Open Access Article
Open Access ArticleModelling the effects of E/Z photoisomerization of a cyclocurcumin analogue on the properties of cellular lipid membranes†
Anastasiia
Delova
a,
Raul
Losantos
 ab,
Jérémy
Pecourneau
c,
Maxime
Mourer
c,
Andreea
Pasc
ab,
Jérémy
Pecourneau
c,
Maxime
Mourer
c,
Andreea
Pasc
 c and
Antonio
Monari
c and
Antonio
Monari
 *a
*a
aUniversité Paris Cité and CNRS, ITODYS, F-75006 Paris, France. E-mail: Antonio.monari@u-paris.fr
bUniversidad de La Rioja, Departamento de Química, Centro de Investigación en Síntesis Química, 26006 Logroño, Spain
cUniversité de Lorraine CNRS, L2CM UMR 7053, F-54000 Nancy, France
First published on 17th July 2023
Abstract
The use of photosensitive molecules capable of isomerizing under light stimuli, and thus induce perturbation in biological systems, is becoming increasingly popular for potential light-activated chemotherapeutic purposes. We recently show that a cyclocurcumin derivative (CCBu), may be suitable for light-activated chemotherapy and may constitute a valuable alternative to traditional photodynamic therapy, due to its oxygen-independent mechanism of action, which allows the treatment of hypoxic solid tumors. In particular, we have shown that the E/Z photoisomerization of CCBu correlates with strong perturbations of model lipid bilayers. In this work, we perform all-atom classical molecular dynamics for a more complex bilayer, whose composition is, thus, much closer to eukaryotic outer cell membranes. We have evidenced important differences in the interaction pathway between CCBu and the complex lipid bilayer as compared to previous models, concerning both the membrane penetration capacity and the isomerization-induced perturbations. While we confirm that structural perturbations of the lipid membrane are induced by isomerization, we also show how the use of a simplified membrane model can result in an oversimplification of the system and hinder key physical and biological phenomena. Although, CCBu may be considered as a suitable candidate for light-activated chemotherapy, we also underline how the inclusion of bulkier substituents, inducing larger perturbations upon photoisomerization, may enhance its efficiency.
Introduction
Cancer remains a complex and devastating disease that affects millions of individuals. Cancer treatments such as surgery, chemotherapy, and radiation therapy have shown significant advancements in recent years, allowing for impressive progresses in therapeutic protocols thus increase of survival rates and improve life-conditions for the patients. Yet, conventional anticancer approaches often come with severe side effects and limited effectiveness. Amongst the different cancer treatments, photodynamic therapy (PDT) appears as a promising alternative,1–5 to increase therapeutic efficiency and selectivity. Indeed, PDT is based on the synergistic effect of a photo-active drug and its activation by an external light stimulus.6,7 Differently from conventional chemotherapeutics, the drug is activated only in the body regions exposed to light, hence avoiding, or limiting, systemic toxicity. From a photophysical point of view PDT golden standard relies on the activation of the photosensitizers (PS) using a specific light wavelength to produce singlet oxygen (1O2) and other reactive oxygen species (ROS) that can disrupt cellular macromolecular systems, such as nucleic acids, proteins, and lipid membranes, and thus initiate cancer cell death. Interestingly, PDT is not only restricted to cancer treatment and can find useful applications also for antibacterial or antiviral purposes, or for food processing.8–10 To assure 1O2 efficient production, the PS should usually show a facile intersystem crossing leading to a high population of its triplet manifold and the subsequent energy transfer to the molecular oxygen to produce 1O2 or other ROS. To increase the intersystem crossing probability, heavy metal having high spin–orbit coupling elements may also be used.11 Even if it is highly promising, PDT may show some drawbacks in particular related to limited selectivity, leading to adverse secondary effects also caused by poor metabolism and metal accumulation in the body.12 Furthermore, efficient PS should absorb in the red or near infrared region of the electromagnetic spectrum, in particular covering the therapeutic windows, to assure the maximum wavelength penetration in the tissues.10,13 While active research lines are operative to decorate PS with peripheral groups specifically recognizing receptors overexpressed in cancer cells to maximize accumulation,14 drug delivering strategies may also be pursued to counteract the usual poor solubility of PDT agents,15–18 which may lead to a globally poor bioavailability. Furthermore, PDT is relying on the activation of molecular oxygen and thus its efficiency is strongly limited in the case of solid tumors under hypoxia.19–21 Therefore, alternative cancer treatments need to be explored. In this context light-activated chemotherapy (LAC) is a promising approach,22,23 as it is oxygen-independent while maintaining the activation of the drugs by a luminous stimulus. Different strategies for LAC have been reported, for instance the light-activated release of ROS by organometallic,24,25 usually Ru-based, compounds have been proposed recently.26,27 A slightly different strategy, which we have also explored in the past, relies in the use of molecular photoswitches, such as E/Z isomerizable molecules.23,28–30 Indeed, upon exposure to light of a specific wavelength, the PS may undergo photoisomerization leading to a significant change in its geometry. If the PS is at the same time interacting with a biological macromolecule, the light-induced large structural change may lead to its perturbation and change its properties, eventually triggering apoptotic signals. For instance, if a photoswitch is interacting with a lipid bilayer, its isomerization may drastically change the membrane properties, causing its permeabilization and rupture, eventually followed by the cell death, without the mediation of singlet oxygen.31 As a matter of fact, the photoisomerization strategy offers several advantages in light-activated chemotherapy, including reduced toxicity and improved efficacy also reducing the risk of adverse effects by limiting the activation of the photosensitizer to specific regions or types of cells. Furthermore, by modifying the structure of photosensitizers, it is possible to change their absorption properties and tune the illumination wavelength required to activate them. Globally, photoswitches comprise several classes of molecules as azobenzenes,32 spiropyrans,33,34 diarylethenes,33 in which light is used to convert the double carbon bond from E- to Z-conformation or to perform photocyclization reactions where the closed-ring specie converts to the open-ring isomer causing changes in the mechanical properties of its environment. The use of non-toxic, natural available, or bio-analogues switches is also particularly attractive to limit general toxicity and side-effects.31,35,36Turmeric extracts have been often employed for a number of therapeutic objectives including treating allergy and facilitate healing.37 The pharmacological properties of turmeric extracts have been further supported by modern scientific study, with a focus on its antibacterial, antioxidant, and anti-inflammatory activities.38 The main component of turmeric, i.e. curcumin, has revealed excellent ADMET (Absorption, Distribution, Metabolism, Excretion, and Toxicity) properties and a high degree of bioavailability,39 it is metabolized primarily in the liver and excreted via the biliary route. Importantly, curcumin demonstrates a favorable toxicity profile, with low levels of toxicity observed across various studies. A secondary component of turmeric, cyclocurcumin, may also present a favourable pharmacological potential, yet its characterization is much scarcer.
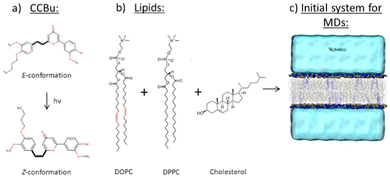
In our previous work31,40,41 we described, thanks to a combination of experimental and molecular modeling tools, the potentiality of a biomimetic cyclocurcumin derivative (CCBu) for use in light-activated chemotherapy (Fig. 1(a)). Indeed, upon light-absorption, CCBu undergoes a structural change from the more stable E-configuration to the bulkier Z-conformation.42 In particular we have shown that our biomimetic analogues present superior properties over the natural cyclocurcumin, both in term of isomerization quantum yield and interaction with a model lipid membrane, the experimental results having been rationalized via multiscale molecular modeling and simulation.35,42 However, both the experiments and modeling were conducted considering a DPPC lipid bilayer membrane only,31,40 which is a widely used model to study lipid bilayer interactions. Notably, we have rationalized both the interactions between CCBu and the lipid bilayer, as well as the effects of E/Z-photoisomerization and different CCBu concentrations on these interactions.40 Specifically, we have shown that CCBu may penetrate the lipid bilayer and remain inside the membrane near the lipid's polar groups stabilized by hydrogen bond mediated interactions. We have also shown that the isomerization of CCBu is indeed altering the membrane properties, three different phases, characterized by different order parameters may be observed depending on the PS concentration.40
In the present work, we aimed to go a step further and investigate the interactions of CCBu in its E/Z-conformations with a more complex and flexible membrane, which closely mimics the composition of eukaryotic outer membranes.43,44 To achieve this, we used a mixed membrane containing three different types of lipids: DPPC, DOPC, and cholesterol, which were chosen due to their abundance and importance in mamalian cell membranes. Indeed, while saturated fatty acids like DPPC are widely recognized for their stiff and well-ordered packing in membranes, DOPC, an unsaturated fatty acid, is in a liquide disordered phase and leads to more fluid membranes. Finally, steroid such as cholesterol have a crucial role in cell membranes especially for preserving their shape and functionality, controlling the fluidity and permeability of the membrane and assuming a role in cell signaling.45
As it will be highlited in the following, we show that the interaction of CCBu strongly depends on the membrane composition, and even more stringently, the magnitude of the light-induced perturbation may vary from the model membrane system to the more biological relevant systems.
Computational methodology
Equilibrium all-atom molecular dynamics
The initial model systems consisting of lipid bilayer, water, and physiological NaCl concentration were generated using the CHARMM-GUI software.46–48 The simulation box contained a lipid bilayer consisting of 3 different types of lipids, namely DOPC, DPPC, and cholesterol (with a molar fraction ratio of 75%, 20%, and 5%, respectively). A total of 200 lipids for each leaflet have been considered and the bilayer is surrounded by a water buffer of 40 Å (81 water molecules per each lipid) including Na+ and Cl− ions at a concentration of 0.15 M to properly model membrane cell. CCBu was also inserted in the water bulk and the total size of the final system amounted to 115 Å × 115 Å × 120 Å (Fig. 1).All the MD simulations were performed using NAMD code49,50 and analyzed and visualized with VMD software.51 The lipids were modeled with the Amber14 lipid force field,52 water was modeled with TIP3P.53 CCBu was modeled using the general AMBER force field (GAFF)54 and atomic point charges were acquired through the restricted electrostatic potential fitting procedure, coherently with the protocol used in our previous contributions.31,40,41 Hydrogen mass repartitions55 has been consistently used, allowing, in combination with Rattle and Shake,56 to use a time step of 4.0 fs to integrate the Newton equations of motion and propagate the MD simulation. Prior to production all the systems have been submitted to minimization followed by thermalization and equilibration progressively removing positional constraints on non-water heavy atoms for a total of 6 ns. The initial temperature was set at 300 K and held constant throughout the entire MD simulation time, assuring a liquid phase for the membrane system. Equilibration was carried on in the isothermal and isobaric (NPT) ensemble to assure the adjustment of the density. Conversely, production was performed in the isothermal NVT ensemble. Conservation of temperature and pression was enforce using Langevin thermostat57 and barostat,58 respectively. Three initial model lipid bilayer systems have been constructed: one system, i.e. the control, involved no CCBu sensitizers, in the second system one CCBu molecule in E-configuration was added in the water bulk, and finally in the third system the Z-isomer of CCBu was added to the bulk. Equilibrium MD simulations reaching the 660–1000 ns time scale was performed, and the stability of the aggregates was checked through the root-mean-square deviation (RMSD) analysis.
To study the effect of CCBu concentrations on the model membranes, systems involving 5, 10, 15, 20, 30, 40 and 50 CCBu in E-conformation have been constructed. Afterwards, steered MD (SMD)59 was performed to pull all molecules separately inside the lipid bilayer from the water bulk by applying a harmonic force of 10 kcal mol−1 over 200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps with timestep of 4 fs. The center of mass of each CCBu molecule and the center of the membrane were used as collective variables. We note that each molecule has reached the lipid core without causing the rupture of the membrane. After SDM, unconstrained equilibrium MD simulations (650–1200 ns) has been run to assure the equilibration of the whole system.
000 steps with timestep of 4 fs. The center of mass of each CCBu molecule and the center of the membrane were used as collective variables. We note that each molecule has reached the lipid core without causing the rupture of the membrane. After SDM, unconstrained equilibrium MD simulations (650–1200 ns) has been run to assure the equilibration of the whole system.
Free energy profile
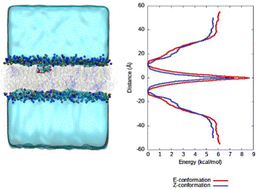
In order to comprehend and quantify the process of CCBu internalizing into the membrane, enhanced sampling methods allowing the calculation of the related free energy profiles has been performed. In particular, and coherently with the strategy previously used we relied to the combination of extended adaptive biased force (eABF)60 with metadynamics61 leading to the meta-eABF strategy62,63 which allows an efficient and faster exploration of complex and rough free energy potentials. As a collective variable (CV) we chose, for the system comprising only one CCBu unit, the distance along the Z-axis between the centers of mass of the CCBu and the lipid bilayer as our CV. The highest boundary of the system (55.0 Å on Oz axis) and the center of the lipid bilayer (0.0 Å on Oz axis) were used as limits for the meta-eABF procedure. The meta-eABF was propagated for 660 ns to ensure proper sampling of the conformational space and obtain a well-converged potential of mean force, also checking that the full collective variable has been explored. Free energy calculations have been performed using NAMD and the Colvar module.64Simulation of E/Z-photoisomerization
Additionally, SMD was used to enforce the effects of the E/Z-photoisomerization of CCBu. This was done considering the equilibrated systems involving 5, 10, 15, 20, 30, 40, and 50 CCBu in E-conformation embedded in the lipid bilayer. Such number of chromophores correspond to a CCBu/lipids molar ratio of 1.25, 2.5, 3.75, 5, 7.5, 10, 12.5%, respectively. Additionally, the CCBu molar concentrations can be estimated to 0.5 × 10−4, 1.1 × 10−4, 1.7 × 10−4, 2.3 × 10−4, 3.4 × 10−4, 4.6 × 10−4, 5.7 × 10−4 M.To enforce the isomerization a 5 kcal mol−1 harmonic potential was applied to change the dihedral angle around the isomerizable carbon–carbon double bond from 179° (E-isomer) to 7.9° (Z-isomer). The harmonic potential was applied for 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 steps using a time step of 0.5 fs to avoid numerical instabilities. After the dihedral angle change, the force fields of CCBu was switched to the one describing its Z-conformation and unconstrained equilibrium MD simulation performed for 1130 ns.
000 steps using a time step of 0.5 fs to avoid numerical instabilities. After the dihedral angle change, the force fields of CCBu was switched to the one describing its Z-conformation and unconstrained equilibrium MD simulation performed for 1130 ns.
Analysis of membrane properties
To study the effects of different concentrations of CCBu (i.e. different numbers of CCBu molecules in the lipid bilayer) and the effects of the forced E/Z-isomerization on the lipid bilayer properties, we have explicitly calculated membrane structural parameters such as area per lipid chain metric. The deuterium order parameter (-Scd) and the membrane thickness have also been obtained and are reported in ESI.† Membrane structural parameters have been obtained by using the MEMBPLUGIN tool65 extension of VMD. Notably, in this work we have calculated the averaged values of the area per lipid chain for each lipid in the system (DPPC, DOPC) along the equilibrated MD trajectories.Results and discussion
In our previous work, we simulated the behaviour of CCBu with a pure DPPC lipid bilayer. More specifically, the free energy profiles (FEP) for CCBu in both E- and Z-configuration were calculated and it was found that only the E-conformer presented an absolute minimum inside the membrane and close to the lipid polar head groups. On the contrary, the Z-isomer only presented a metastable state when internalized in the membrane, and a free energy barrier needed to be overcome to allow the interaction with the polar heads. Coherently with those results during the equilibrium MD simulations only spontaneous penetration of the E-isomer was observed.31Conversely, in the present case (see ESI†) we observe spontaneous penetration of both isomers inside the mixed membrane and their positioning in the vicinity of the polar head region. Furthermore, meta-eABF was used to calculate the FEP for one CCBu molecule in both E- and Z-configurations penetrating inside a complex membrane composed of DPPC, DOPC and cholesterol and it was observed that both isomers can enter the mixed lipid bilayer unimpeded (Fig. 2). Furthermore, both isomers present a global free energy minimum at the interface between polar heads and lipid tails, as can also be appreciated by the snapshots reported in Fig. 2. Interestingly, a free energy gain of about 6 kcal mol−1 can be appreciated for both isomers, compared to the situation in which CCBu is in the water bulk, and at the global minimum CCBu is interacting with the polar groups of the DPPC and DOPC lipids. This situation represents a first very crucial difference experienced by the more complex membrane under study here. The easiness of penetration of CCBu may be ascribed to the membrane higher fluidity and flexibility, which most notably enables even the sterically bulkier Z-isomer to penetrate the membrane, while the more rigid DPPC-only membrane made penetration of Z-CCBu impossible and energetically unfavorable due to the larger energetic barrier.
We have also identified the main interactions stabilizing CCBu inside the lipid membrane. Our findings reveal that hydrogen bonds (HBs) between CCBu and the lipid polar heads play the most important role. Specifically, we found that HBs involving either the hydroxyl group of the phenyl ring and the carbonyl oxygen of the pyron ring (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) are the most important occurrences and takes place with both DPPC and DOPC polar moieities, as shown in Fig. 3. Interestingly, by analysing the MD trajectories we evidenced that distances shorter than 3 Å between CCBU and the polar heads occurred for 6% of the whole simulation span while distances shorter than 4 Å were populated for 59% of the simulation span. Furthermore, we also observed for 58% of the MDs trajectory the establishment of HBs (with a distance of 3 Å) between CCBu and water molecules (Fig. 3(c)). We also evidenced that weak hydrophobic interactions between the hydrophobic tail of DPPC and DOPC lipids and the butyl chain of CCBu contribute to the overall stability of the membrane aggregate.
O) are the most important occurrences and takes place with both DPPC and DOPC polar moieities, as shown in Fig. 3. Interestingly, by analysing the MD trajectories we evidenced that distances shorter than 3 Å between CCBU and the polar heads occurred for 6% of the whole simulation span while distances shorter than 4 Å were populated for 59% of the simulation span. Furthermore, we also observed for 58% of the MDs trajectory the establishment of HBs (with a distance of 3 Å) between CCBu and water molecules (Fig. 3(c)). We also evidenced that weak hydrophobic interactions between the hydrophobic tail of DPPC and DOPC lipids and the butyl chain of CCBu contribute to the overall stability of the membrane aggregate.
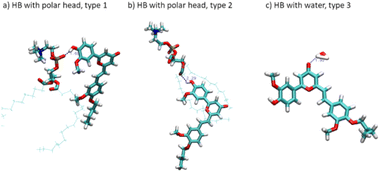 | ||
| Fig. 3 Representation of the hydrogen bonds (HB) stabilizing CCBu at the polar head/water interface. | ||
We also considered the effects of CCBu orientation inside the lipid membrane. To accomplish this, we constructed two extreme configurations obtained after full equilibration of CCBu in E-conformation and situated within the lipid bilayer. In the first system, CCBu is positioned perpendicular to the lipid's hydrophobic chains, i.e. the membrane axis, while in the second system, it is oriented parallel to them. To investigate the effects of these orientations, we conducted MD simulations for a total of 100 ns for both systems. We observed that for the first system CCBu remains in a perpendicular orientation for the whole duration of the simulation, the average angle between the membrane Oz axis and the CCBu main axis stabilizing at 120°. Conversely, in the second system CCBu rapidly reorients to a more perpendicular arrangement after 50 ns. Thus, it appears that the slightly perpendicular orientation is the most favourable arrangement, at least when considering only one ligand. This result is not unexpected, since this conformation allows to maximize at the same time HBs with the polar head and hydrophobic interactions with the lateral chains.
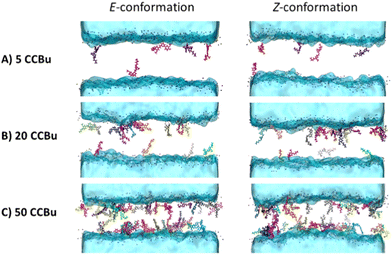
Having assessed that both isomers are indeed penetrating inside the lipid membrane, we then turn to consider the impact of various concentrations of E- and Z-conformers on the membrane structural properties. It is important to note that in this work we investigate the concomitant isomerization of the whole ensemble of chromophores in the lipid bilayer. The study of partially isomerized systems would be much more expensive and would require a large number of initial structures leading to a combinatorial complexity. Nevertheless, we have previously assessed the partial isomerization of CCBu in a single-lipid membrane environment. A strong correlation between the membrane perturbation observed for fully and partial isomerized systems justify the choice done here to consider only the full isomerization. Furthermore, having simulated the isomerization by Steered MD only, all the fast or ultrafast (ps-scale) details are clearly not well described. However, the full study of photoisolerization would require exploring the excited state potential energy surface with high-level quantum chemistry approaches which would dramatically limit the accessible time-scales. Indeed, in this contribution, we focus more on the non-equilibrium response of the membrane to the isomerization which requires sampling longer time-scales.
Because of the more complex composition of the membrane, we computed the average area per lipid chain separately for each lipid in the system and the results are presented in Fig. 4. Once again, we may observe a striking difference with the situation observed for the pure DPPC membrane,40 which was showing the coexistence of three different phases, characterized by different ordered and disordered arrangement of CCBu. As reported in Fig. 4, in the case of the mixed membrane studied here we may observe an almost perfectly linear increase of the area per lipid as a function of the concentration, whatever the isomer considered.
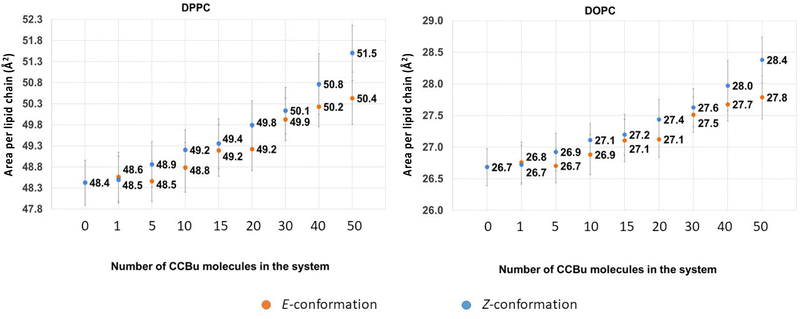 | ||
| Fig. 4 The average values for area per lipid chain for (A) DPPC and (B) DOPC as a function of varying concentrations of CCBu in both E and Z conformation. | ||
Interestingly, the same global behavior is maintained for the two amphiphilic lipids, i.e. DPPC and DOPC, while cholesterol presents a more intricate behavior as shown in ESI.† Interestingly, the constant increase of the area per lipid leads to a grow of area per lipid chain of about 4% for DPPC and 7% for DOPC for 50 CCBu molecules.
Unsurprisingly, we also observe a globally stronger perturbation induced by the bulkier and more compact Z-isomer, indeed the average area per lipid chain after the forced E/Z isomerization and ensuing equilibration are systematically higher than in the case of the E-isomer. Although, the perturbation appears relatively small, the observed differences are statistically significant, and thus indicative of an effective perturbation. Nevertheless, it is also important to note that the difference appears slightly lower than the one observed for concentrations compatible with the ordered phases in the case of the pure membrane. This difference can again be ascribed to the higher flexibility of the mixed membrane, which is thus, more efficient in accommodating the structural perturbations brought by the isomerization. Yet, as we already observed, it appears that the inclusion of bulkier peripheral substituents could be beneficial to maximize the membrane perturbation.
Similar results can also be appreciated for the evolution of membrane interdigitation and the deuterium order parameters which are shown in ESI.†
The evolution of the membrane structural properties with the concentration shows no discontinuity and thus points to the absence of specific phase transitions and concentration dependent specific arrangements. This can also be pictorially appreciated by the representative snapshots reported in Fig. 5 for the E- and Z-isomer, respectively. The transbilayer density profiles of CCBu in both E- and Z-conformations have been obtained and are reported in ESI.† CCBU predominantly resides within the lipid bilayer in the vicinity of the polar heads. Notably, the density profile of CCBu remains unaffected after the photoisomerization. Furthermore, the increase in the concentration of CCBU is not altering its equilibrium position which remains close to the polar heads. Indeed, a globally disordered arrangement is always and consistently observed, without the propensity of the CCBu moieties to orient parallelly to each other as it was observed for the pure membrane system. Interestingly, the same disordered pattern is observed for the planar E- and the more globular Z-isomer, coherently with the linear behavior of the area per lipid evolution as reported in Fig. 4. Furthermore, and even at slightly high concentration we may notice the absence of clustering of CCBu suggesting that the hydrophobic interactions developed at the interface with the lipid tails are actually preventing aggregation, which is instead highly favorable in aqueous environment.
Next, we analysed the orientation of CCBu within the lipid bilayer with respect to the membrane axis. We observed that CCBu in the E-conformation orient at angles of 50° and 130° relative to the Oz axis, i.e. the membrane axis, whatever the concentration (Fig. 6). This observation is also in agreement with the results previously discussed and obtained for a single ligand embedded in the membrane. Upon photoisomerization, we observed that the Z-CCBu retains the same global behavior, with however a slightly more perpendicular orientation the angle peaking, at 60°. Interestingly, for both isomers, the distributions are quite large and the orientation spans a wide allowed region. Once again, this observation is coherent with the more fluid nature of the mixed membrane, as compared to the simple model.
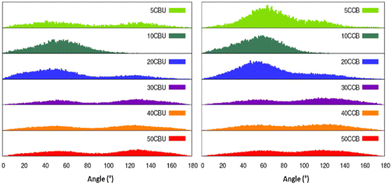 | ||
| Fig. 6 Distribution of the orientation of the CCBu molecules with respect to the Oz axis, i.e. the membrane axis. | ||
Conclusions
We have performed a long-time scale MD simulation to unravel the behavior of a cyclocurcumin derivative, CCBu, interacting with a model lipid bilayer, whose composition mimics a eukaryotic membrane. Equilibrium MD simulation achieving the 660–1200 ns time-range and free energy methods have been considered throughout. We have evidenced some specific differences in the behavior of the mixed membrane compared to a simpler DPPC model. In particular, we found that in the case of the more flexible mixed membrane penetration of CCBu is more favorable and stable states in which the sensitizers are placed at the polar head/lipid tail interface exist for both isomers. Furthermore, and differently from the rigid DPPC model,31,40 we also found that no concentration dependent phase transition can be observed. Instead, CCBu always assumes a disordered arrangement inside the lipid bilayers, whatever the concentration, a situation which translates in a linear dependence of the average area per lipid metric with the CCBu concentration.The use of CCBu for LAC purposes relies on its capacity to induce membrane perturbations upon photoisomerization. Indeed, we have shown that the bulkier Z-isomer consistently leads to larger values of the area per lipid, which represent a statistically significant trend. Yet, the perturbation induced are relatively small, and are even less important than those observed in the case of the more rigid DPPC membrane. Therefore, we confirm that the inclusion of bulky and rigid peripheral substituents could represent a most valuable strategy to increase the ability of CCBu to perturb lipid bilayers.
In the future, we plan to extend the work presented here considering on the one side the use of longer peripheral chains which should favor even more the penetration inside the membrane of CCBu and the perturbation induced by the photoisomerization. Furthermore, the effects of partial isomerization will also be taken into account to propose an even more complete model.
Author contributions
A. D. performed all the MD simulations, A. M. and A. P. conceptualized and planned the study. Results have been discussed by all the authors. The manuscript was written by the contribution of all the authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors thank GENCI and Explor computing centers for computational resources. A. D. thanks Université Paris Cité for her PhD fellowships. R. L. thanks Universidad de La Rioja and Ministerio de Universidades for his “Margarita Salas” grant. Support from the European Union European Regional Development Funds (Programme opérationnel FEDER-FSE Lorraine et Massif des Vosges 2014-2020 “Fire Light” project: “Photo-bio-active molecules and nanoparticles”) is also acknowledged. A. M. thanks ANR and CGI for their financial support of this work through Labex SEAM ANR 11 LABX 086, ANR 11 IDEX 05 02. The support of the IdEx “Université Paris 2019” ANR-18-IDEX-0001 and of the Platform P3MB is gratefully acknowledged.Notes and references
- R. L. Yanovsky, D. W. Bartenstein, G. S. Rogers, S. J. Isakoff and S. T. Chen, Photodermatol., Photoimmunol. Photomed., 2019, 35, 295–303 CrossRef PubMed.
- J. F. Algorri, M. Ochoa, P. Roldán-Varona, L. Rodríguez-Cobo and J. M. López-Higuera, Cancers, 2021, 13, 4447 CrossRef CAS PubMed.
- R. R. Allison and K. Moghissi, Photodiagn. Photodyn. Ther., 2013, 10, 331–341 CrossRef CAS PubMed.
- J. Zhang, C. Jiang, J. P. Figueiró Longo, R. B. Azevedo, H. Zhang and L. A. Muehlmann, Acta Pharm. Sin. B, 2018, 8, 137–146 CrossRef PubMed.
- T. C. Pham, V. N. Nguyen, Y. Choi, S. Lee and J. Yoon, Chem. Rev., 2021, 121, 13454–13619 CrossRef CAS PubMed.
- A. B. Ormond and H. S. Freeman, Materials, 2013, 6, 817–840 CrossRef CAS PubMed.
- A. Master, M. Livingston and A. Sen Gupta, J. Controlled Release, 2013, 168, 88–102 CrossRef CAS PubMed.
- E. Koshi, A. Mohan, S. Rajesh and K. Philip, J. Indian Soc. Periodontol., 2011, 15, 323 CrossRef PubMed.
- T. Dai, Y. Y. Huang and M. R. Hamblin, Photodiagn. Photodyn. Ther., 2009, 6, 170–188 CrossRef CAS PubMed.
- W. Fan, P. Huang and X. Chen, Chem. Soc. Rev., 2016, 45, 6488–6519 RSC.
- L. B. Josefsen and R. W. Boyle, Met.-Based Drugs, 2008, 2008, 276109 Search PubMed.
- S. Ren, X. Cheng, M. Chen, C. Liu, P. Zhao, W. Huang, J. He, Z. Zhou and L. Miao, ACS Appl. Mater. Interfaces, 2017, 9, 31509–31518 CrossRef CAS PubMed.
- Z. Li and K. B. Grant, RSC Adv., 2016, 6, 24617–24634 RSC.
- K. Li, W. Dong, Q. Liu, G. Lv, M. Xie, X. Sun, L. Qiu and J. Lin, J. Photochem. Photobiol., B, 2019, 190, 1–7 CrossRef CAS PubMed.
- I. Yakavets, I. Yankovsky, M. Millard, L. Lamy, H. P. Lassalle, A. Wiehe, V. Zorin and L. Bezdetnaya, Int. J. Pharm., 2017, 529, 568–575 CrossRef CAS PubMed.
- I. Yankovsky, E. Bastien, I. Yakavets, I. Khludeyev, H. P. Lassalle, S. Gräfe, L. Bezdetnaya and V. Zorin, Eur. J. Pharm. Sci., 2016, 91, 172–182 CrossRef CAS PubMed.
- I. Yakavets, H. P. Lassalle, D. Scheglmann, A. Wiehe, V. Zorin and L. Bezdetnaya, Nanomaterials, 2018, 8, 1–15 CrossRef PubMed.
- B. Aslanoglu, I. Yakavets, V. Zorin, H. P. Lassalle, F. Ingrosso, A. Monari and S. Catak, Phys. Chem. Chem. Phys., 2020, 22, 16956–16964 RSC.
- J. M. Brown, Cancer Biol. Ther., 2002, 1, 453–458 CrossRef PubMed.
- J. M. Brown, Methods Enzymol., 2007, 435, 297–321 CAS.
- B. Chen, B. W. Pogue, X. Zhou, J. A. O’Hara, N. Solban, E. Demidenko, P. J. Hoopes and T. Hasan, Clin. Cancer Res., 2005, 11, 720–727 CrossRef CAS PubMed.
- X. Li, J. F. Lovell, J. Yoon and X. Chen, Nat. Rev. Clin. Oncol., 2020, 17, 657–674 CrossRef PubMed.
- P. Dunkel and J. Ilaš, Cancers, 2021, 13, 3237 CrossRef CAS PubMed.
- A. C. Jung, F. Moinard-Butot, C. Thibaudeau, G. Gasser and C. Gaiddon, Pharmaceutics, 2021, 13, 1788 CrossRef CAS PubMed.
- M. N. Pinto and P. K. Mascharak, J. Photochem. Photobiol., C, 2020, 42, 100341 CrossRef CAS.
- A. Busemann, I. Flaspohler, X. Q. Zhou, C. Schmidt, S. K. Goetzfried, V. H. S. van Rixel, I. Ott, M. A. Siegler and S. Bonnet, J. Biol. Inorg. Chem., 2021, 26, 667–674 CrossRef CAS PubMed.
- X.-Q. Zhou, P. Wang, V. Ramu, L. Zhang, S. Jiang, X. Li, S. Abyar, P. Papadopoulou, Y. Shao, L. Bretin, M. A. Siegler, F. Buda, A. Kros, J. Fan, X. Peng, W. Sun and S. Bonnet, Nat. Chem., 2023, 15, 980–987 CrossRef CAS PubMed.
- J. Broichhagen, J. A. Frank and D. Trauner, Acc. Chem. Res., 2015, 48, 1947–1960 CrossRef CAS PubMed.
- M. M. Lerch, M. J. Hansen, G. M. van Dam, W. Szymanski and B. L. Feringa, Angew. Chem., Int. Ed., 2016, 55, 10978–10999 CrossRef CAS PubMed.
- W. Szymański, J. M. Beierle, H. A. V. Kistemaker, W. A. Velema and B. L. Feringa, Chem. Rev., 2013, 113, 6114–6178 CrossRef PubMed.
- J. Pecourneau, R. R. Losantos, A. Delova, Y. Bernhard, S. S. Parant, M. Mourer, A. Monari and A. Pasc, Langmuir, 2022, 38, 15642–15655 CrossRef CAS PubMed.
- H. M. D. Bandara and S. C. Burdette, Chem. Soc. Rev., 2012, 41, 1809–1825 RSC.
- Z. L. Pianowski, Chem. – Eur. J., 2019, 25, 5128–5144 CrossRef CAS PubMed.
- A. V. Chernyshev, A. A. Guda, A. Cannizzo, E. V. Solov’Eva, N. A. Voloshin, Y. Rusalev, V. V. Shapovalov, G. Smolentsev, A. V. Soldatov and A. V. Metelitsa, J. Phys. Chem. B, 2019, 123, 1324–1331 CrossRef CAS PubMed.
- M. Marazzi, A. Francés-Monerris, M. Mourer, A. Pasc and A. Monari, Phys. Chem. Chem. Phys., 2020, 22, 4749–4757 RSC.
- R. Losantos, A. Pasc and A. Monari, Phys. Chem. Chem. Phys., 2021, 23, 24757–24764 RSC.
- M. C. P. Araujo, L. M. G. Antunes and C. S. Takahashi, Teratog., Carcinog., Mutagen., 2001, 21, 175–180 CrossRef CAS PubMed.
- S. C. Gupta, S. Patchva, W. Koh and B. B. Aggarwal, Clin. Exp. Pharmacol. Physiol., 2012, 39, 283–299 CrossRef CAS PubMed.
- D. Shanmugarajan, P. P., B. R. P. Kumar and B. Suresh, RSC Adv., 2020, 10, 31385–31399 RSC.
- A. Delova, R. Losantos, J. Pecourneau, Y. Bernhard, M. Mourer, A. Pasc and A. Monari, J. Chem. Inf. Model., 2023, 63, 299–307 CrossRef CAS PubMed.
- R. Losantos, J. Pecourneau, M. Mourer, S. Parant, A. Pasc and A. Monari, Phys. Chem. Chem. Phys., 2021, 23, 12842–12849 RSC.
- J. Pecourneau, R. Losantos, A. Monari, S. Parant, A. Pasc and M. Mourer, J. Org. Chem., 2021, 86, 8112–8126 CrossRef CAS PubMed.
- D. Casares, P. V. Escribá and C. A. Rosselló, Int. J. Mol. Sci., 2019, 20, 2167 CrossRef CAS PubMed.
- G. Van Meer, D. R. Voelker and G. W. Feigenson, Nat. Rev. Mol. Cell Biol., 2008, 9, 112–124 CrossRef CAS PubMed.
- D. S. Schade, L. Shey and R. P. Eaton, Endocr. Pract., 2020, 26, 1514–1523 CrossRef PubMed.
- S. Jo, J. B. Lim, J. B. Klauda and W. Im, Biophys. J., 2009, 97, 50–58 CrossRef CAS PubMed.
- S. Jo, T. Kim, V. G. Iyer and W. Im, J. Comput. Chem., 2008, 29, 1859–1865 CrossRef CAS PubMed.
- E. L. Wu, X. Cheng, S. Jo, H. Rui, K. C. Song, E. M. Dávila-Contreras, Y. Qi, J. Lee, V. Monje-Galvan, R. M. Venable, J. B. Klauda and W. Im, J. Comput. Chem., 2014, 35, 1997–2004 CrossRef CAS PubMed.
- J. C. Phillips, R. Braun, W. Wang, J. Gumbart, E. Tajkhorshid, E. Villa, C. Chipot, R. D. Skeel, L. Kalé and K. Schulten, J. Comput. Chem., 2005, 26, 1781–1802 CrossRef CAS PubMed.
- J. C. Phillips, D. J. Hardy, J. D. C. Maia, J. E. Stone, J. V. Ribeiro, R. C. Bernardi, R. Buch, G. Fiorin, J. Hénin, W. Jiang, R. McGreevy, M. C. R. Melo, B. K. Radak, R. D. Skeel, A. Singharoy, Y. Wang, B. Roux, A. Aksimentiev, Z. Luthey-Schulten, L. V. Kalé, K. Schulten, C. Chipot and E. Tajkhorshid, J. Chem. Phys., 2020, 153, 044130 CrossRef CAS PubMed.
- W. Humphrey, A. Dalke and K. Schulten, J. Mol. Graphics, 1996, 14, 33–38 CrossRef CAS PubMed.
- J. A. Maier, C. Martinez, K. Kasavajhala, L. Wickstrom, K. E. Hauser and C. Simmerling, J. Chem. Theory Comput., 2015, 11, 3696–3713 CrossRef CAS PubMed.
- P. Mark and L. Nilsson, J. Phys. Chem. A, 2001, 105, 9954–9960 CrossRef CAS.
- J. Wang, R. M. Wolf, J. W. Caldwell, P. A. Kollman and D. A. Case, J. Comput. Chem., 2004, 25, 1157–1174 CrossRef CAS PubMed.
- C. W. Hopkins, S. Le Grand, R. C. Walker and A. E. Roitberg, J. Chem. Theory Comput., 2015, 11, 1864–1874 CrossRef CAS PubMed.
- S. Miyamoto and P. A. Kollman, J. Comput. Chem., 1992, 13, 952–962 CrossRef CAS.
- R. L. Davidchack, R. Handel and M. V. Tretyakov, J. Chem. Phys., 2009, 130, 234101 CrossRef PubMed.
- S. E. Feller, Y. Zhang, R. W. Pastor and B. R. Brooks, J. Chem. Phys., 1995, 103, 4613–4621 CrossRef CAS.
- S. Izrailev, S. Stepaniants, B. Isralewitz, D. Kosztin, H. Lu, F. Molnar, W. Wriggers and K. Schulten, J. Phys. Chem. B, 2011, 115, 1211–1219 CrossRef PubMed.
- T. Zhao, H. Fu, T. Lelièvre, X. Shao, C. Chipot and W. Cai, J. Chem. Theory Comput., 2017, 13, 1566–1576 CrossRef CAS PubMed.
- A. Barducci, M. Bonomi and M. Parrinello, Wiley Interdiscip. Rev.: Comput. Mol. Sci., 2011, 1, 826–843 CAS.
- H. Fu, H. Zhang, H. Chen, X. Shao, C. Chipot and W. Cai, J. Phys. Chem. Lett., 2018, 9, 4738–4745 CrossRef CAS PubMed.
- H. Fu, X. Shao, W. Cai and C. Chipot, Acc. Chem. Res., 2019, 52, 3254–3264 CrossRef CAS PubMed.
- G. Fiorin, M. L. Klein and J. Hénin, Mol. Phys., 2013, 111, 3345–3362 CrossRef CAS.
- R. Guixà-González, I. Rodriguez-Espigares, J. M. Ramírez-Anguita, P. Carrió-Gaspar, H. Martinez-Seara, T. Giorgino and J. Selent, Bioinformatics, 2014, 30, 1478–1480 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Radial distribution function, membrane thickness, evolution of the collective variable during SDM, RMSD for different concentration of E- and Z CCBu, transbilayer density profiles, force field parameters. See DOI: https://doi.org/10.1039/d3cp02955g |
| This journal is © the Owner Societies 2023 |