Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Assembly and optically triggered disassembly of lipid–DNA origami fibers†
Sofia
Julin
 a,
Nadine
Best
a,
Nadine
Best
 abc,
Eduardo
Anaya-Plaza
abc,
Eduardo
Anaya-Plaza
 a,
Eeva
Enlund
a,
Veikko
Linko
a,
Eeva
Enlund
a,
Veikko
Linko
 ad and
Mauri A.
Kostiainen
ad and
Mauri A.
Kostiainen
 *ae
*ae
aBiohybrid Materials, Department of Bioproducts and Biosystems, Aalto University, 00076 Aalto, Finland. E-mail: mauri.kostiainen@aalto.fi
bTechnische Universität Darmstadt, 64289 Darmstadt, Germany
cFraunhofer Institute for Microengineering and Microsystems IMM, 55129 Mainz, Germany
dInstitute of Technology, University of Tartu, 50411 Tartu, Estonia
eLIBER Center of Excellence, Aalto University, 00076 Aalto, Finland
First published on 9th November 2023
Abstract
The co-assembly of lipids and other compounds has recently gained increasing interest. Here, we report the formation of stimuli-responsive lipid–DNA origami fibers through the electrostatic co-assembly of cationic lipids and 6-helix bundle (6HB) DNA origami. The photosensitive lipid degrades when exposed to UV-A light, which allows a photoinduced, controlled release of the 6HBs from the fibers. The presented complexation strategy may find uses in developing responsive nanomaterials e.g. for therapeutics.
Lipid self-assembly is important for the formation of many cellular compartments in biological systems, but lipids can also co-assemble with other molecular entities into hierarchical structures with different structural morphologies.1 Nucleic acids, for example, form highly ordered supramolecular assemblies (lipoplexes) when complexed with cationic lipids.2,3 Lipoplexes have potential applications in nanomedicine3–5 as well as in optoelectronics and synthetic chemistry.5 We have recently demonstrated that lipids could also co-assemble with larger DNA origami nanostructures and that DNA origami may serve as nucleation templates for the growth of multilamellar lipid assemblies.6 DNA origami are well-defined two- and three-dimensional (2D and 3D) DNA-based nanostructures prepared by folding a long-single-stranded DNA “scaffold” strand by a set of shorter oligonucleotides (“staples”).7–9 Versatile and custom-shaped DNA origami can be utilized in, for example, drug delivery,10–12 synthetic cells,13,14 nanorobotics,15–17 and bottom-up nanofabrication.18,19 In addition, the phosphate groups in the DNA backbone give the DNA origami an overall negative charge, and therefore, DNA origami can direct formation of ordered assemblies from positively charged compounds merely via electrostatic interactions.20–23
Currently, the research interests are increasingly focusing on functional nanomaterials that can respond to external stimuli.24 Responsive nanomaterials that are able to release encapsulated compounds on demand would be advantageous especially for therapeutics.25 In our previous work, we showed that DNA origami could be released from the multilamellar lipid assemblies by adding competitive polyanions.6 However, for practical applications this disassembly approach is rather unfeasible and it would be advantageous to explore other stimuli-responsive disassembly methods. Light as a stimulus is non-invasive and straightforward to apply with high spatiotemporal control.26 Therefore, we designed and synthesized a cationic photosensitive lipid, spermine-hydroxyethyl photolinker-oleic ester (SpllO, Fig. 1a), with the ability to co-assemble with DNA origami (Fig. 1b) into highly ordered lipid–DNA origami fibers through electrostatic and hydrophobic interactions (Fig. 1c). SpllO contains an o-nitrobenzyl group that undergoes photolytic degradation when exposed to UV-A light (λ ∼ 350 nm),27 thus cleaving the oleic ester tail from the spermine head group (inset in Fig. 1a). The spermine group alone has low affinity to DNA, and by removing the lipid tail, the self-assembled multivalency of SpllO driven by hydrophobic interactions, is lost, and the SpllO-DNA origami fibers are disassembled.28–30
The photosensitive lipid SpllO was synthesized and characterized using standard methods (ESI,† Scheme S1 and Fig. S1, S2, S25–S35). Briefly, a photolabile o-nitrobenzyl linking group (pll) was functionalized with a tert-butyloxycarbonyl protected spermine (BOC-spermine, 1) by an amide bond.27 In a subsequent step, the BOC-spermine-pll (2) was coupled with the oleic acid in a Steglich esterifcation. Finally, the water-soluble SpllO was obtained by a deprotection of the BOC-protected spermine group by acidic hydrolysis. For the co-assembly with SpllO, we used a long, rod-shaped 6-helix bundle (6HB) DNA origami31 as a model structure (Fig. 1b and ESI,† Fig. S3–S6). The negatively charged DNA origami interacts with the positively charged amines in SpllO, which facilitates the electrostatic co-assembly (
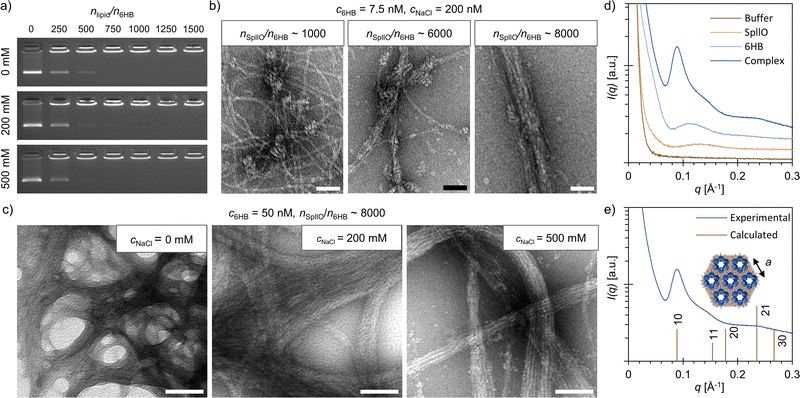
SpllO–6HB complexes were formed by mixing the two compounds together in 6HB folding buffer (FOB; 1× Tris-acetate-EDTA (TAE) and 12.5 mM MgCl2) supplemented with different amounts of NaCl. Initially, the electrostatic co-assembly of SpllO and 6HBs was studied using an agarose gel electrophoretic mobility shift assay (EMSA, Fig. 2a). The binding of SpllO and the subsequent formation of complexes immobilize the 6HBs in the gel, which is observed as a gradual decrease in the intensity of the free 6HB band with increasing stoichiometric ratios, nSpllO/n6HB, between SpllO and 6HBs. High concentrations of free ions in the assembly buffer typically prevent assembly formation by screening the electrostatic interactions between the charged compounds.22,32 However, in this case, the electrostatic co-assembly was not significantly affected by the NaCl concentration of the assembly buffer, and a nSpllO/n6HB ∼ 1000 was sufficient for complexation even at high NaCl concentrations (Fig. 2a and ESI,† Fig. S7). This further confirms that the co-assembly is driven not only by electrostatic but also hydrophobic interactions.
To gain additional insights into the assembly, we used transmission electron microscopy (TEM) to visualize the SpllO–6HB assemblies (c6HB = 7.5 nM) formed in 1× FOB supplemented with 200 mM NaCl (Fig. 2b and ESI,† Fig. S8–S10). At nSpllO/n6HB ∼ 1000, the 6HBs were, as also suggested by agarose gel EMSA, highly aggregated, although mostly uncoated. Small lipid-like structures were visible on the 6HB interfaces, which indicates that SpllO acts as a supramolecular “glue” bringing the structures together. By increasing the nSpllO/n6HB considerably, the amount of uncoated 6HBs decreased, and a nSpllO/n6HB ∼ 8000 was found to be required to entirely encapsulate the 6HBs with SpllO. Furthermore, the TEM images revealed that the structural morphology of the SpllO–6HB complexes changed from less ordered aggregates towards more fiber-like structures with increasing nSpllO/n6HB.
The initial co-assembly, shown in Fig. 2a and b, was carried out using relatively low 6HB concentrations (c6HB = 7.5 nM), but for further characterization, we formed SpllO–6HB complexes at remarkably higher concentrations (c6HB ≥ 50 nM, ESI,† Fig. S11–S16). As for the assemblies formed at low 6HB concentrations, a nSpllO/n6HB ∼ 8000 was found to be sufficient for the assembly (ESI,† Fig. S12–S14). At high concentrations (c6HB = 50 nM), we also studied the effect of the NaCl concentration on the assembly formation more thoroughly. TEM shows that large, micrometer-sized fibers are formed for all three NaCl concentrations, but that the most well-aligned fibers are formed at cNaCl = 200 mM (Fig. 2c and ESI,† Fig. S11, S14, S15). In the absence of NaCl the fibers tend to aggregate, whereas at high NaCl concentrations (cNaCl = 500 mM) the fibers are smaller and the 6HBs are more loosely packed. This is in line with the small-angle X-ray scattering (SAXS) data measured for SpllO–6HB assemblies formed at different NaCl concentrations, which indicates that 200 mM NaCl would be the optimum salt concentration for the assembly (ESI,† Fig. S17).
SAXS measurements were conducted on both the SpllO–6HB complexes and the individual components to confirm the observations from TEM (Fig. 2e and ESI,† Fig. S18). The SpllO–6HB complexes had a clearly different scattering profile than the individual building blocks. Additional analysis of the SAXS data for the SpllO–6HB complex further suggests that the complexes form a 2D hexagonal lattice. The peak positions found at q = 0.089, 0.148 and 0.235 Å−1 fit approximately with the reflections from [10], [11] and [21] planes for a 2D hexagonal lattice with a lattice constant of a = 7.1 nm. The lattice constant is slightly larger than the 6HB diameter (6 nm), and therefore matches the dimensions of the building blocks. Previously, similar fiber-like structures have been reported when 6HBs were co-assembled with positively charged phthalocyanines.22
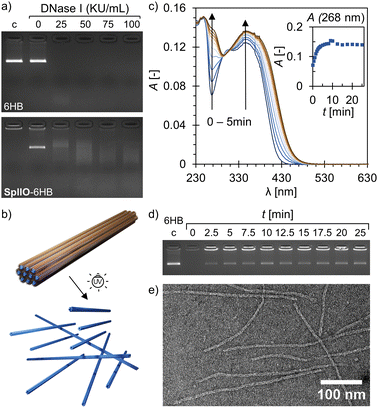
Plain DNA origami are typically susceptible to digestion by nucleases, such as DNase I,33 but the structural stability against nucleases can be enhanced by applying different coatings or by encapsulating the structures.34,35 Co-assembly with SpllO (c6HB = 7.5 nM, nSpllO/n6HB ∼ 8000) was also observed to shield the 6HBs and slow down the DNase I digestion. The plain 6HBs were completely digested when exposed to DNase I at a concentration of 25 Kunitz units (KU) mL−1 for 1 h at RT, while the 6HBs co-assembled with SpllO were only partially digested (Fig. 3a). The DNase I was inactivated with sodium dodecyl sulfate (SDS) before running the gel, which disassembled the SpllO–6HB complexes and released the 6HBs. The 6HBs embedded in the fibers formed at high concentrations (c6HB = 50 nM) withstand DNase I at least to the same extent as the 6HBs co-assembled with SpllO at low concentrations (ESI,† Fig. S19).
The responsiveness of the assembly was demonstrated by exposing the SpllO–6HB complexes to UV-A light (λ ∼ 350 nm). Irradiation with UV-A light cleaves the photosensitive o-nitrobenzyl group of SpllO, which facilitates a controlled, optically triggered release of the 6HBs from the assemblies (Fig. 3b). The photolytic degradation of plain SpllO was confirmed by exposing an aqueous SpllO solution to UV-A light and following the degradation over time using UV/Vis spectroscopy (Fig. 3c). UV-A irradiation changed the absorbance spectra significantly, and clear increases in the absorbance at 268 nm and 347 nm were observed, which is typical for the proposed photolytic degradation.27,36 Moreover, after ∼8 min of irradiation, the SpllO degradation leveled off and reached a plateau. Further irradiation after that resulted in a decrease again of the absorbance at 347 nm (ESI,† Fig. S20). In addition, also TEM demonstrated that SpllO is degraded by UV-A light (ESI,† Fig. S21). The disassembly of the complexes (c6HB = 7.5 nM, nSpllO/n6HB ∼ 8000) and the subsequent release of the 6HBs upon irradiation was studied using an agarose gel EMSA (Fig. 3d and ESI,† Fig. S22). A band with similar electrophoretic mobility as the plain 6HB appeared in the gel after only 2.5 min of UV-A exposure, and maximum 6HB recovery of ∼ 30% was observed after ∼ 10 min. The co-assembly with SpllO entangles the 6HBs (as shown in Fig. 2b), and therefore some 6HBs will remain aggregated also after the UV-A exposure although the supramolecular multivalency effect is destroyed. DNA origami are known to maintain their structural integrity even upon high-dose UV-A irradiation,37 and TEM further confirmed that intact 6HBs are released from the complexes (Fig. 3e and ESI,† Fig. S23). The 6HBs could also be recovered from the larger and more well-aligned fibers formed at high 6HB concentrations (c6HB = 50 nM), but a longer irradiation time was required (ESI,† Fig. S24).
In conclusion, we have demonstrated a straightforward and scalable approach for constructing well-aligned and photo-responsive fibers by the co-assembly of a cationic photosensitive lipid SpllO and 6HB DNA origami. The lipid co-assembly provided protection for the 6HBs against DNase I digestion, and the SpllO–6HB complexes could be controllably disassembled and the 6HBs released through UV-A light exposure. Lipid nanoparticles are already widely used for mRNA delivery,38 but stimuli-responsive lipid-based systems for the delivery of larger DNA origami could find intriguing uses in therapeutics. The high addressability of the DNA origami allows a variety of drugs, therapeutic proteins and antibody fragments to be precisely positioned onto the structure,39 and genes folded within DNA origami have been successfully expressed in cells.40
This work was financially supported by the Academy of Finland (no. 314671 and 341057), ERA Chair MATTER under the EU's H2020 research and innovation programme (no. 856705), Aalto University School of Chemical Engineering, Finnish Cultural Foundation, Sigrid Jusélius Foundation, Emil Aaltonen Foundation and Jane and Aatos Erkko Foundation. The work was carried out under the Academy of Finland Centers of Excellece Programme (2022–2029) in Life-Inspired Hybrid Materials (LIBER), no. 346110. The authors thank S. Nummelin, D. Langerreiter, I. Seitz and K. Malinen for technical assistance, and acknowledge the provision of facilities and technical support by Aalto University Bioeconomy Facilities and Ota Nano – Nanomicroscopy Center (Aalto-NMC).
Conflicts of interest
The authors declare no conflict of interest.References
- G. Tresset, PMC Biophys., 2009, 2, 3 CrossRef PubMed.
- C. R. Safinya, Curr. Opin. Struct. Biol., 2001, 11, 440–448 CrossRef CAS PubMed.
- L. Wasungu and D. Hoekstra, J. Controlled Release, 2006, 116, 255–264 CrossRef CAS PubMed.
- C. T. de Ilarduya, Y. Sun and N. N. Düzgüneş, Eur. J. Pharm. Sci., 2010, 40, 159–170 CrossRef PubMed.
- K. Liu, L. Zheng, C. Ma, R. Göstl and A. Herrmann, Chem. Soc. Rev., 2017, 46, 5147–5172 RSC.
- S. Julin, Nonappa, B. Shen, V. Linko and M. A. Kostiainen, Angew. Chem., Int. Ed., 2021, 60, 827–833 CrossRef CAS PubMed.
- P. W. K. Rothemund, Nature, 2006, 440, 297–302 CrossRef CAS PubMed.
- S. M. Douglas, H. Dietz, T. Liedl, B. Högberg, F. Graf and W. M. Shih, Nature, 2009, 459, 414–418 CrossRef CAS PubMed.
- S. Dey, C. Fan, K. V. Gothelf, J. Li, C. Lin, L. Liu, N. Liu, M. A. D. Nijenhuis, B. Saccà, F. C. Simmel, H. Yan and P. Zhan, Nat. Rev. Methods Primers, 2021, 1, 13 CrossRef CAS.
- A. Keller and V. Linko, Angew. Chem., Int. Ed., 2020, 59, 15818–15833 CrossRef CAS PubMed.
- J. Weiden and M. M. C. Bastings, Curr. Opin. Colloid Interface Sci., 2021, 52, 101411 CrossRef CAS.
- S. Jiang, Z. Ge, S. Mou, H. Yan and C. Fan, Chem, 2021, 7, 1156–1179 CAS.
- H. Shen, Y. Wang, J. Wang, Z. Li and Q. Yuan, ACS Appl. Mater. Interfaces, 2019, 11, 13859–13873 CrossRef CAS PubMed.
- K. Jahnke and K. Göpfrich, Interface Focus, 2023, 13, 20230028 CrossRef PubMed.
- S. Nummelin, B. Shen, P. Piskunen, Q. Liu, M. A. Kostiainen and V. Linko, ACS Synth. Biol., 2020, 9, 1923–1940 CrossRef CAS PubMed.
- M. DeLuca, Z. Shi, C. E. Castro and G. Arya, Nanoscale Horiz., 2020, 5, 182–201 RSC.
- P. Pitikultham, Z. Wang, Y. Wang, Y. Shang, Q. Jiang and B. Ding, ChemMedChem, 2022, 17, e202100635 CrossRef CAS PubMed.
- A. Heuer-Jungemann and V. Linko, ACS Cent. Sci., 2021, 7, 1969–1979 CrossRef CAS PubMed.
- P. Zhan, A. Peil, Q. Jiang, D. Wang, S. Mousavi, Q. Xiong, Q. Shen, Y. Shang, B. Ding, C. Lin, Y. Ke and N. Liu, Chem. Rev., 2023, 123, 3976–4050 CrossRef CAS PubMed.
- T. Jiang, T. A. Meyer, C. Modlin, X. Zuo, V. P. Conticello and Y. Ke, J. Am. Chem. Soc., 2017, 139, 14025–14028 CrossRef CAS PubMed.
- S. Julin, A. Korpi, Nonappa, B. Shen, V. Liljeström, O. Ikkala, A. Keller, V. Linko and M. A. Kostiainen, Nanoscale, 2019, 11, 4546–4551 RSC.
- A. Shaukat, E. Anaya-Plaza, S. Julin, V. Linko, T. Torres, A. de la Escosura and M. A. Kostiainen, Chem. Commun., 2020, 56, 7341–7344 RSC.
- I. Seitz, S. Saarinen, E.-P. Kumpula, D. McNeale, E. Anaya-Plaza, V. Lampinen, V. P. Hytönen, F. Sainsbury, J. J. L. M. Cornelissen, V. Linko, J. T. Huiskonen and M. A. Kostiainen, Nat. Nanotechnol., 2023, 18, 1205–1212 CrossRef CAS PubMed.
- X. Zhang, L. Chen, K. H. Lim, S. Gonuguntla, K. W. Lim, D. Pranantyo, W. P. Yong, W. J. T. Yam, Z. Low, W. J. Teo, H. P. Nien, Q. W. Loh and S. Soh, Adv. Mater., 2019, 31, 1804540 CrossRef PubMed.
- S. Mura, J. Nicolas and P. Couvreur, Nat. Mater., 2013, 12, 991–1003 CrossRef CAS PubMed.
- M. A. Kostiainen, O. Kasyutich, J. J. L. M. Cornelissen and R. J. M. Nolte, Nat. Chem., 2010, 2, 394–399 CrossRef CAS PubMed.
- M. A. Kostiainen, D. K. Smith and O. Ikkala, Angew. Chem., Int. Ed., 2007, 46, 7600–7604 CrossRef CAS PubMed.
- A. Barnard and D. K. Smith, Angew. Chem., Int. Ed., 2012, 51, 6572–6581 CrossRef CAS PubMed.
- N. P. Gabrielson and J. Cheng, Biomaterials, 2010, 31, 9117–9127 CrossRef CAS PubMed.
- L. E. Fechner, B. Albanyan, V. M. P. Vieira, E. Laurini, P. Posocco, S. Pricl and D. K. Smith, Chem. Sci., 2016, 7, 4653–4659 RSC.
- H. Bui, C. Onodera, C. Kidwell, Y. Tan, E. Graugnard, W. Kuang, J. Lee, W. B. Knowlton, B. Yurke and W. L. Hughes, Nano Lett., 2010, 10, 3367–3372 CrossRef CAS PubMed.
- M. A. Kostiainen, P. Hiekkataipale, A. Laiho, V. Lemieux, J. Seitsonen, J. Ruokolainen and P. Ceci, Nat. Nanotechnol., 2013, 8, 52–56 CrossRef CAS PubMed.
- V. Linko and A. Keller, Small, 2023, 19, 2301935 CrossRef CAS PubMed.
- H. Bila, E. E. Kurisinkal and M. M. C. Bastings, Biomater. Sci., 2019, 7, 532–541 RSC.
- A. R. Chandrasekaran, Nat. Rev. Chem., 2021, 5, 225–239 CrossRef CAS PubMed.
- M. Smet, L.-X. Liao, W. Dehaen and D. V. McGrath, Org. Lett., 2000, 2, 511–513 CrossRef CAS PubMed.
- H. Chen, R. Li, S. Li, J. Andréasson and J. H. Choi, J. Am. Chem. Soc., 2017, 139, 1380–1383 CrossRef CAS PubMed.
- X. Hou, T. Zaks, R. Langer and Y. Dong, Nat. Rev. Mater., 2021, 6, 1078–1094 CrossRef CAS PubMed.
- I. Seitz, H. Ijäs, V. Linko and M. A. Kostiainen, ACS Appl. Mater. Interfaces, 2022, 14, 38515–38524 CrossRef CAS PubMed.
- J. A. Kretzmann, A. Liedl, A. Monferrer, V. Mykhailiuk, S. Beerkens and H. Dietz, Nat. Commun., 2023, 14, 1017 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3cc04677j |
| This journal is © The Royal Society of Chemistry 2023 |