Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Design strategies for countering the effect of fluorophore-quencher labelling on DNA hairpin thermodynamics†
Yan Shan
Ang
 and
Lin-Yue Lanry
Yung
and
Lin-Yue Lanry
Yung
 *
*
Department of Chemical & Biomolecular Engineering, National University of Singapore, Singapore 117585, Singapore. E-mail: cheyly@nus.edu.sg
First published on 12th October 2023
Abstract
We report the impact of fluorophore-quencher labelling on the thermodynamics of hairpin opening by testing five fluorophores and two quenchers labelled at the end and/or internal positions. Two counter strategies were introduced, i.e. label the hairpin probe at an internal position or append an external hairpin stem on the trigger strand to promote coaxial stacking hybridization. The observations remained valid for complex hairpin opening operations such as hybridization chain reaction.
The predictability of Watson–Crick base-pairing has enabled the rational design of nucleic acid probes and machineries for diverse applications, including molecular probes for PCR, DNA circuits for biosensing and DNA barcodes for multiplexed bio-imaging.1,2 A common approach for generating a readout signal is via the labelling of oligonucleotides (oligo) with fluorophores and/or quenchers either in the duplex or hairpin structure, herein termed probes.3,4 The presence of the dye, often with complex ring structures, introduces additional molecular interactions such as π–π stacking, hydrogen bonding and intercalation to the double helix structure.5,6 Several experimental studies have confirmed the stabilization effect induced by the dyes modified on the ends of the probes7 and even destabilization when the dyes are modified in the internal regions of the probes.8
Dye-induced deviations from the predictable Watson–Crick base pairing thermodynamics can manifest in a non-negligible difference in the annealing temperature up to 7.5 °C.6 Such variation depends on multiple parameters such as the fluorophore identity, and their position relative to one another and within the oligo. The effects are challenging to model predictively and can only be elucidated through deliberate experimental studies but often go unnoticed in a typical experimental workflow. For example, the experimental feasibility of a DNA circuit design is often checked via gel electrophoresis using a high concentration of unmodified oligos before transiting to fluorescence measurement using a relatively lower concentration of the modified probes.9–11 There is an inherent yet inaccurate assumption that the unmodified oligos and modified probes are equivalent as recently studied by Li et al.12 In their study of toehold-mediated DNA strand displacement involving duplex substrates, the end modification of duplexes slows the strand displacement kinetics, while modification on the toehold domains accelerates the kinetics.
In this work, we studied the impact of different configurations of site-specific fluorophore-quencher modifications in both duplex and hairpin structures (Fig. 1). Within the combinations tested, hairpin structures with end modifications consistently suppressed the equilibrium signal generated to the greatest extent due to the additional stabilization effect of the fluorophore-quenchers at the terminals. Such hairpin structures are often used as molecular beacon probes or monomers facilitating isothermal amplification such as in hybridization chain reaction (HCR)13,14 and catalytic hairpin assembly (CHA).15,16
To overcome the unintended stabilization induced by the dye, a simple strategy of using an external hairpin stem structure at the end of the invading trigger strand to provide an additional base stacking effect, or otherwise known as coaxial stacking hybridization,17–19 was introduced to energetically favour the forward strand displacement reaction. A similar strategy has been used to improve hybridization to short sequences such as for microRNA detection20–23 but has yet to be applied in this context of overcoming the unintended effect of dye modifications to nucleic acid structures.
Four different direct trigger (DT) strands were designed, i.e. single stranded DT with a 9 nt toehold (DT9) and three hairpin-primed DT with a 4 nt, 6 nt and 10 nt hairpin stem length respectively (DT9-HP4, DT9-HP6, DT9-HP10). They were each reacted with probes of different fluorophore-quencher configurations (Section S2, ESI†), and the various reaction products are shown in Fig. S2 (ESI†).
We first established the baseline observation using an FAM-quencher duplex probe, wherein the equilibrium fluorescence signal increased only marginally when reacted with hairpin-primed DT of longer stem length (Fig. 2a). When the FAM-quencher duplex was replaced with a hairpin structure of identical domain sequences and fluorophore-quencher combination, the fluorescence signal generated using the single stranded DT9 trigger strand dropped drastically which demonstrated the significant impact of dye modification on hairpin opening (Fig. 2b). The contribution from the hairpin structure outweighed the positional effect of the dye in a duplex probe (Fig. S3, ESI†). This could be due to the stronger intra-hydrogen bonding at the stem region of the hairpin structure favouring its closed state as compared to inter-hydrogen bonding in the duplex structure.24,25 The invading strand had to form a stronger intermediate complex at the toehold domain for the forward strand displacement reaction to proceed.18 By adding a hairpin stem to the trigger strand (DT9-HP4/6/10), the equilibrium fluorescence signal increased substantially due to the coaxial base stacking contributed by the hairpin stem.
 | ||
| Fig. 2 Evolution of fluorescence signal when 20 nM of the respective trigger strands (DT) were incubated with (a) FAM-Q duplex, as well as hairpins with (b) end-modified FAM-Q, (c) end-modified Alexa Fluor 555 (AF555), (d) internally modified AF555 (int), (e) end-modified Alexa Fluor 647 (AF647), (f) internally modified AF647 (int), (g) internally modified mFluor Blue 580 (MF580 int) and (h) internally modified mFluor Blue 630 (MF630 int). The y-axis is expressed in thousand units; note that fluorescence intensity was measured in arbitrary units and the absolute numerical value cannot be compared directly across different excitation/emission combinations. Refer to Fig. S4 (ESI†) for plots with error bars (n = 3). (i) Comparison of the fold-change of the signal intensity when the respective probes were reacted with hairpin-primed DT (DT-HP4) versus single-stranded DT. All data are shown as mean ± standard deviation (n = 3). | ||
DNA hairpins, particularly those modified with a fluorophore-quencher configuration, are frequently used in DNA circuit designs26 and biosensing applications.3 To test the generality of our observation, two other fluorophores of different colours, Alexa Fluor 555 (AF555, Fig. 2c) and 647 (AF647, Fig. 2e), were conjugated onto the hairpin probe which generated similar trends to that of FAM, i.e. a modest signal generated with single stranded DT9, a huge signal boost with hairpin-primed DT and the marginal-to-negligible increase in signal with a longer hairpin stem. This was within expectation as the terminal modification of oligos with a fluorophore and quencher is known to enhance its annealing temperature due to the effects of base stacking stabilization and intercalation of the dye to the nucleic acids.9
The next parameter studied was the position of the fluorophore and quencher. By shifting the AF555 or AF647 and Black Hole Quencher 2 to an internal position within the stem of HP1, a significantly higher fluorescence signal was generated using a DT9 trigger compared to the end-modified configuration (Fig. 2d and f). This can be attributed to the thermodynamically destabilizing effect of having internally modified dyes which locally disrupt the usual Watson–Crick base pairing interactions.8 As such, DT9 could successfully initiate strand displacement and open the hairpin efficiently. The strategy of modifying the hairpin at internal rather than end positions to overcome the additional base stacking effect of hairpin probes remained effective when tested with two other non-Alexa Fluor family dyes, mFluor580 (MF580, Fig. 2g) and mFluor630 (MF630, Fig. 2h). A similar observation was made with modification at another position along the stem (nearer the loop) (Fig. S5, ESI†).
A common observation across all dye-modified probes was that a short hairpin-primed trigger of stem length 4 nt sufficed in restoring the opening of hairpin probes efficiently. As such there was marginal-to-no incremental benefit of having longer stems for the endpoint fluorescence measurement and analysis from this point onwards was only performed with DT9 and DT9-HP4. Nonetheless, we observed that the melting temperature of the hairpin-trigger complexes increased when DT9-HP”n” trigger with longer stem lengths was used, which was direct evidence of the coaxial stacking effect in stabilizing the desired product and favouring the forward hybridization reaction (Section S5, ESI†).
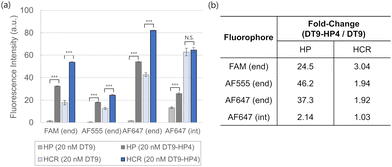
We calculated the fold-change of the equilibrium fluorescence intensity obtained using DT9-HP4 to that using DT9 and observed significant differences across the eight fluorophore/quencher combinations tested (Fig. 2i). The signal generated was restored significantly (>20 fold-change) using the proposed hairpin-primed DT strategy for all end-modified hairpins. The magnitude of change depended on the specific pairing of the fluorophore and quencher due to differences in their chemical structures and hence molecular interactions between the pair, and with the nucleic acid backbone. Previously, Zimmers et al. have reported an increase in annealing temperature from 4.0 °C (for FAM) to 7.5 °C (for rhodamine 101) depending on the fluorophore identity.6 Alternatively, if the use of a conventional single stranded trigger strand were preferred, a consistently effective strategy for restoring the hairpin opening thermodynamics was to shift the dye position from end to internal.
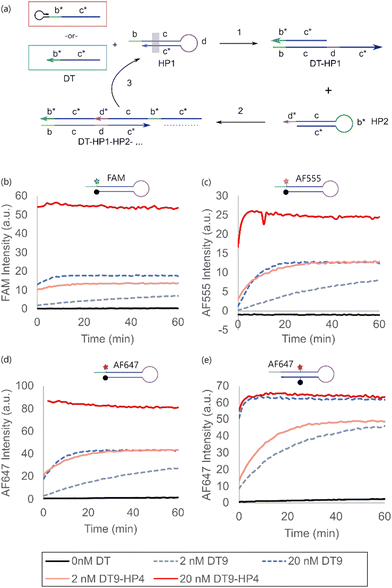
The impact of dye modification on more complex molecular hairpin operations was characterized using hybridization chain reaction (HCR) as a model system. HCR is a signal amplification mechanism involving the cascaded opening of a pair of hairpin monomers (HP1 and HP2) upon initiation by a trigger strand (DT),13,14 commonly used for biosensing and spatial imaging applications.27–29 HP1 was modified with the four sets of fluorophore-quencher pairings selected from the previous section, while HP2 was unmodified (Fig. 3a).
For all end-modified fluorophore-quencher pairs, the hairpin-primed direct trigger (DT9-HP4) outperformed that of using the single stranded DT (DT9) (Fig. 3b–d). Interestingly, the equilibrium fluorescence signal of 20 nM DT9 was generally close to that generated by 2 nM DT9-HP4, suggesting that the former had only ca. 10% HCR triggering efficiency on the molar basis of the trigger strand concentration. This represented significant inefficiency of trigger utilization, which would otherwise go undetected in typical experiments.
Similar to the case of a single hairpin opening, the internal modification of HP1 destabilized the hairpin sufficiently such that the single stranded trigger (DT9) was approximately just as efficient as the hairpin-primed trigger (DT9-HP4) in activating HCR (Fig. 3d). In spite of the apparent destabilizing effect, there was no significant increase in the background leakage and a high signal-to-noise ratio was still achieved. The practical implication is that one can simply overcome the inefficient utilization of the single stranded trigger strand by adopting an internally modified hairpin design instead of adding a hairpin-primed structure to the trigger strand in scenarios where this is not possible, e.g. if the trigger were a naturally occurring sequence, or the sequence design gets too complicated due to the longer trigger length.
Finally, the single stranded trigger was more effective in generating a signal in HCR versus the opening of a single hairpin (Fig. 4a). This was likely due to the combined effect of both DT9 and HP2 in stabilizing the opened state of HP1 in HCR, on top of the cascaded amplification contribution by HCR. This was in line with our previous report that the melting temperature of complexes formed via HCR was higher than that of a single DT-HP1 duplex due to the co-stabilization effect by HP2.30 The fold-change of the equilibrium fluorescence intensity obtained using DT9-HP4 to that using DT9 across all designs was compared in Fig. 4b. Generally, the effect of the hairpin-primed trigger was less pronounced in the HCR system compared to the direct opening of a single hairpin, such as molecular beacon design.
In this work, we studied the effect of fluorophore modifications on DNA hybridization thermodynamics with a focus on hairpin opening. We observed an extremely low hybridization efficiency of a single stranded trigger strand with end-modified hairpins. The hybridization efficiency improved drastically, i.e. up to ca. 40-folds, by incorporating an external hairpin stem structure on the single stranded trigger strand to promote coaxial stacking hybridization. An alternative counter strategy was to modify the dye internally in the hairpin stem to destabilize its closed state. Lastly, the effect of dye modifications on hairpin opening was similarly observed in the more complex molecular operation of HCR. Overall, the study affirmed that the careful choice of fluorophore-quencher pair, aside from their spectral properties, and their strategic placement should be accounted for in the design of oligo probes, which is particularly crucial for probes with a hairpin structure.
This work was supported by research funding from the Singapore Ministry of Education Academic Research Fund Tier 1 (A-0009534-01-00) and Tier 2 (MOE2019-T2-1-116 and MOE-T2EP50120-0018).
Conflicts of interest
There are no conflicts to declare.References
- D. Y. Zhang and G. Seelig, Nat. Chem., 2011, 3, 103–113 CrossRef CAS PubMed.
- S. K. Saka, Y. Wang, J. Y. Kishi, A. Zhu, Y. Zeng, W. Xie, K. Kirli, C. Yapp, M. Cicconet, B. J. Beliveau, S. W. Lapan, S. Yin, M. Lin, E. S. Boyden, P. S. Kaeser, G. Pihan, G. M. Church and P. Yin, Nat. Biotechnol., 2019, 37, 1080–1090 CrossRef CAS PubMed.
- N. Bidar, M. Amini, F. Oroojalian, B. Baradaran, S. S. Hosseini, M.-A. Shahbazi, M. Hashemzaei, A. Mokhtarzadeh, M. R. Hamblin and M. de la Guardia, TrAC, Trends Anal. Chem., 2021, 134, 116143 CrossRef CAS.
- M. K. Johansson, H. Fidder, D. Dick and R. M. Cook, J. Am. Chem. Soc., 2002, 124, 6950–6956 CrossRef CAS PubMed.
- B. G. Moreira, Y. You, M. A. Behlke and R. Owczarzy, Biochem. Biophys. Res. Commun., 2005, 327, 473–484 CrossRef CAS PubMed.
- Z. A. Zimmers, N. M. Adams, W. E. Gabella and F. R. Haselton, Anal. Methods, 2019, 11, 2862–2867 RSC.
- B. G. Moreira, Y. You and R. Owczarzy, Biophys. Chem., 2015, 198, 36–44 CrossRef CAS PubMed.
- W. Lee, P. H. von Hippel and A. H. Marcus, Nucleic Acids Res., 2014, 42, 5967–5977 CrossRef CAS PubMed.
- J. Wang, D.-X. Wang, J.-Y. Ma, Y.-X. Wang and D.-M. Kong, Chem. Sci., 2019, 10, 9758–9767 RSC.
- Y. Chu, S.-J. Xiao and J.-J. Zhu, Anal. Chem., 2023, 95, 4317–4324 CrossRef CAS PubMed.
- Y. Jiang, B. Li, X. Chen and A. Ellington, Molecules, 2012, 17, 13211–13220 CrossRef CAS PubMed.
- C. Li, Z. Li, W. Han, X. Yin, X. Liu, S. Xiao and H. Liang, Chem. Commun., 2022, 58, 5849–5852 RSC.
- R. M. Dirks and N. A. Pierce, Proc. Natl. Acad. Sci. U. S. A., 2004, 101, 15275–15278 CrossRef CAS PubMed.
- Y. S. Ang and L.-Y. L. Yung, Chem. Commun., 2016, 52, 4219–4222 RSC.
- P. Yin, H. M. T. Choi, C. R. Calvert and N. A. Pierce, Nature, 2008, 451, 318–322 CrossRef CAS PubMed.
- J. Liu, Y. Zhang, H. Xie, L. Zhao, L. Zheng and H. Ye, Small, 2019, 15, 1902989 CrossRef CAS PubMed.
- V. A. Vasiliskov, Nucleic Acids Res., 2001, 29, 2303–2313 CrossRef CAS PubMed.
- N. Srinivas, T. E. Ouldridge, P. Šulc, J. M. Schaeffer, B. Yurke, A. A. Louis, J. P. K. Doye and E. Winfree, Nucleic Acids Res., 2013, 41, 10641–10658 CrossRef CAS PubMed.
- D. Y. Zhang and E. Winfree, J. Am. Chem. Soc., 2009, 131, 17303–17314 CrossRef CAS PubMed.
- H. Liu, T. Tian, Y. Zhang, L. Ding, J. Yu and M. Yan, Biosens. Bioelectron., 2017, 89, 710–714 CrossRef PubMed.
- W. Liu, X. Zhou and D. Xing, Biosens. Bioelectron., 2014, 58, 388–394 CrossRef CAS PubMed.
- C. Chen, Nucleic Acids Res., 2005, 33, e179 CrossRef PubMed.
- D. Duan, K. Zheng, Y. Shen, R. Cao, L. Jiang, Z. Lu, X. Yan and J. Li, Nucleic Acids Res., 2011, 39, e154 CrossRef CAS PubMed.
- Y. Gao, L. K. Wolf and R. M. Georgiadis, Nucleic Acids Res., 2006, 34, 3370–3377 CrossRef CAS PubMed.
- J. S. Schreck, T. E. Ouldridge, F. Romano, P. Šulc, L. P. Shaw, A. A. Louis and J. P. K. Doye, Nucleic Acids Res., 2015, 43, 6181–6190 CrossRef CAS PubMed.
- H. Peng, A. M. Newbigging, M. S. Reid, J. S. Uppal, J. Xu, H. Zhang and X. C. Le, Anal. Chem., 2020, 92, 292–308 CrossRef CAS PubMed.
- S. Bi, S. Yue and S. Zhang, Chem. Soc. Rev., 2017, 46, 4281–4298 RSC.
- Y. S. Ang, P. S. Lai and L.-Y. L. Yung, Anal. Chem., 2020, 92, 11164–11170 CrossRef CAS PubMed.
- Y. S. Ang, J. J. Li, P.-J. Chua, C.-T. Ng, B.-H. Bay and L.-Y. L. Yung, Anal. Chem., 2018, 90, 6193–6198 CrossRef CAS PubMed.
- Y. S. Ang, T. Bando, H. Sugiyama and L. L. Yung, ChemBioChem, 2020, 21, 2912–2915 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedures, DNA sequence design and fluorophore/quencher pair used, and data representing the reproducibility of observations. See DOI: https://doi.org/10.1039/d3cc02427j |
| This journal is © The Royal Society of Chemistry 2023 |