Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The history of genome editing: advances from the interface of chemistry & biology
Daisuke
Matsumoto
ab and
Wataru
Nomura
 *abc
*abc
aGraduate School of Biomedical and Health Sciences, Hiroshima University, 1-2-3 Kasumi Minami-ku, Hiroshima, 734-8553, Japan. E-mail: wnomura@hiroshima-u.ac.jp
bSchool of Pharmaceutical Sciences, Hiroshima University, 1-2-3 Kasumi Minami-ku, Hiroshima, 734-8553, Japan
cResearch Institute for Nanodevices, Hiroshima University, 1-4-2 Kagamiyama, Higashi-Hiroshima 739-8527, Japan
First published on 24th May 2023
Abstract
Genome editing had a long history before the appearance of CRISPR. Although a decade has passed since the initial use of CRISPR with mammalian cells, the first attempts at gene editing occurred in the 1980's. Subsequently, many researchers tried to develop methods to edit specific genes. Here, we review the history of genome editing and improvements in genome editing tools. In the last two decades, genome editing tools have been applied in basic sciences, the bio-industry, and therapeutics. We provide examples in which genome editing tools have been applied to various tasks. Recently, new CRISPR-Cas techniques, such as base and prime editing and anti-CRISPR proteins, have attracted considerable interest. Accordingly, these topics are also reviewed.
1. Introduction
All living organisms possess genetic information, which provides instructions for their development, function, and reproduction. This genetic information, subdivided into functional units called genes, is composed of deoxyribonucleic acid (DNA), a polymeric molecule composed of four types of nucleotides. Genes are sequences of nucleotides that encode ribonucleic acids (RNA) and proteins. However, external stimuli such as UV irradiation and oxidative stress, as well as internal stimuli, such as replication errors, can alter nucleotide sequences. These changes, called mutations, disrupt the functions of cells and induce diseases such as cancer. This means that it may be possible to treat these diseases by restoring mutations to their original DNA sequences. In addition, this ability to rewrite genetic information at will is very useful in understanding the functions of genes in all living organisms.Genome editing is one of the most important inventions in biomedical sciences in the last two or three decades. CRISPR-Cas9 and the Cas family of proteins have been its primary drivers. Not only site-directed mutagenesis in eukaryotic cells, but also the DNA-binding properties of Cas9 or nuclease-null Cas9 (dCas9) have been applied to epigenetic modifications, gene regulation, fluorescent imaging of genome dynamics, etc. There are many other technologies related to gene mutagenesis or gene regulation, such as triple helix-forming oligonucleotides (TFOs), antisense-oligonucleotides, recombinases (Cre1,2 or phiC313), and mega-nucleases such as I-SceI.4 Current concepts of genome editing/engineering technologies are mainly based on zinc-finger enzymes,5–8 which recognize specific DNA sequences, gene-9–12 or protein-modification enzymes,13 or effector domains for transcription regulation.14,15
A historic view of current genome editing from the discovery of zinc-finger domains and their applications is the starting point for our discussion (Fig. 1). To regulate protein functions, chemical compounds or photochemicals often provide efficient output. Chemistry-driven methods of genome editing, such as precise regulation of the catalytic activity of enzymes or efficient delivery of component proteins or plasmid DNA into cells, have expanded the scope and application of this technology.
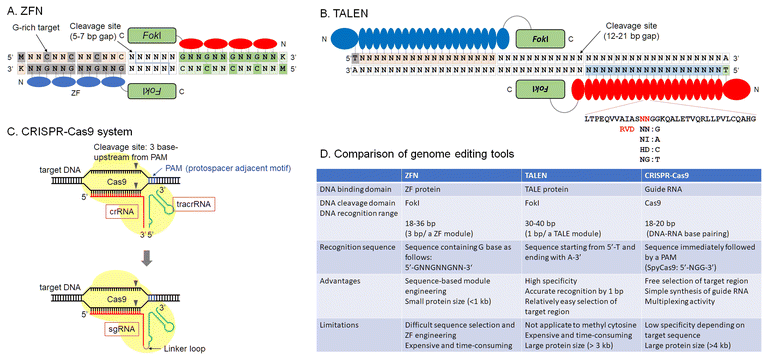 | ||
| Fig. 1 Overview of current genome editing tools, ZFN (A), TALEN (B), CRISPR-Cas9 system (C), and a comparison table of these tools (D). | ||
This review emphasizes current chemistry-driven methods for genome editing or engineering and epigenetic regulation. In addition, future directions of the development and applications of genome editing will be discussed.
2. A view of genome editing technologies from history
As mentioned above, rewriting DNA sequences allows us to understand how those genes work, paving the way for gene therapy. Restriction enzymes derived from bacteria were the first enzymes used to manipulate DNA. Most restriction enzymes recognize short palindromic DNA sequences to cleave a target DNA. In mammalian cells, it is difficult to modify DNA because restriction enzymes recognize short sequences, cleaving many sites. For example, a restriction enzyme that recognizes a 6-base-pair DNA sequence can cleave approximately 7.5 × 106 sites in 3 × 109 base pairs of human genomic DNA. On the other hand, a meganuclease that recognizes 18 non-palindromic base pairs, such as Sce-I, derived from mitochondria in Saccharomyces cerevisiae, can computationally cleave only one site in human genomic DNA. However, the recognition sequence of Sce-I is fixed. This makes it difficult to edit arbitrary DNA sequences using such natural enzymes and highlights the importance of developing DNA-cleaving techniques for long, non-palindromic sequences. In this section, the development of technology for creating DNA double-strand breaks is discussed. The main milestones of DNA double-strand break technology, currently called genome editing, are zinc finger nucleases (ZFNs), transcription activator-like effector nuclease (TALEN), and CRISPR-Cas9. However, efforts to develop reagents including protein or nucleic acid-based enzymes and small organic compounds for DNA double-strand breaks have continued.Zinc-finger proteins (ZFs) were one of the pioneering platforms for genome engineering/editing (Fig. 1A). The first zinc-finger protein, TFIIIA, was discovered by Klug et al. in Xenopus laevis.16,17 In the early 1990's, accelerated development of X-ray crystallographic techniques helped to reveal protein structures.18–21 Complex structures of DNA-binding proteins and their target DNAs were the main targets of these projects.22 These structures provided detailed insights into the DNA recognition mode of zinc-finger proteins. At about that same time, phage display was developed to engineer potent monoclonal antibodies.23–25 Using methods of molecular evolution, customization of zinc-finger domains for target sequence recognition became possible.26–30 Customized zinc-finger domains were utilized for various applications, such as artificial transcription regulators or DNA-modification enzymes. In particular, ZFN, fusions with FokI catalytic domains, can be used to introduce indels into mammalian cell genomes. Their use constituted a major early milestone in the development of genome editing technologies.5,7,31
Zinc-finger domains recognize triplet nucleotides or codons. Minimum binding affinity required for DNA recognition results from a pair of ZF domains, whereas artificially designed single ZF domains with cationic polypeptide chains or tail structures based on the GATA-1 zinc finger can bind to target sequences.32 The rule of three base pairs per ZF module has made the construction of programmable ZF domains difficult because ZF–DNA-base interactions are not truly modular, but there is another important interaction with the complimentary strand having Asp in the −2 position in the alpha helix of ZF modules.
TALE domains were regarded as a promising option for genome editing tools (Fig. 1B).33,34 DNA recognition by TALE domains is simple because single RVD domains recognize single base pairs. This more accessible technology expanded genome editing research.
After the report of CRISPR-Cas9,35,36 the genome editing community underwent a large expansion (Fig. 1C). Designable and programmable sgRNAs for any target sequences in the genome reduced time and cost for gene knockout or knockdown, and sequence conversion. CRISPR-Cas9 and other Cas family proteins have been developed for efficient genome editing or gene modification. They require only template sgRNAs to target sequences; thus, the time from experimental design until results was greatly shortened. As multiple sgRNA templates can be encoded in a single plasmid, multiplex genome editing in a single cell is easily performed.37,38
DNA double-stranded break technology can be traced back to the pre-ZFN period. DNA cleaving reagents were important tools for functional analysis of antibiotics or anticancer reagents39–41 and they led to the development of artificial restriction enzymes.42,43 Analysis of DNA binding and cleaving reagents were important topics in the 1980's in bioorganic and bioinorganic chemistry. Once structural and DNA-selective properties of zinc-finger domains were known, sequence selectivity to reduce cytotoxicity became manageable. Zinc-finger domains are very small and are expected to be non-immunogenic when used in cells, because of the abundance of zinc finger family proteins in eukaryotes. In addition, zinc finger domains can autonomously penetrate mammalian cells.44 Although the Cas9/sgRNA complex can be delivered in ribonucleoprotein form by electroporation of cells or embryos, direct delivery of ZF proteins will offer large advantages, even in this CRISPR era.
3. Programmable genome editing or gene regulation by chemical compounds
Genome editing tools can be used to edit desired DNA sequences in genomic DNA. However, these tools are still not perfect and sometimes cut unintended DNA sequences that are similar to the target DNA sequence. Such unintended cleavages induce mutations at unexpected sites and are known as off-target effects. In many cases, off-target effects are caused by high levels of active nucleases in the nucleus. This section describes the state-of-the-art in chemical control of genome editing and gene regulation. Although this technology is protein-based, activity can be specifically controlled by binding of naturally occurring or designed small molecules. Control of enzymatic activity related to genome editing is important so as to avoid off-target effects. Double-strand breaks in DNA effected with nucleases engage a repair pathway called non-homologous end joining (NHEJ), which leads to indel mutagenesis at target sites. Enzymatic activity can be regulated chemically at some stages, such as enzyme translocation to the nucleus, protein folding, or more generally, protein production stages.45–48 For protein function regulation, the time lag between chemical stimulation and activation of protein function should be small. Therefore, regulation of translation and protein folding should be favored over regulation of protein expression. Regulation of translocation was reported by Barbas et al. using activation of a hormone receptor called estrogen receptor T2 (ERT2), leading to the translocation of fusion proteins into the nucleus (Fig. 2A).49 This system was also applied to Cas9.50,51 ERT2 is kept outside the nucleus by heat shock protein 90 (HSP90. After the addition of 4-hydroxytamoxifen (4-OHT), HSP90 binding disappears and the ERT2 fusion nuclease rapidly relocates into the nucleus. Inhibition of nuclease activity by altering folding could include insertion of external sequences, such as intein, which are excised by the addition of 4-OHT52 or division of protein sequences into split fragments, that can be fused using chemically inducible dimerization.53–56 Chemical limitation of protein activity can avoid the constitutive activity of nucleases, lowering the probability of off-target cleavage. Another approach is stabilization of nucleases using fused degradation domains.57–60 In this case, the degradation domain is stabilized by the addition of chemicals, such as Shield, for DD domains (Clontech), which utilize fast degradation of mutated FKBP protein. Limiting the duration of stabilized nucleases can control the catalytic activity of enzymes, reducing off-target effects (Fig. 2A).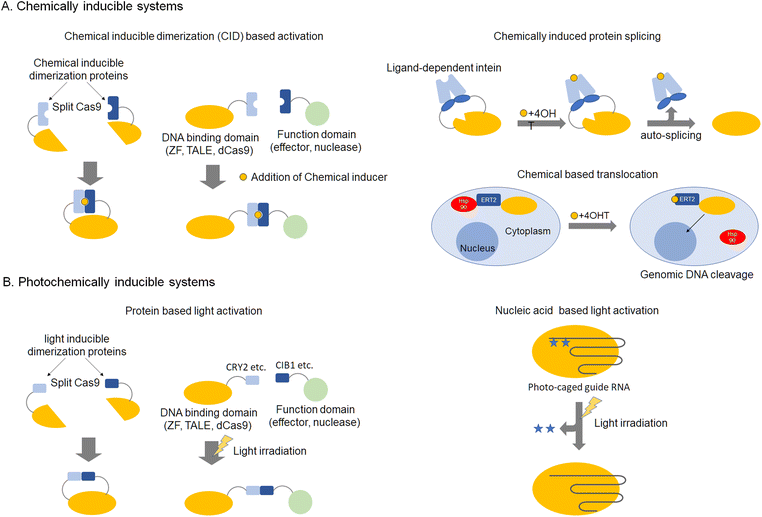 | ||
| Fig. 2 Inducible genome editing technologies, chemically inducible systems (A) and photochemically inducible systems (B). | ||
4. Programmable genome editing or gene regulation by photochemistry
This section describes the application of artificial regulation of protein function by photochemistry to genome editing and gene regulation. While the previous section discussed chemical control, photochemical control has the advantage that the scope of action can be limited to the cell or used partially in vivo. Association of photo-responsive proteins as a switch to turn on/off activity of nucleases with UV irradiation or visible light, can control the association and dissociation of protein domains, respectively (Fig. 2B). As these reactions are reversible, they can be utilized for optical control of gene expression by ZF-, TALE-, and dCas9-based artificial transcription factors.61–63 For a genomic anchor, a customizable DNA-binding domain (DBD) fused to a light-sensitive cryptochrome 2 (CRY2) protein from Arabidopsis thaliana (DBD-CRY2) is utilized. The interacting partner of CRY2, CIB1, is fused to a desired effector domain (CIB1-effector). In the absence of light, DBD-CRY2 binds the promoter region of the target gene, while the CIB1-effector remains free in the nuclear compartment. Illumination with UV light triggers a conformational change of CRY2, which subsequently recruits a CIB1-effector.61,62,64,65 Other light inducible domains such as pMag-nMag63,66 and trCIB1-CRY2PHR64 have also been reported. Light activation of protein function avoids cytotoxicity, and areas for activation in tissues or cells can be precisely regulated.Another photo-responsive genome editing method is the use of caged-amino acids incorporated into the nuclease domain. In the case of ZFN, modification of the hydroxyl group of tyrosine in the catalytic core of the nuclease domain with an ortho-nitrobenzyl group was reported.67 Incorporation of a caged amino acid was also demonstrated for Cas9.68 The protecting group inhibits catalysis by the enzyme, but photolysis of the protecting group with UV irradiation activates the nuclease, resulting in DNA double-strand breaks at the target sequences. A light-activated system can also be constructed by incorporating photocleavable oligonucleotides that complement the target regions of the sgRNA in the absence of Cas9 modifications69–71 (Fig. 2B).
5. Efficient protein delivery by chemically-modified or designed vehicles for genome editing
Delivery of expression plasmids or nuclease proteins into nuclei is a key step for efficient genome editing and reduction of off-target effects. The probability of off-target effects can be reduced by controlling the degradation rates and amounts of functional nucleases in cells. As mentioned above, direct protein-delivery is one of the most promising approaches to optimize the amount of plasmid or protein in cells (Fig. 3). For ZF-based enzymes, autonomous cell penetration is observed because of the cationic charges of amino acid side chains of ZF (Fig. 3A).44 Cationic charges of protein surfaces are important for cell penetration and stabilization, as demonstrated by the construction of super-charged EGFP.72 Cre recombinase or TALEN, on the other hand, can fuse with negatively super-charged EGFP to reduce its net charge and to form a complex with cationic lipids for delivery. In the case of Cas9, complex sgRNAs have enough negative charges to form complexes with cationic polymers.73 There are several reports of Cas9 protein as a platform for delivery. The first approach used to deliver Cas9 protein directly into cells was electroporation (Fig. 3B).74 Recombinant Cas9 is complexed with in vitro transcribed sgRNA. Direct delivery of Cas9–sgRNA ribonucleoprotein (RNP) edits target genes with nearly the same efficiency as plasmid delivery and with fewer off-target effects. Gold nanoparticles would also be a viable delivery option (Fig. 3C).75,76 Gold nanoparticles appear to be a safe drug delivery method in vivo because they are biocompatible and non-toxic. However, the release of proteins from particles after delivery into cells could be a major hurdle to optimizing the technology. Rotello et al. demonstrated that optimized surface modification is the key for protein delivery, utilizing the Cas9–sgRNA complex.75 A negatively charged tag called E-tag was attached to Cas9, enabling it to bind with Arginine modified gold nanoparticles via electrostatic interaction. Efficient delivery of Cas9 into culture cells has been achieved by delivery design. In addition, there are some reports of Cas9 protein delivery using nanotechnologies, such as cell squeezing with microfluidics,77,78 acoustoporation,79 and nano-needles80,81 (Fig. 3D). A common feature of these methods is instantaneous physical disruption of the cell membrane. Such direct delivery, which is not mediated by endocytosis, can efficiently deliver biomolecules into the cytoplasm because there is no need for them to escape from endosomes. Although these methods still need improvements, such as better delivery efficiency and reduced cytotoxicity, these problems will be solved in the near future because of the rapid development of nanomaterials in recent years.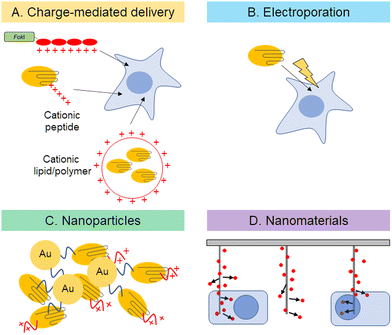 | ||
| Fig. 3 Description of direct delivery of genome editing tools into cells by charge-mediated delivery (A), electroporation (B), nanoparticles (C), and nanomaterials (D). | ||
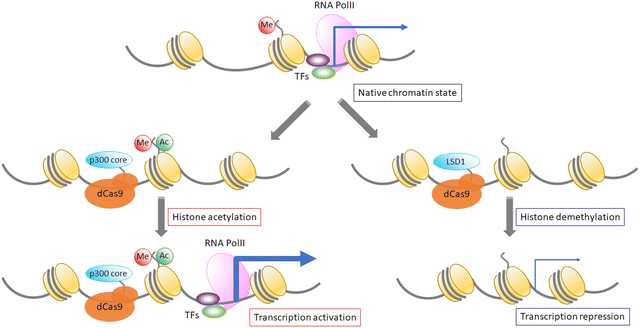
6. Epigenome editing by modification of DNA or proteins using “EpiEffectors”
Epigenetic modification of DNA or proteins is important to control gene functions. Variable modifications of histone core proteins can synergistically affect the outcome of gene regulation. Histone modification proteins such as acetyltransferases, deacetylases, methyltransferases, and demethylases are used by fusing them with dCas9 protein. An acetyltransferase, p300, fused with dCas9 can induce site-specific acetylation of H3K27 at the hypersensitive site 2 (HS2) enhancer region in the IL1RN, MYOD, and OCT4 promoters, resulting in drastic enhancement of gene expression in human cells.82 Moreover, LSD1 histone methyltransferase fused with a TALE domain83 or dCas984 represses target gene expression, and it has been shown that dCas9-LSD1 works more efficiently than dCas9-fused Krüppel-associated box (KRAB), which is a common transcription repressor. On the other hand, repressive histone modifications are not sufficient for gene repression because such repression is not correlated with the deposition of either H3K27me3 or H3K27me3.85 Further analysis and development are required for precise control of targeted gene expression. It has been estimated that about 70% of CpG sequences are methylated in mammalian cells. DNA methylation promotes histone deacetylation, and heterochromatin formation represses gene expression.86,87 As DNA methylation patterns are maintained after cell division, DNA methylation is important to maintain gene expression patterns in cells, and artificial changes of methylation patterns could induce heritable changes of expression patterns. Recently, the pathway of cytosine demethylation has been determined, and the functions of TET family proteins, which catalyze demethylation reactions, have been addressed.88,89 The Jaenisch group showed that dCas9-Tet1 (for DNA demethylation) and dCas9-Dnmt3a (for DNA methylation) control the methylation level of a specific promoter and the expression level of the target gene (Fig. 4).90 Another way to apply DNA methylase to target methylation is to use split DNA methylase. Two split fragments of methylase are fused with a ZF domain and methylase fragments are reassembled after target sequence binding by ZF domains,9,10 resulting in tight control of DNA methylation. This approach can control the CpG methylation pattern with single-nucleotide resolution, which is more precise than direct fusion of full-length methylase with DNA-binding domains, such as ZF, TALE, and dCas9 (Fig. 4).7. Next-generation genome editing tools, the base editor and the prime editor
This section describes breakthroughs in genome editing technology that explore the possibility of alternative sequences for target genes other than DNA double-strand breaks. Most human genetic variants associated with disease result from single point mutations.91,92 The safety of clinical applications could be improved if it were possible to edit point mutations without DNA cleavage. The Liu group first reported a base editor that mediates the direct conversion of cytidine to uridine (Fig. 5A).93 Nishida et al. also reported a different cytidine deaminase called PmCDA1 from sea lamprey fused with nickase Cas9 (nCas9) called “Target-AID” (Fig. 5A).94 They showed that the combination of DNA nicking activity and deamination by Target-AID was efficient in yeast and the use of uracil DNA glycosylase inhibitor improved the efficiency. Liu et al. added an adenine base editor that can convert A-T base pairs into G-C base pairs to their repertoire (Fig. 5B).95,96 TadA* was obtained by molecular evolution of an adenosine deaminase called TadA in E. coli, which is usually fused with nCas9 along with wild type TadA. Now, base editors can mediate all four possible transition mutations. Several groups improved these base editors and their applications for medical uses, as in base substitution in human cells97–101 and bio-industrial uses, such as base substitution in bacteria102–114 and plants.115–123 Activity controlling methods described above, such as chemical or photo-activation and exogenous small molecules, increase the editing specificity and advance the use of base editors in various fields. Base editors cannot currently introduce transversion point mutations. The Liu group reported a new method to edit genes, called prime editor, by combining Cas9 nickase with reverse transcriptase and using prime editing guide RNA (pegRNA), which links single-guide RNA (sgRNA) with a primer template for reverse transcription at the 3’ end of sgRNA (Fig. 5C).124 With this method, the RNA primer template region of pegRNA hybridizes with a nicked non-targeted DNA and the target DNA serves as a primer for the reverse transcriptase, inserting a mutation by reverse transcription of the pegRNA template site, which contains a targeted mutation for editing. After the introduction of a mutation in one strand of the target DNA, a corresponding mutation is inserted in the other strand by the DNA repair pathway. Liu et al. showed that the prime editor can introduce transversion point mutations in mammalian cells. This unique property of prime editing makes it a promising tool for genome modification in various species.125–127 As the prime editor complements single point mutations of the base editor, next-generation gene editors could be powerful tools in clinical applications to treat genetic diseases caused by single point mutations.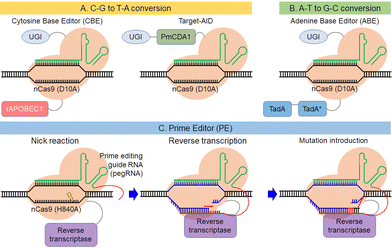 | ||
| Fig. 5 Description of the base editors, C-G to T-A conversion (A) and A-T to G-C conversion (B), and the prime editor (C). | ||
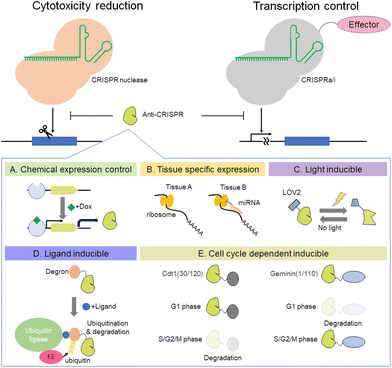
8. Use of anti-CRISPR for regulation of Cas activity
Recently, the existence of anti-CRISPR molecules was discovered.128,129 This section focuses on the functions of anti-CRISPR proteins, mainly inhibition of CRISPR-Cas activity and its use to improve the accuracy of genome editing. For inhibition of SpCas9, the anti-CRISPR from Listeria monocytogenes prophages is very effective.130 The crystal structure of AcrIIA4–SpCas9–sgRNA shows that AcrIIA4 binds to the SpCas9–sgRNA complex through the protospacer adjacent motif (PAM) interaction site and the RuvC domain.131–134 AcrIIA4 strongly binds to SpCas9 as an sgRNA complex (KD ∼ 0.6 nM), whereas interaction with apo-SpCas9 is very weak (KD ∼ 4.8 μM).134 The utility of AcrIIA4 inhibitory activity against SpCas9 has been shown in the field of synthetic biology (Fig. 6). The Lei group has demonstrated that combinations of anti-CRISPR, including AcrII4 and transcription mediators, such as dCas9-VPR and dCas9-KRAB, can control gene expression precisely in yeast and mammalian cells.135 In addition, Davidson et al. found that transcription of anti-CRISPR proteins is regulated by an anti-CRISPR-associated gene (aca) that encodes an anti-CRISPR operon-binding protein.136 By using an anti-CRISPR control system that utilizes spontaneous transcription in conjunction with a gene expression regulator, it is possible to develop more complex logic gates. Another way to use anti-CRISPR is to control Cas9 cleavage activity for precise editing. The Niopek group reported that fusion proteins of SpCas9 with AcrIIA4 variants D14A/G38A, ins5, and N39A reduce the off-target mutation rate while maintaining target gene editing efficiency.137 The equilibrium between the bound and unbound states of the AcrIIA4 variant to Cas9 regulates Cas9 activity autonomously. Anti-CRISPR-based regulation of Cas9 activity is applicable not only to this equilibrium-based method, but also to chemical expression control (Fig. 6A), tissue-specific expression by miRNAs138 (Fig. 6B), and optogenetic control139 (Fig. 6C and D). Moreover, we recently reported that cell cycle-dependent expression of AcrIIA4 fused with human Cdt1 can make genome editing with SpCas9 more precise (Fig. 6E).140 This Cdt1-based anti-CRISPR expression regulation is also applicable to AcrIIA5, and when combined with Cas9-Geminin, it shows a synergetic effect on precise genome editing.141 Studies utilizing anti-CRISPR for precision genome editing by combining it with light- or chemically inducible methods or nucleic acid-mediated control systems such as aptamer and miRNA will enhance the possibility of anti-CRISPR applications with CRISPR-Cas systems. New anti-CRISPRs against many types of Cas protein have been reported continuously since the appearance of the first anti-CRISPR.142–145 For example, Type II anti-CRISPRs, such as the AcrII-A subtype146–151 and the AcrII-C subtype152–154 inhibit Cas9, and Type V anti-CRISPRs, such as the AcrV-A subtype, inhibit Cas12a.144,155,156 The use of anti-CRISPRs and Cas protein will continue to spread not only for the purposes of DNA cleavage, but also RNA cleavage, in concert with base editors, prime editor, and Epi-effectors.9. Conclusions
The CRISPR-Cas9 system has made genome editing more active and competitive. Not only CRISPR-Cas9, but also other Cas orthologs are being applied to gene editing of eukaryotic cells. Moreover, the development of various CRISPR-Cas-based tools, such as gene regulator, epi-effector, base editor, and prime editor, will continue to develop variations of CRISPR-Cas platforms for applications in other research fields. To solve the remaining problems or to enhance the functions of genome editing tools, such as improving target editing efficiency, reducing off-target effects, and improving the efficiency of tissue-specific delivery methods, one of the most important ways could be collaborations between physicians and chemists. Now that we have technologies to edit genomes of many organisms, including humans, we need to carefully consider how to use this technology properly, from ethical and political viewpoints. Moreover, it is important that scientists continue to improve genome editing technology in keeping with governmental and ethical restrictions.Author contributions
D. M. and W. N. wrote and edited the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
We thank Dr Steven D. Aird for English language editing. This work was supported in part by the New Energy and Industrial Technique Development Organization (NEDO) of Japan, the Japan Society for the Promotion of Science (JSPS) KAKENHI (JP22H02201, JP19H02827 and JP20K21253 to WN, JP21K14740 to DM), and Grants-in-Aid for Scientific Research on Innovative Areas (JP16H01420 (Resonance Bio) to WN), funding from Core Research for Organelle Diseases at Hiroshima University (the MEXT program to enhance Japanese research universities), HIRAKU-Global Program which is funded by MEXT's “Strategic Professional Development Program for Young Researchers” (to DM), funding from Takeda Science Foundation, the Naito Foundation, the Uehara Memorial Foundation, the Mochida Memorial Foundation, and the Suzuken Memorial Foundation (to WN).References
- B. Sauer, Mol. Cell. Biol., 1987, 7, 2087–2096 CAS.
- N. Sternberg, B. Sauer, R. Hoess and K. Abremski, J. Mol. Biol., 1986, 187, 197–212 CrossRef CAS PubMed.
- A. M. Campbell, Adv. Genet., 1962, 11, 101–145 Search PubMed.
- A. Jacquier and B. Dujon, Cell, 1985, 41, 383–394 CrossRef CAS PubMed.
- Y. G. Kim, J. Cha and S. Chandrasegaran, Proc. Natl. Acad. Sci. U. S. A., 1996, 93, 1156–1160 CrossRef CAS PubMed.
- M. Bibikova, M. Golic, K. G. Golic and D. Carroll, Genetics, 2002, 161, 1169–1175 CrossRef CAS PubMed.
- M. Bibikova, K. Beumer, J. K. Trautman and D. Carroll, Science, 2003, 300, 764 CrossRef CAS PubMed.
- D. Carroll, Gene Ther., 2008, 15, 1463–1468 CrossRef CAS PubMed.
- W. Nomura and C. F. Barbas, J. Am. Chem. Soc., 2007, 129, 8676–8677 CrossRef CAS PubMed.
- G. E. Meister, S. Chandrasegaran and M. Ostermeier, Biochem. Biophys. Res. Commun., 2008, 377, 226–230 CrossRef CAS PubMed.
- G. E. Meister, S. Chandrasegaran and M. Ostermeier, Nucleic Acids Res., 2010, 38, 1749–1759 CrossRef CAS PubMed.
- B. Chaikind and M. Ostermeier, PLoS One, 2014, 9, e96931 CrossRef PubMed.
- M. R. Grimmer, S. Stolzenburg, E. Ford, R. Lister, P. Blancafort and P. J. Farnham, Nucleic Acids Res., 2014, 42, 10856–10868 CrossRef CAS PubMed.
- R. R. Beerli, D. J. Segal, B. Dreier and C. F. Barbas, Proc. Natl. Acad. Sci. U. S. A., 1998, 95, 14628–14633 CrossRef CAS PubMed.
- D. J. Segal and C. F. Barbas, 3rd, Curr. Opin. Biotechnol, 2001, 12, 632–637 CrossRef CAS PubMed.
- J. Miller, A. D. McLachlan and A. Klug, EMBO J., 1985, 4, 1609–1614 CrossRef CAS PubMed.
- A. Klug and D. Rhodes, Cold Spring Harbor Symp. Quant. Biol., 1987, 52, 473–482 CrossRef CAS PubMed.
- W. H. Bragg, Nature, 1913, 90, 360–361 CrossRef.
- W. H. Bragg and W. L. Bragg, Proc. R. soc. Lond. Ser. A-Contain. Pap. Math., 1913, 88, 428 CAS.
- W. L. Bragg and W. H. Bragg, Proc. R. soc. Lond. Ser. A-Contain. Pap. Math., 1913, 89, 248–277 CAS.
- J. M. Thomas, Angew. Chem., Int. Ed., 2012, 51, 12946–12958 CrossRef CAS PubMed.
- N. P. Pavletich and C. O. Pabo, Science, 1991, 252, 809–817 CrossRef CAS PubMed.
- G. P. Smith, Science, 1985, 228, 1315–1317 CrossRef CAS PubMed.
- J. K. Scott and G. P. Smith, Science, 1990, 249, 386–390 CrossRef CAS PubMed.
- D. Corp, Chem. Biol., 1997, 4, 977–978 CrossRef PubMed.
- Y. Choo and A. Klug, Proc. Natl. Acad. Sci. U. S. A., 1994, 91, 11168–11172 CrossRef CAS PubMed.
- E. J. Rebar and C. O. Pabo, Science, 1994, 263, 671–673 CrossRef CAS PubMed.
- H. Wu, W. P. Yang and C. F. Barbas, 3rd, Proc. Natl. Acad. Sci. U. S. A., 1995, 92, 344–348 CrossRef CAS PubMed.
- E. J. Rebar, H. A. Greisman and C. O. Pabo, Methods Enzymol., 1996, 267, 129–149 CAS.
- S. A. Wolfe, H. A. Greisman, E. I. Ramm and C. O. Pabo, J. Mol. Biol., 1999, 285, 1917–1934 CrossRef CAS PubMed.
- M. H. Porteus and D. Baltimore, Science, 2003, 300, 763 CrossRef PubMed.
- J. G. Omichinski, C. Trainor, T. Evans, A. M. Gronenborn, G. M. Clore and G. Felsenfeld, Proc. Natl. Acad. Sci. U. S. A., 1993, 90, 1676–1680 CrossRef CAS PubMed.
- J. Boch, H. Scholze, S. Schornack, A. Landgraf, S. Hahn, S. Kay, T. Lahaye, A. Nickstadt and U. Bonas, Science, 2009, 326, 1509–1512 CrossRef CAS PubMed.
- M. J. Moscou and A. J. Bogdanove, Science, 2009, 326, 1501 CrossRef CAS PubMed.
- G. Gasiunas, R. Barrangou, P. Horvath and V. Siksnys, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E2579–2586 CrossRef CAS PubMed.
- M. Jinek, K. Chylinski, I. Fonfara, M. Hauer, J. A. Doudna and E. Charpentier, Science, 2012, 337, 816–821 CrossRef CAS PubMed.
- L. Cong, F. A. Ran, D. Cox, S. L. Lin, R. Barretto, N. Habib, P. D. Hsu, X. B. Wu, W. Y. Jiang, L. A. Marraffini and F. Zhang, Science, 2013, 339, 819–823 CrossRef CAS PubMed.
- T. Sakuma, A. Nishikawa, S. Kume, K. Chayama and T. Yamamoto, Sci. Rep., 2014, 4, 5400 CrossRef CAS PubMed.
- P. G. Schultz and P. B. Dervan, Proc. Natl. Acad. Sci. U. S. A., 1983, 80, 6834–6837 CrossRef CAS PubMed.
- M. W. Van Dyke, R. P. Hertzberg and P. B. Dervan, Proc. Natl. Acad. Sci. U. S. A., 1982, 79, 5470–5474 CrossRef CAS PubMed.
- Y. Sugiura, Y. Uesawa, Y. Takahashi, J. Kuwahara, J. Golik and T. W. Doyle, Proc. Natl. Acad. Sci. U. S. A., 1989, 86, 7672–7676 CrossRef CAS PubMed.
- Y. Sugiura and T. Suzuki, J. Biol. Chem., 1982, 257, 10544–10546 CrossRef CAS PubMed.
- P. B. Dervan and B. F. Baker, Ann. N. Y. Acad. Sci., 1986, 471, 51–59 CrossRef CAS PubMed.
- T. Gaj, J. Guo, Y. Kato, S. J. Sirk and C. F. Barbas, Nat. Methods, 2012, 9, 805–807 CrossRef CAS PubMed.
- F. Gonzalez, Z. Zhu, Z. D. Shi, K. Lelli, N. Verma, Q. V. Li and D. Huangfu, Cell Stem Cell, 2014, 15, 215–226 CrossRef CAS PubMed.
- T. Wang, J. J. Wei, D. M. Sabatini and E. S. Lander, Science, 2014, 343, 80–84 CrossRef CAS PubMed.
- B. J. Aubrey, G. L. Kelly, A. J. Kueh, M. S. Brennan, L. O'Connor, L. Milla, S. Wilcox, L. Tai, A. Strasser and M. J. Herold, Cell Rep., 2015, 10, 1422–1432 CrossRef CAS PubMed.
- L. E. Dow, J. Fisher, K. P. O'Rourke, A. Muley, E. R. Kastenhuber, G. Livshits, D. F. Tschaharganeh, N. D. Socci and S. W. Lowe, Nat. Biotechnol., 2015, 33, 390–394 CrossRef CAS PubMed.
- R. R. Beerli, U. Schopfer, B. Dreier and C. F. Barbas, 3rd, J. Biol. Chem., 2000, 275, 32617–32627 CrossRef CAS PubMed.
- K. I. Liu, M. N. Ramli, C. W. Woo, Y. Wang, T. Zhao, X. Zhang, G. R. Yim, B. Y. Chong, A. Gowher, M. Z. Chua, J. Jung, J. H. Lee and M. H. Tan, Nat. Chem. Biol., 2016, 12, 980–987 CrossRef CAS PubMed.
- D. P. Nguyen, Y. Miyaoka, L. A. Gilbert, S. J. Mayerl, B. H. Lee, J. S. Weissman, B. R. Conklin and J. A. Wells, Nat. Commun., 2016, 7, 12009 CrossRef CAS PubMed.
- K. M. Davis, V. Pattanayak, D. B. Thompson, J. A. Zuris and D. R. Liu, Nat. Chem. Biol., 2015, 11, 316–318 CrossRef CAS PubMed.
- B. Zetsche, S. E. Volz and F. Zhang, Nat. Biotechnol., 2015, 33, 139–142 CrossRef CAS PubMed.
- J. Lu, C. Zhao, Y. Zhao, J. Zhang, Y. Zhang, L. Chen, Q. Han, Y. Ying, S. Peng, R. Ai and Y. Wang, Nucleic Acids Res., 2018, 46, e25 CrossRef PubMed.
- W. Nomura, D. Matsumoto, T. Sugii, T. Kobayakawa and H. Tamamura, Biochemistry, 2018, 57, 6452–6459 CrossRef CAS PubMed.
- D. Matsumoto, H. Tamamura and W. Nomura, Biochemistry, 2020, 59, 197–204 CrossRef CAS PubMed.
- S. M. Pruett-Miller, D. W. Reading, S. N. Porter and M. H. Porteus, PLoS Genet., 2009, 5, e1000376 CrossRef PubMed.
- D. Balboa, J. Weltner, S. Eurola, R. Trokovic, K. Wartiovaara and T. Otonkoski, Stem Cell Rep., 2015, 5, 448–459 CrossRef CAS PubMed.
- B. Maji, C. L. Moore, B. Zetsche, S. E. Volz, F. Zhang, M. D. Shoulders and A. Choudhary, Nat. Chem. Biol., 2017, 13, 9–11 CrossRef CAS PubMed.
- S. Senturk, N. H. Shirole, D. G. Nowak, V. Corbo, D. Pal, A. Vaughan, D. A. Tuveson, L. C. Trotman, J. B. Kinney and R. Sordella, Nat. Commun., 2017, 8, 14370 CrossRef CAS PubMed.
- L. R. Polstein and C. A. Gersbach, J. Am. Chem. Soc., 2012, 134, 16480–16483 CrossRef CAS PubMed.
- S. Konermann, M. D. Brigham, A. Trevino, P. D. Hsu, M. Heidenreich, L. Cong, R. J. Platt, D. A. Scott, G. M. Church and F. Zhang, Nature, 2013, 500, 472–476 CrossRef CAS PubMed.
- Y. Nihongaki, F. Kawano, T. Nakajima and M. Sato, Nat. Biotechnol., 2015, 33, 755–760 CrossRef CAS PubMed.
- Y. Nihongaki, S. Yamamoto, F. Kawano, H. Suzuki and M. Sato, Chem. Biol., 2015, 22, 169–174 CrossRef CAS PubMed.
- L. R. Polstein and C. A. Gersbach, Nat. Chem. Biol., 2015, 11, 198–200 CrossRef CAS PubMed.
- F. Kawano, H. Suzuki, A. Furuya and M. Sato, Nat. Commun., 2015, 6, 6256 CrossRef CAS PubMed.
- C. Chou and A. Deiters, Angew. Chem., Int. Ed., 2011, 50, 6839–6842 CrossRef CAS PubMed.
- J. Hemphill, E. K. Borchardt, K. Brown, A. Asokan and A. Deiters, J. Am. Chem. Soc., 2015, 137, 5642–5645 CrossRef CAS PubMed.
- P. K. Jain, V. Ramanan, A. G. Schepers, N. S. Dalvie, A. Panda, H. E. Fleming and S. N. Bhatia, Angew. Chem., Int. Ed., 2016, 55, 12440–12444 CrossRef CAS PubMed.
- E. V. Moroz-Omori, D. Satyapertiwi, M. C. Ramel, H. Hogset, I. K. Sunyovszki, Z. Q. Liu, J. P. Wojciechowski, Y. Y. Zhang, C. L. Grigsby, L. Brito, L. Bugeon, M. J. Dallman and M. M. Stevens, ACS Cent. Sci., 2020, 6, 695–703 CrossRef CAS PubMed.
- W. Y. Zhou, W. Brown, A. Bardhan, M. Delaney, A. S. Ilk, R. R. Rauen, S. I. Kahn, M. Tsang and A. Deiters, Angew. Chem., Int. Ed., 2020, 59, 8998–9003 CrossRef CAS PubMed.
- B. R. McNaughton, J. J. Cronican, D. B. Thompson and D. R. Liu, Proc. Natl. Acad. Sci. U. S. A., 2009, 106, 6111–6116 CrossRef CAS PubMed.
- J. A. Zuris, D. B. Thompson, Y. Shu, J. P. Guilinger, J. L. Bessen, J. H. Hu, M. L. Maeder, J. K. Joung, Z. Y. Chen and D. R. Liu, Nat. Biotechnol., 2015, 33, 73–80 CrossRef CAS PubMed.
- S. Kim, D. Kim, S. W. Cho, J. Kim and J. S. Kim, Genome Res., 2014, 24, 1012–1019 CrossRef CAS PubMed.
- R. Mout, M. Ray, G. Yesilbag Tonga, Y. W. Lee, T. Tay, K. Sasaki and V. M. Rotello, ACS Nano, 2017, 11, 2452–2458 CrossRef CAS PubMed.
- P. Wang, L. M. Zhang, Y. Z. Y. Xie, N. X. Wang, R. B. Tang, W. F. Zheng and X. Y. Jiang, Adv. Sci., 2017, 4, 1700175 CrossRef PubMed.
- A. Sharei, J. Zoldan, A. Adamo, W. Y. Sim, N. Cho, E. Jackson, S. Mao, S. Schneider, M. J. Han, A. Lytton-Jean, P. A. Basto, S. Jhunjhunwala, J. Lee, D. A. Heller, J. W. Kang, G. C. Hartoularos, K. S. Kim, D. G. Anderson, R. Langer and K. F. Jensen, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 2082–2087 CrossRef CAS PubMed.
- J. Yen, M. Fiorino, Y. Liu, S. Paula, S. Clarkson, L. Quinn, W. R. Tschantz, H. Klock, N. Guo, C. Russ, V. W. C. Yu, C. Mickanin, S. C. Stevenson, C. Lee and Y. Yang, Sci. Rep., 2018, 8, 16304 CrossRef PubMed.
- S. Yoon, P. Wang, Q. Peng, Y. Wang and K. K. Shung, Sci. Rep., 2017, 7, 5275 CrossRef PubMed.
- J. W. Sessions, C. S. Skousen, K. D. Price, B. W. Hanks, S. Hope, J. K. Alder and B. D. Jensen, SpringerPlus, 2016, 5, 1521 CrossRef PubMed.
- A. Yamagishi, D. Matsumoto, Y. Kato, Y. Honda, M. Morikawa, F. Iwata, T. Kobayashi and C. Nakamura, Appl. Sci., 2019, 9, 965 CrossRef CAS.
- I. B. Hilton, A. M. D'Ippolito, C. M. Vockley, P. I. Thakore, G. E. Crawford, T. E. Reddy and C. A. Gersbach, Nat. Biotechnol., 2015, 33, 510–517 CrossRef CAS PubMed.
- E. M. Mendenhall, K. E. Williamson, D. Reyon, J. Y. Zou, O. Ram, J. K. Joung and B. E. Bernstein, Nat. Biotechnol., 2013, 31, 1133–1136 CrossRef CAS PubMed.
- N. A. Kearns, H. Pham, B. Tabak, R. M. Genga, N. J. Silverstein, M. Garber and R. Maehr, Nat. Methods, 2015, 12, 401–403 CrossRef CAS PubMed.
- H. O'Geen, C. H. Ren, C. M. Nicolet, A. A. Perez, J. Halmai, V. M. Le, J. P. Mackay, P. J. Farnham and D. J. Segal, Nucleic Acids Res., 2017, 45, 9901–9916 CrossRef PubMed.
- A. Bird, Genes Dev., 2002, 16, 6–21 CrossRef CAS PubMed.
- R. Jaenisch and A. Bird, Nat. Genet., 2003, 33, 245–254 CrossRef CAS PubMed.
- K. D. Rasmussen and K. Helin, Genes Dev., 2016, 30, 733–750 CrossRef CAS PubMed.
- P. Melamed, Y. Yosefzon, C. David, A. Tsukerman and L. Pnueli, Front. Cell Dev. Biol., 2018, 6, 22 CrossRef PubMed.
- X. S. Liu, H. Wu, X. Ji, Y. Stelzer, X. B. Wu, S. Czauderna, J. Shu, D. Dadon, R. A. Young and R. Jaenisch, Cell, 2016, 167, 233–247 CrossRef CAS PubMed.
- M. J. Landrum, J. M. Lee, G. R. Riley, W. Jang, W. S. Rubinstein, D. M. Church and D. R. Maglott, Nucleic Acids Res., 2014, 42, D980–D985 CrossRef CAS PubMed.
- M. J. Landrum, J. M. Lee, M. Benson, G. Brown, C. Chao, S. Chitipiralla, B. Gu, J. Hart, D. Hoffman, J. Hoover, W. Jang, K. Katz, M. Ovetsky, G. Riley, A. Sethi, R. Tully, R. Villamarin-Salomon, W. Rubinstein and D. R. Maglott, Nucleic Acids Res., 2016, 44, D862–868 CrossRef CAS PubMed.
- A. C. Komor, Y. B. Kim, M. S. Packer, J. A. Zuris and D. R. Liu, Nature, 2016, 533, 420–424 CrossRef CAS PubMed.
- K. Nishida, T. Arazoe, N. Yachie, S. Banno, M. Kakimoto, M. Tabata, M. Mochizuki, A. Miyabe, M. Araki, K. Y. Hara, Z. Shimatani and A. Kondo, Science, 2016, 353, aaf8729 CrossRef PubMed.
- N. M. Gaudelli, A. C. Komor, H. A. Rees, M. S. Packer, A. H. Badran, D. I. Bryson and D. R. Liu, Nature, 2017, 551, 464–471 CrossRef CAS PubMed.
- N. M. Gaudelli, A. C. Komor, H. A. Rees, M. S. Packer, A. H. Badran, D. I. Bryson and D. R. Liu, Nature, 2018, 559, E8 CrossRef CAS PubMed.
- A. C. Chadwick, X. Wang and K. Musunuru, Arterioscler., Thromb., Vasc. Biol., 2017, 37, 1741–1747 CrossRef CAS PubMed.
- G. Li, Y. Liu, Y. Zeng, J. Li, L. Wang, G. Yang, D. Chen, X. Shang, J. Chen, X. Huang and J. Liu, Protein Cell, 2017, 8, 776–779 CrossRef CAS PubMed.
- P. Liang, C. Ding, H. Sun, X. Xie, Y. Xu, X. Zhang, Y. Sun, Y. Xiong, W. Ma, Y. Liu, Y. Wang, J. Fang, D. Liu, Z. Songyang, C. Zhou and J. Huang, Protein Cell, 2017, 8, 811–822 CrossRef CAS PubMed.
- C. Zhou, M. Zhang, Y. Wei, Y. Sun, Y. Sun, H. Pan, N. Yao, W. Zhong, Y. Li, W. Li, H. Yang and Z. J. Chen, Protein Cell, 2017, 8, 772–775 CrossRef CAS PubMed.
- Y. Zeng, J. Li, G. Li, S. Huang, W. Yu, Y. Zhang, D. Chen, J. Chen, J. Liu and X. Huang, Mol. Ther., 2018, 26, 2631–2637 CrossRef CAS PubMed.
- W. Chen, Y. Zhang, Y. Zhang, Y. Pi, T. Gu, L. Song, Y. Wang and Q. Ji, iScience, 2018, 6, 222–231 CrossRef CAS PubMed.
- T. N. Gu, S. Q. Zhao, Y. S. Pi, W. Z. Chen, C. Y. Chen, Q. Liu, M. Li, D. L. Han and Q. J. Ji, Chem. Sci., 2018, 9, 3248–3253 RSC.
- Y. Wang, Y. Liu, J. Liu, Y. M. Guo, L. W. Fan, X. M. Ni, X. M. Zheng, M. Wang, P. Zheng, J. B. Sun and Y. H. Ma, Metab. Eng., 2018, 47, 200–210 CrossRef CAS PubMed.
- K. Zheng, Y. Wang, N. Li, F. F. Jiang, C. X. Wu, F. Liu, H. C. Chen and Z. F. Liu, Commun. Biol., 2018, 1, 32 CrossRef PubMed.
- L. G. Huang, H. Z. Dong, J. W. Zheng, B. Wang and L. Pan, Microbiol. Res., 2019, 223, 44–50 CrossRef PubMed.
- Q. Li, F. M. Seys, N. P. Minton, J. Yang, Y. Jiang, W. Jiang and S. Yang, Biotechnol. Bioeng., 2019, 116, 1475–1483 CrossRef CAS PubMed.
- J. Tan, F. Zhang, D. Karcher and R. Bock, Nat. Commun., 2019, 10, 439 CrossRef CAS PubMed.
- Y. J. Tong, C. M. Whitford, H. L. Robertsen, K. Blin, T. S. Jorgensen, A. K. Klitgaard, T. Gren, X. L. Jiang, T. Weber and S. Y. Lee, Proc. Natl. Acad. Sci. U. S. A., 2019, 116, 20366–20375 CrossRef CAS PubMed.
- Y. Wang, Y. Liu, J. W. Li, Y. Yang, X. M. Ni, H. J. Cheng, T. Huang, Y. M. Guo, H. W. Ma, P. Zheng, M. Wang, J. B. Sun and Y. H. Ma, Biotechnol. Bioeng., 2019, 116, 3016–3029 CrossRef CAS PubMed.
- Y. Wang, Z. P. Wang, Y. Chen, X. T. Hua, Y. S. Yu and Q. J. Ji, Cell Chem. Biol., 2019, 26, 1732–1742 CrossRef CAS PubMed.
- S. J. Bae, B. G. Park, B. G. Kim and J. S. Hahn, Biotechnol. J., 2020, 15, e1900238 CrossRef PubMed.
- L. Cheng, D. Min, R. L. He, Z. H. Cheng, D. F. Liu and H. Q. Yu, Biotechnol. Bioeng., 2020, 117, 2389–2400 CrossRef CAS PubMed.
- Y. Luo, M. Ge, B. Wang, C. Sun, J. Wang, Y. Dong and J. J. Xi, Microb. Cell Fact., 2020, 19, 93 CrossRef CAS PubMed.
- J. Y. Li, Y. W. Sun, J. L. Du, Y. D. Zhao and L. Q. Xia, Mol. Plant, 2017, 10, 526–529 CrossRef CAS PubMed.
- Y. M. Lu and J. K. Zhu, Mol. Plant, 2017, 10, 523–525 CrossRef CAS PubMed.
- Z. Shimatani, S. Kashojiya, M. Takayama, R. Terada, T. Arazoe, H. Ishii, H. Teramura, T. Yamamoto, H. Komatsu, K. Miura, H. Ezura, K. Nishida, T. Ariizumi and A. Kondo, Nat. Biotechnol., 2017, 35, 441–443 CrossRef CAS PubMed.
- Y. Zong, Y. P. Wang, C. Li, R. Zhang, K. L. Chen, Y. D. Ran, J. L. Qiu, D. W. Wang and C. X. Gao, Nat. Biotechnol., 2017, 35, 438–440 CrossRef CAS PubMed.
- X. S. Li, Y. Wang, Y. J. Liu, B. Yang, X. Wang, J. Wei, Z. Y. Lu, Y. X. Zhang, J. Wu, X. X. Huang, L. Yang and J. Chen, Nat. Biotechnol., 2018, 36, 324–327 CrossRef CAS PubMed.
- B. Ren, F. Yan, Y. J. Kuang, N. Li, D. W. Zhang, X. P. Zhou, H. H. Lin and H. B. Zhou, Mol. Plant, 2018, 11, 623–626 CrossRef CAS PubMed.
- S. W. Tian, L. J. Jiang, X. X. Cui, J. Zhang, S. G. Guo, M. Y. Li, H. Y. Zhang, Y. Ren, G. Y. Gong, M. Zong, F. Liu, Q. J. Chen and Y. Xu, Plant Cell Rep., 2018, 37, 1353–1356 CrossRef CAS PubMed.
- F. Yan, Y. J. Kuang, B. Ren, J. W. Wang, D. W. Zhang, H. H. Lin, B. Yang, X. P. Zhou and H. B. Zhou, Mol. Plant, 2018, 11, 631–634 CrossRef CAS PubMed.
- F. Veillet, L. Perrot, L. Chauvin, M. P. Kermarrec, A. Guyon-Debast, J. E. Chauvin, F. Nogue and M. Mazier, Int. J. Mol. Sci., 2019, 20, 402 CrossRef PubMed.
- A. V. Anzalone, P. B. Randolph, J. R. Davis, A. A. Sousa, L. W. Koblan, J. M. Levy, P. J. Chen, C. Wilson, G. A. Newby, A. Raguram and D. R. Liu, Nature, 2019, 576, 149–157 CrossRef CAS PubMed.
- Q. Lin, Y. Zong, C. Xue, S. Wang, S. Jin, Z. Zhu, Y. Wang, A. V. Anzalone, A. Raguram, J. L. Doman, D. R. Liu and C. Gao, Nat. Biotechnol., 2020, 38, 582–585 CrossRef CAS PubMed.
- M. Marzec and G. Hensel, Trends Plant Sci., 2020, 25, 722–724 CrossRef CAS PubMed.
- J. Yan, A. Cirincione and B. Adamson, Mol. Cell, 2020, 77, 210–212 CrossRef CAS PubMed.
- A. L. Borges, A. R. Davidson and J. Bondy-Denomy, Annu. Rev. Virol., 2017, 4, 37–59 CrossRef CAS PubMed.
- S. Y. Stanley and K. L. Maxwell, Annu. Rev. Genet., 2018, 52, 445–464 CrossRef CAS PubMed.
- B. J. Rauch, M. R. Silvis, J. F. Hultquist, C. S. Waters, M. J. McGregor, N. J. Krogan and J. Bondy-Denomy, Cell, 2017, 168, 150–158.e10 CrossRef CAS PubMed.
- D. Dong, M. Guo, S. Wang, Y. Zhu, S. Wang, Z. Xiong, J. Yang, Z. Xu and Z. Huang, Nature, 2017, 546, 436–439 CrossRef CAS PubMed.
- J. Shin, F. Jiang, J. J. Liu, N. L. Bray, B. J. Rauch, S. H. Baik, E. Nogales, J. Bondy-Denomy, J. E. Corn and J. A. Doudna, Sci. Adv., 2017, 3, e1701620 CrossRef PubMed.
- H. Yang and D. J. Patel, Mol. Cell, 2017, 67(117–127), e115 Search PubMed.
- I. Kim, M. Jeong, D. Ka, M. Han, N. K. Kim, E. Bae and J. Y. Suh, Sci. Rep., 2018, 8, 3883 CrossRef PubMed.
- M. Nakamura, P. Srinivasan, M. Chavez, M. A. Carter, A. A. Dominguez, M. La Russa, M. B. Lau, T. R. Abbott, X. Xu, D. Zhao, Y. Gao, N. H. Kipniss, C. D. Smolke, J. Bondy-Denomy and L. S. Qi, Nat. Commun., 2019, 10, 194 CrossRef PubMed.
- S. Y. Stanley, A. L. Borges, K. H. Chen, D. L. Swaney, N. J. Krogan, J. Bondy-Denomy and A. R. Davidson, Cell, 2019, 178, 1452–1464.e13 CrossRef CAS PubMed.
- S. Aschenbrenner, S. M. Kallenberger, M. D. Hoffmann, A. Huck, R. Eils and D. Niopek, Sci Adv, 2020, 6, eaay0187 CrossRef CAS PubMed.
- M. Hirosawa, Y. Fujita and H. Saito, ACS Synth. Biol., 2019, 8, 1575–1582 CrossRef CAS PubMed.
- F. Bubeck, M. D. Hoffmann, Z. Harteveld, S. Aschenbrenner, A. Bietz, M. C. Waldhauer, K. Borner, J. Fakhiri, C. Schmelas, L. Dietz, D. Grimm, B. E. Correia, R. Eils and D. Niopek, Nat. Methods, 2018, 15, 924–927 CrossRef CAS PubMed.
- D. Matsumoto, H. Tamamura and W. Nomura, Commun. Biol., 2020, 3, 601 CrossRef CAS PubMed.
- D. Matsumoto, K. Kishi, E. Matsugi, Y. Inoue, K. Nigorikawa and W. Nomura, FEBS Lett., 2023, 597, 985–994 CrossRef CAS PubMed.
- J. Bondy-Denomy, A. Pawluk, K. L. Maxwell and A. R. Davidson, Nature, 2013, 493, 429–432 CrossRef CAS PubMed.
- N. D. Marino, J. Y. Zhang, A. L. Borges, A. A. Sousa, L. M. Leon, B. J. Rauch, R. T. Walton, J. D. Berry, J. K. Joung, B. P. Kleinstiver and J. Bondy-Denomy, Science, 2018, 362, 240–242 CrossRef CAS PubMed.
- K. E. Watters, C. Fellmann, H. B. Bai, S. M. Ren and J. A. Doudna, Science, 2018, 362, 236–239 CrossRef CAS PubMed.
- K. G. Wandera, S. P. Collins, F. Wimmer, R. Marshall, V. Noireaux and C. L. Beisel, Methods, 2020, 172, 42–50 CrossRef CAS PubMed.
- A. P. Hynes, G. M. Rousseau, M. L. Lemay, P. Horvath, D. A. Romero, C. Fremaux and S. Moineau, Nat. Microbiol., 2017, 2, 1374–1380 CrossRef CAS PubMed.
- A. P. Hynes, G. M. Rousseau, D. Agudelo, A. Goulet, B. Amigues, J. Loehr, D. A. Romero, C. Fremaux, P. Horvath, Y. Doyon, C. Cambillau and S. Moineau, Nat. Commun., 2018, 9, 2919 CrossRef PubMed.
- D. Ka, S. Y. An, J. Y. Suh and E. Bae, Nucleic Acids Res., 2018, 46, 485–492 CrossRef CAS PubMed.
- K. J. Forsberg, I. V. Bhatt, D. T. Schmidtke, K. Javanmardi, K. E. Dillard, B. L. Stoddard, I. J. Finkelstein, B. K. Kaiser and H. S. Malik, eLife, 2019, 8, e46540 CrossRef PubMed.
- L. Liu, M. Yin, M. Wang and Y. Wang, Mol. Cell, 2019, 73, 611–620.e3 CrossRef CAS PubMed.
- R. V. Uribe, E. van der Helm, M. A. Misiakou, S. W. Lee, S. Kol and M. O. A. Sommer, Cell Host Microbe, 2019, 26, 702 CrossRef CAS PubMed.
- J. Lee, A. Mir, A. Edraki, B. Garcia, N. Amrani, H. E. Lou, I. Gainetdinov, A. Pawluk, R. Ibraheim, X. D. Gao, P. Liu, A. R. Davidson, K. L. Maxwell and E. J. Sontheimer, mBio, 2018, 9, e02321-18 CrossRef PubMed.
- Y. Kim, S. J. Lee, H. J. Yoon, N. K. Kim, B. J. Lee and J. Y. Suh, FEBS J., 2019, 286, 4661–4674 CrossRef CAS PubMed.
- A. Thavalingam, Z. Cheng, B. Garcia, X. Huang, M. Shah, W. Sun, M. Wang, L. Harrington, S. Hwang, Y. Hidalgo-Reyes, E. J. Sontheimer, J. Doudna, A. R. Davidson, T. F. Moraes, Y. Wang and K. L. Maxwell, Nat. Commun., 2019, 10, 2806 CrossRef PubMed.
- L. Y. Dong, X. Y. Guan, N. N. Li, F. Zhang, Y. W. Zhu, K. Ren, L. Yu, F. X. Zhou, Z. F. Han, N. Gao and Z. W. Huang, Nat. Struct. Mol. Biol., 2019, 26, 308–314 CrossRef CAS PubMed.
- G. J. Knott, B. W. Thornton, M. J. Lobba, J. J. Liu, B. Al-Shayeb, K. E. Watters and J. A. Doudna, Nat. Struct. Mol. Biol., 2019, 26, 315–321 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2023 |