Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Supramolecular localization in liquid–liquid phase separation and protein recruitment in confined droplets†
Hiroki
Obayashi
 a,
Rie
Wakabayashi
a,
Rie
Wakabayashi
 *a,
Noriho
Kamiya
*a,
Noriho
Kamiya
 ab and
Masahiro
Goto
ab and
Masahiro
Goto
 ab
ab
aDepartment of Applied Chemistry, Graduate School of Engineering, Kyushu University, 744 Motooka, Nishi-ku, Fukuoka 819-0395, Japan. E-mail: wakabayashi.rie.122@m.kyushu-u.ac.jp
bCenter for Future Chemistry, Kyushu University, 744 Motooka, Nishi-ku, Fukuoka 819-0395, Japan
First published on 5th December 2022
Abstract
This study investigated the localization of artificial peptide supramolecular fibers in liquid–liquid phase separation (LLPS). Hierarchical organization led to the localization of supramolecules in LLPS droplets. Moreover, proteins were recruited into confined droplets by the physical adsorption of proteins on the supramolecules, enabling an enhanced cascade reaction.
Intracellular environments contain millions of biomolecules, resulting in crowded conditions.1 To achieve highly regulated intracellular chemical reactions in such a crowded cellular environment, each component must encounter the corresponding partner, such as enzymes and substrates, with appropriate timing and location.2,3 Recent studies have shown that cells utilize liquid–liquid phase separation (LLPS) to dynamically form liquid droplets that confine specific biomolecules, which regulates intracellular reactions for gene expression, metabolic regulation, signal transmission, and stress adaptation.4–6 This regulation is considered to be enabled not only by increased local biomolecule concentrations, but also by changes in the surrounding local environment of biomolecules in droplets, which affects the biophysical properties and/or interactions of biomolecules.
In addition to improving the understanding of naturally formed droplets, recent attempts to construct droplets that can localize specific biomolecules in cells have focused on controlling cellular functions.7,8 The formation of these droplets is driven by the association of biomolecules through oligomerization or coiled-coil interactions by introducing these associative motifs to biomolecules, and controlled recruitment and/or release of client biomolecules has been demonstrated.9,10 In parallel, the phase selectivity of biomolecules has been studied in vitro in LLPS systems using natural/synthetic polymers, such as polyethylene glycol (PEG) and dextran (Dex), to form cell-free droplets.11,12 In addition to the physicochemical properties, supramolecule formation by biomolecules, such as the polymerization of actin proteins and duplex formation of DNA, has been reported to affect the phase selectivity of biomolecules in LLPS, with higher-order structures, such as filamented actins and duplexed DNAs, favoring the Dex-rich phase over the PEG-rich phase.13,14 This suggests the possibility of controlling biomolecule localization by changing their organization and properties. However, whether this phenomenon is limited to biological supramolecules with defined structures and regulated organizations remains to be determined.
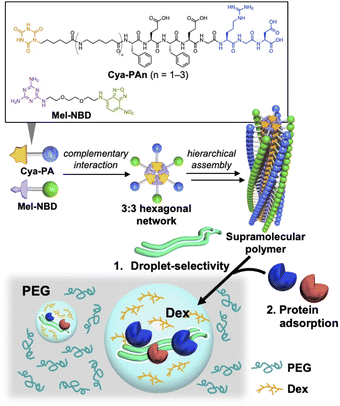
This study used artificial supramolecules based on peptide amphiphiles (PAs) to demonstrate that control of the phase selectivity through supramolecule formation can also be applied to an artificial system. Although the formation of LLPS at the very initial stage of peptide supramolecular polymerization,15 or the co-existence of peptide supramolecular fibers in LLPS droplets,16 has recently been reported using artificial peptides as a droplet component, this research uses macromolecular droplets formed by PEG and Dex to study the supramolecular organization and phase selectivity of artificial supramolecules independently from droplet formation. For this purpose, we used supramolecules formed by co-assembly of cyanuric acid (Cya)-introduced PAs (Cya-PAs) and melamine (Mel)-modified fluorescent molecule nitrobenzofurazan (Mel-NBD).17 Supramolecular formation is initiated by the 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 complementary hydrogen bond network between Cya and Mel, called a rosette, which leads to a hierarchical assembly to form supramolecular polymers.18 We previously reported that the sizes of the supramolecules were controlled by the number of C6 alkyl chains of Cya-PAs.17 Therefore, we studied the formation of supramolecules and size-dependent control of supramolecular localization in the LLPS system. In addition, the recruitment of native proteins into droplets was demonstrated through the physical adsorption of proteins on the supramolecules (Fig. 1).
3 complementary hydrogen bond network between Cya and Mel, called a rosette, which leads to a hierarchical assembly to form supramolecular polymers.18 We previously reported that the sizes of the supramolecules were controlled by the number of C6 alkyl chains of Cya-PAs.17 Therefore, we studied the formation of supramolecules and size-dependent control of supramolecular localization in the LLPS system. In addition, the recruitment of native proteins into droplets was demonstrated through the physical adsorption of proteins on the supramolecules (Fig. 1).
We first confirmed the formation of supramolecules in the absence of LLPS. Three Cya-PAs with different numbers of C6 alkyl chains between Cya and peptide (FEFEGRGD) were prepared (Cya-PAn: n = 1–3) and mixed with Mel-NBD (Fig. 1). Cya-PA1/Mel-NBD formed rods of several hundreds of nanometers long, while Cya-PA2/Mel-NBD and Cya-PA3/Mel-NBD formed micron-scale fibrous assemblies (Fig. S1, ESI†). Compared with Cya-PA2/Mel-NBD, Cya-PA3/Mel-NBD formed larger-sized, bundled assembles, which was evidenced by confocal laser scanning microscopy (CLSM) observation (Fig. S1i, ESI†).
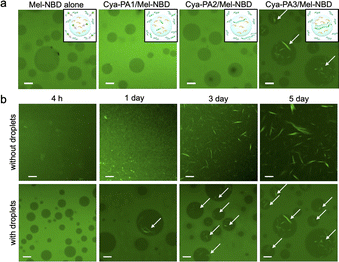
We then investigated the formation and localization of supramolecules in the LLPS system using CLSM. Cya-PA and Mel-NBD were added to an LLPS solution containing 5 wt% PEG and Dex to form Dex-rich droplets and a PEG-rich continuous phase, and the solutions were incubated at 37 °C. After 5 d, homogeneous green fluorescence was observed in both the PEG and Dex phases, with a higher relative intensity in the PEG phase, for Mel-NBD alone and co-assemblies with Cya-PA1 and Cya-PA2 (Fig. 2a). When partition measurement using liquid chromatography was applied to the mixture of Cya-PA1 and Mel-NBD, an approx. 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio of Cya-PA1 and Mel-NBD was detected in both the PEG and Dex phases, indicating the formation of a Cya-PA1/Mel-NBD co-assembly (Fig. S2, ESI†). Therefore, the homogeneous green fluorescence might stem from the smaller supramolecule size, which was difficult to observe by CLSM (Fig. S1g, ESI†). In contrast, fibrous assemblies of Cya-PA3/Mel-NBD were clearly localized in Dex droplets (Fig. 2a and Fig. S3, ESI†), regardless of the molecular weights of PEG/Dex (Fig. S4, ESI†). These results suggest that not the formation of fibrous structures but larger-sized, bundled assembly formation might lead to localization in the Dex phase. To support this hypothesis, we examined the correlation between the formation of well-developed fibrous structures and the phase selectivity through time-dependent observation. In our co-assembly system, the complementary interaction between Cya and Mel occurred at the initial stage of co-assembly, while hierarchical fibrous assembly formation occurred with aging (Fig. S5, ESI†). After 4 h, when the assembly was at the initial stage, little fibrous objects were detected in the absence of LLPS (Fig. 2b, top) and relatively high green fluorescence was observed in the PEG phase in the presence of LLPS (Fig. 2b, bottom). Assemblies of Cya-PA3/Mel-NBD started to be detected in PBS solution after 1 d and continued to grow with aging. Interestingly, localization of the fibrous structures in Dex droplets was also detected after 1 d. These results suggested that the formation of large assemblies was necessary for localization in the Dex phase.
1 molar ratio of Cya-PA1 and Mel-NBD was detected in both the PEG and Dex phases, indicating the formation of a Cya-PA1/Mel-NBD co-assembly (Fig. S2, ESI†). Therefore, the homogeneous green fluorescence might stem from the smaller supramolecule size, which was difficult to observe by CLSM (Fig. S1g, ESI†). In contrast, fibrous assemblies of Cya-PA3/Mel-NBD were clearly localized in Dex droplets (Fig. 2a and Fig. S3, ESI†), regardless of the molecular weights of PEG/Dex (Fig. S4, ESI†). These results suggest that not the formation of fibrous structures but larger-sized, bundled assembly formation might lead to localization in the Dex phase. To support this hypothesis, we examined the correlation between the formation of well-developed fibrous structures and the phase selectivity through time-dependent observation. In our co-assembly system, the complementary interaction between Cya and Mel occurred at the initial stage of co-assembly, while hierarchical fibrous assembly formation occurred with aging (Fig. S5, ESI†). After 4 h, when the assembly was at the initial stage, little fibrous objects were detected in the absence of LLPS (Fig. 2b, top) and relatively high green fluorescence was observed in the PEG phase in the presence of LLPS (Fig. 2b, bottom). Assemblies of Cya-PA3/Mel-NBD started to be detected in PBS solution after 1 d and continued to grow with aging. Interestingly, localization of the fibrous structures in Dex droplets was also detected after 1 d. These results suggested that the formation of large assemblies was necessary for localization in the Dex phase.
To further investigate the localization mechanism of Cya-PA3/Mel-NBD co-assembly in the droplets, we observed co-assembly formation in the presence of either PEG or Dex polymer. After 1 d, when assemblies started to be detected in PBS, fibrous objects were observed both in 5 wt% PEG and Dex solutions (Fig. S6, ESI†). The assembly size continued increasing in PBS and 5 wt% PEG until day 5. In contrast, in 5 wt% Dex solution, almost complete growth was observed after 1 d. These results suggested that the formation of hierarchical assemblies was accelerated in 5 wt% Dex. However, the presence of 5 wt% PEG neither inhibited the formation nor co-existence of hierarchical assemblies, suggesting that accelerated fibrous formation in the Dex phase was not the direct reason for their localization in Dex droplets. Next, we checked the phase selectivity of the pre-formed Cya-PA3/Mel-NBD co-assembly in the two-phase system. When imaged soon after mixing the co-assembly and LLPS solutions, complete localization of the fibers in the droplets was observed (Fig. S7, ESI†). Furthermore, the partition of each assembly component (Cya-PA3 or Mel-NBD) showed either non-selectivity (Cya-PA3) or selectivity for the PEG phase (Mel-NBD) (Fig. S8, ESI†), indicating the lower possibility of selective co-assembly formation in the Dex phase. Altogether, the droplet localization of Cya-PA3/Mel-NBD supramolecules was due to the higher order organization of the assemblies. Similar droplet selectivity depending on assembly maturation has been reported for intracellular supramolecular protein polymers formed with actin or FtsZ, as explained by the high affinity of the developed supramolecular structures for the coarser structure of Dex phase than for the PEG phase.13,14
Intracellular supramolecular cytoskeletons like actin fibers have been reported to exhibit non-covalent adsorption of biomolecules on their surface to regulate biological processes.19 Similarly, some synthetic supramolecular polymers composed of simple peptides physically adsorb proteins.20–22 If our novel synthetic supramolecular polymers also showed a protein-binding property, they would become useful materials to recruit proteins into a compartmentalized space, namely droplets, without modification of the proteins.
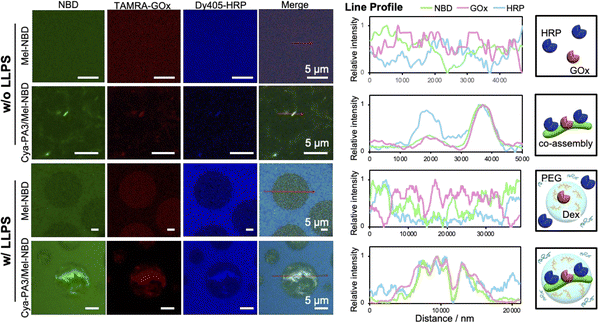
To verify the feasibility of this concept, we examined the interaction between proteins and the co-assemblies. Various proteins, including fluorescent protein mDsRed, and cationic (lysozyme) and anionic (ovalbumin) proteins, interacted with the Cya-PA3/Mel-NBD co-assembly as evidenced by the overlapped red (proteins) and green (NBD) fluorescence observed by CLSM (Fig. S9, ESI†). We also examined whether the simultaneous adsorption of two proteins on the supramolecule could be achieved. Glucose oxidase (GOx) and horseradish peroxidase (HRP), which are representative cascade enzymes, were used for this purpose. CLSM observation of the Cya-PA3/Mel-NBD co-assembly after mixing with labeled TAMRA-GOx and Dy405-HRP showed overlapped green (NBD), red (TAMRA-GOx), and blue (Dy405-HRP) fluorescence, indicating simultaneous retention of the two proteins on the supramolecular polymers (Fig. 3, top). Since both the Cya-PA3/Mel-NBD co-assembly and proteins are negatively charged (Fig. S10, ESI†) and protein adsorption was not inhibited when 0.5 M NaCl was added (Fig. S11, ESI†), non-covalent interactions other than electrostatic interaction should be operating. Bio-layer interferometry measurements showed that co-assembly formation was necessary to adsorb proteins because their components (Cya-PA3 or Mel-NBD alone) did not show apparent adsorptions (Fig. S12, ESI†).
We then evaluated the recruitment of proteins into droplets using our supramolecules. The localization of proteins is dependent on their physicochemical properties and the LLPS conditions. Under the LLPS conditions in the present study, mDsRed, ovalbumin, and GOx were localized in Dex droplets, while lysozyme and HRP were in the PEG phase. However, protein localization was influenced by the Cya-PA3/Mel-NBD assemblies and supramolecules in the droplets adsorbing the proteins, regardless of their original localizations (Fig. 3, bottom, Fig. S13 and S14, ESI†). This result suggested that the two properties of the co-assemblies, namely, adsorption of proteins and droplet-selectivity, did not mutually affect and enable the recruitment of proteins across phases. Furthermore, it was suggested that the non-covalent adsorption was strong enough to convert the phase-selectivity of the proteins.
The recruitment of cascade enzymes into a confined space enables proximate positioning of the enzymes, which would accelerate the reaction rate. Our supramolecule was able to bind and bring several types of enzymes into the droplets, making it a great candidate material for non-covalent compartmentalization of cascade enzymes. The enzymatic cascade activity was evaluated using the production rate of a final product of the GOx-HRP catalytic system, the ABTS diradical, using glucose and ABTS as substrates for GOx and HRP, respectively. When Cya-PA3/Mel-NBD co-existed in the absence of droplets, the production rate was not increased (Fig. 4 and Fig. S15, ESI†). However, under LLPS conditions, the co-existence of the co-assembly significantly increased the cascade reaction. As the presence of LLPS intrinsically decreases the reaction rate, presumably owing to the separated localizations of GOx and HRP, the increased cascade reaction rate in the presence of both droplets and the co-assembly should result from the recruitment of both enzymes onto the structures in the droplets (Fig. 3, bottom and Fig. S16, ESI†). This was also supported by the little increase in the reaction rate when a Cya-PA1/Mel-NBD co-assembly showing little localization in the droplets was added (Fig. S17, ESI†). Further investigation of the influence of the local environment around the co-assembly, and interaction of the macromolecular polymers with enzymes, substrates, and co-assemblies, will provide deeper insight into the mechanism of change in the cascade reaction and provide a better supramolecule design.
In summary, we have demonstrated that artificial supramolecular polymers show localization in a specific phase in an LLPS system depending on the hierarchical organization of supramolecules, where highly developed supramolecules showed localization in Dex droplets. The co-assembly non-covalently adsorbed proteins on the structure, leading to the recruitment of proteins into the droplets without modification. Furthermore, these features of the co-assembly enabled the confinement of several enzymes in droplets, as demonstrated by the increased cascade reaction rate. The significant achievements of the present study were as follows: (i) the unique supramolecular system, which allowed the controlled organization that enabled the relationship between hierarchical organizations of supramolecules and phase selectivity in LLPS to be investigated; and (ii) achieving artificial manipulation of the spatial control of biomolecules without modification. This study not only provides a fundamental understanding of supramolecule behavior in an LLPS system, but also expands the potential utilization of LLPS/supramolecular systems in the bioengineering field.
This work was supported by the Japan Society for the Promotion of Science (JSPS) KAKENHI, grant numbers JP22H04552 and JP21H05227. R. W. thanks The Kurata Grants from The Hitachi Global Foundation for funding support. H. O. is grateful to the JSPS Research Fellowship for Young Scientists (Grant No. JP20J20799) and Kyushu University Program for Leading Graduate Schools, Advanced Graduate Course on Molecular Systems for Devices, for a scholarship. The authors thank the Center of Advanced Instrumental Analysis, Kyushu University, and the Advanced Research Infrastructure for Materials and Nanotechnology in Japan for facility support.
Conflicts of interest
There are no conflicts to declare.Notes and references
- R. Milo, BioEssays, 2013, 35, 1050–1055 CrossRef CAS PubMed.
- M. C. Good, J. G. Zalatan and W. A. Lim, Science, 2011, 332, 680–686 CrossRef CAS PubMed.
- S. F. Banani, H. O. Lee, A. A. Hyman and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2017, 18, 285–298 CrossRef CAS PubMed.
- B. Wang, L. Zhang, T. Dai, Z. Qin, H. Lu, L. Zhang and F. Zhou, Signal Transduction Targeted Ther., 2021, 6, 1–16 CrossRef PubMed.
- A. S. Lyon, W. B. Peeples and M. K. Rosen, Nat. Rev. Mol. Cell Biol., 2021, 22, 215–235 CrossRef CAS PubMed.
- S. Alberti, A. Gladfelter and T. Mittag, Cell, 2019, 176, 419–434 CrossRef CAS PubMed.
- M. v Garabedian, W. Wang, J. B. Dabdoub, M. Tong, R. M. Caldwell, W. Benman, B. S. Schuster, A. Deiters and M. C. Good, Nat. Chem. Biol., 2021, 17, 998–1007 CrossRef CAS PubMed.
- M. Yoshikawa and S. Tsukiji, Biochemistry, 2021, 60, 3273–3276 CrossRef CAS PubMed.
- M. Yoshikawa, T. Yoshii, M. Ikuta and S. Tsukiji, J. Am. Chem. Soc., 2021, 143, 6434–6446 CrossRef CAS PubMed.
- C. D. Reinkemeier and E. A. Lemke, Curr. Opin. Chem. Biol., 2021, 64, 174–181 CrossRef CAS PubMed.
- Y. Chao and H. C. Shum, Chem. Soc. Rev., 2020, 49, 114–142 RSC.
- C. D. Keating, Acc. Chem. Res., 2012, 45, 2114–2124 CrossRef CAS PubMed.
- B. Monterroso, S. Zorrilla, M. Sobrinos-Sanguino, C. D. Keating and G. Rivas, Sci. Rep., 2016, 6, 1–13 CrossRef PubMed.
- N. Nakatani, H. Sakuta, M. Hayashi, S. Tanaka, K. Takiguchi, K. Tsumoto and K. Yoshikawa, ChemBioChem, 2018, 19, 1370–1374 CrossRef CAS PubMed.
- C. Yuan, A. Levin, W. Chen, R. Xing, Q. Zou, T. W. Herling, P. K. Challa, T. P. J. Knowles and X. Yan, Angew. Chem., Int. Ed., 2019, 58, 18116–18123 CrossRef CAS PubMed.
- A. Jain, S. Kassem, R. S. Fisher, B. Wang, T. de Li, T. Wang, Y. He, S. Elbaum-Garfinkle and R. v Ulijn, J. Am. Chem. Soc., 2022, 144, 15002–15007 CrossRef CAS PubMed.
- R. Wakabayashi, H. Obayashi, R. Hashimoto, N. Kamiya and M. Goto, Chem. Commun., 2019, 55, 6997–7000 RSC.
- B. Adhikari, X. Lin, M. Yamauchi, H. Ouchi, K. Aratsu and S. Yagai, Chem. Commun., 2017, 53, 9663–9683 RSC.
- H. Hu, A. Juvekar, C. A. Lyssiotis, E. C. Lien, J. G. Albeck, D. Oh, G. Varma, Y. P. Hung, S. Ullas, J. Lauring, P. Seth, M. R. Lundquist, D. R. Tolan, A. K. Grant, D. J. Needleman, J. M. Asara, L. C. Cantley and G. M. Wulf, Cell, 2016, 164, 433–446 CrossRef CAS PubMed.
- R. Jain, V. K. Pal and S. Roy, Biomacromolecules, 2020, 21, 4180–4193 CrossRef CAS PubMed.
- N. Kapil, A. Singh and D. Das, Angew. Chem., Int. Ed., 2015, 54, 6492–6495 CrossRef CAS PubMed.
- A. Chatterjee, C. Mahato and D. Das, Angew. Chem., Int. Ed., 2021, 60, 202–207 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Materials and methods, morphological observation of co-assemblies, time-dependent hierarchical assembly formation of Cya-PA3/Mel-NBD, and physical adsorption of proteins on the assemblies. See DOI: https://doi.org/10.1039/d2cc05910j |
| This journal is © The Royal Society of Chemistry 2023 |