Open Access Article
Open Access ArticleIncreasing the functional density of threose nucleic acid†
Biju
Majumdar‡
 a,
Daisy
Sarma‡
a,
Daisy
Sarma‡
 a,
Yutong
Yu
a,
Yutong
Yu
 a,
Adriana
Lozoya-Colinas
a,
Adriana
Lozoya-Colinas
 a and
John C.
Chaput
a and
John C.
Chaput
 *abcd
*abcd
aDepartment of Pharmaceutical Sciences, University of California, Irvine, CA 92697-3958, USA. E-mail: jchaput@uci.edu; Tel: +1 949-824-8149
bDepartment of Chemistry, University of California, Irvine, CA 92697-3958, USA
cDepartment of Molecular Biology and Biochemistry, University of California, Irvine, CA 92697-3958, USA
dDepartment of Chemical and Biomolecular Engineering, University of California, Irvine, CA 92697-3958, USA
First published on 25th October 2023
Abstract
Chemical strategies that augment genetic polymers with amino acid residues that are overrepresented on the paratope surface of an antibody offer a promising route for enhancing the binding properties of nucleic acid aptamers. Here, we describe the chemical synthesis of α-L-threofuranosyl cytidine nucleoside triphosphate (tCTP) carrying either a benzyl or phenylpropyl side chain at the pyrimidine C-5 position. Polymerase recognition studies indicate that both substrates are readily incorporated into a full-length α-L-threofuranosyl nucleic acid (TNA) product by extension of a DNA primer-template duplex with an engineered TNA polymerase. Similar primer extension reactions performed using nucleoside triphosphate mixtures containing both C-5 modified tCTP and C-5 modified tUTP substrates enable the production of doubly modified TNA strands for a panel of 20 chemotype combinations. Kinetic measurements reveal faster on-rates (kon) and tighter binding affinity constants (Kd) for engineered versions of TNA aptamers carrying chemotypes at both pyrimidine positions as compared to their singly modified counterparts. These findings expand the chemical space of evolvable non-natural genetic polymers by offering a path for improving the quality of biologically stable TNA aptamers for future clinical applications.
Introduction
Crystal structures of antibody–antigen complexes have long shown that protein epitope recognition is driven by the presence of aromatic side chains found on the paratope surface, which are five times more abundant in this region than the rest of the antibody surface.1,2 In an effort to improve the binding properties of aptamers,3,4 nucleic acid sequences that mimic antibodies by folding into structures with ligand binding pockets,5 numerous groups have sought to emulate nature by augmenting nucleic acid scaffolds with diversity-enhancing functional groups.6–10 The simplest approach to this problem involves employing copper(I)-catalyzed alkyne–azide cycloaddition reactions (click chemistry) to link azide containing compounds to libraries that have been prepared by replacing thymidine with C5-ethynyl-2′-deoxyuridine.11 While this approach provides access to regions of chemical space that lie outside the boundary of what is permitted by the structural requirements of natural or engineered polymerases, sub-quantitative coupling leads to the formation of complex heterogenous mixtures of base-modified strands. Despite this potential weakness, click chemistry has been used to modify genetic polymers with a range of functional groups, including amino acids, carbohydrates, and dyes, and subsequent in vitro selection experiments have yielded aptamers against targets that were previously refractory to in vitro selection.12–16The preparation of uniformly modified libraries requires synthetic strategies that introduce new chemical functionality directly into the oligonucleotide sequence during the library generation step of the selection. One approach involves the concept of genetic alphabet expansion, which increases the sequence diversity of the starting library through the incorporation of a third Watson–Crick base pair.17,18 Using this strategy, the Benner and Hirao laboratories have evolved several examples of aptamers that contain more than four genetic letters in their DNA sequence, some of which function with solution binding affinity constants (Kd) in the picomolar range.19–22 Functionally enhanced libraries prepared using nucleoside triphosphates that are modified to include additional functionality at the C-5 position of pyrimidines and N-7 or C-8 positions of purines offer a different path for improving aptamer activity.14,23–27 Slow off-rate modified aptamers (SOMAmers), for example, utilize a highly versatile palladium-catalyzed carboxyamidation reaction to install diversity enhancing functional groups at the C-5 position of pyrimidines.28 This approach revolutionized the field of aptamer-based diagnostics by increasing the success rate of aptamer selections from ∼30% for standard base libraries to 84% for functionally enhanced libraries.29 Close inspection of aptamer–protein co-crystal structures reveals striking similarities in the recognition of protein epitopes by SOMAmers and antibodies, showing the ability for base-modified aptamers to effectively mimic the paratope surface of an antibody.30
Recent advances in polymerase engineering have enabled the development of laboratory evolved polymerases that can replicate synthetic genetic polymers (XNAs) with backbone structures that are distinct from those found in nature.31 These enzymes, along with similar improvements in nucleic acid chemistry, have permitted the evolution of aptamers and catalysts from libraries of XNA sequences.32–36 Given the success of functionally enhanced DNA aptamers, base modifications were viewed as a critical next step in the development of therapeutic aptamers that function with high biological stability and slow off-rate binding kinetics. Threomers, functionally-enhanced aptamers based on an α-L-threofuranosyl nucleic acid (TNA) backbone architecture (Fig. 1), were previously shown to function with enhanced binding affinity.37 Here, we explore the potential for generating threomers with increased functional density using nucleoside triphosphate mixtures carrying chemical modifications on both pyrimidine residues. We suggest that this approach will help drive the development of highly functionalized XNA aptamers for future clinical applications.
Results
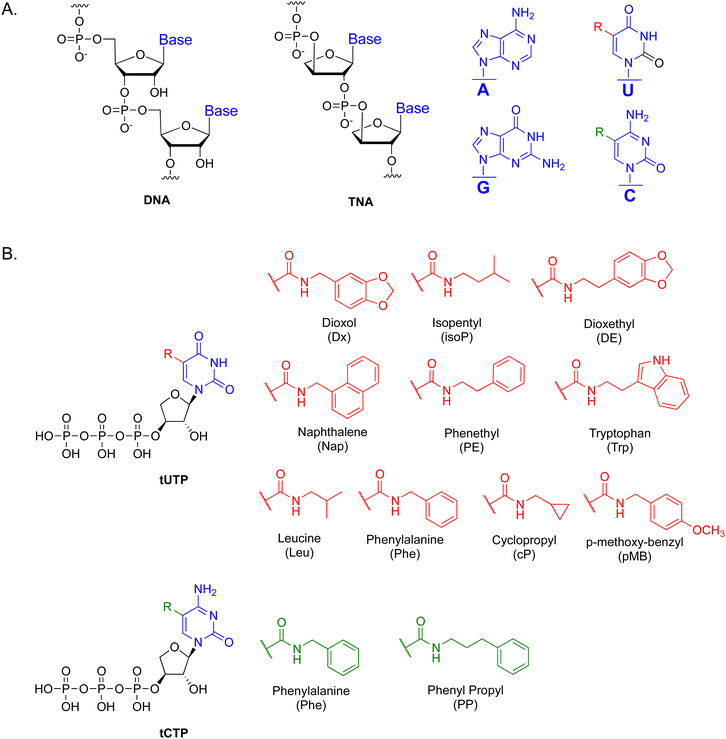
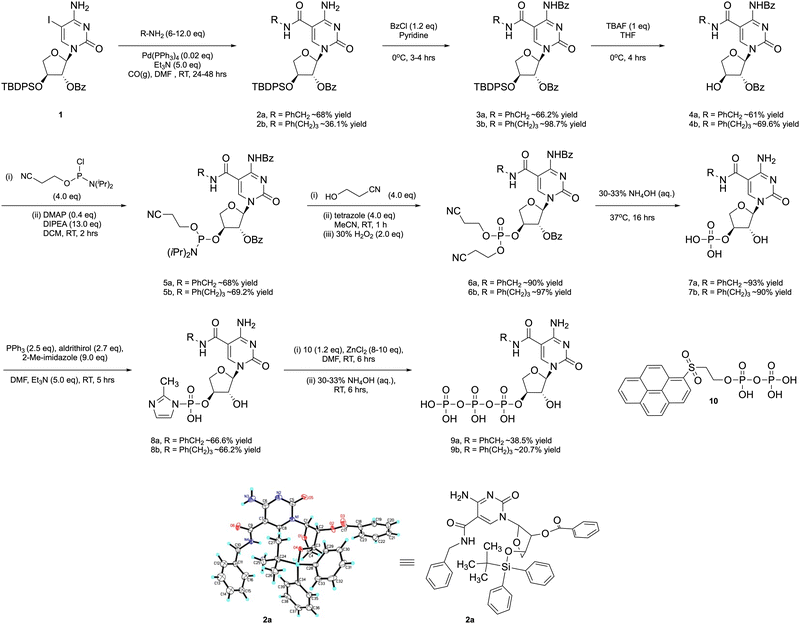
The side chains analyzed in this study included phenylalanine (Phe) and phenyl propyl (PP), viewed here as representative aromatic groups (Fig. 1(B)) with properties that are expected to be similar to the side chains found on the paratope surface of the antigen binding domain of an antibody.38 We considered this initial proof-of-concept study as a possible generalizable route to modified tCTP analogs with diverse chemotypes that could be used in future aptamer selections. However, before such systems could be explored by in vitro selection, it was first necessary to demonstrate the impact of increased functional density toward TNA aptamer function through the synthesis and polymerase recognition of modified tCTPPhe or tCTPPP substrates in the presence of known tUTP-modified analogs.39The starting 5-iodo-1-(2′-O-benzoyl-α-L-threofuranosyl)-cytosine nucleoside 1 was prepared in 8 steps from L-ascorbic acid using a Vorbrüggen reaction to conjugate 5-iodo-cytosine to an orthogonally protected threose sugar (Scheme S1, ESI†). Subsequent conjugation of the side chains to the nucleobase was accomplished by heating the aryl amines with TNA nucleoside 1 in the presence of 10 mol% Pd(PPh3)4, and CO (Scheme 1). The stereochemistry of the modified nucleoside was unambiguously determined by solving a small molecule X-ray crystal structure for the phenylalanine derivative 2a. The aromatic side chain is linked to the C-5 position of the cytosine nucleobase via an amide bond with the correct stereochemistry observed at the C1′, C2′, and C3′ sugar positions (Scheme 1). After purification by silica gel chromatography, nucleosides 2a and 2b were converted to the desired nucleoside-3′-triphosphates 9a and 9b. The exocyclic amines were first protected with benzoyl chloride to afford 3a and 3b, which were treated with TBAF to remove the TBDPS protecting groups and afford nucleosides 4a and 4b.
 | ||
| Scheme 1 Synthetic strategy for preparing C-5 modified tCTP analogs with crystallographic validation of nucleoside 2a. | ||
Conversion of the free 3′ OH groups of 4a and 4b to the desired nucleoside-3′-triphosphates 9a and 9b required the use of a specialized pyrene pyrophosphate reagent as previous studies have shown that the Yoshikawa40 and Ludwig–Eckstein41 methods commonly used to prepare DNA nucleoside triphosphates are incompatible with TNA.42 This problem is likely due to the reduced nucleophilicity and steric compression of the 3′ OH found in TNA as compared to the 5′ OH of DNA. Accordingly, 4a and 4b were converted to the 3′-O-phsophoramidite derivatives 5a and 5b by treatment with 2-cyanoethoxy-N,N-diisopropylchorophosphine in the presence of Hunig's base and DMAP. Intermediates 5a and 5b were converted to their trialkyl phosphite derivatives by replacing the N,N-diisopropyl amino group with a base-labile cyanoethoxy group and oxidized to their phosphate derivatives 6a and 6b. Deprotection of the alkyl phosphate chains and both benzoyl groups yielded the 3′-monophosphates 7a and 7b, which were activated with 2-methyl-imidazole (8a and 8b) and converted to their fully protected α-L-threofuranosyl cytidine-5′-triphosphates upon treatment with ZnCl2 and pyrene pyrophosphate.42 Following purification, the triphosphate intermediates were deprotected with concentrated ammonium hydroxide (33% aq.) and precipitated as a sodium salt to afford the desired nucleoside-3′-triphosphates 9a and 9b (Fig. S1, ESI†).
In addition to the C-5 modified tCTP analogs, this study also required the preparation of all four TNA triphosphates with the natural bases of adenine (tATP), cytosine (tCTP), thymine (tTTP), and guanine (tGTP), which were each synthesized in 14 steps from L-ascorbic acid.42,43 Additionally, ten C-5 modified tUTP analogs were prepared in 15 steps from L-ascorbic acid.39 Together, these molecules represent 16 different TNA triphosphates that can be used as substrates in polymerase-mediated TNA synthesis reactions by coping DNA templates into TNA with a laboratory-engineered TNA polymerase.44
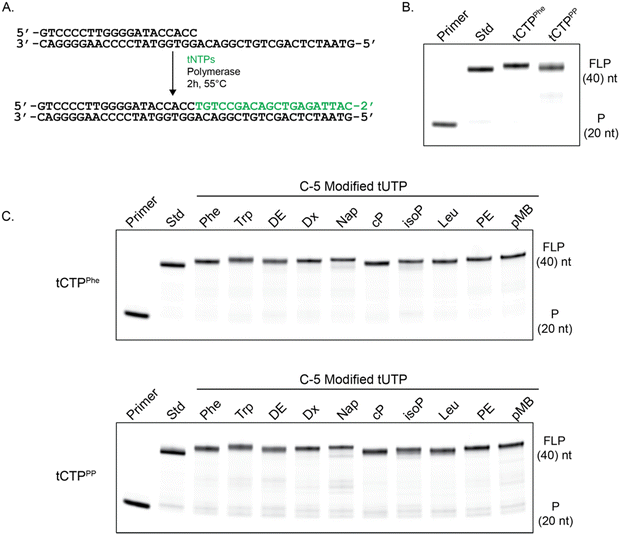
To confirm that the C-5 modified tCTP analogs 9a and 9b were viable substrates for enzymatic synthesis, primer extension assays were performed using a laboratory-engineered TNA polymerase. Accordingly, a 5′-IR680 dye-labeled DNA primer annealed to a DNA template containing an unpaired region of 20 nucleotides (nt) was challenged to extend the DNA primer using a mixture of TNA triphosphates (tNTPs) that contained either standard bases only or a mixture of tNTPs in which the tCTP substrate was replaced with either of the newly synthesized analogs tCTPPhe or tCTPPP (Fig. 2(A)). Similar modifications were previously evaluated using modified tUTP substrates to install the desired chemotypes at sequence-defined positions.39 The primer extension reactions were incubated at 55 °C for 2 hours in ThermoPol buffer [20 mM tris (pH 8.8), 10 mM KCl, 10 mM (NH4)2SO4, and 20 mM MgSO4] and product formation was analyzed by 12% denaturing polyacrylamide gel electrophoresis (PAGE). As illustrated in Fig. 2(B), formation of the full-length product was observed in each case, indicating that the TNA polymerase is able to recognize and incorporate the newly synthesized tCTP substrates. The incorporation of a base-modified nucleotide into the TNA product was confirmed by MALDI-ToF mass spectrometry for a model template that was more amenable to ionization than a longer, fully modified sequence (Fig. S2, ESI†).
Encouraged by the ability of our polymerase to synthesize TNA carrying amino acid-like side chains at the C-5 position of cytidine, we explored the potential for this enzyme to recognize tNTP mixtures in which both pyrimidine substrates were modified with function-enhancing chemotypes. Primer extension reactions were performed using the newly synthesized tCTPPhe or tCTPPP substrates and a set of ten previously synthesized C-5 modified tUTP substrates.39 Analysis of a panel of 20 combinations of doubly modified TNA synthesis reactions reveal that full-length product formation is observed in all cases with the product bands migrating with similar electrophoretic mobilities as the standard base control reaction (Fig. 2(C)). Importantly, this result demonstrates that many of the synthetic challenges previously overcome for base-modified DNA substrates can now be extended to base-modified XNAs, like TNA. This includes the challenge of identifying polymerases with the ability to incorporate nucleoside triphosphates carrying modifications on both the backbone and base moieties of the nucleic acid structure.
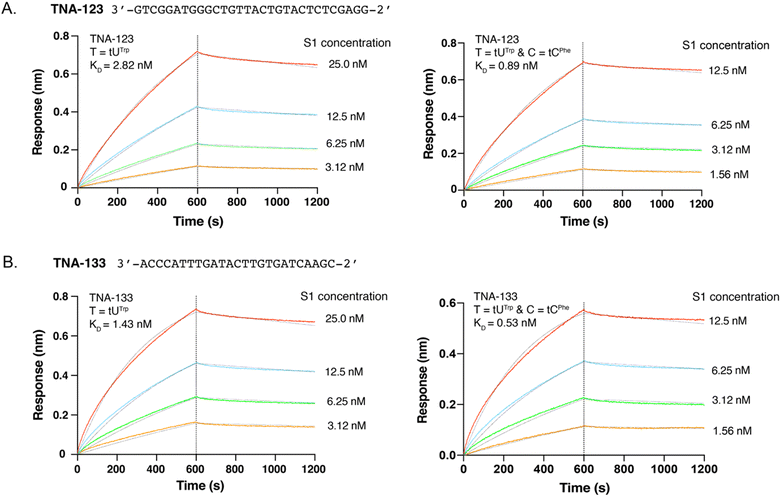
To explore the potential for modified cytidine analogs to improve the binding properties of TNA aptamers, we chose to evaluate the binding properties of two threomers previously evolved to recognize the S1 subunit of the SARS-CoV-2 spike protein.37 TNA aptamers S1-123 and S1-133 were prepared in their original Trp chemotype forms using a tNTP mixture that contained the tUTPTrp substrate in place of tTTP. Additionally, the doubly modified forms of both aptamers were prepared using the tUTPTrp and tCTPPhe substrates to install the indole and benzyl chemotypes at sequence-defined positions of the uracil and cytosine bases, respectively. The TNA extended regions encode aptamers that are 25 and 30 nts in length and carry base-modified residues at ∼30% of the positions (Fig. 3). All four aptamers were generated by extension of a 5′-biotinylated DNA primer that allowed for kinetic binding assays to be performed using a streptavidin-coated biosensor tip of a bio-layer interferometry (BLI) instrument. Following PAGE purification and electroelution, the binding kinetics of the single and doubly modified threomers were evaluated by BLI, a label-free optical approach for precisely measuring the binding kinetics of aptamer–protein complexes.45 Analysis of the resulting sensorgrams indicates that the doubly modified threomers achieve ∼3-fold tighter binding than their singly modified counterparts (Fig. 3). The increase in binding affinity is primarily due to faster association kinetics (kon) of the aptamer–protein complex, which may be attributed to favorable hydrophobic interactions with the S1 protein and possibly an improved pre-folded state of the aptamer. This interpretation is consistent with control experiments, which show that protein binding affinity is abrogated when the aptamers are prepared with either standard bases only or as singly modified versions that carry the tCPhe modification (Fig. S3, ESI†). It also confirms that the tUTrp modifications previously discovered by in vitro selection provide critical hydrophobic interactions that drive aptamer binding to the protein target.
Discussion
Antibodies have long been recognized as the gold standard in affinity reagent technology.46 Driven by strong shape complementarity resulting from an extensive network of non-covalent interactions in the antigen binding site, antibodies have been shown to bind their cognate targets with high affinity and high specificity.1 However, despite their routine use in biomedical applications, antibodies are limited by several factors, including their high cost of discovery and manufacturing and their tendency for batch-to-batch variability, which can negatively impact assay reproducibility.47 Aptamers, nucleic acid sequences that fold into shapes with target binding affinity, offer a possible alternative to antibodies.48 Most aptamers are discovered in vitro and chemically synthesized, which reduces their discovery and production costs, and allows for minimal batch-to-batch variability as compared to antibodies. Although aptamer technology has improved significantly over time,49 the next generation of aptamers will need to be more biologically stable, function with slower dissociation constants, and have a discovery platform that is more amenable to high throughput screening.50Inspired by SOMAmers, slow off-rate binding DNA aptamers,51 we initiated a program of study aimed at generating functionally enhanced XNA aptamers based on a biologically stable framework of TNA.52 In an earlier study, we showed that functionally enhanced TNA aptamers, referred to as threomers, can be readily generated using uracil nucleotides carrying amino acid-like side chains at the C-5 position.37 Much like their DNA counterparts,51 these reagents functioned with higher affinity and slower dissociation rates than their unmodified standard base sequences. In the current study, we examined the hypothesis that threomers activity could be further improved through increased chemical diversity. We tested this hypothesis by first chemically synthesizing two prototypical C-5 modified cytosine nucleoside triphosphates and then evaluating their incorporation into TNA using a laboratory-engineered TNA polymerase. Primer extension assays showed that it is possible to enzymatically synthesize TNA sequences in which both pyrimidine nucleotides are modified with a different aromatic side chain, which was a nontrivial result given the potential for engineered polymerases to stall during XNA synthesis.53 We then used this methodology to test our hypothesis by comparing the binding properties to two previously discovered threomers prepared in their original Trp-modified form and their new doubly-modified Trp and Phe forms. Kinetic titration assays indicate that both doubly-modified threomers function with tighter binding affinity than their singly-modified forms, suggesting that it should be possible to evolve new examples of threomers from doubly-modified libraries that carry an assortment of functional groups at the pyrimidine positions.
As these studies continue, we anticipate that the expansion of chemical space through the synthesis of additional TNA nucleoside triphosphate building blocks will lead to the discovery of new types of aptamers with superior binding properties. Such libraries could, for example, be used to target proteins that have previously been refractory to standard base libraries or resulted in aptamers with weaker binding affinity. It may also allow for platform technologies to be created that accelerate the discovery process by making it easier for aptamers to be isolated by in vitro selection. Finally, functionally-enhanced aptamers based on non-natural backbone structures, like TNA, offer a viable route for developing therapeutic aptamers towards potential clinical applications, as they are invisible to the enzymes that degrade DNA and RNA.52
In conclusion, we have shown that chemical density has the potential to increase the functional activity of in vitro selected TNA aptamers. This finding expands the chemical space of evolvable non-natural genetic polymers by offering a path for improving the quality of biologically stable TNA aptamers for future diagnostic and therapeutic applications.
Author contributions
All authors conceived the project and designed the experiments. BM, DS, ALC, and YY performed the experiments. JC wrote the paper with input from all of the authors.Conflicts of interest
JC is a consultant for X, The Moonshot Factory.Acknowledgements
We wish to thank members of the Chaput lab for helpful comments and suggestions on the manuscript. This work was supported by a sponsored research project from X, The Moonshot Factory.References
- T. Ramaraj, T. Angel, E. A. Dratz, A. J. Jesaitis and B. Mumey, Antigen-antibody interface properties: composition, residue interactions, and features of 53 non-redundant structures, Biochim. Biophys. Acta, 2012, 1824, 520–532 CrossRef CAS PubMed.
- J. V. Kringelum, M. Nielsen, S. B. Padkjaer and O. Lund, Structural analysis of B-cell epitopes in antibody:protein complexes, Mol. Immunol., 2013, 53, 24–34 CrossRef CAS PubMed.
- A. D. Ellington and J. W. Szostak, In vitro selection of RNA molecules that bind specific ligands, Nature, 1990, 346, 818–822 CrossRef CAS PubMed.
- C. Tuerk and L. Gold, Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase, Science, 1990, 249, 505–510 CrossRef CAS PubMed.
- T. Hermann and D. J. Patel, Adaptive recognition by nucleic acid aptamers, Science, 2000, 287, 820–825 CrossRef CAS PubMed.
- J. A. Latham, R. Johnson and J. J. Toole, The application of a modified nucleotide in aptamer selection: novel thrombin aptamers containing 5-(1-pentynyl)-2′-deoxyuridine, Nucleic Acids Res., 1994, 22, 2817–2822 CrossRef CAS PubMed.
- D. M. Perrin, T. Garestier and C. Helene, Expanding the catalytic repertoire of nucleic acid catalysts: simultaneous incorporation of two modified deoxyribonucleoside triphosphates bearing ammonium and imidazolyl functionalities, Nucleosides Nucleotides, 1999, 18, 377–391 CrossRef CAS.
- N. K. Vaish, R. Larralde, A. W. Fraley, J. W. Szostak and L. W. McLaughlin, A novel, modification-dependent ATP-binding aptamer selected from an RNA library incorporating a cationic functionality, Biochemistry, 2003, 42, 8842–8851 CrossRef CAS PubMed.
- S. Jager, G. Rasched, H. Kornreich-Leshem, M. Engeser, O. Thum and M. Famulok, A versatile toolbox for variable DNA functionalization at high density, J. Am. Chem. Soc., 2005, 127, 15071–15082 CrossRef.
- T. R. Battersby, D. N. Ang, P. Burgstaller, S. C. Jurczyk, M. T. Bowser, D. D. Buchanan, R. T. Kennedy and S. A. Benner, Quantitative analysis of receptors for adenosine nucleotides obtained via in vitro selection from a library incorporating a cationic nucleotide analog, J. Am. Chem. Soc., 1999, 121, 9781–9789 CrossRef CAS PubMed.
- F. Tolle, M. Rosenthal, F. Pfeiffer and G. Mayer, Click Reaction on Solid Phase Enables High Fidelity Synthesis of Nucleobase-Modified DNA, Bioconjugate Chem., 2016, 27, 500–503 CrossRef CAS.
- J. S. Temme, I. S. MacPherson, J. F. DeCourcey and I. Krauss, High temperature SELMA: evolution of DNA-supported oligomannose clusters which are tightly recognized by HIV bnAB 2G12, J. Am. Chem. Soc., 2014, 136, 1726–1729 CrossRef CAS.
- F. Tolle, G. M. Brandle, D. Natzner and G. Mayer, A versatile approach towards nucleobase-modified aptamers, Angew. Chem., Int. Ed., 2015, 54, 10971–10974 CrossRef CAS PubMed.
- Y. W. Cheung, P. Rothlisberger, A. E. Mechaly, P. Weber, F. Levi-Acobas, Y. Lo, A. W. C. Wong, A. B. Kinghorn, A. Haouz, G. P. Savage, M. Hollenstein and J. A. Tanner, Evolution of abiotic cubane chemistries in a nucleic acid aptamer allows selective recognition of a malaria biomarker, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 16790–16798 CrossRef CAS.
- K. B. Wu, C. J. A. Skrodzki, Q. Su, J. Lin and J. Niu, “Click handle”-modified 2′-deoxy-2′-fluoroarabino nucleic acid as a synthetic genetic polymer capable of post-polymerization functionalization, Chem. Sci., 2022, 13, 6873–6881 RSC.
- C. K. L. Gordon, D. Wu, A. Pusuluri, T. A. Feagin, A. T. Csordas, M. S. Eisenstein, C. J. Hawker, J. Niu and H. T. Soh, Click-Particle Display for Base-Modified Aptamer Discovery, ACS Chem. Biol., 2019, 14, 2652–2662 CrossRef CAS.
- K. H. Lee, K. Hamashima, M. Kimoto and I. Hirao, Genetic alphabet expansion biotechnology by creating unnatural base pairs, Curr. Opin. Biotechnol., 2018, 51, 8–15 CrossRef CAS PubMed.
- S. Hoshika, N. A. Leal, M. J. Kim, M. S. Kim, N. B. Karalkar, H. J. Kim, A. M. Bates, N. E. Watkins Jr., H. A. SantaLucia, A. J. Meyer, S. DasGupta, J. A. Piccirilli, A. D. Ellington, J. SantaLucia Jr., M. M. Georgiadis and S. A. Benner, Hachimoji DNA and RNA: A genetic system with eight building blocks, Science, 2019, 363, 884–887 CrossRef CAS.
- M. Kimoto, R. Yamashige, K.-I. Matsunaga, S. Yokoyama and I. Hirao, Generation of high affinity DNA aptamers using an expanded genetic alphabet, Nat. Biotechnol., 2013, 31, 453–457 CrossRef CAS.
- K. I. Matsunaga, M. Kimoto and I. Hirao, High-Affinity DNA Aptamer Generation Targeting von Willebrand Factor A1-Domain by Genetic Alphabet Expansion for Systematic Evolution of Ligands by Exponential Enrichment Using Two Types of Libraries Composed of Five Different Bases, J. Am. Chem. Soc., 2017, 139, 324–334 CrossRef CAS.
- K. Sefah, Z. Yang, K. M. Bradley, S. Hoshika, E. Jimenez, L. Zhang, G. Zhu, S. Shanker, F. Yu, D. Turek, W. Tan and S. A. Benner, In vitro selection with artificial expanded genetic information systems, Proc. Natl. Acad. Sci. U. S. A., 2014, 111, 1449–1454 CrossRef CAS.
- L. Q. Zhang, Z. Y. Yang, T. L. Trinh, I. T. Teng, S. Wang, K. M. Bradley, S. Hoshika, Q. F. Wu, S. Cansiz, D. J. Rowold, C. McLendon, M. S. Kim, Y. Wu, C. Cui, Y. Liu, W. J. Hou, K. Stewart, S. Wan, C. Liu, S. A. Benner and W. H. Tan, Aptamers against Cells Overexpressing Glypican 3 from Expanded Genetic Systems Combined with Cell Engineering and Laboratory Evolution, Angew. Chem., Int. Ed., 2016, 55, 12372–12375 CrossRef CAS PubMed.
- J. Niu, R. Hili and D. R. Liu, Enzyme-free translation of DNA into sequence-defined synthetic polymers structurally unrelated to nucleic acids, Nat. Chem., 2013, 5, 282–292 CrossRef CAS.
- H. Cahova, A. Panattoni, P. Kielkowski, J. Fanfrlik and M. Hocek, 5-Substituted Pyrimidine and 7-Substituted 7-Deazapurine dNTPs as Substrates for DNA Polymerases in Competitive Primer Extension in the Presence of Natural dNTPs, ACS Chem. Biol., 2016, 11, 3165–3171 CrossRef CAS PubMed.
- Z. Chen, P. A. Lichtor, A. P. Berliner, J. C. Chen and D. R. Liu, Evolution of sequence-defined highly functionalized nucleic acid polymers, Nat. Chem., 2018, 10, 420–427 CrossRef CAS PubMed.
- M. Ondrus, V. Sykorova, L. Bednarova, R. Pohl and M. Hocek, Enzymatic synthesis of hypermodified DNA polymers for sequence-specific display of four different hydrophobic groups, Nucleic Acids Res., 2020, 48, 11982–11993 CrossRef CAS.
- C. Mulholland, I. Jestrabova, A. Sett, M. Ondrus, V. Sykorova, C. L. Manzanares, O. Simoncik, P. Muller and M. Hocek, The selection of a hydrophobic 7-phenylbutyl-7-deazaadenine-modified DNA aptamer with high binding affinity for the Heat Shock Protein 70, Commun. Chem., 2023, 6, 65 CrossRef CAS.
- J. D. Vaught, C. Bock, J. Carter, T. Fitzwater, M. Otis, D. Schneider, J. Rolando, S. Waugh, S. K. Wilcox and B. E. Eaton, Expanding the chemistry of DNA for in vitro selection, J. Am. Chem. Soc., 2010, 132, 4141–4151 CrossRef CAS PubMed.
- L. Gold, et al., Aptamer-Based Multiplexed Proteomic Technology for Biomarker Discovery, Plos One, 2010, 5, e15004 CrossRef CAS PubMed.
- A. D. Gelinas, D. R. Davies and N. Janjic, Embracing proteins: structural themes in aptamer–protein complexes, Curr. Opin. Struct. Biol., 2016, 36, 122–132 CrossRef CAS PubMed.
- A. Nikoomanzar, N. Chim, E. J. Yik and J. C. Chaput, Engineering polymerases for applications in synthetic biology, Q. Rev. Biophys., 2020, 53, e8 CrossRef CAS.
- H. Yu, S. Zhang and J. C. Chaput, Darwinian evolution of an alternative genetic system provides support for TNA as an RNA progenitor, Nat. Chem., 2012, 4, 183–187 CrossRef CAS PubMed.
- V. B. Pinheiro, A. I. Taylor, C. Cozens, M. Abramov, M. Renders, S. Zhang, J. C. Chaput, J. Wengel, S. Y. Peak-Chew, S. H. McLaughlin, P. Herdewijn and P. Holliger, Synthetic genetic polymers capable of heredity and evolution, Science, 2012, 336, 341–344 CrossRef CAS PubMed.
- A. I. Taylor, V. B. Pinheiro, M. J. Smola, A. S. Morgunov, S. Peak-Chew, C. Cozens, K. M. Weeks, P. Herdewijn and P. Holliger, Catalysts from synthetic genetic polymers, Nature, 2015, 518, 427–430 CrossRef CAS.
- Y. Wang, A. K. Ngor, A. Nikoomanzar and J. C. Chaput, Evolution of a general RNA-cleaving FANA enzyme, Nat. Commun., 2018, 9, 5067 CrossRef PubMed.
- Y. Wang, Y. Wang, D. Song, X. Sun, Z. Li, J. Y. Chen and H. Yu, An RNA-cleaving threose nucleic acid enzyme capable of single point mutation discrimination, Nat. Chem., 2022, 14, 350–359 CrossRef.
- C. M. McCloskey, Q. Li, E. J. Yik, N. Chim, A. K. Ngor, E. Medina, I. Grubisic, L. Co Ting Keh, R. Poplin and J. C. Chaput, Evolution of Functionally Enhanced alpha-L-Threofuranosyl Nucleic Acid Aptamers, ACS Synth. Biol., 2021, 10, 3190–3199 CrossRef CAS PubMed.
- J. C. Rohloff, A. D. Gelinas, T. C. Jarvis, U. A. Ochsner, D. J. Schneider, L. Gold and N. Janjic, Nucleic Acid Ligands With Protein-like Side Chains: Modified Aptamers and Their Use as Diagnostic and Therapeutic Agents, Mol. Ther.–Nucleic Acids, 2014, 3, e201 CrossRef CAS PubMed.
- Q. Li, V. A. Maola, N. Chim, J. Hussain, A. Lozoya-Colinas and J. C. Chaput, Synthesis and Polymerase Recognition of Threose Nucleic Acid Triphosphates Equipped with Diverse Chemical Functionalities, J. Am. Chem. Soc., 2021, 143, 17761–17768 CrossRef CAS PubMed.
- M. Yoshikawa, T. Kato and T. Takenishi, A novel method for phosphorylation of nucleosides to 5′-nucleotides, Tetrahedron Lett., 1967, 8, 5065–5068 CrossRef PubMed.
- J. Ludwig and F. Eckstein, Rapid and efficient synthesis of nucleoside 5′-0-(1-thiotriphosphates), 5′-triphosphates and 2′,3′-cyclophosphorothioates using 2-chloro-4H-1,3,2-benzodioxaphosphorin-4-one, J. Org. Chem., 1989, 54, 631–635 CrossRef CAS.
- J.-Y. Liao, S. Bala, A. K. Ngor, E. J. Yik and J. C. Chaput, P(V) Reagents for the Scalable Synthesis of Natural and Modified Nucleoside Triphosphates, J. Am. Chem. Soc., 2019, 141, 13286–13289 CrossRef CAS PubMed.
- S. P. Sau, N. E. Fahmi, J.-Y. Liao, S. Bala and J. C. Chaput, A scalable synthesis of α-L-threose nucleic acid monomers, J. Org. Chem., 2016, 81, 2302–2307 CrossRef CAS.
- A. Nikoomanzar, D. Vallejo, E. J. Yik and J. C. Chaput, Programmed allelic mutagenesis of a DNA polymerase with single amino acid resolution, ACS Synth. Biol., 2020, 9, 1873–1881 CrossRef CAS PubMed.
- X. Lou, M. Egli and X. Yang, Determining Functional Aptamer–Protein Interaction by Biolayer Interferometry, Curr. Protoc. Nucleic Acid Chem., 2016, 67, 7251–72515 Search PubMed.
- R. M. Lu, Y. C. Hwang, I. J. Liu, C. C. Lee, H. Z. Tsai, H. J. Li and H. C. Wu, Development of therapeutic antibodies for the treatment of diseases, J. Biomed. Sci., 2020, 27, 1 CrossRef CAS.
- M. Baker, Reproducibility crisis: Blame it on the antibodies, Nature, 2015, 521, 274–276 CrossRef CAS PubMed.
- S. D. Jayasena, Aptamers: An emerging class of molecules that rival antibodies in diagnostics, Clin. Chem., 1999, 45, 1628–1650 CrossRef CAS.
- M. R. Dunn, R. M. Jimenez and J. C. Chaput, Analysis of aptamer discovery and technology, Nat. Rev. Chem., 2017, 1, 0076 CrossRef CAS.
- J. Zhou and J. Rossi, Aptamers as targeted therapeutics: current potential and challenges, Nat. Rev. Drug Discovery, 2017, 16, 181–202 CrossRef CAS PubMed.
- F. Pfeiffer, M. Rosenthal, J. Siegl, J. Ewers and G. Mayer, Customised nucleic acid libraries for enhanced aptamer selection and performance, Curr. Opin. Biotechnol., 2017, 48, 111–118 CrossRef CAS PubMed.
- M. C. Culbertson, K. W. Temburnikar, S. P. Sau, J.-Y. Liao, S. Bala and J. C. Chaput, Evaluating TNA stability under simulated physiological conditions, Bioorg. Med. Chem. Lett., 2016, 26, 2418–2421 CrossRef CAS PubMed.
- E. Medina, E. J. Yik, P. Herdewijn and J. C. Chaput, Functional Comparison of Laboratory-Evolved XNA Polymerases for Synthetic Biology, ACS Synth. Biol., 2021, 10, 1429–1437 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: The experimental methods and compound characterization. See DOI: https://doi.org/10.1039/d3cb00159h |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2024 |