Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
A precise design strategy for a cell-derived extracellular matrix based on CRISPR/Cas9 for regulating neural stem cell function†
Yuanxin
Zhai
a,
Lingyan
Yang
 *a,
Wenlong
Zheng
b,
Quanwei
Wang
a,
Zhanchi
Zhu
a,
Fang
Han
a,
Ying
Hao
a,
Sancheng
Ma
*b and
Guosheng
Cheng
*ac
*a,
Wenlong
Zheng
b,
Quanwei
Wang
a,
Zhanchi
Zhu
a,
Fang
Han
a,
Ying
Hao
a,
Sancheng
Ma
*b and
Guosheng
Cheng
*ac
aCAS Key Laboratory of Nano-Bio Interface, Suzhou Institute of Nano-Tech and Nano-Bionics. Chinese Academy of Sciences, Suzhou, Jiangsu 215123, China. E-mail: lyy2013@sinano.ac.cn; gscheng2006@sinano.ac.cn
bSuzhou Kowloon Hospital, Shanghai Jiaotong University Medical School, Suzhou, Jiangsu 215123, China. E-mail: sanchengma@hotmail.com
cGuangdong Institute of Semiconductor Micro-Nano Manufacturing Technology, Guangdong 528200, China
First published on 15th March 2023
Abstract
The extracellular matrix (ECM) is a natural microenvironment pivotal for stem cell survival, as well as proliferation, differentiation and metastasis, composed of a variety of biological molecular complexes secreted by resident cells in tissues and organs. Heparan sulfate proteoglycan (HSPG) is a type of ECM protein that contains one or more covalently attached heparan sulfate chains. Heparan sulphate chains have high affinity with growth factors, chemokines and morphogens, acting as cytokine-binding domains of great importance in development and normal physiology. Herein, we constructed endogenous HSPG2 overexpression in mouse embryonic fibroblasts based on the CRISPR/Cas9 synergistic activation mediator system and then fabricated a cell-derived HSPG2 functional ECM (ECMHSPG2). The ECMHSPG2 is capable of enriching basic fibroblast growth factor (bFGF), which binds more strongly than the negative control ECM. With a growing bFGF concentration, ECMHSPG2 could better maintain neural stem cell (NSCs) stemness and promote NSC proliferation and differentiation in culture. These findings provide a precise design strategy for producing a specific cell-derived ECM for biomaterials in research and regenerative medicine.
Introduction
Stem cell survival, maintenance, proliferation, differentiation and metastasis are regulated by the microenvironment that they reside in.1–3 The microenvironment regulates stem cell fate through specific niche components, including supporting cells, growth factors and the extracellular matrix (ECM).4,5 The ECM is the natural microenvironment for all cells in the body, produced and secreted by resident cells in tissues or organs.6 The complicated relationship between stem cells and their surrounding ECM is a crucial control mechanism for stem cell function. The ECM derived from human umbilical vein endothelial cells could promote osteogenic differentiation of human bone marrow mesenchymal stem cells.7 It has been shown that the ECM is a vital regulator of epidermal stem cell fate, and destructive ECM components could impair epidermal stem cell homeostasis and morphogenesis.8 A cell-derived ECM with suitable hydrophilic properties could promote neural progenitor cell adhesion, proliferation and differentiation into basal forebrain cholinergic neurons.9 The ECM could be a favourable cell culture carrier for neural progenitor cell attachment and has great application potential in cell therapy for Alzheimer's disease.Although these basic functions are well defined, currently little is known about ECM individual components and their roles in stem cell fate regulation. One of the major challenges in the fields of tissue engineering and regenerative medicine is unravelling this mystery and designing biomaterials to repair or replace damaged tissues and organs. Nowadays, most ECM proteins have been well described and characterized. Complex ECM proteins can be classified into two main types: structural matrix proteins and proteoglycans.10 Collagens, laminin, fibronectin and elastin are major structural proteins that provide the structural support and biomechanical stability for tissues.11 Proteoglycans, which consist of a core protein and one or more covalently bound glycosaminoglycan chains, play many essential roles in biological processes, including development and disease progression.12,13 Heparan sulfate proteoglycan (HSPG) is one kind of proteoglycan consisting of a core protein with several covalently attached heparan sulfate chains, and it is widely distributed on the cell surface and in the ECM, where it interacts with a variety of ligands to regulate cellular function.14 Over the last decade, the binding properties of these chains have attracted more attention. Heparan sulfate chains have high affinity for heparin-binding growth factors, cytokines, chemokines and morphogens, and act as cytokine-binding domains, which increases the local concentration and protects them against proteolysis.15 Owing to the great capacity of HSPG for holding water and binding growth factors, it has been reported to participate in many kinds of biological activities and processes.16
HSPGs in the ECM can maintain endogenous stem cells or serve as a platform for receiving and directing the integration of transplanted stem cells. The development of gene editing tools allows humans to precisely edit target genes by the deletion, insertion, or site-directed mutagenesis of specific DNA segments.17 However, the transcript of HSPG2 is more than 13![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 bp, which exceeds the delivery capacity of traditional virus vectors, including lentiviral, adenovirus and adeno-associated virus vectors. Because of the convenient operation and efficient gene editing ability of the CRISPR/Cas9 system, it has been widely used by researchers in the field of genome editing for species including humans, rats, mice, fish, fruit flies, pigs, rice, wheat and bacteria and exhibits great application potential in disease prevention and intervention, animal and crop breeding, animal disease model development, and other fields.18,19
000 bp, which exceeds the delivery capacity of traditional virus vectors, including lentiviral, adenovirus and adeno-associated virus vectors. Because of the convenient operation and efficient gene editing ability of the CRISPR/Cas9 system, it has been widely used by researchers in the field of genome editing for species including humans, rats, mice, fish, fruit flies, pigs, rice, wheat and bacteria and exhibits great application potential in disease prevention and intervention, animal and crop breeding, animal disease model development, and other fields.18,19
In this study, we designed and fabricated a specific HSPG2-enriched ECM (ECMHSPG2) based on the CRISPR/Cas9 synergistic activation mediator (SAM) system. The endogenous HSPG2 was activated and overexpressed in mouse embryonic fibroblasts (MEFs). Additionally, the multiple functions of the ECMHSPG2 were evaluated, including the loading of the endogenous basic fibroblast growth factor (bFGF), which is a pivotal factor for neural stem cell (NSC) survival and development and also the promotion of NSC proliferation and differentiation into neurons in vitro. This ECM engineering strategy gives an example of manipulating cell-derived ECM and holds great promise for stem-cell based therapy and tissue regeneration.
Results
Endogenous HSPG2 overexpressing MEFs
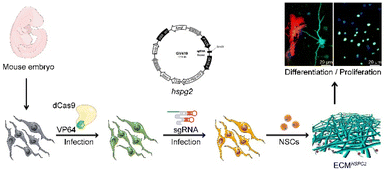
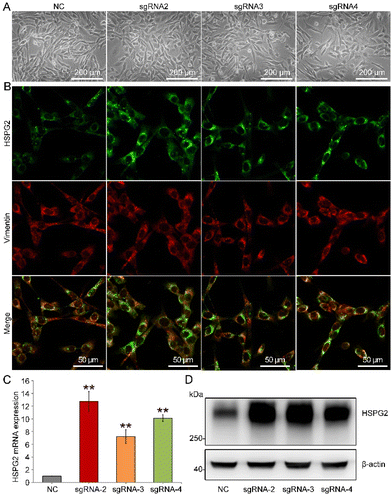
The strategic overview of our experiment is presented in Fig. 1. HSPG2-overexpressing MEFs were constructed to increase the content level of HSPG2 in a cell-derived ECM. In an SAM system, the RNA-guided nuclease-null Cas9 (dCas9) is fused to a transcriptional activator, VP64, which is reengineered as a programmable transcriptional factor that can activate the transcriptional activity of target genes through the binding of single-guide RNAs (sgRNAs) to the promoter regions.20Herein, we designed three sgRNAs (named sgRNA-2, sgRNA-3 and sgRNA-4) targeting three different loci within the HSPG2 promoter. Immunofluorescence staining, real-time PCR and western blot results showed that the expression of HSPG2 was significantly increased in MEFs (Fig. 2 and Fig. S1†). Different sgRNA groups showed about 12.71-, 11.48- and 11.08-fold higher levels of transcription activation than the negative sgRNA infection control (NC) (Fig. 2C), separately, and the sgRNA-2 group was selected in the subsequent experiments, due to its highest expression in both mRNA and protein levels (Fig. 2D). These results indicated that endogenous transcriptional activation of HSPG2 in MEFs had been achieved through the SAM system.
Fabrication and characterization of cell-derived ECM
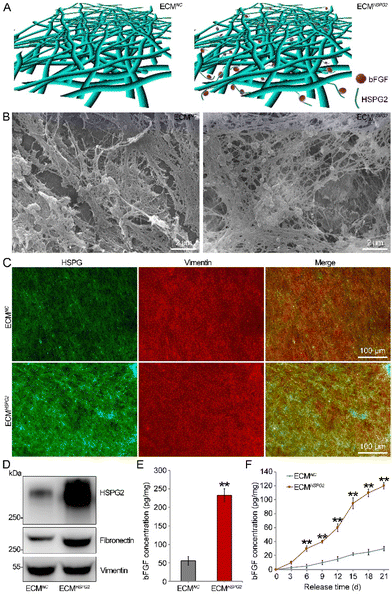
The NC or HSPG2 transcriptional activated MEFs fabricated cell-derived ECMs, named ECMNC or ECMHSPG2, respectively, as described in Fig. 3A. Upon stimulation with L-ascorbic acid and sodium ascorbate, the cells were embedded in the deposited ECM secreted by themselves and interwoven together. After decellularization, the remaining ECMNC or ECMHSPG2 consisting of a large number of interconnected micro- or nanoscale filaments was able to form a highly porous network, as shown by scanning electron microscopy (Fig. 3B). Then, the composition of the cell-derived ECM was identified by immunofluorescence staining and western blot analysis. It was shown that HSPG2 protein content in the ECMHSPG2 was significantly higher than that in the ECMNC from both immunofluorescence (Fig. 3C) and western blot results (Fig. 3D). Fibronectin, another important ECM structural protein, was also heavily deposited in the ECMHSPG2 compared to in ECMNC without changing its mRNA level. Meanwhile, there was no significant difference in the level of vimentin between these two kinds of cell-derived ECMs (Fig. 3D and Fig. S2, S3, S4†).bFGF is a heparin-binding growth factor which possesses mitogenic and cell-survival promoting functions involved in a variety of biological processes.21 It has been proved that bFGF is a crucial factor for NSC proliferation and differentiation.22 According to the results of ELISA, ECMHSPG2 could significantly enrich the endogenous bFGF to a level of 232.68 ± 18.56 pg mg−1 of total ECM protein, while the enrichment level of endogenous bFGF was 55.26 ± 12.43 pg mg−1 in the ECMNC group (Fig. 3E). We also detected the other two HSPG2-overexpressing MEFs constructed by sgRNA-3 and sgRNA-4, and they all could enrich the endogenous bFGF, significantly, without obvious changes to the Fgf2 mRNA (Fig. S2†). Moreover, the amount of bFGF released from ECMHSPG2 was significantly more than that from ECMNC at all the time points detected (Fig. 3F). These results showed that MEF-derived ECMHSPG could not only enrich endogenous bFGF, but also promote the release of bFGF to the surrounding microenvironment, providing it with promising application potential for regulating the NSC cell fate.
NSC proliferation capacity on the cell-derived ECM
The NSCs were initially cultured under serum-free conditions in the presence of bFGF to promote the proliferation of primary neurospheres (Fig. S5A†). The neurospheres were characterized by immunofluorescence staining with protein markers for NSCs including nestin and Sox2 (Fig. S5B†). As investigated by immunofluorescence staining with an anti-nestin antibody, ECMNC and ECMHSPG2 both effectively maintained the stemness properties after NSC adherent culture for 7 d (Fig. 4).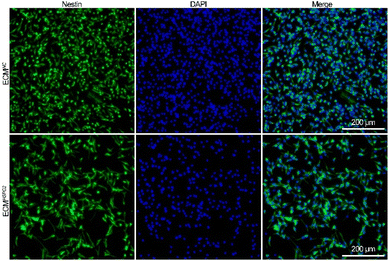 | ||
| Fig. 4 Immunofluorescence staining against nestin images of NSCs cultured on ECMNC or ECMHSPG2 for 7 d. | ||
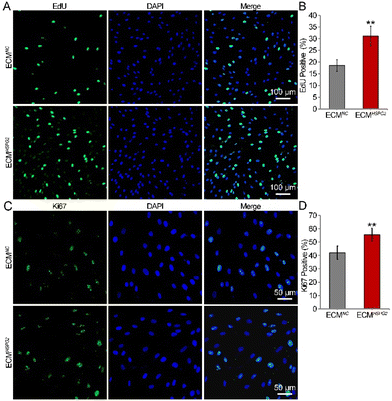
The proliferation capacity of NSCs cultured on ECMNC or ECMHSPG2 was examined by measuring the ratio of EdU-positive cells after NSCs were maintained in the proliferation medium for 7 d. The EdU/DAPI immunofluorescence results showed that ECMHSPG2 promoted NSC proliferation to a significantly higher level than ECMNC (Fig. 5A). The percentage of EdU-positive cells in the ECMHSPG2 group was significantly higher than that of the ECMNC group (31 ± 4% vs. 18 ± 2%, Fig. 5B). The NSC proliferation capacity was also verified by measuring the ratio of Ki67-positive cells (Fig. 5C), and similar to the EdU incorporation result, the percentage of Ki67-positive cells in the ECMHSPG2 group was significantly higher than that of the ECMNC group (55 ± 4% vs. 42 ± 5%, Fig. 5D). These results indicated that ECMHSPG2 could promote the proliferation of NSCs, probably through the enrichment and release of bFGF.
NSC differentiation capacity on cell-derived ECM
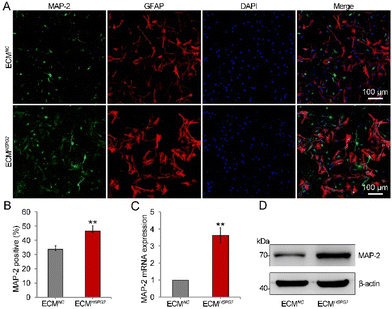
In the presence of EGF and bFGF, NSCs remain in a relatively undifferentiated state. With the addition of serum, B27 or N2 supplement and the removal of cytokines to the culture media, NSCs may be induced to differentiate into neurons, astrocytes, and oligodendrocytes.23 In order to stimulate the proliferation of NSCs before differentiation, bFGF can be added to the medium at the early stage of the differentiation process. However, an alternative differentiation method, which leverages the stimulation of bFGF, could induce increased numbers of neurons. Since ECMHSPG2 could significantly enrich and release bFGF (Fig. 3E and F), we next investigated the influence of it on the differentiation of NSCs through culturing NSCs on ECMNC or ECMHSPG2 without cytokines. After 14 d of differentiation, immunofluorescence staining showed a vast increase in the number of cells positive for microtubule-associated protein 2 (MAP-2) from 33 ± 2% on ECMNC to 46 ± 3% on ECMHSPG2 (Fig. 6A and B). Meanwhile, there was no significant difference between ECMNC and ECMHSPG2 groups regarding the number of GFAP-positive cells (Fig. S6A†). For further quantitative analysis, cells were harvested and subjected to real-time PCR and western blot assays. As shown in Fig. 6C and D, compared with the cells on ECMNC, a significant increase in the expression of MAP-2 was achieved in the cells cultured on ECMHSPG2. We also did not observe any significant differences in GFAP mRNA and protein expression between the two conditions (Fig. S6B and C†). These results suggested that ECMHSPG2 could increase the differentiation capacity of NSCs, probably through the stimulation of bFGF. Furthermore, owing to its proliferation- and differentiation-promoting properties, this ECM engineering strategy represents a prospective tool for cell therapy and even tissue repair in the future.Discussion
ECM-mimicking substrates and matrices improve cell culture conditions and focus on tissue engineering applications, establishing superior substrates to remodel and repair diseased tissues and organs.24 In order to improve the efficiency of drug delivery and the function of a drug in an environment mimicking the in situ cellular niche, more materials are being developed to simulate the ECM.25 To this end, electrostatic spinning, hydrogels and three-dimensional bio-printing technologies have been used to produce scaffolds from synthetic materials or a limited set of natural polymer blends, but none of these have adequately mimicked the complex morphology and composition of natural ECM.26,27 Meanwhile, the application of tissue-derived ECM scaffolds also has limitations, such as limited donors, potential pathogen transmission and uncontrollable degradation. Therefore, cell-derived ECM has become a promising biomaterial for tissue engineering and regenerative medicine, mainly due to its ability to establish a bionic microenvironment that provides biochemical and physical cues for cells.28,29 Purified individual ECM components, including fibrin, laminin, fibronectin, collagen and hyaluronan etc., have no significant effects on cell proliferation and differentiation. In previous work, it has been shown that cell-derived ECM presented a more suitable hydrophilicity than laminin, which is favourable for cell adhesion.9 The appropriate hydrophilicity surfaces could adsorb more proteins and transmit signals to the cells through cell adhesion receptors and thereby affect cell survival, growth and differentiation. Various mesenchymal cells or fibroblasts produce cell-derived ECM when stimulated by L-ascorbic acid and 15% fetal bovine serum (FBS), which provide adequate nutrition for the cells. However, the field is progressing towards the expansion/differentiation of NSCs under chemically defined and xeno-free conditions. The composition of FBS is uncertain, nor it is clear whether the composition of FBS could affect the composition of cell-derived ECM deposition. Therefore, the use of serum-free components, such as N2 and B27, to stimulate cell-derived ECM formation, or the use of different batches or even different brands of FBS tests, should be further validated in the future.In addition, cell-derived ECM with specific functions can be designed and fabricated by altering the expression of specific ECM protein components in the originating cells. The cell-derived ECM enriched with a collagen-binding domain fused with membrane metalloendopeptidase or an insulin-degrading enzyme could reduce the aggregation of Aβ peptides. This prevents the formation of amyloid plaques and also suppresses the phosphorylation of Tau protein, providing a cell-derived ECM that can be potentially used in the treatment of Alzheimer's disease.30 Farhang N et al.31 demonstrated the increased expression and deposition of aggrecan and collagen II without exogenous growth factors in ECM pellet culture. Fibronectin knockout by CRISPR/Cas9 in human infrapatellar fat pad-derived stem cells (IPFSCs) and the derived decellularized ECM could inhibit the proliferation of high-passage IPFSCs along with a decline in CDH2 expression.32
The SAM system enables the robust transcriptional activation of endogenous genes targeted by sgRNAs that bind within 200 bp upstream of the transcription start site.33 This system fuses RNA-guided dCas9 with a well-characterized transcription regulatory domain, using predesigned sgRNAs to direct the binding of the transcriptional complex to the target sites. Using nuclease-null dCas9 protein, the complex can be targeted to specific sites without cutting or altering the genomic DNA. After dCas9 binds the targeted DNA sequence, the fused transcription regulatory domains recruit inhibitory or activating effectors to modify gene expression.34 Therefore, we could apply the SAM system to precisely design and develop cell-derived ECM with encapsulated growth factors.
Indeed, various stem cell types adapt to different specific ECM proteins, and some ECM proteins have specialized functions that are restricted to distinct stem cell niches, for stem cell survival and self-renewal in vivo and in vitro. For example, tenascin C and osteopontin are specifically present in hematopoietic stem cell niches, whereas β-1 integrins are predominantly expressed in epidermal stem cell niches.35 Fibronectin induces the differentiation of either myoblasts into skeletal muscle cells or adipose tissue-derived stem cells into endothelial cells.36,37 Proteoglycans can be considered as one of the first critical ECM components for maintaining normal cell function and tissue development.38 With multiple biological functions in development and disease, HSPG2 is a key proteoglycan.39 Importantly, HSPG2 regulates cellular behaviour and biological processes by binding cytokines, growth factors and chemokines, such as bFGF.15,40
Like other cytokines, bFGF is unstable under physiological conditions and is likely to be broken down by proteolytic degradation when in an unbound state. When injected in vivo, the biological functions of bFGF were reduced due to diffusion, enzymolysis, and weak binding with co-receptors. However, HSPG2 protects bFGF from proteolytic degradation, thanks to its physical structure, and it can also induce bFGF biological activities by acting as a co-receptor.16 In addition to bFGF, other cytokines that possess binding affinity to HSPG2, such as VEGF, BMP-9, EGF, FGF8 and SHH, are also considered to be pivotal for stem cell fate and function.14,16,41,42 Our designed ECMHSPG2 is an ideal delivery carrier that enriches the endogenous and exogenous bFGF, which can be further distributed at the injury site and degraded by proteolysis under normal physiological conditions. In this study, with enriched bFGF, ECMHSPG2 promotes NSC proliferation and their differentiation into the neurons, which signifies that ECMHSPG2 has the potential to become an agent for stem cell transplantation.
As a major component of ECM in the basement membrane, HSPG also binds with many other ECM proteins to form the skeleton of the ECM, making HSPG a link molecule targeting ECM proteins. Herein, we showed that HSPG2 is capable of facilitating ECM remodeling and the deposition of structural matrix proteins, such as fibronectin, which plays an essential role in cell adhesion, migration and the development of embryonic vascular structures.43 Thus, we suggest that using this ECM engineering strategy will benefit not only fundamental research about the ECM function, but also clinical applications of ECM in cell therapy and tissue repair.
Experimental
All animal procedures were performed in accordance with the Guidelines for the Care and Use of Laboratory Animals of Suzhou Institute of Nano-Tech and Nano-Bionics, Chinese Academy of Sciences, and approved by the Animal Ethics Committee of Suzhou Institute of Nano-Tech and Nano-Bionics, Chinese Academy of Sciences. Efforts were made to minimize the number of animals used and any discomfort experienced. Detailed information on essential materials used in this study can be found in the ESI.†Isolation and culture of MEFs
Skin from an E12.5 fetal C57BL/6 mouse was collected in a bacteria-free environment and then cut into small pieces and spread onto a culture dish. The pieces were continuously cultured for 4–7 d in high-glucose DMEM supplemented with 10% fetal bovine serum (FBS). The MEFs were purified using the trypsin digestion-adherence method.Construction of endogenous HSPG2 transcriptionally activated MEFs
CRISPR plasmids and lentiviral vectors for dCAS9-VP64-Puro and gRNA LV-sgRNA-MS2-P65-HSF1-Neo (GV419) were constructed by Genechem (Shaihai, China). MEFs infected with dCAS9-VP64-Puro and negative gRNA were used as the negative control. The sequences of sgRNAs used in this article are listed in Table S1.† MEFs were seeded in 6-well plates at a density of 1 × 104 cells per cm2 and transduced at 10 MOI with dCAS9-VP64-Puro. Cells were subcultured and selected with 2 μg mL−1 puromycin. After two weeks of selection culture, cells were seeded in 6-well plates at a density of 1 × 104 cells per cm2 and infected with 10 MOI Lenti-MS2-p65-HSF1. Cells were subcultured and selected with 10 μg mL−1 G418. After stable cell lines were obtained, the HSPG2 expression level was assessed by real-time PCR, western blot assay and immunofluorescence staining.Production of cell-derived ECMNC and ECMHSPG2
Cell-derived ECMs were fabricated by stimulating the MEFs or HSPG2 transcriptionally activated MEFs, according to our previously published work.9 In brief, the culture substrates were pre-treated by incubating with 0.5% gelatin at 37 °C overnight and exposed to ultraviolet rays for 2 h to enhance crosslinking. Cells were seeded on the substrates at the same density of 5 × 104 cells per cm2. After the cells were cultured to 100% confluence, the medium was changed to DMEM with 15% FBS and 50 mg mL−1L-ascorbic acid and 100 mg mL−1 sodium ascorbate for 10 d. After the stimulation of ECM formation, the cells were washed with deionized water for 20 min, followed by treatments with 0.5% Triton X-100 plus 1% deoxycholate for 10 min and with 100 U mL−1 DNase I for 30 min at 37 °C.Characterization of the binding and release of bFGF
The bFGF binding and release assay was performed using an FGF2 (bFGF) Mouse ELISA Kit according to the manufacturer's instructions. Freshly prepared cell-derived ECMs were dissociated in RIPA lysis buffer with a protease inhibitor, and the supernatant was obtained by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g. The concentration of ECM-bound bFGF was normalized to the total ECM protein content, detected using a BCA kit. To measure the amount of bFGF contained in ECM, cell-derived ECMs were placed at 37 °C for 0, 3, 6, 9, 12, 15, 18, and 21 d, and the supernatant was collected and tested using the ELISA kit as described above.
000g. The concentration of ECM-bound bFGF was normalized to the total ECM protein content, detected using a BCA kit. To measure the amount of bFGF contained in ECM, cell-derived ECMs were placed at 37 °C for 0, 3, 6, 9, 12, 15, 18, and 21 d, and the supernatant was collected and tested using the ELISA kit as described above.
Isolation and culture of NSCs
The cerebral cortex was dissected from the brain of fetal mice at E14 and then digested with trypsin at 37 °C for 2 min. After gentle dissociation into small pieces using a pipette, single cells were acquired by centrifugation. The cells were re-suspended and cultured in a proliferation medium containing NeuroCult Basal Medium with 10% NeuroCult Proliferation Supplement, 1% PS, 20 ng mL−1 of human recombinant epidermal growth factor (EGF), and 10 ng mL−1 of human recombinant bFGF. The cells were expanded as spheres, and the medium was half-refreshed every 3 d. In this study, passage 3 and 5 NSCs were used for further experiments.Proliferation and differentiation of NSCs
The NSCs were cultured in the proliferation medium as previously mentioned. For the differentiation study, the NSC spheres were digested into single cells with Accutase and seeded on the ECM at a density of 2 × 105 cells per cm2 in the proliferation medium for 12 h, and then the medium was changed to the differentiation medium containing NeuroCult Basal Medium with 2% B27, 2 μM glutamine, and 1% PS for 14 d. The differentiation medium was replaced every 3 d.Immunofluorescence staining
MEFs, ECM and NSCs were fixed in 4% paraformaldehyde at room temperature for 20 min, washed with PBS and blocked with 5% normal goat serum containing 0.1% Triton X-100 at room temperature for 1 h. Subsequently, the cells were incubated with primary antibodies specific to HSPG2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), vimentin (1
200), vimentin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), Nestin (1
200), Nestin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), Sox-2 (1
200), Sox-2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200), Ki67 (1
200), Ki67 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) and MAP-2 (1
200) and MAP-2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200) at 4 °C overnight. After washing, the cells were incubated with Alexa Fluor 488 or 594 labelled secondary antibodies in the dark at room temperature for 2 h. The cell nuclei were counterstained with 5 μg mL−1 DAPI. Fluorescence microscopy images were collected using a confocal laser scanning microscope with the same magnification and the same fluorescence intensity (Olympus, Japan). The percentages of EdU-, Ki67- and MAP2-positive cells were calculated from ten random fields, and each experiment was performed in triplicate.
200) at 4 °C overnight. After washing, the cells were incubated with Alexa Fluor 488 or 594 labelled secondary antibodies in the dark at room temperature for 2 h. The cell nuclei were counterstained with 5 μg mL−1 DAPI. Fluorescence microscopy images were collected using a confocal laser scanning microscope with the same magnification and the same fluorescence intensity (Olympus, Japan). The percentages of EdU-, Ki67- and MAP2-positive cells were calculated from ten random fields, and each experiment was performed in triplicate.
Real-time PCR
Total RNA was isolated from MEFs and NSCs with TRIzol reagent, and the concentration was quantified using a Nano100 spectrophotometer (Thermo, USA). First-strand cDNA was synthesized from 1 μg total RNA (A260/280: 1.8–2.0, A260/230: 2.0–2.2) using a first-strand cDNA synthesis kit. Thereafter, real-time PCR was carried out using the obtained cDNA with SYBR Mixture for 40 cycles. The 2−ΔΔCt method was used to calculate the target gene relative mRNA expression. The primers were synthesized by Genewiz (Suzhou, China), and the sequences are listed in Table S1.† GAPDH was used as an endogenous control, and each experiment was performed in triplicate.Western blot
MEFs, ECM and NSCs were lysed in an ice-cold RIPA buffer with a protease inhibitor. The lysates were then centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 20 min to collect the supernatant. The protein concentration was determined with a BCA assay kit and denaturised in boiling water for 5 min. Equal amounts of proteins were separated by SDS-PAGE and transferred to the PVDF membrane. After being blocked for 1 h at room temperature, the membrane was then incubated with primary antibodies specific to HSPG2 (1
000g for 20 min to collect the supernatant. The protein concentration was determined with a BCA assay kit and denaturised in boiling water for 5 min. Equal amounts of proteins were separated by SDS-PAGE and transferred to the PVDF membrane. After being blocked for 1 h at room temperature, the membrane was then incubated with primary antibodies specific to HSPG2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), fibronectin (1
1000), fibronectin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), vimentin (1
1000), vimentin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000), MAP-2 (1
1000), MAP-2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) and β-actin (1
1000) and β-actin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) at 4 °C overnight. The membranes were washed with TBST three times and then incubated with HRP-conjugated secondary antibodies (1
1000) at 4 °C overnight. The membranes were washed with TBST three times and then incubated with HRP-conjugated secondary antibodies (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000) at room temperature for 2 h. The protein bands were visualized using an ECL substrate under an LAS4000 imaging system (Fuji Film, Japan). β-Actin was used as an endogenous control, and each experiment was performed in triplicate.
1000) at room temperature for 2 h. The protein bands were visualized using an ECL substrate under an LAS4000 imaging system (Fuji Film, Japan). β-Actin was used as an endogenous control, and each experiment was performed in triplicate.
Scanning electron microscopy imaging
Fresh prepared ECM samples were washed three times with PBS and fixed with 4% paraformaldehyde for 20 min at room temperature, followed by post-fixation in 2.5% glutaraldehyde for 30 min at 4 °C. The samples were dehydrated in 50%, 70%, 80%, 90%, and 100% ethanol for 10 min, respectively. After being replaced with 100% tert-butanol, they were frozen at −20 °C for 30 min and lyophilized in a vacuum dryer. The ECM samples were coated with a gold film before observation under a scanning electron microscope (Quanta 400 FEG, FEI, USA).Statistical analysis
All experiments were performed in triplicate, and the data are shown as the means ± SD of three separate experiments. Statistical analysis was performed using one-way analysis of variance followed by Dunnett's two-tailed test, and then Student's independent samples t-test was performed to compare the difference between the two groups. Probability values of **p < 0.01 were considered significant.Conclusions
Although various artificial materials have been exploited to mimic the ECM regulation of stem cell function, there were almost no reports on how to make natural cell-derived ECM that is more appropriate for stem cell study and application. In this study, we fabricated a specific functional cell-derived ECM based on the SAM system. In addition, the ECM protein can also be used to enrich bFGF suggesting that ECMHSPG2 is a promising biomaterial candidate for the stem cell niche. ECMHSPG2 promoted NSC proliferation and differentiation into neurons in vitro. This research will provide a precise design strategy to develop a functional cell-derived ECM for stem cell-based biomaterials research and regenerative medicine. However, the mechanism by which HSPG2 increases the fibronectin deposition and bFGF binding efficiency requires further study.Author contributions
Y. L. Y. and Z. Y. X.: conceptualization and writing – original draft; Z. Y. X., and Z. W. L.: data curation; W. Q. W., Z. Z. C., H. F. and H. Y.: investigation; and M. S. C. and C. G. S.: funding acquisition and writing – review & editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by grants from the National Natural Science Foundation of China (31700831) and Guangdong Province Joint Fund (2019B1515120090).References
- D. T. Scadden, Nature, 2006, 441, 1075 CrossRef CAS PubMed.
- D. Lucas, Adv. Exp. Med. Biol., 2017, 1041, 5 CrossRef CAS PubMed.
- G. M. Crane, E. Jeffery and S. J. Morrison, Nat. Rev. Immunol., 2017, 17, 573 CrossRef CAS PubMed.
- M. Votteler, P. J. Kluger, H. Walles and K. Schenke-Layland, Macromol. Biosci., 2010, 10, 1302 CrossRef CAS PubMed.
- Y. Xu, J. Zhou, C. Liu, S. Zhang, F. Gao, W. Guo, X. Sun, C. Zhang, H. Li, Z. Rao, S. Qiu, Q. Zhu, X. Liu, X. Guo, Z. Shao, Y. Bai, X. Zhang and D. Quan, Biomaterials, 2021, 268, 120596 CrossRef CAS PubMed.
- S. Yi, F. Ding, L. Gong and X. Gu, Curr. Stem Cell Res. Ther., 2017, 12, 233 CrossRef CAS PubMed.
- Y. Kang, S. Kim, J. Bishop, A. Khademhosseini and Y. Yang, Biomaterials, 2012, 33, 6998 CrossRef CAS PubMed.
- E. Chermnykh, E. Kalabusheva and E. Vorotelyak, Int. J. Mol. Sci., 2018, 19, 1003 CrossRef PubMed.
- L. Yang, Z. Jiang, L. Zhou, K. Zhao, X. Ma and G. Cheng, RSC Adv., 2017, 7, 45587 RSC.
- A. D. Theocharis, S. S. Skandalis, C. Gialeli and N. K. Karamanos, Adv. Drug Delivery Rev., 2016, 97, 4 CrossRef CAS PubMed.
- J. Halper and M. Kjaer, Adv. Exp. Med. Biol., 2014, 802, 31 CrossRef CAS PubMed.
- L. M. Jenkins, B. Horst, C. L. Lancaster and K. Mythreye, Cytokine Growth Factor Rev., 2018, 39, 124 CrossRef CAS PubMed.
- N. K. Karamanos, Z. Piperigkou, A. D. Theocharis, H. Watanabe, M. Franchi, S. Baud, S. Brézillon, M. Götte, A. Passi, D. Vigetti, S. Ricard-Blum, R. D. Sanderson, T. Neill and R. V. Iozzo, Chem. Rev., 2018, 118, 9152 CrossRef CAS PubMed.
- M. Xie and J. P. Li, Cell. Signalling, 2019, 54, 115 CrossRef CAS PubMed.
- S. Sarrazin, W. C. Lamanna and J. D. Esko, Cold Spring Harbor Perspect. Biol., 2011, 3(7), a004952 Search PubMed.
- J. Shi, C. Fan, Y. Zhuang, J. Sun, X. Hou, B. Chen, Z. Xiao, Y. Chen, Z. Zhan, Y. Zhao and J. Dai, Biomater. Sci., 2019, 7, 5438 RSC.
- F. Akram, U. H. Ikram, Z. Ahmed, H. Khan and M. S. Ali, Protein Pept. Lett., 2020, 27, 931 CrossRef CAS PubMed.
- C. Lyu, J. Shen, R. Wang, H. Gu, J. Zhang, F. Xue, X. Liu, W. Liu, R. Fu, L. Zhang, H. Li, X. Zhang, T. Cheng, R. Yang and L. Zhang, Stem Cell Res. Ther., 2018, 9, 92 CrossRef CAS PubMed.
- C. Jiang, L. Meng, B. Yang and X. Luo, Clin. Genet., 2020, 97, 73 CrossRef CAS PubMed.
- L. A. Gilbert, M. H. Larson, L. Morsut, Z. Liu, G. A. Brar, S. E. Torres, N. Stern-Ginossar, O. Brandman, E. H. Whitehead, J. A. Doudna, W. A. Lim, J. S. Weissman and L. S. Qi, Cell, 2013, 154, 442 CrossRef CAS PubMed.
- D. Nawrocka, M. A. Krzyscik, L. Opalinski, M. Zakrzewska and J. Otlewski, Int. J. Mol. Sci., 2020, 21, 4108 CrossRef CAS PubMed.
- C. Li, L. H. Che, L. Shi and J. L. Yu, J. Cell. Biochem., 2017, 118, 3875 CrossRef CAS PubMed.
- N. Li, Qi. Zhang, S. Gao, R. Huang, L. Wang, L. Liu, J. Dai, M. Tang and G. Cheng, Sci. Rep., 2013, 3, 1604 CrossRef PubMed.
- H. Lu, T. Hoshiba, N. Kawazoe, I. Koda, M. Song and G. Chen, Biomaterials, 2011, 32, 9658 CrossRef CAS PubMed.
- J. Hwang, M. O. Sullivan and K. L. Kiick, Front. Bioeng. Biotechnol., 2020, 8, 69 CrossRef PubMed.
- M. Keshvardoostchokami, S. S. Majidi and P. Huo, Nanomaterials, 2020, 11, 21 CrossRef PubMed.
- J. Wang, Y. Yu, J. Guo, W. Lu, Q. Wei and Y. Zhao, Adv. Biosyst., 2020, 4(2), e1900238 CrossRef PubMed.
- Y. Yang, H. Lin, H. Shen, B. Wang, G. Lei and R. S. Tuan, Acta Biomater., 2018, 69, 71–82 CrossRef CAS PubMed.
- J. C. Silva, M. S. Carvalho, K. Xia, J. M. S. Cabral, C. L. da Silva, F. C. Ferreira, D. Vashishth and R. J. Linhardt, Methods Cell Biol., 2020, 156, 85 CAS.
- S. Zhang, T. Xiao, Y. Yu, Y. Qiao, Z. Xu, J. Geng, Y. Liang, Y. Mei, Q. Dong, B. Wang, J. Wei and G. Suo, J. Tissue Eng. Regener. Med., 2019, 13, 1759 CrossRef CAS PubMed.
- N. Farhang, B. Davis, J. Weston, M. Ginley-Hidinger, J. Gertz and R. D. Bowles, Tissue Eng., Part A, 2020, 26, 1169 CrossRef CAS PubMed.
- Y. Wang, Y. Fu, Z. Yan, X. B. Zhang and M. Pei, Front. Bioeng. Biotechnol., 2019, 7, 321 CrossRef PubMed.
- S. Konermann, M. D. Brigham, A. E. Trevino, J. Joung, O. O. Abudayyeh, C. Barcena, P. D. Hsu, N. Habib, J. S. Gootenberg, H. Nishimasu, O. Nureki and F. Zhang, Nature, 2015, 517, 583 CrossRef CAS PubMed.
- P. Mali, J. Aach, J. H. Lee, D. Levner, L. Nip and G. M. Church, Nat. Methods, 2013, 10, 403 CrossRef CAS PubMed.
- S. Stier, Y. Ko, R. Forkert, C. Lutz, T. Neuhaus, E. Grünewald, T. Cheng, D. Dombkowski, L. M. Calvi, S. R. Rittling and D. T. Scadden, J. Exp. Med., 2005, 201, 1781 CrossRef CAS PubMed.
- S. Heydarkhan-Hagvall, K. Schenke-Layland, J. Q. Yang, S. Heydarkhan, Y. Xu, P. A. Zuk, W. R. MacLellan and R. E. Beygui, Cells Tissues Organs, 2008, 187, 263–274 CrossRef PubMed.
- M. A. Lan, C. A. Gersbach, K. E. Michael, B. G. Keselowsky and A. J. Garcia, Myoblast proliferation and differentiation on fibronectin-coated self assembled monolayers presenting different surface chemistries, Biomaterials, 2005, 26, 4523–4531 CrossRef CAS PubMed.
- N. Hassan, B. Greve, N. A. Espinoza-Sanchez and M. Gotte, Cell. Signalling, 2021, 77, 109822 CrossRef CAS PubMed.
- H. Kaneko, M. Ishijima, I. Futami, N. Tomikawa-Ichikawa, K. Kosaki, R. Sadatsuki, Y. Yamada, H. Kurosawa, K. Kaneko and E. Arikawa-Hirasawa, Matrix Biol., 2013, 32, 178 CrossRef CAS PubMed.
- J. R. Bishop, M. Schuksz and J. D. Esko, Nature, 2007, 446, 1030 CrossRef CAS PubMed.
- D. C. Kraushaar, S. Rai, E. Condac, A. Nairn, S. Zhang, Y. Yamaguchi, K. Moremen, S. Dalton and L. Wang, J. Biol. Chem., 2012, 287, 22691 CrossRef CAS PubMed.
- A. Wade, A. McKinney and J. J. Phillips, Biochim. Biophys. Acta, 2014, 1840, 2520 CrossRef CAS PubMed.
- M. Marchand, C. Monnot, L. Muller and S. Germain, Semin. Cell Dev. Biol., 2019, 89, 147 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: The detailed information for essential reagents, sequences of the sgRNA and real-time PCR primers, mRNA expression and protein expression of HSPG2 in three sgRNAs transfected MEFs. See DOI: https://doi.org/10.1039/d2bm01466a |
| This journal is © The Royal Society of Chemistry 2023 |