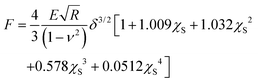A study of macrophage mechanical properties and functional modulation based on the Young's modulus of PLGA-PEG fibers†
Bokai
Zhang‡
 *abc,
Massimiliano
Galluzzi‡
*abc,
Massimiliano
Galluzzi‡
 c,
Guoqiao
Zhou
c and
Haoyang
Yu
*ab
c,
Guoqiao
Zhou
c and
Haoyang
Yu
*ab
aDGENE (Dongjin) Bighealth (Shenzhen) Co., Ltd, P.R. China. E-mail: sonicgoboy@gmail.com
bBenHealth Biopharmaceutical (Shenzhen) Co., Ltd, P.R. China
cMaterials and Interfaces Center, Shenzhen Institute of Advanced Technology, Chinese Academy of Science, Shenzhen 518055, Guangdong, China
First published on 14th November 2022
Abstract
The immune response of macrophages plays an important role in defending against viral infection, tumor deterioration and repairing of contused tissue. Macrophage functional differentiation induced by nanodrugs is the leading edge of current research, but nanodrugs have toxic side effects, and the influence of their physical properties on macrophages is not clear. Here we create an alternative way to modulate macrophage function through PLGA-PEG fibers’ Young's modulus. Previously, we revealed that by controlling the Young's modulus of the fibers from kPa to MPa, all the fibers entered murine macrophage cells (RWA 264.7) in a similar manner, and based on that, we found that macrophages’ mechanical properties were affected by the fibers’ Young's modulus, that is, hard fibers with a Young's modulus of ∼1 MPa increased the cell average Young's modulus, but did not affect the cell shape, while soft fibers with a Young's modulus of ∼100 kPa decreased the cell average Young's modulus and modulated the cell shape to a more spherical one. On the other hand, only the soft fibers induced proinflammatory cytokine secretion, indicating an M1 macrophage functional modulation by low Young's modulus fibers. This study explored the mechanical properties of the interactions between PLGA-PEG fibers and cells, in particular, when guiding the direction of the modulation of macrophage function, which is of great significance for the applications of material biology in the biomedical field.
1. Introduction
Macrophages are the most versatile cells of the immune system; they are found in all tissues and show great functional diversity.1 They play important roles in pathogen immunity,2 tissue development,3 homeostasis4 and damage.3 To achieve such functions, their functional modulation becomes important. For instance, the M1 pro-inflammatory phenotype increases the expression of pro-inflammatory cytokines and the production of reactive oxygen species, the M2 pro-healing phenotype, has anti-inflammatory and wound healing functions.5 At present, the understanding of macrophage function regulation is mostly limited to how soluble chemicals such as cytokines and chemokines affect differentiation, which brings cytotoxic side effects.6 According to such concerns, the application of nanoparticles as nanodrugs for immune modulation was proposed due to their low cytotoxicity upon controlled release.7 However, the nanoparticles cannot fully avoid cytotoxicity because they are still using the embedded/conjugated chemicals, which upon release still cause cytotoxic side effects, even though they are slighter than those by pure chemical means.8 Thus, finding another way to stimulate macrophage function differentiation in a safer way becomes important.The complexity of the macrophage microenvironment emphasizes that it is necessary to consider other factors that may affect macrophage function differentiation. Recent studies have shown that physical and mechanical signals can regulate the mechanical, differentiation, proliferation and migration behaviors of many cell types by modulating the integrin–ligand interaction and cytoskeleton reorganization (cell mechanics) of macrophages.9 For example, the surface morphology, geometry, Young's modulus and roughness of the pericellular environment can regulate the different behavior types of macrophages.10 Among them, cell mechanics was found to provide information about many physiological phenomena closely related to the cell structure and function.11 At the macro-scale level, alveolar cells (i.e. epithelial cells and macrophages) do not rupture in the continuous cyclic extension under controlled mechanical stress, while receiving mechanical signals from the environment and physiologically responding.12 Macrophages in the diseased lung tissue are affected by the high tensile stress of surrounding tissues (hardened cell microenvironment), and their mechanical state is also changed accordingly as a warning signal of lesions.13 The effects of these mechanical factors on macrophages are at the micro-scale level; however, whether these properties play a role in inducing macrophages to differentiate into the required phenotype remains unclear.
On the other hand, macrophages coordinate their behaviors during various biological processes largely depending on the mechanical properties of the external environment, providing us the possibility to apply materials’ mechanical properties to modulate macrophage function. Previously, authors successfully synthesized PLGA-PEG fibers with the Young's modulus ranging from kPa to MPa and found that fibers with high apparent Young's modulus (average value >240 kPa) maintained their original shape upon phagocytosis, while their counterparts with low apparent Young's moduli (average value <150 kPa) curled in cells. The observed deformation of soft fibers in cells coincided with abnormal intracellular actin translocation, the absence of lysosome/phagosome fusion in macrophages, and the resulting more rounded cell morphology, which is one of the physical indicators of the M1 phenotype;14 however, there is still a lack of evidence for the modulation of macrophage function by PLGA-PEG fibers’ Young's modulus, especially immune differentiation. To verify it, the M1 markers, such as iNOs,15 TNF-α and IFN-γ,16 and M2 markers, such as, Arg1,17 IL4 and IL10,16 are well accepted biochemical indicators. Thus, to complement the picture of the mechanical properties of materials and the mechano-related cellular pathways affecting cell physiology, the mechanical responses and physiological changes of macrophages induced by the fibers’ mechanical modulus are necessary. Other recent works show the efficacy of PLGA-PEG-based nanoparticles as drug carriers for tissue regeneration18 or inducing macrophage apoptosis.19 Although some achievements were made in biomedical applications, the underlying mechanical interaction was ignored. Still a deep understanding of the interactions between macrophages and the shapes of PLGA-PEG fibers from a biomechanical perspective is missing.
In this study, we selected two PLGA-PEG fibers with the highest (≈1 MPa) and lowest (≈100 kPa) Young's moduli according to the authors’ previous work to further investigate their influence on murine macrophage RAW 264.7 cells by mechanical means. Firstly, an AFM/fluorescence microscopy system was used to co-locally investigate the cell mechanics distribution, morphology and intracellular location of fibers; then cryo-TEM was used to study the ultra-fine cellular structure affected by the fibers, supplementing with the 3D confocal images which revealed the mechanical structure-actin affected by the fibers; and finally biochemical methods were applied to investigate the relevance between the changes of cell nanomechanics and phenotype differentiation induced by the fibers.
2. Experimental
2.1. Preparation of the PLGA-PEG fibers
Needle-shaped PLGA-PEG fibers were synthesized by a two-step process as reported previously.20 Firstly, the PLGA-PEG polymers (Akina, Inc.) were synthesized as spherical shaped particles via the precipitation/solvent diffusion method.21 Briefly, 50 mg of PLGA-PEG (and 0.5 mg of Nile Red (Invitrogen), only for the CLSM experiment) was dissolved in 1.25 mL of DCM (Sigma-Aldrich) for at least 3 h. The polymer solution was directly added to 5 mL of 5% PVA (Mw 31![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000–50
000–50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 Da, 87–89% hydrolyzed, Sigma) solution. The mixture was then homogenized for 1 min by using a probe sonicator at 10 W to generate an oil-in-water (O/W) emulsion. The formed emulsion was added to 25 mL of ice-cold DI water and stirred for 3 h at room temperature to evaporate DCM. After that, the particles were collected and washed by centrifugation at 15
000 Da, 87–89% hydrolyzed, Sigma) solution. The mixture was then homogenized for 1 min by using a probe sonicator at 10 W to generate an oil-in-water (O/W) emulsion. The formed emulsion was added to 25 mL of ice-cold DI water and stirred for 3 h at room temperature to evaporate DCM. After that, the particles were collected and washed by centrifugation at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500g at 4 °C, three times, and then passed through a 1.2 μm filter, resulting in ∼450 nm diameter spherical PLGA-PEG particles.
500g at 4 °C, three times, and then passed through a 1.2 μm filter, resulting in ∼450 nm diameter spherical PLGA-PEG particles.
Secondly, the synthesized spherical PLGA-PEG particles were stretched into needle-shaped particles via the reported stretching method.22 Briefly, the spherical PLGA-PEG particles were dispersed in a solution containing 10% PVA and 1% glycerol to a concentration up to 0.1% wt/vol. 15 mL of the solution was dried on a 12 × 16 cm2 flat surface to form an 80 μm thick film. The film was cut into sections and stretched in one direction at a temperature higher than 70 °C in air by using a custom-made apparatus. The finished films were dissolved in ice-cold DI water. The stretched fibers were collected by centrifugation at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g, and washed with the same solution at least 5 times to remove PVA from their surface. The fibers were finally freeze dried, weighed and stored at −20 °C for further use.
000g, and washed with the same solution at least 5 times to remove PVA from their surface. The fibers were finally freeze dried, weighed and stored at −20 °C for further use.
2.2. Introduction of PLGA-PEG fibers to cells
RAW 264.7 (ATCC®TIB-71 TM) murine macrophage cells were used in this study. The cells were cultured with Dulbecco's modified Eagle's media (Life technology) and supplemented with 10% fetal bovine serum (Life technology). The cells were grown in a standard cell culture incubator at 37 °C with 5% CO2 in a humidified atmosphere. The cells were allowed to be incubated for 24 h before the needle-shaped PLGA-PEG fibers were introduced. All the fibers were sterilized using UV light for 15 min before feeding. The feeding concentration of the fibers was always kept at 100 μg mL−1 unless otherwise specified.2.3. AFM mechanical measurements
Topographic and mechanical imaging were performed using an AFM model MFP3D-Bio from Asylum Research in the Force Mapping mode, acquiring a force vs. indentation curve (force curve, FC) for each point of a regular square grid on the surface.23 The cells were firstly seeded on a glass slide supported in a culture dish for 24 hours, followed by incubation with fibers for 6, 14 and 24 hours. All the experiments were performed in PBS solutions at a constant temperature of 37 °C, maintained using a BioHeater stage from Asylum Research.The complete protocol and methodology for nanomechanical measurements on soft matter, specifically living cells, is described in a series of methodological studies from our group.23–25 Briefly, we used spherical colloidal probes (Novascan) of borosilicate glass having a typical nominal spring constant k = 0.2 N m−1 and radius R = 5 μm. The radius of the sphere was characterized by means of AFM reverse imaging of the colloidal probe on a spiked grating (TGT1 from NT-MDT).26 The spring constant and lever sensitivity were calibrated by acquiring force curves on a hard surface (glass) and an integrated thermal noise routine from Asylum Research. The mechanical properties of the fibers were obtained by using standard sharp probes AC240 (Asylum Research). The acquisition was performed with the following parameters for each FC: ramp size 8 μm, force setpoint FMAX ≈ 5–10 nN, approaching velocity v = 32 μm s−1, and a ramp rate of 2 Hz. Using spherical probes and relatively slow speed force spectroscopy, allowed us to neglect the viscosity effects and focus on actin cytoskeleton network mechanics. The setpoint force was selected in order to obtain roughly 50% of the curve in contact with the cell and 50% non-contact. A map with 32 × 32 = 1024 FCs was typically acquired for a population of 10 cells under each condition for statistical purposes, while 1–2 cells were at high resolution (64 × 64). The compressed topographic maps of the cells were built using the local z-position corresponding to the maximum setpoint force.
The contact part of each FC was individuated by binning the force axis and producing a histogram. The non-contact part is determined as a sharply defined Gaussian distribution that peaked at zero force. The region of the FC above the width of the Gaussian distribution is considered as the indentation for the fitting procedure. The contact part is then analyzed using the modified Hertz model for finite thickness samples:27,28
Here F is the applied force, δ is the indentation, ν is the Poisson's ratio, E is the effective Young's modulus of the cell, and R is the radius of the spherical probe and the adimensional parameter is 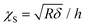 . The correction by Dimitriadis et al. was designed for two extreme boundary conditions: the bound layer (i.e. well adherent) and the free-to-move layer.29 Since the cells are alive, they can dynamically move and adhere on the substrate using focal adhesion points; therefore, we used a boundary condition between the bound and not bound limits by employing the coefficients of the χS power-law as the arithmetic means between the bound and not-bound states.
. The correction by Dimitriadis et al. was designed for two extreme boundary conditions: the bound layer (i.e. well adherent) and the free-to-move layer.29 Since the cells are alive, they can dynamically move and adhere on the substrate using focal adhesion points; therefore, we used a boundary condition between the bound and not bound limits by employing the coefficients of the χS power-law as the arithmetic means between the bound and not-bound states.
For each condition, the mean Young's modulus value E standard deviation of the mean σE was calculated by summing in quadrature the average uncertainty of a single measurement (propagated uncertainty in the probe calibration and the variability of Young's modulus on the cell area) and the standard deviation from the cell population.
2.4. AFM fluorescence colocalization microscopy
Colocalization experiments between optical fluorescence and AFM are performed by using an MFP3Dbio AFM mounted on an inverted optical microscope (Nikon Eclipse Ti2). A mercury lamp (Nikon) and a TRITC (tetramethylrhodamine isothiocyanate) filter allow the excitation of Nile red during the AFM experiments. The optical output is recorded using a colour-sensitive DS-Fi3 camera (Nikon). All the optical images are post-processed using NIS Element AR (Nikon, ver: 5.02.03).2.5. TEM study
For a transmission electron microscopy study, the fibers were diluted in ethanol, put on the TEM grid drop wisely, allowed to dry in air, and observed using TEM with a cryo sample holder (FEI TS12). The fibers fed cells were fixed using typical procedures30 at the end of their incubation with NPs. Then, the cells were dehydrated in a graded series of ethanol and embedded in Spurr resin. The resin blocks were sectioned by using an ultramicrotome (Leica) and the 90 nm thickness sections were transferred onto a TEM grid (TED Pella Inc. USA). The grids were further stained with uranyl acetate and lead acetate solutions. The cell samples were also observed using a TEM with a cryo sample holder (FEI TS12).2.6. Confocal microscopy study
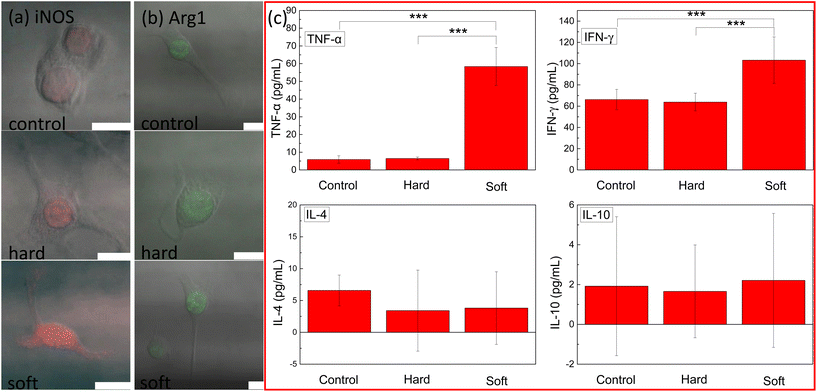
Confocal laser scanning microscopy (CLSM, Leica SP5TCS II) was used to study the actin revolution influenced by the fibers. Cells were seeded in a slide glass for 24 hours and then fed with fibers. The fiber fed cells were collected at different time intervals, fixed with 4% formaldehyde, permeabilized with 0.1% Triton X-100, stained with Phalloidin (Invitrogen), and then observed by a CLSM (Ex: 488 nm, Em: 516 nm for Phalloidin, 650 for fibers).The phenotype differentiation was also revealed by confocal microscopy. Firstly, cells were seeded in confocal dishes (35 mm, Ibidi, Gräfelfing, Germany) for 24 hours, followed by incubation with fibers for 24 hours. After that, the cells were fixed with 4% PFA for 10 min; permeabilized with 0.1% Triton X-100 for 10 min; blocked with 1% BSA for 1 hour, Rt; labeled with iNOS antibody (Thermo, PA5-16524) at 2 μg mL−1, or Arg1 antibody (Thermo PA5-29645) at 3 μg mL−1 in 0.1% BSA for 2 hours; labeled with Goat anti-Rabbit IgG (H + L) secondary antibody at a dilution of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 for 45 min; and finally observed with a CLSM (Ex: 498, Em: 520).
2000 for 45 min; and finally observed with a CLSM (Ex: 498, Em: 520).
2.7. Immune response study
The immune response induced by the PLGA-PEG fibers was evaluated by determining the cytokine profile (TNF-α, INF-γ, IL-4 and IL-10). Briefly, RAW 264.7 cells were seeded at 5 × 103 per well in a 96 well plate for 24 hours. Fibers dispersed in FBS-free DMEM at 100 μg mL−1 were fed to cells for another 24 hours. The supernatants were collected, and the cytokine secretion levels were determined by using a mouse-specific ELISA Kit (Thermo, BMS607-3, EM40RB, BMS613INST, BMS614INST) according to the manufacturer's instructions. Finally, the absorbance was recorded at 450 nm with a reference of 550 nm.3. Results
3.1. Cell morphology and mechanical properties affected by PLGA-PEG fibers
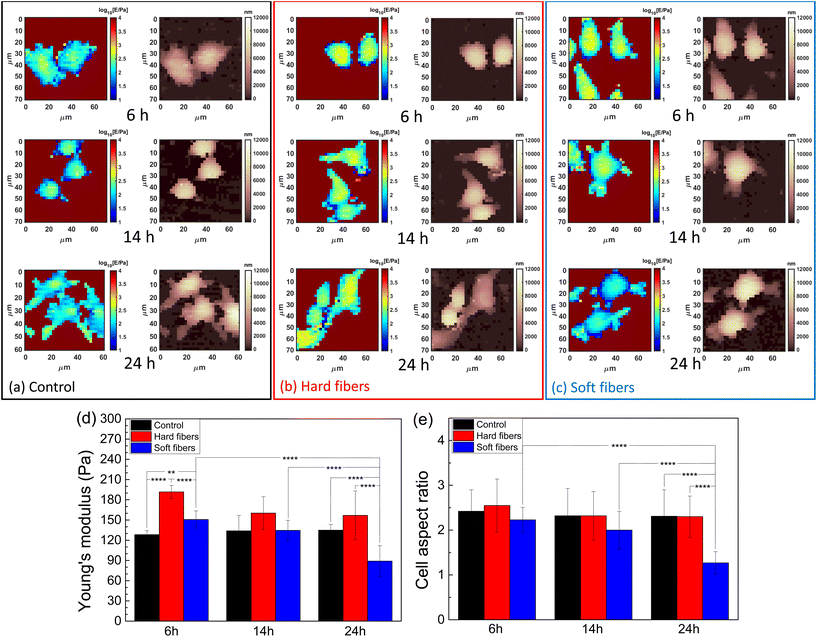
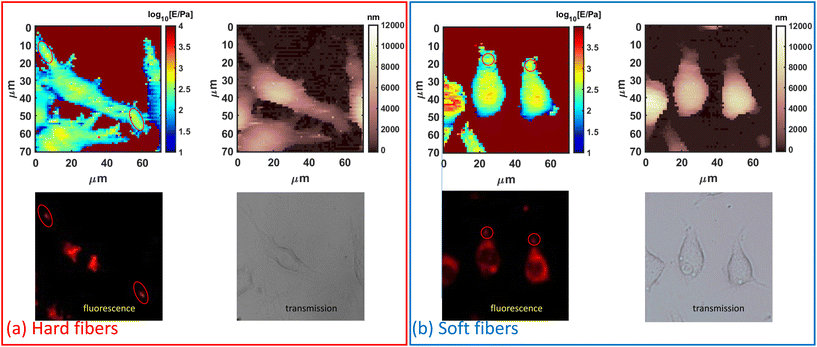
According to the authors’ previous work, the macrophages’ actin distribution was affected by the Young's modulus of PLGA-PEG fibers in a time-dependent way,20 as actin was well accepted to be the intracellular mechanical structure, which indicated the macrophage mechanics could be modulated by PLGA-PEG fibers’ Young's modulus. Thus, we selected PLGA-PEG fibers with the highest (≈1 MPa) and the lowest (≈100 kPa) Young's moduli as measured by AFM mechanics using a sharp probe in liquid (Fig. S1†). The needle-shaped morphology was further confirmed by using the TEM images (Fig. S2†) as a model system,20 and incubation periods of 6, 14 and 24 hours as the representative time points correspond to the highest, lowest and final (equilibrium) quantity of actin coating the fibers. Such time points, used for AFM nanomechanics, were selected by analyzing the actin fluorescence signal in the confocal images shown in Fig. S3.† At each time point, the cells had different Young's modulus behaviors when fed with different fibers (Fig. 1a–c). i.e., for the hard fiber-fed cells, their Young's modulus was higher than those of the control cells and the soft fiber-fed cells at each time point; for the soft fiber-fed cells, the Young's modulus was higher than those of the control cells at 6 hours, but much lower at 24 hours. The averaged Young's moduli of the cell populations are shown in Fig. 1(d); at 6 hours of feeding time, the hard fibers induced a significant increase in cell Young's modulus when compared to soft fiber fed cells and control cells; at 14 hours feeding time, all cases showed similar cell Young's modulus; and at 24 hours feeding time, the soft fiber fed cells showed a significant decrease in Young's modulus compared to hard fiber fed cells and control cells, all of which matched the mechanical observation by force mapping. These results confirmed the authors’ hypothesis that macrophage mechanics could be modulated by controlling PLGA-PEG fibers’ Young's modulus. Another effect induced by the PLGA-PEG fibers was the modification of cell shape, which was represented by the cell aspect ratio in the study. Cells fed with hard fibers had similar aspect ratios to controls at all the selected time points, while the cells fed with soft fibers showed decreasing aspect ratios, reaching the lowest value at 24 hours, which was nearly half of the control and hard fiber fed cells, meaning the cell shape was rounder. This dramatic shape change was coincident with the lowered Young's modulus, and therefore we hypothesized that cell morphology causes a serious decrease in cell mechanical properties.Moreover, an AFM has enough resolution to distinguish heterogeneity in the scale length of probe contact radius (≈1 μm for typical indentation and spherical probes). The fibers’ locations can be distinguished by a local increase in Young's modulus caused by the intracellular distribution of the fiber. This effect was verified by AFM-fluorescence co-localized microscopy. From the AFM-optical fluorescence images (Fig. 2), the locally increased cell Young's modulus matched the fibers’ distribution. In most cases (Fig. S4†), the cells had increased Young's modulus at the protrusions and such locations were coincident with the fibers’ locations, indicating an increased local mechanical property induced by the fibers.
The overall modification of Young's modulus revealed by AFM was dependent on actin fluctuation and redistribution of the external cortical layers of actin where AFM is more sensitive. As studied by confocal microscopy (Fig. S3†), the soft fibers were firstly internalized and located at the cells’ protrusions with actin coat and then deeply located in the cytosol near the nucleus after 14 hours, being less visible to a spherical probe and less contributing to the Young's modulus of the whole cell. This effect is also visible in the cells’ retracting peripheral regions (rich in actin) while assuming rounder shapes. In contrast, hard fibers are difficult to move after internalization and are preferentially located in the external periphery causing an average increase in Young's modulus.
3.2. Intracellular structure affected by PLGA-PEG fibers
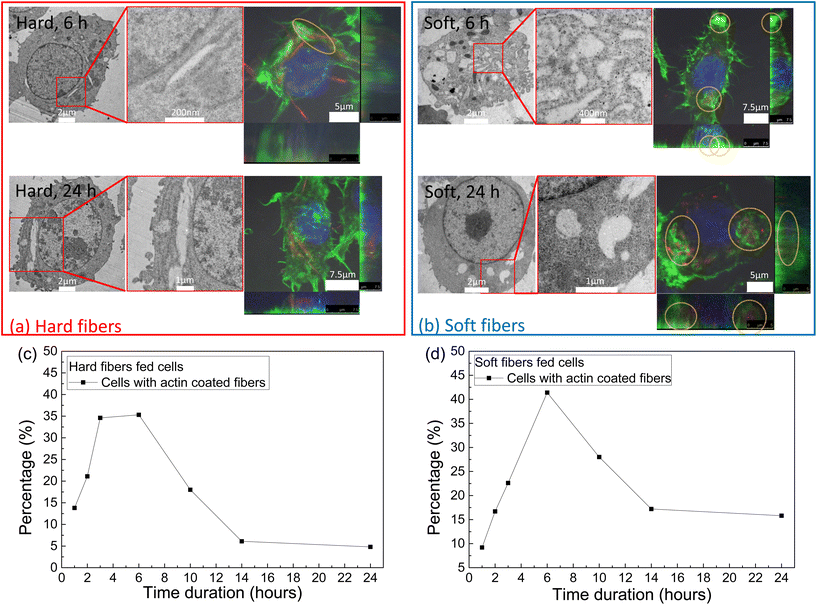
During the first 6 h of incubation, both the hard and soft fibers induced needle-like vesicles inside the cells as indicated by both TEM and confocal images (Fig. 3a and b). After 6 hours, the hard fibers maintained their shape associated with the needle-like vesicles upto 24 hours of incubation, while some enlarged spherical-shaped vesicles appeared in the group of soft fiber-fed cells after 6 hours and the number of such spherical vesicles increased (Fig. S5†). On the other hand, needle-like vesicles can always be found during the whole experiment from both the TEM and confocal images (Fig. S5–S7†). All the fiber-induced vesicle shapes fully matched the fibers’ shapes observed by confocal images. When the percentage of cells with actin-coated fibers was counted (Fig. 3c and d), we found that both the hard and soft fiber groups reached their peak at 6 hours, which was further verified by confocal images that the most actin coats on the vesicles containing fibers at the same time. After that, the number of cells with actin coated fibers decreased until 14 hours and then remained constant. We noticed that the actin signal for the soft fiber case was higher than that of the hard fiber case, explaining the observation that even though actin depleted at 24 hours, some of it still coated the soft fiber containing vesicles.3.3. Immune response study
It was reported that the shape of a macrophage is associated with the phenotype.14 iNOS and Arg1 expression, which represented the M1 and M2 macrophage phenotypes, were determined by confocal images. From Fig. 4a and b, obvious iNOS expression (red) was observed in the soft fiber case, while the hard fiber case was similar to that of the control; Arg1 expression was negligible in all the cases. Such results indicated that soft fibers induced M1 macrophage differentiation. Cytokines are inflammatory mediators secreted along with an immunological response.31 The increased secretion of cytokines indicated the stimulation of the immune system.32 The different physiological responses (actin evolution, mechanical variation, and morphological change) suggested a possible difference in the immune response stimulated by the fibers with different Young's moduli. In this set of experiments, RAW 264.7 cells were fed with hard and soft PLGA-PEG fibers for 24 hours and the cytokine secretion was measured by enzyme-linked immunosorbent assay (ELISA). Significant enhancement in cytokine secretion (TNF-α and IFN-γ, which were associated with inflammation encouragement) was observed when the cells were fed with the soft fibers, while the secretions of IL-4 and IL-10 (which were associated with the pro-healing response) were negligible (Fig. 4c), indicating that only soft fibers could induce M1 macrophage phenotype differentiation.4. Discussion
Research has shown that the phenotypic differentiation of macrophages is related to their physiological state. For example, the morphology of macrophages tended to increase roundness and reduce the extension area in the M1 phenotype and tended to become long in shape in the M2 phenotype;33 for the mechanical properties, the macrophage of the M1 phenotype was hard and the M2 phenotype was soft.34 In addition, in the extracellular environment of macrophages, such as cardiopulmonary tissue, the macrophages bore the mechanical force of continuous circulation and the levels of these mechanical forces were also changed by the influence of the histopathological state and further acted on macrophages. For example, in the pathological state, the Young's modulus of the microenvironment where the macrophages were located increased, the mechanical force that the macrophages receive increased, and the macrophages’ Young's modulus also increased, and finally pro-inflammatory cytokines were secreted.35Being underestimated in most biophysical research studies, the mechanical effect on cell physiological change is gathering more and more attention. The experimental observations were that (1) hard PLGA-PEG fibers increased the macrophage Young's modulus and retained the cell shape, while the soft fibers decreased the macrophage Young's modulus but had rounded cells; (2) both hard and soft fibers increased cell Young's modulus at the protrusions (less protrusion when using soft fibers), which coincided with the fibers’ locations; (3) the long-lasting actin presence on vesicles containing soft fibers (24 hours) was associated with the vesicle shape change from the needle shape to the sphere shape and the quick actin depletion (6 hours) of hard fibers yielded needle-shaped vesicles; and (4) only the soft fibers induced immune response: the M1 phenotype differentiation. The results suggested a picture of PLGA-PEG fibers’ Young's modulus modulated macrophage physiological variations.
Whether the immune cells would respond to mechanical stimulations is an important topic. It had been found that stiffer substrates increase the secretion of pro-inflammatory and anti-inflammatory cytokines36 and also cause differentiation in macrophage phenotypes.14 Nevertheless, all previous works focused on the intercellular but not the intracellular microenvironment of the studied cells. In the present work, pro-inflammatory response (the secretion of INF-α and IFN-γ) was observed when the soft fibers were internalized and associated with cell softening, rounding, and actin evolution.37,38 A possible correlation among the fibers’ Young's modulus, the cell mechanics, morphology, actin evolution, and the immune response exists. However, the exact signaling pathway for the observed immune response associated with the soft fibers’ internalization remained unclear and needed further exploration.
5. Conclusions
This work showed that PLGA-PEG fibers’ mechanical properties impacted a lot of biophysical responses and controlling the fibers’ Young's modulus is a viable strategy to modulate biological interactions. Specifically, this work showed that the soft PLGA-PEG fibers decreased the cell average Young's modulus, modulated the cell shape to be more spherical, caused an intracellular vesicle shape change from a needle to spherical shape with a long-lasting actin coat, and induced proinflammatory cytokine secretion, while the hard PLGA-PEG fibers induced an actin-related cell average Young's modulus increase but without changing the cell shape, and retained the needle-shaped vesicles along with actin depletion. All of these emphasized the influence of PLGA-PEG fibers’ Young's modulus on macrophage physiological modulation and revealed a corresponding correlation among the cell morphology, mechanical properties, actin distribution and phenotype differentiation, thus suggesting a method based on mechanics in order to modulate macrophage differentiation. However, it still lacks the signaling cascade according to such modulation at the biomolecular level, arousing further investigation of the underlying mechanism.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the support from the National Natural Science Foundation of China (Grants No. 32071318 and 21950410518), Shenzhen Science and Technology Innovation Committee (JCYJ20190807163007525), Youth Innovation Promotion Association of the Chinese Academy of Sciences (2022363), and the National Natural Science Foundation of China (11872369).References
- T. A. Wynn, A. Chawla and J. W. Pollard, Nature, 2013, 496, 445–455 CrossRef CAS.
- S. Gordon and P. R. Taylor, Nat. Rev. Immunol., 2005, 5, 953–964 CrossRef CAS.
- L. C. Davies, S. J. Jenkins, J. E. Allen and P. R. Taylor, Nat. Immunol., 2013, 14, 986–995 CrossRef CAS PubMed.
- D. A. Ovchinnikov, genesis, 2008, 46, 447–462 CrossRef PubMed.
- Y. Oishi and I. Manabe, Int. Immunol., 2018, 30, 511–528 CrossRef CAS PubMed.
- L. Cassetta and J. W. Pollard, Nat. Rev. Drug Discovery, 2018, 17, 887–904 CrossRef CAS PubMed.
- R. Parmar, R. Misra and S. Mohanty, Colloids Surf., B, 2015, 129, 198–205 CrossRef CAS.
- D. Reichel, M. Tripathi and J. M. Perez, Nanotheranostics, 2019, 3, 66–88 CrossRef.
- B. Ladoux and R.-M. Mège, Nat. Rev. Mol. Cell Biol., 2017, 18, 743–757 CrossRef CAS PubMed.
- O. Veiseh, J. C. Doloff, M. Ma, A. J. Vegas, H. H. Tam, A. R. Bader, J. Li, E. Langan, J. Wyckoff, W. S. Loo, S. Jhunjhunwala, A. Chiu, S. Siebert, K. Tang, J. Hollister-Lock, S. Aresta-Dasilva, M. Bochenek, J. Mendoza-Elias, Y. Wang, M. Qi, D. M. Lavin, M. Chen, N. Dholakia, R. Thakrar, I. Lacík, G. C. Weir, J. Oberholzer, D. L. Greiner, R. Langer and D. G. Anderson, Nat. Mater., 2015, 14, 643–651 CrossRef CAS PubMed.
- N. Jain, J. Moeller and V. Vogel, Annu. Rev. Biomed. Eng., 2019, 21, 267–297 CrossRef CAS.
- S. Féréol, R. Fodil, G. Pelle, B. Louis and D. Isabey, Respir. Physiol. Neurobiol., 2008, 163, 3–16 CrossRef PubMed.
- R. D. Hubmayr, J. Appl. Physiol., 2005, 99, 384–385 CrossRef PubMed.
- F. Y. McWhorter, T. Wang, P. Nguyen, T. Chung and W. F. Liu, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 17253–17258 CrossRef CAS.
- S. B. Weisser, K. W. McLarren, E. Kuroda and L. M. Sly, in Basic Cell Culture Protocols, ed. C. D. Helgason and C. L. Miller, Humana Press, Totowa, NJ, 2013, vol. 946, pp. 225–239 Search PubMed.
- F. O. Martinez and S. Gordon, F1000Prime Rep, 2014, 6, 12703 Search PubMed.
- L. Lisi, G. M. P. Ciotti, D. Braun, S. Kalinin, D. Currò, C. Dello Russo, A. Coli, A. Mangiola, C. Anile, D. L. Feinstein and P. Navarra, Neurosci. Lett., 2017, 645, 106–112 CrossRef CAS PubMed.
- P. Ghandforoushan, J. Hanaee, Z. Aghazadeh, M. Samiei, A. M. Navali, A. Khatibi and S. Davaran, Drug Delivery Transl. Res., 2022, 12, 2960–2978 CrossRef CAS.
- J. Hou, J. Zhou, M. Chang, G. Bao, J. Xu, M. Ye, Y. Zhong, S. Liu, J. Wang, W. Zhang, H. Ran, Z. Wang, Y. Chen and D. Guo, Bioact. Mater., 2022, 16, 120–133 CrossRef CAS PubMed.
- B. Zhang, M. Zhu, Z. Li, P. Lung, W. Chrzanowski, C. T. Kwok, J. Lu and Q. Li, Acta Biomater., 2020, 112, 182–189 CrossRef CAS PubMed.
- P. Xu, E. Gullotti, L. Tong, C. B. Highley, D. R. Errabelli, T. Hasan, J.-X. Cheng, D. S. Kohane and Y. Yeo, Mol. Pharm., 2009, 6, 190–201 CrossRef CAS PubMed.
- J. A. Champion and S. Mitragotri, Proc. Natl. Acad. Sci. U. S. A., 2006, 103, 4930–4934 CrossRef CAS PubMed.
- M. Galluzzi, G. Tang, C. S. Biswas, J. Zhao, S. Chen and F. J. Stadler, Nat. Commun., 2018, 9, 3584 CrossRef.
- M. Galluzzi, C. S. Biswas, Y. Wu, Q. Wang, B. Du and F. J. Stadler, NPG Asia Mater., 2016, 8, e327–e327 CrossRef CAS.
- L. Puricelli, M. Galluzzi, C. Schulte, A. Podestà and P. Milani, Rev. Sci. Instrum., 2015, 86, 033705 CrossRef PubMed.
- M. Indrieri, A. Podestà, G. Bongiorno, D. Marchesi and P. Milani, Rev. Sci. Instrum., 2011, 82, 023708 CrossRef CAS.
- G. Tang, M. Galluzzi, B. Zhang, Y.-L. Shen and F. J. Stadler, Langmuir, 2019, 35, 7578–7587 CrossRef CAS PubMed.
- M. Galluzzi, C. Schulte, P. Milani and A. Podestà, Langmuir, 2018, 34, 12452–12462 CrossRef CAS PubMed.
- E. K. Dimitriadis, F. Horkay, J. Maresca, B. Kachar and R. S. Chadwick, Biophys. J., 2002, 82, 2798–2810 CrossRef CAS PubMed.
- F. Peng, M. I. Setyawati, J. K. Tee, X. Ding, J. Wang, M. E. Nga, H. K. Ho and D. T. Leong, Nat. Nanotechnol., 2019, 14, 279 CrossRef CAS PubMed.
- M. D. Turner, B. Nedjai, T. Hurst and D. J. Pennington, Biochim. Biophys. Acta, Mol. Cell Res., 2014, 1843, 2563–2582 CrossRef CAS PubMed.
- G. Arango Duque and A. Descoteaux, Front. Immunol., 2014, 5, 491 Search PubMed.
- N. Jain and V. Vogel, Nat. Mater., 2018, 17, 1134–1144 CrossRef CAS PubMed.
- N. R. Patel, M. Bole, C. Chen, C. C. Hardin, A. T. Kho, J. Mih, L. Deng, J. Butler, D. Tschumperlin, J. J. Fredberg, R. Krishnan and H. Koziel, PLoS One, 2012, 7, e41024 CrossRef CAS PubMed.
- V. S. Meli, P. K. Veerasubramanian, H. Atcha, Z. Reitz, T. L. Downing and W. F. Liu, J. Leukocyte Biol., 2019, 106, 283–299 CrossRef CAS PubMed.
- A. K. Blakney, M. D. Swartzlander and S. J. Bryant, J. Biomed. Mater. Res., Part A, 2012, 100, 1375–1386 CrossRef.
- B. M. Barth, S. Stewart-Smeets and T. B. Kuhn, Mol. Cell. Neurosci., 2009, 41, 274–285 CrossRef CAS.
- T. Kihara, T. Matsumoto, Y. Nakahashi and K. Tachibana, Hum. Cell, 2021, 34, 1709–1716 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2bm01351g |
| ‡ These authors contributed equally. |
| This journal is © The Royal Society of Chemistry 2023 |