Open Access Article
Open Access ArticleCapillary driven microfluidic sequential flow device for point-of-need ELISA: COVID-19 serology testing†
Cody
Carrell‡
a,
Ilhoon
Jang‡
 ab,
Jeremy
Link
a,
James S.
Terry
d,
Zachary
Call
a,
Yosita
Panraksa
e,
Orawon
Chailapakul
e,
David S.
Dandy
ab,
Jeremy
Link
a,
James S.
Terry
d,
Zachary
Call
a,
Yosita
Panraksa
e,
Orawon
Chailapakul
e,
David S.
Dandy
 cf,
Brian J.
Geiss
cf,
Brian J.
Geiss
 df and
Charles S.
Henry
df and
Charles S.
Henry
 *acf
*acf
aDepartment of Chemistry, Colorado State University, CO 80523, USA. E-mail: Chuck.Henry@colostate.edu
bDepartment of Mechanical Engineering, Hanyang University, Seoul, 04763, Korea
cDepartment of Chemical and Biological Engineering, Colorado State University, CO 80523, USA
dDepartment of Microbiology, Immunology and Pathology, Colorado State University, CO 80523, USA
eProgram in Biotechnology, Faculty of Science, Chulalongkorn University, Bangkok, 10330, Thailand
fSchool of Biomedical Engineering, Colorado State University, CO 80523, USA
First published on 18th April 2023
Abstract
A capillary-driven microfluidic sequential flow device, designed for eventual at-home or doctor's office use, was developed to perform an enzyme-linked immunosorbent assay (ELISA) for serology assays. Serology assays that detect SARS-CoV-2 antibodies can be used to determine prior infection, immunity status, and/or individual vaccination status and are typically run using well-plate ELISAs in centralized laboratories, but in this format SARs-CoV-2 serology tests are too expensive and/or slow for most situations. Instead, a point-of-need device that can be used at home or in doctor's offices for COVID-19 serology testing would provide critical information for managing infections and determining immune status. Lateral flow assays are common and easy to use, but lack the sensitivity needed to reliably detect SARS-CoV-2 antibodies in clinical samples. This work describes a microfluidic sequential flow device that is as simple to use as a lateral flow assay, but as sensitive as a well-plate ELISA through sequential delivery of reagents to the detection area using only capillary flow. The device utilizes a network of microfluidic channels made of transparency film and double-sided adhesive combined with paper pumps to drive flow. The geometry of the channels and storage pads enables automated sequential washing and reagent addition steps with two simple end-user steps. An enzyme label and colorimetric substrate produce an amplified, visible signal for increased sensitivity, while the integrated washing steps decrease false positives and increase reproducibility. Naked-eye detection can be used for qualitative results or a smartphone camera for quantitative analysis. The device detected antibodies at 2.8 ng mL−1 from whole blood, while a well-plate ELISA using the same capture and detection antibodies could detect 1.2 ng mL−1. The performance of the capillary-driven immunoassay (CaDI) system developed here was confirmed by demonstrating SARS-CoV-2 antibody detection, and we believe that the device represents a fundamental step forward in equipment-free point-of-care technology.
1. Introduction
Serology assays are traditionally performed in laboratory settings using enzyme-linked immunosorbent assays (ELISA) or at the point of care using lateral flow assays (LFAs).1,2 While traditional ELISAs have superior analytical performance versus LFAs, they require expensive equipment and samples must be shipped to a centralized laboratory for testing. The volume of serology tests needed in the US alone for a robust serosurvey of different populations can overwhelm clinical laboratory resources and samples may take days or weeks to process for COVID-19 and other diseases. Additionally, at >$100 per sample the cost of a test represents a significant financial burden to patients.3 Furthermore, when access to clinical laboratories is limited, e.g., in low and middle income countries (LMICs), laboratory ELISAs are not practical. The World Health Organization has outlined four key target product profiles for SARS-related infections, and one is a point of care serology assay that takes <2 h to perform, can be used by an untrained person, and costs <$12.4 Traditional LFAs meet these requirements, but lack the analytical performance needed for this critical assay.LFAs use a series of porous membranes to wick sample across a detection zone. Target analyte is bound to a nitrocellulose membrane by a capture agent and is typically labeled with nanoparticles that form a colored line for naked-eye detection.5 LFAs can be performed in a matter of minutes, require no external instrumentation, cost ∼$5–15 per test, and can be used outside of a centralized laboratory. Several LFAs have been developed to detect SARS-CoV-2 antibodies but results from these devices are not trusted in large scale surveys because of poor clinical sensitivity and specificity.6 LFAs will inherently perform more poorly than ELISAs for two reasons: (1) they cannot wash excess reagent and/or sample from the detection zone to mitigate non-specific adsorption, and (2) nanoparticle labels cannot amplify colorimetric signal like enzyme reporters. Previous attempts to create simplified ELISAs for use at the point-of-need have been successful, but require multiple timed steps,7,8 have inefficient washing,9 are inconsistent because of end-user intervention, and can take over one hour,10 limiting their practicality in at-home settings.7,11–13 There is a pressing need for a POC device that is as easy to use as a one-step LFA with the sensitivity and specificity of an ELISA.
This article describes a capillary-driven immunoassay (CaDI) that passively performs sequential washing and reagent addition steps using capillary-driven flow in hydrophilic channels and paper pumps for realizing point-of-need colorimetric ELISA. Prior work from our group demonstrated a similar concept using electrochemical detection but that method required screen-printed electrodes, a potentiostat, and a mobile phone user interface.14 The CaDI reported here uses a recently developed pump-free microfluidic device to achieve automated and sequential sample transport, plasma separation from whole-blood, reagent delivery, washing steps, and visual detection 20 min after sample addition.15 Individual assay steps are controlled through simple but highly effective valving and flow control structures in a laminate device with hydrophilic channels for capillary-driven fluid transport previously reported.15,16 The channel geometry in the laminate device enables programmable flow of sample, washing buffer, and reagents for sequential delivery to a detection zone on a nitrocellulose membrane. These processes are performed with only two end-user steps: sample addition (whole blood) followed by buffer addition. Sequential flow of reagents and washing buffer stands in contrast to an LFA where all steps occur within the sample matrix without washing. The device is made of a combination of hydrophilic transparency films, pressure sensitive adhesive, and nitrocellulose. As a result, devices are inexpensive (∼$3 in materials and reagents) and can be mass produced. Flow control methods embedded in the CaDI along with the design and optimization of the colorimetric assay are discussed. To compare device performance to an ELISA, Anti-SARS-CoV-2 IgG nucleocapsid protein (anti-N protein) was detected using recombinant N protein as a capture probe and a horseradish peroxidase detection antibody (HRP-Ab) as the label. The chromogenic substrate 3,3′-diaminobenzidine (DAB) was used for colorimetric detection without the need for equipment if desired. The performance of the disposable ELISA assay was compared to standard well-plate ELISA and antibodies were detected from whole blood, a common matrix for serology testing. Although the application of the device in this manuscript is serology testing for COVID-19, the technology will open the possibility of equipment free at-home and/or low-resource setting ELISA testing using whole blood as a sample matrix.
2. Materials and methods
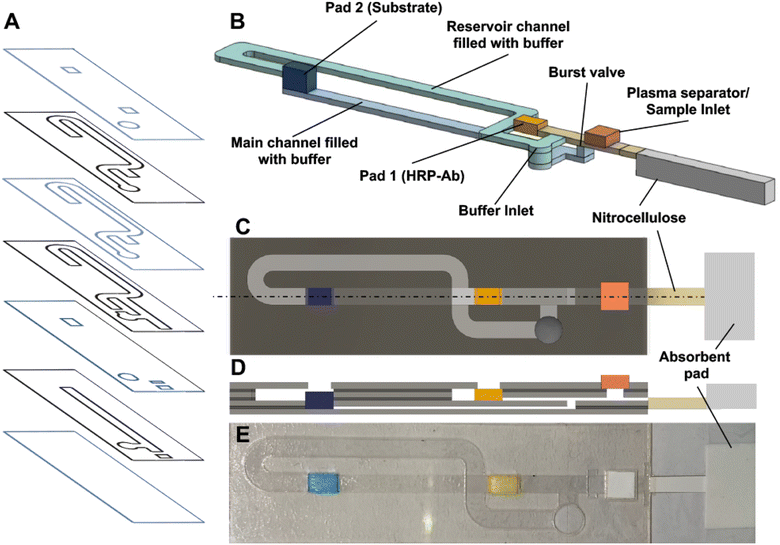
2.1. Device construction
The fluidic channels in the device are constructed from 3M™ 9962 diagnostic microfluidic hydrophilic film, which is 99 μm thick and coated with a proprietary hydrophilic coating on both sides. Four layers of the 9962 film are cut using a CO2 laser cutter (Epilog, Zing 1000) to form channels for flow. These layers were designed in CorelDRAW X4. 3M™ MP467 double-sided adhesive is used between each piece of 9962 film to bind the films together and to form a gap between the layers. Each piece of double-sided adhesive is 50 μm thick and is patterned with the same CO2 laser cutter. Three layers of double-sided adhesive are laminated between the layers of 9962 film to create the final device as shown in Fig. 1. Layers are shown in more detail in Fig. S1.† 3 × 5 mm2 glass fiber pads (Millipore Sigma, GXDX203000) were used as conjugate release membranes and the detection antibody and substrate were dried and stored on these pads. The pads are inserted into the device before the final 9962 film layer is sealed on top. A plasma separation membrane (Vivid GX Membrane, Pall Corporation) was integrated above the main channel between the buffer inlet and the nitrocellulose. Finally, a 3 × 15 mm2 nitrocellulose membrane (GE FF120) striped with capture antigen is inserted into the outlet of the channel and a waste pad made of GE CF4 membrane is placed at the end of the nitrocellulose. To demonstrate sequential delivery and washing in the device tartrazine (yellow dye, 1870 μM) and erioglaucine (blue dye, 800 μM) were used.162.2. Anti-SARS-CoV-2 assay
The anti-SARS-CoV-2 immunoassay used SARS-CoV-2 nucleocapsid protein (N protein AA133-419) as the capture antigen. Recombinant SARS-CoV-2 N protein was produced as previously described.17 Briefly, a bacteria-codon optimized gBlock was cloned into a pET28a bacterial expression with a C-terminal 6xHis tag. Recombinant protein was expressed in BL21(DE3) pLysS E. coli and purified by nickel affinity and size exclusion chromatography in 50 mM HEPES buffer (pH 7.4) and 500 mM NaCl throughout purification to reduce aggregation. Protein purity and quality was verified by SDS-PAGE gel electrophoresis. N protein was striped onto the nitrocellulose strip with a reagent dispenser (Claremont Bio). The striping solution contained 45 mM trehalose, 4.5% glycerol, and 0.5 mg mL−1 N protein. The trehalose and glycerol were used to improve storage stability. Roughly 120 ng of N protein was added to each 3 mm nitrocellulose strip. The detection antibody was an anti-mouse-IgG conjugated to horse radish peroxidase (HRP) (Abcam ab97040, lot no. 3327554). The antibody was diluted to 5.0 μg mL−1 in a solution of 0.01 M FeSO4–EDTA, 4% trehalose, and 0.1% BSA to improve long term storage.18 Two 5.00 μL aliquots of the detection antibody solution (50 ng) were sequentially dried onto a 3 × 5 glass fiber pad. The colorimetric substrate used was 3,3′-diaminobenzidine (DAB). Pierce™ DAB Substrate kit from ThermoFisher, which included a 10× solution of DAB and a peroxide buffer, was used for the substrate and washing buffer. 15.00 μL of the 10× DAB solution was added to a 3 × 5 glass fiber pad in three 5.00 μL aliquots and air dried at 37 °C for 20 min. The peroxide buffer was adjusted to a pH of 6.5 with sodium hydroxide and Tween-80 (Fisher Scientific) was added to a concentration of 0.1%.To start the assay, 10.0 μL of sample is added to the Vivid plasma separation membrane (sample pad). The target analyte used was SARS-CoV-2 nucleocapsid antibody (GeneTex Cat. No. GTX632269 lot No. 43936). The target antibody was diluted in phosphate buffer pH 7.2 or commercially obtained single donor human whole blood (Innovative Research) to concentrations ranging from 0.01 ng mL−1 to 100.0 μg mL−1. All blood samples were performed in compliance with all relevant guidelines. After the sample is added to the sample pad, 75.00 μL of wash buffer is added to the buffer inlet (Fig. 1). Once buffer has been added, the waste pad pumps the buffer through the system for 20 min at which point the nitrocellulose membrane is imaged using a Motorola One smartphone. The color intensity at the test line is quantified using NIH ImageJ. Here, the image of the test line is converted to grayscale and inverted. The grayscale intensity across the test line is divided by the intensity adjacent to the test line to obtain a “gray ratio” for quantification. The ratio is used to reduce any error caused by variation in lighting. Lighting for this project was also stabilized with a 16 LED light box for consistency.7,19
2.3. Well-plate ELISA
The analytical performance of the CaDI was compared to a traditional well-plate ELISA using the same capture antigen, target, and detection antibody. In the well-plate ELISA, polystyrene plates were coated with 50 μL of 0.20 μg mL−1 N protein overnight on a shaker at 4 °C and then blocked with 4% dry non-fat milk powder in 1× PBS and 0.1% Tween-20 at RT. 50.0 μL of the target antibody solution (GeneTex Cat. No. GTX632269 lot No. 43936) was incubated in the wells for 1 h at RT on a shaker followed by washing 3× with 0.1% Tween-20 in PBS (200.0 μL). The detection antibody is the same used in the CaDI (Abcam ab97040), and 50.00 μL of 0.10 μg mL−1 was incubated in each well for 1 h. The wells were washed 3× before addition of the 50.00 μL 1-step Ultra TMB substrate (Thermo Fisher Scientific, 34028). After 4 min the reaction was quenched with 1 M H2SO4 and absorbance readings were taken with a PerkinElmer Victor X5 plate reader at 450 nm.3. Results and discussion
3.1. Design of CaDI
The CaDI is composed of porous material as well as a hydrophilic film layer and can perform sequential delivery of reagents for a point-of-need assay. The device is composed of three sections: the laminate hydrophilic flow channels for programmable reagent release and washing that generates sequential flow; a nitrocellulose membrane strip where the sandwich immunoassay readout is performed; and a waste pad to draw the sample through the nitrocellulose (Fig. 1a). The nitrocellulose membrane provides a zone for instrument free detection. The CaDI uses capillary action of the membrane and waste pad to drive flow just like an LFA, but the flow channels placed upstream of the membrane can provide more advanced functions than an LFA alone. The flow channel was designed in a multi-layered format to achieve sequential delivery of the sample, buffer, and reagents to the nitrocellulose membrane after two end-user steps.The flow channel is divided into two sections: the main channel of 50 μm height and the reservoir channel of 200 μm height. Two main channels are placed between each glass pad and nitrocellulose fiber and designed as a straight channel to make reagent flows uniform in the channel. Therefore, two main channels have different vertical positions and are merged into a single layer channel prior to the sample inlet. The sample and reagent inlets were placed in order of delivery to the detection zone along the main straight channel (Fig. 1b), and the timing of the sample and reagent delivery was controlled by adjusting the distance between the nitrocellulose and each reagent inlet. The sample is added to the main channel through a plasma separation membrane while the reagents dried in the glass pad are rehydrated with buffer for delivery through the main channel. The reservoir channel, which connects upstream of each reagent pad, supplies the buffer solution to rehydrate reagents and generate flow in the main channel. Although the buffer channels are connected upstream of the glass pads, the buffer inlet is placed downstream of the glass pads so the buffer can fill both the main channel and the reservoir channel upon buffer addition. This channel geometry also makes the device compact. Since the two reagent channels have different vertical positions, buffer can fill the main channel without being affected by the glass pads (Fig. 1c). The opening above the glass fiber pads prevents air bubbles from forming in the main and/or reservoir channels and helps maintain consistent flow, but the flow should reach the glass pad upstream and downstream at the same time to realize this function. Therefore, the length of the main and reservoir channels was designed based on a previous study of flow rate in a capillary-driven lamination device.20 Finally, a burst valve was implemented under the separation membrane, which releases the sample and buffer into the main channel at the same time without air bubbles.16 The burst valve initiates flow in a channel only after two separate channels meet to ensure proper flow timing.
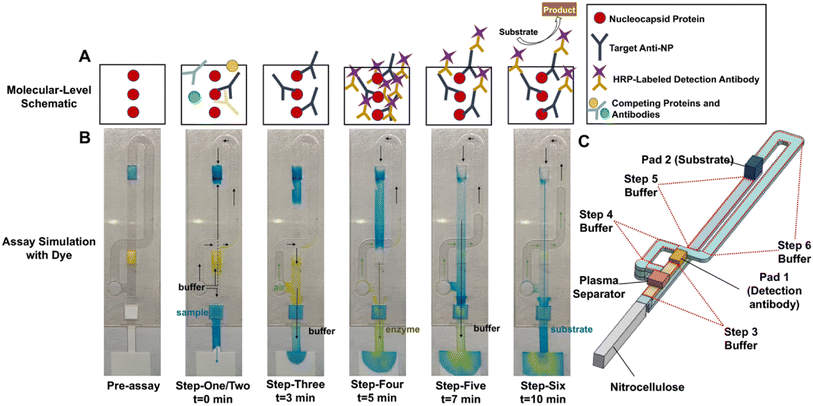
3.2. Sequential flow for ELISA
To visualize the automated sequential steps of ELISA, food dye was used in place of immunoassay reagents (Fig. 2). The assay steps are as follows: (1) sample addition by user; (2) buffer addition by user; (3) sample is washed through nitrocellulose membrane with washing buffer via capillary-driven flow and target is conjugated to proteins on detection zone; (4) rehydrated detection antibody flows through the nitrocellulose membrane strip and conjugates to captured target; (5) wash buffer removes excess detection antibody from the nitrocellulose membrane strip; and (6) rehydrated substrate flows through the nitrocellulose membrane strip, reacts with enzymatic label and produces a visible-by-eye color change. In addition to the still images in Fig. 2b, a video showing the flow is included in the ESI.†In Video S1† blue dye is added to the sample pad, yellow dye is dried on glass pad 1 to simulate the detection antibody, and blue dye is dried on glass pad 2 to simulate the substrate. After sample and buffer addition (steps 1 and 2) all channels are filled via capillary action, and the sample, detection antibody, and substrate are connected via buffer. Once the channels are filled, the nitrocellulose strip and waste pad pull the sample, buffer, and immunoassay reagents by the negative pressure of capillary force. Because the sample is placed at the front of the device it is programmed to pass through the nitrocellulose membrane first and the target is captured on the test line. Next, the buffer between the sample inlet and pad 1 (yellow) will flow through the nitrocellulose membrane and wash away excess sample constituents that might interfere with the remaining assay (step 3). Interestingly, although the main channel has a short connection to the buffer inlet, air does not enter the main channel until air fills the reservoir channel. This is because the interface area between air and buffer solution in the reservoir channel is larger than the main channel. Therefore, the flow could be maintained without air bubble problems. The pressure at pad one (yellow) and two (blue) is atmospheric because of the opening above each pad. However, the pressure difference from pad one to the nitrocellulose membrane is larger than that of the difference between the pad two (blue) and the nitrocellulose membrane, so the rehydrated enzyme label from the yellow pad flows through next and any target analyte on the test line will capture the enzyme label (step 4). Once the buffer behind pad one is depleted, the flow from the blue pad to the nitrocellulose begins. The rehydrated substrate stored on pad two will be preceded by a plug of buffer between pads 1 and 2 in the main channel (step 5). Once the buffer has washed away excess label, thereby preventing non-specific signal from arising, the substrate reaches the test line and reacts with the enzyme label to produce a visible color change (step 6). Unlike previous work from our group, which used a similar channel geometry, the CaDI in this study was designed for automated two-reagent delivery without additional user steps after buffer addition. After flow stops (∼20 min), the color change is detected with by naked eye for qualitative detection or imaged (smartphone camera) for quantitative information. The volume of buffer used to wash the sample or reagents through the nitrocellulose membrane can be controlled by changing the length of channels for a given assay. In the current device design, the total volume of washing buffer was 75.00 μL and the assay takes 20 min to run.
3.3. Anti-SARS-CoV assay optimization
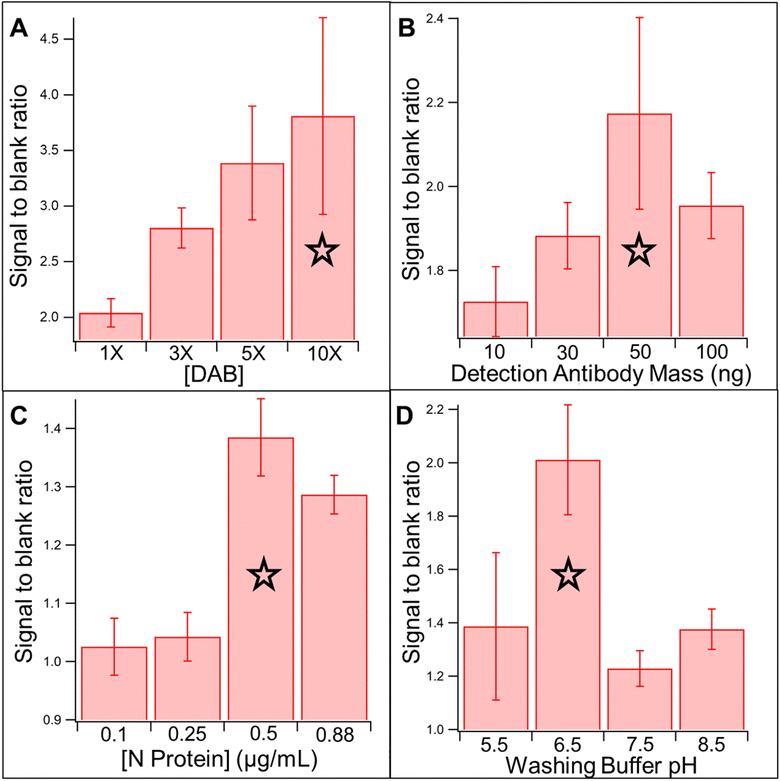
Once sequential reagent delivery and washing steps were confirmed, an immunoassay for anti-SARS-CoV-2 antibodies was performed using the CaDI device. Separate glass fiber pads were used to store the detection antibody and substrate. A drying buffer (Section 2.2) was used to dilute the detection antibody to improve storage capabilities.18 Sequential flow could not be confirmed through observation during the assay, but a colored signal on the test line, with no background color in the nitrocellulose, indicates that indeed sequential addition of the HRP-labeled antibody, wash buffer, and substrate has occurred. In future iterations of the device a control line will also be incorporated.The assay parameters described in Section 2.2 were optimized using positive signal![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) blank signal ratio, and the key results for washing buffer pH, capture antigen concentration, detection antibody concentration, and DAB concentration are shown in Fig. 3 with a blue star indicating the conditions chosen for the final assay. Separated positive and blank signals are shown in Fig. S2.† One of the struggles of running a sandwich immunoassay in the CaDI versus a well-plate ELISA is that assay buffer conditions are constant during all steps instead of optimized for each individual sample addition or washing step. For example, a commercial peroxide buffer was used as the washing buffer to improve the activity of HRP with DAB, but the buffer was adjusted from a pH of 5.5 to 6.5 for the final assay. While changing from 5.5 to 6.5 decreased the HRP activity, it improved the antibody–antigen binding, which had a larger impact on assay sensitivity. 1 μg mL−1 target antibody was used during the optimization experiments as that concentration falls within the linear range of the assay.
blank signal ratio, and the key results for washing buffer pH, capture antigen concentration, detection antibody concentration, and DAB concentration are shown in Fig. 3 with a blue star indicating the conditions chosen for the final assay. Separated positive and blank signals are shown in Fig. S2.† One of the struggles of running a sandwich immunoassay in the CaDI versus a well-plate ELISA is that assay buffer conditions are constant during all steps instead of optimized for each individual sample addition or washing step. For example, a commercial peroxide buffer was used as the washing buffer to improve the activity of HRP with DAB, but the buffer was adjusted from a pH of 5.5 to 6.5 for the final assay. While changing from 5.5 to 6.5 decreased the HRP activity, it improved the antibody–antigen binding, which had a larger impact on assay sensitivity. 1 μg mL−1 target antibody was used during the optimization experiments as that concentration falls within the linear range of the assay.
3.4. Assay from whole-blood
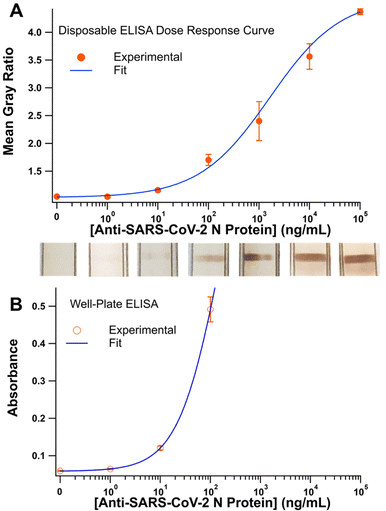
Whole blood was used to evaluate the ability of CaDI to handle real samples. Plasma from whole blood is the most common matrix for serology testing and in point-of-need or at-home settings blood can be obtained with a lancet to prick a finger and draw a drop of blood. To ensure that red blood cells do not interfere with the colorimetric readout on the nitrocellulose membrane, they must be removed using a plasma separation membrane. In this device, a plasma separation membrane is integrated just upstream of the nitrocellulose membrane (Fig. 1). After adding 10.00 μL of sample to the plasma separation membrane, red blood cells are captured in the membrane and plasma flows into the device and through the nitrocellulose. 10.00 μL was chosen because it is similar to the volume of blood obtained with a standard finger prick.21,22 Video S2 in the ESI† captures this process along with the rest of the assay. Note that the video depicts the formation of a test line, but this version of the assay does not include a control line, so only a single line is developed. A potential problem in using blood as a sample type is that finger prick volumes have a large variation (10–150 μL) and when applying the blood to the device, end users will not have access to a pipette, so volumes applied by simply pressing a finger with a drop of blood to the device will vary widely. Explicit instructions will be important to ensure sufficient volume is distributed sample pad (e.g. add until sample pad is saturated), and a siphon pad that removes excess blood will need to be incorporated to ensure the plasma separation membrane does not flood. A dose-response curve was generated using blood spiked with anti-N protein antibody (Fig. 4a). Images of the test line at the various concentrations show the ability to visually observe signal below 10 ng mL−1. The data were fit to a four-parameter logistic curve as is common for sandwich immunoassays (eqn S(1)†).23 The detection limit using the CaDI in blood was 2.8 ng mL−1, which is sensitive enough to detect anti-N protein IgG from clinical samples (>10 μg mL−1).24 The detection limit was calculated using eqn S(2).† Importantly, when real blood samples with large amounts of potentially interfering proteins and antibodies were used, non-specific interactions were not observed in any blanks, which indicates the robust washing capabilities of the CaDI. Future studies will focus on using the CaDI with real patient samples to establish a clinical sensitivity and specificity for the device.We next compared CaDI to a well-plate ELISA using the same antigen and detection antibody. The results for the well-plate ELISA are shown in Fig. 4b. The well-plate ELISA dose-response data were once again fit with a 4-parameter logistic curve and a detection limit of 1.2 ng mL−1 was calculated. The detection limit of the CaDI (2.8 ± 0.4 ng mL−1) is comparable to the detection limit in the ELISA (1.2 ± 0.2 ng mL−1). These results exceeded expectations for the CaDI considering that a well-plate ELISA uses absorbance values from a plate reader, while our device quantifies results using images from a smartphone. The advantages of the CaDI over a traditional well-plate assay include faster assay time (20 min versus 2.5 h), decreased end-user steps (2 versus 13), increased portability, decreased cost, and increased ease of use. Hands-on time and preparation time are also significantly lower in the CaDI than the well-plate ELISA. Most importantly, the CaDI is a true at-home test that can be run outside of a centralized laboratory by an untrained end-user. These advantages are common for POC assays but are typically offset by sacrificing analytical performance. This is not the case for the CaDI, which has the sensitivity of a well-plate ELISA and is as easy to use as an LFA.
4. Conclusion and future directions
This article describes a fluidic system that enables sequential reagent delivery and washing for automated, pump-free, ELISA. The CaDI demonstrated ELISA-level performance in a disposable and easy-to-use device. The laminated channels of CaDI were fabricated with hydrophilic film and double-sided adhesive layers. The multi-layered geometry enabled several flow control methods such as burst valves so that the CaDI can deliver sample, wash buffer, detection antibody, and substrate to the detection zone on the nitrocellulose membrane. The sequential flow was designed to automate ELISA in a single use format and was confirmed using blue and yellow dyes. The CaDI detected anti-SARS-CoV-2 antibodies from whole blood to demonstrate assay performance in real samples. Although additional specificity challenges will likely exist when detecting human antibodies from human blood, (instead of recombinant mouse antibodies spiked into human blood) the goal of this experiment was to showcase the ability of the CaDI to process complex samples such as whole blood through simple and inexpensive means. The CaDI can achieve ELISA-level sensitivity with a colorimetric read-out because an enzyme label is used to amplify the signal and sequential washing and reagent addition steps are performed automatically after sample and buffer addition. Qualitative detection can be performed with the naked eye, or a smartphone can be used to capture quantitative information. The detection limit of CaDI was 2.8 ng mL−1, nearly the same as a traditional well-plate ELISA (1.2 ng mL−1). Several retrospective studies indicate that the 2.8 ng mL−1 LOD for antibodies in CaDI devices is solidly within the concentration ranges of SARS-CoV-2 specific antibodies found in COVID-19 patients (1–10 ng mL−1 based on well-plate ELISA assays),25,26 indicating that the serological CaDI assay can provide potentially clinically useful data. Future iterations of the device will use electrochemical sensing as a more quantitative alternative to colorimetric detection. Eventually, we envision this technology will be used in resource-limited settings to detect influenza, tuberculosis, or other pathogens, biomarkers for chronic illness such as heart failure or cancer, and/or other antibodies at levels that were previously undetectable outside a centralized laboratory.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was financially supported by a National Research Foundation of Korea (NRF) grant funded by the Korea government (No. 2018R1A6A3A11043917). In addition, this work was supported by grants from the National Science Foundation (NSF STTR 2032222) and the National Institutes of Health (UO1-HL152405 and 1R01AI132668).References
- H. Flinck, A. Rauhio, B. Luukinen, T. Lehtimaki, A. M. Haapala, T. Seiskari and J. Aittoniemi, Diagn. Microbiol. Infect. Dis., 2021, 99, 115197 CrossRef CAS PubMed.
- M. A. C. Huergo and N. T. K. Thanh, Analyst, 2021, 146, 382–402 RSC.
- Q. Diagnostics, COVID-19 test price information, https://www.questdiagnostics.com/business-solutions/health-plans/covid-19/pricing, accessed March 13, 2022 Search PubMed.
- COVID-19 Target product profiles for priority diagnostics to support response to the COVID-19 pandemic v.0.1, World Health Organization, 2020 Search PubMed.
- G. A. Posthuma-Trumpie, J. Korf and A. van Amerongen, Anal. Bioanal. Chem., 2009, 393, 569–582 CrossRef CAS PubMed.
- F. Krammer and V. Simon, Science, 2020, 368, 1060–1061 CrossRef CAS.
- C. S. Carrell, R. M. Wydallis, M. Bontha, K. E. Boehle, J. R. Beveridge, B. J. Geiss and C. S. Henry, RSC Adv., 2019, 9, 29078–29086 RSC.
- M. S. Verma, M.-N. Tsaloglou, T. Sisley, D. Christodouleas, A. Chen, J. Milette and G. M. Whitesides, Biosens. Bioelectron., 2018, 99, 77–84 CrossRef CAS PubMed.
- A. Apilux, Y. Ukita, M. Chikae, O. Chailapakul and Y. Takamura, Lab Chip, 2013, 13, 126–135 RSC.
- G. E. Fridley, H. Le and P. Yager, Anal. Chem., 2014, 86, 6447–6453 CrossRef CAS PubMed.
- S.-G. Jeong, J. Kim, S. H. Jin, K.-S. Park and C.-S. Lee, Korean J. Chem. Eng., 2016, 33, 2761–2770 CrossRef CAS.
- P. Preechakasedkit, W. Siangproh, N. Khongchareonporn, N. Ngamrojanavanich and O. Chailapakul, Biosens. Bioelectron., 2018, 102, 27–32 CrossRef CAS PubMed.
- A. Yakoh, S. Chaiyo, W. Siangproh and O. Chailapakul, ACS Sens., 2019, 4, 1211–1221 CrossRef CAS PubMed.
- K. M. Clark, M. S. Schenkel, T. W. Pittman, I. C. Samper, L. B. R. Anderson, W. Khamcharoen, S. Elmegerhi, R. Perera, W. Siangproh, A. J. Kennan, B. J. Geiss, D. S. Dandy and C. S. Henry, ACS Meas. Sci. Au, 2022, 2(6), 584–594 CrossRef CAS PubMed.
- I. Jang, D. B. Carrão, R. F. Menger, A. R. Moraes de Oliveira and C. S. Henry, ACS Sens., 2020, 5, 2230–2238 CrossRef CAS PubMed.
- J. Ilhoon, D. S. David, G. J. Brian, H. Charles, K. Hyunwoong and S. Simon, Flow Control in a Laminate Capillary-Driven Microfluidic Device, 2020 Search PubMed.
- A. Fagre, J. Lewis, M. Eckley, S. Zhan, S. M. Rocha, N. R. Sexton, B. Burke, B. Geiss, O. Peersen, R. Kading, J. Rovnak, G. D. Ebel, R. B. Tjalkens, T. Aboellail and T. Schountz, bioRxiv, 2020, preprint, DOI:10.1101/2020.08.07.241810.
- S. Ramachandran, E. Fu, B. Lutz and P. Yager, Analyst, 2014, 139, 1456–1462 RSC.
- K. E. Boehle, E. Doan, S. Henry, J. R. Beveridge, S. L. Pallickara and C. S. Henry, Anal. Methods, 2018, 10, 5282–5290 RSC.
- I. Jang, H. Kang, S. Song, D. S. Dandy, B. J. Geiss and C. S. Henry, Analyst, 2021, 146, 1932–1939 RSC.
- H. Fruhstorfer, G. Schmelzeisen-Redeker and T. Weiss, Eur. J. Pain, 1999, 3, 283–286 CrossRef.
- J. Hauser, G. Lenk, J. Hansson, O. Beck, G. Stemme and N. Roxhed, Anal. Chem., 2018, 90, 13393–13399 CrossRef CAS PubMed.
- T. K. Christopoulos and E. P. Diamandis, Immunoassay, 1996, 25–50, DOI:10.1016/b978-012214730-2/50004-2.
- H. Ma, W. Zeng, H. He, D. Zhao, Y. Yang, D. Jiang, P. Zhou, Y. Qi, W. He, C. Zhao, R. Yi, X. Wang, B. Wang, Y. Xu, Y. Yang, A. J. Kombe Kombe, C. Ding, J. Xie, Y. Gao, L. Cheng, Y. Li, X. Ma and T. Jin, medRxiv, 2020, preprint, DOI:10.1101/2020.04.17.20064907.
- A. Sukumaran, R. E. Thomas, R. A. Krishnan, T. Thomas, R. Thomas, D. K. Vijayan, J. K. Paul and D. Vasudevan, Indian J. Clin. Biochem., 2022, 37, 349–355 CrossRef CAS PubMed.
- P. Chellamuthu, A. N. Angel, M. A. MacMullan, N. Denny, A. Mades, M. Santacruz, R. Lopez, C. Bagos, J. G. Casian and K. Trettner, Front. Immunol., 2021, 12, 777858 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3ay00225j |
| ‡ C. C. and I. J. contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |