Open Access Article
Open Access ArticleMolecularly imprinted polymers as effective capturing receptors in a pseudo-ELISA immunoassay for procalcitonin detection in veterinary species†
Federica
Battaglia
 *ab,
Francesca
Bonelli
b,
Micaela
Sgorbini
b,
Luigi
Intorre
b,
Maria
Minunni
a,
Simona
Scarano
a and
Valentina
Meucci
*b
*ab,
Francesca
Bonelli
b,
Micaela
Sgorbini
b,
Luigi
Intorre
b,
Maria
Minunni
a,
Simona
Scarano
a and
Valentina
Meucci
*b
aDepartment of Chemistry “Ugo Schiff”, University of Florence, 50019 Sesto Fiorentino, FI, Italy. E-mail: federica.battaglia@unifi.it
bDepartment of Veterinary Science, University of Pisa, 56122 Via Livornese, PI, Italy. E-mail: valentina.meucci@unipi.it
First published on 29th November 2022
Abstract
In this study, a new sandwich-type immunoenzymatic assay, based on a molecularly imprinted polymer (MIP) as an artificial antibody (pseudo-ELISA), was developed for the determination of procalcitonin (PCT) in veterinary species. The quantification of PCT in human medicine represents the state of the art for the diagnosis of sepsis; instead the clinical studies on the relevance of PCT as a sepsis predictor in veterinary patients are few, likely due to the total absence of validated assays. MIPs have been widely used as antibody mimics for important applications, and MIP-based sandwich assays have emerged as promising analytical tools for the detection of disease biomarkers. Herein, a polynorepinephrine (PNE)-based imprinted film was directly synthesized on the well surface of a 96-well plate. Subsequently, based on a commercial ELISA kit, the PCT quantification was accomplished via a colorimetric sandwich assay by replacing the capture antibody of the kit with the PNE-based MIP. This method was performed to detect canine and equine PCT in buffer and in plasma samples. Under optimal conditions, the results obtained in plasma samples showed a limit of detection (LOD) of 5.87 ng mL−1 and a reproducibility (CVav%) of 10.0% for canine samples, while a LOD = 4.46 ng mL−1 and CVav% = 7.61% were obtained for equine samples.
Introduction
Immunoassays, conventionally based on the antibody–antigen reaction, are an essential tool widely used in a multitude of bioanalytical fields for the determination of biomarkers for clinical purposes, food safety, environmental monitoring, and forensic analysis.1–4 Among the immunoassays, the so-called enzyme-linked immunosorbent assay (ELISA) is considered the gold standard and it is probably the most commonly used method in clinical routine measurement.5,6 ELISA tests offer significant advantages such as sensitivity and selectivity for the target analyte, are generally convenient for a large sample, and allow the detection of substances from different types of matrices, without pre-treatment and without the need for skilled technicians.7,8 Capturing receptors of conventional ELISA tests are antibodies, and at present still almost the only choice in the field due to their extraordinary ability in expressing specific sites for molecular recognition against a huge variety of target antigens.9,10 However, they have many drawbacks correlated to the high cost and the production, together with the need to use laboratory animals. Furthermore antibodies, as proteins, tend to be sensitive to environmental conditions and are physically, chemically and biochemically unstable.On this basis, molecularly imprinted polymers (MIPs) are excellent versatile tools in different research fields, including biosensing, and have already displayed good performance as an alternative to antibodies capable of binding their target analyte in a specific and selective manner.11–16 Due to the capability of MIPs to create selective recognition sites in synthetic polymers, they offer the specificity and selectivity of naturally occurring receptors with the explicit improvements of ease and speed of preparation, low cost, and good stability to environmental conditions.17–20 MIPs have been applied to a wide range of target analytes from small molecules (e.g. pesticides, drugs, sugars) to macromolecules such as peptides and proteins.21–28 At present, there is an increasing interest in the use of these mimetic receptors for the development of diagnostic antibody-free assay employing a variety of sensitive detection methods such as enzymatic amplification (BELISA); radiodetection; fluorescence and chemiluminescence.29–35 As concerns macromolecule imprinting, the relentless research for new materials for MIP production led to green bio-inspired polymers and in this perspective, the natural neurotransmitter dopamine (DA) and, more recently, norepinephrine (NE), have played a key role. The two catecholamines share the same ability to easily self-polymerize, under alkaline conditions, to form strongly adherent nanofilms on almost any surface such as noble metals, metal oxides, glass, and synthetic polymers.35–41 Thanks to these advantageous features, polydopamine (PDA) has already been applied for the development of MIPs both for small molecules15,31,42–44 and for protein detection.14,45–51 Compared to its PDA analogue, PNE (polynorepinephrine) exhibits a smoother and more hydrophilic surface, due to the presence of intermediate 3,4-dihydroxybenzaldehyde norepinephrine, that reduces the non-specific adsorption, being particularly advantageous for applications in medical and analytical and bioanalytical fields.40,52,53 Based on these considerations we have recently developed two biosensors which utilize PDA- and PNE-based MIPs as capturing receptors for equine and canine procalcitonin (PCT) detection in buffer and plasma. In detail, DA and NE were used as monomers for the synthesis of MIP nanofilms directly on surface plasmon resonance (SPR) gold chips and the imprinting efficiencies of proteins were compared.54 PCT, a prohormone of calcitonin, is encoded by the CALC-1 gene and is synthesized by the C-cells of the thyroid gland. Under normal conditions, PCT is quickly cleaved into three epitopes: (1) an N-terminal region; (2) calcitonin; and (3) katacalcin; therefore, healthy humans commonly have very low levels of plasma PCT (<0.5 μg L−1).55,56 In contrast, in systemic microbial infection, the level of the protein increases up to several thousand-fold, within 6 to 12 hours, and in a correlation manner to the severity of infection. Nowadays, the quantification of PCT in human medicine represents the state of the art for the diagnosis of sepsis, the monitoring of disease and antimicrobial stewardship.57,58 In contrast to the high interest in human procalcitonin, there is no clinical use of PCT in veterinary species and there are very few papers dedicated to its detection.59,60 Among domestic animals, procalcitonin studies are more in horses than in other species. An increase in the circulating level of PCT has been observed in adult horses during pathological conditions caused by bacteria or by the translocation of bacteria and/or their products into the bloodstream.61–64 Moreover, under physiological conditions, PCT levels were traced to be higher in horses than in humans due to the different compositions of intestinal bacterial flora.65 Only about five papers evaluated the plasma levels of canine PCT, showing that septic dogs have higher PCT concentrations than non-septic dogs.66–69 Regarding cattle, high PCT concentrations were observed in neonatal calves with septicaemic colibacillosis and other pathologic conditions such as respiratory and inflammatory diseases and in staphylococcal mastitis. Furthermore, a positive correlation was noted between PCT and other pro-inflammatory cytokines (i.e., TNF-α and INF-γ) and amyloid A protein, the main acute phase protein studied in cattle for the diagnosis of infections.70,71
It is evident that the published studies in the literature for the measurement of PCT principally referred to the human species and include a plethora of papers dedicated to development of innovative analytical methods such as immunological assay and electrochemical and optical biosensors. In the field of veterinary medicine, the search for innovative and appropriate methods for the quantification of PCT is still, today, an ongoing challenge. Moreover, all the published papers dedicated to PCT detection in animals refer to the use of commercially available ELISA kits that are not fully validated in the target species, are expansive, and are useful only for research purposes.72 Notably, no analytical parameters are reported in the literature for equine PCT ELISA kits, and the only ones present referred to the use of human anti-PCT antibodies, due to the current lack of specific anti-equine antibodies.73–75 At the same time, acceptable analytical parameters are reported only for one type of canine PCT ELISA kit.68,69,76 Based on the lack of fully reliable and practical tests, the aim of this work is to develop an innovative analytical assay, for the detection of PCT in veterinary species, by combining the practicality of commercial ELISA kits with the advantages of MIPs. All commonly commercial assays work in a sandwich format in which the first antibody is immobilized on a microplate that serves to capture the antigen, while the secondary antibody (conjugated to an enzyme) is applied as a last step before quantification. The intensity of the signal is directly related to the concentration of the analyte in the sample and involves the horseradish peroxidase (HRP) enzyme as a signal reporter and tetramethylbenzidine (TMB) as a substrate that develops an optical signal in the visible range. Here, commercial ELISA kits were used by replacing the capture antibody with a PNE-based MIP as a capturing receptor for canine and equine PCT detection both in buffer and plasma. For this aim the MIP for equine/canine PCT has been grown on 96-well microplates, obtaining a very sensitive and selective pseudoELISA immunoassay test in a sandwich format with advantages in terms of cost effectiveness and easy preparation of the synthetic receptor.
Experimental
Materials
±-Norepinephrine hydrochloride (NE ≧ 98.0%), tris(hydroxymethyl)aminomethane hydrochloride (Tris–HCl ≧ 99.0%), acetic acid, L-lysine (≧98.0%), L-cysteine hydrochloride monohydrate (≧98.0%) and tris(hydroxymethyl)aminomethane (Tris-base) were purchased from Sigma-Aldrich (Milan, Italy). Recombinant canine procalcitonin (cPCT) and recombinant equine procalcitonin (ePCT) were purchased from Biovendor (Asheville, North Carolina, USA). Polystyrene (PS) 96-well flat-bottom microplates were obtained from Sarstedt (Nümbrecht, Germany).Two different commercial ELISA kits were used:
(1) Recombinant Canine Procalcitonin ELISA kit (Biovendor Asheville, North Carolina, NC, USA).
(2) Human Procalcitonin DuoSet® ELISA (R&D Systems, Inc, Minneapolis, USA).
Water used for all the preparations was obtained from a Milli-Q system.
Preparation of reagents and buffer solutions
All ELISA kits used in this study are sandwich immunoassays. Reagents and all the buffer solutions were prepared according to the manufacturer's instructions. The tests were carried out as reported in the technical data sheet. cPCT was measured with the recombinant canine procalcitonin ELISA kit (Biovendor), while ePCT was measured with the human procalcitonin DuoSet® ELISA kit (R&D Systems). Both the commercial kits included dilution buffers, biotin labelled lyophilized antibody, streptavidin-HRP (S-HRP) solutions, wash buffers, substrate solutions (3,3′,5,5′-tetramethylbenzidine, TMB) and stop solutions (H2SO4, 2 N). The biotin labelled antibody anti-canine PCT was reconstituted and diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 100 just prior to the assay, whereas, for the ePCT detection, the biotinylated sheep anti-human procalcitonin antibody was reconstituted and diluted to a working concentration of 50.0 ng mL−1.
100 just prior to the assay, whereas, for the ePCT detection, the biotinylated sheep anti-human procalcitonin antibody was reconstituted and diluted to a working concentration of 50.0 ng mL−1.
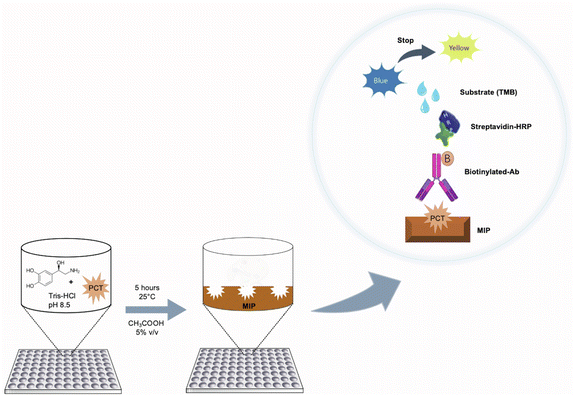
Preparation of imprinted polymers on microplates
PNE-imprinted nanofilms were directly polymerized on the 96-well plate by dropping a monomer solution (100 μL per well) prepared by mixing NE, at a concentration of 5.00 mg mL−1 in 20.0 mmol L−1 Tris–HCl (pH 8.50), in the presence of the template (cPCT/ePCT) at four different concentrations (0.500, 5.00, 50.0 and 500 ng mL−1). For simplicity, in the text, the MIP imprinted with cPCT is renamed cMIP, whereas the one imprinted with ePCT is renamed eMIP. The plates were left upside down and the polymerization was conducted at 25.0 °C for 5 hours. After polymerization was complete, the 96-well plates were washed with deionized water to remove unreacted reagents and then a surface passivation step was performed according to the following procedure: 100 μL of an aqueous solution consisting of 1.00 mmol L−1 cysteine, 1.00 mmol L−1 lysine and 1.00 mmol L−1 tris(hydroxymethyl)aminomethane was dropped onto the MIP surface and left overnight at 25.0 °C by avoiding evaporation. The passivation step is based on the Michael addition reaction and allows the covalent binding of the amines and thiols to the PNE surface in order to minimize possible non-specific interactions of biomolecules present in biological matrices. Finally, the plates were washed with acetic acid (aq. 5.00% v/v, 200 μL per well) three times to remove the template from the MIP, and then with deionized water (200 μL per well, three times).Sandwich assay protocols
Analysis of procalcitonin in plasma samples
The sandwich assays were further tested to detect PCT in spiked horse and dog plasma samples. Blood samples were collected from healthy animals at the Department of Veterinary Science of the University of Pisa. After collection, samples were immediately centrifuged at 3000 rpm for 10 min and plasma aliquots were stored at −80.0 °C from the time of collection and thawed just before use. Hence, the canine/equine plasma samples were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 properly in buffer and spiked respectively, with a known amount of cPCT at concentrations of 25.0, 50.0, 100, 200 and 400 ng mL−1 and ePCT at concentrations of 200, 400, 600, 800 and 1000 ng mL−1. Spiked samples were explored following the same experimental procedure reported above (see paragraph 2.1.4).
10 properly in buffer and spiked respectively, with a known amount of cPCT at concentrations of 25.0, 50.0, 100, 200 and 400 ng mL−1 and ePCT at concentrations of 200, 400, 600, 800 and 1000 ng mL−1. Spiked samples were explored following the same experimental procedure reported above (see paragraph 2.1.4).
Results and discussion
Optimization of the experimental conditions
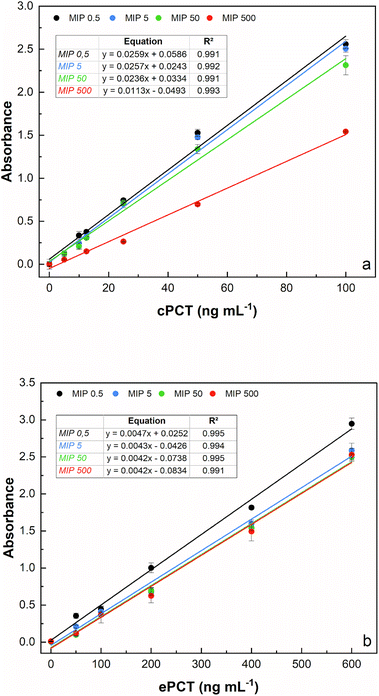
The template concentration (cPCT/ePCT) to be used during PNE imprinting was first optimized to obtain the highest binding performances with the imprinted material. The concentration of the template theoretically reflects the density of binding sites obtained on the MIP surface; therefore, choosing the right concentration is a crucial step of the imprinting process. For both the proteins, the MIPs were synthesized by using four different concentrations of the template (0.500, 5.00, 50.0 and 500 ng mL−1) leaving the starting concentration of the functional monomer (5.00 mg mL−1) unchanged. As shown in Fig. 2a and b, an excellent linearity was obtained for all the tested concentrations for both proteins. In detail, for cPCT (Fig. 2a), a significantly lower absorbance signal was observed at 500 ng mL−1; therefore the highest template concentration was not further considered. Conversely, the best binding ability and the higher absorbance signal were observed for the three lower densities of imprinting (0.500, 5.00 and 50.0 ng mL−1). Among these concentrations, almost overlapping responses were obtained, except for a slight improvement trend that was noted with the decreasing template concentration. For this reason, the subsequent experiments were conducted by using 0.500 ng mL−1 as the template, especially considering that in this study the whole protein was used as a template for MIP synthesis. Instead, for ePCT (Fig. 2b), a different behaviour was observed: similar responses were recorded for template concentrations of 5.00, 50.0 and 500 ng mL−1, whereas an improvement of the absorbance signal was obtained for the lower density of imprinting (0.500 ng mL−1).It is interesting that the absorbance signal increases, for both biomarkers, in an inverse way to the concentration of the template, while it does not increase the absorbance of the negative control (dilution buffer without PCT). This might be attributed to the fact that during the copolymerization, the lower concentration of the template minimizes the steric crowding of the molecules leading to a more efficient creation of the binding cavities for PCT. On the other hand, higher template concentrations could have excessively reduced the amount of norepinephrine available for polymerization, resulting in the formation of insufficient specific cavities for the recognition of the template molecule. On this basis, subsequent experiments were conducted by using 0.500 ng mL−1 as a template concentration for MIP synthesis for canine and equine procalcitonin.
Sandwich assay on MIP-coated microplates
Plasma sample analysis
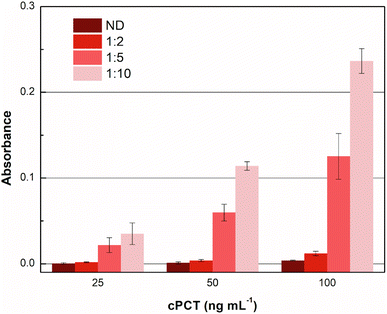
Accordingly, different dilutions of the plasma samples, fortified with cPCT, were compared in order to obtain the maximum output signal and to reduce the residual nonspecific response in plasma. In detail, cPCT was spiked in pooled blank plasma at 25.0, 50.0 and 100 ng mL−1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5, and 1
5, and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 diluted with dilution buffer provided in the kit. Moreover, “as it is” plasma samples fortified with cPCT at the same concentrations were tested. The results reported in Fig. 4 show that the undiluted plasma gave a negligible signal, due to some plasma component that impaired the test function, while the 1
10 diluted with dilution buffer provided in the kit. Moreover, “as it is” plasma samples fortified with cPCT at the same concentrations were tested. The results reported in Fig. 4 show that the undiluted plasma gave a negligible signal, due to some plasma component that impaired the test function, while the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution allowed the best signal. This is likely related to the dilution of some plasma components that tend to unspecifically adsorb to the MIP, reducing the availability of specific recognition sites for the target protein. As a result, after 1
10 dilution allowed the best signal. This is likely related to the dilution of some plasma components that tend to unspecifically adsorb to the MIP, reducing the availability of specific recognition sites for the target protein. As a result, after 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution, the signal significantly increased the dilution of the matrix.
10 dilution, the signal significantly increased the dilution of the matrix.
The 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution was the most favourable condition for the detection of cPCT in plasma samples. This dilution was thus selected for cPCT quantification. For this aim, cPCT was spiked in diluted plasma (1
10 dilution was the most favourable condition for the detection of cPCT in plasma samples. This dilution was thus selected for cPCT quantification. For this aim, cPCT was spiked in diluted plasma (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
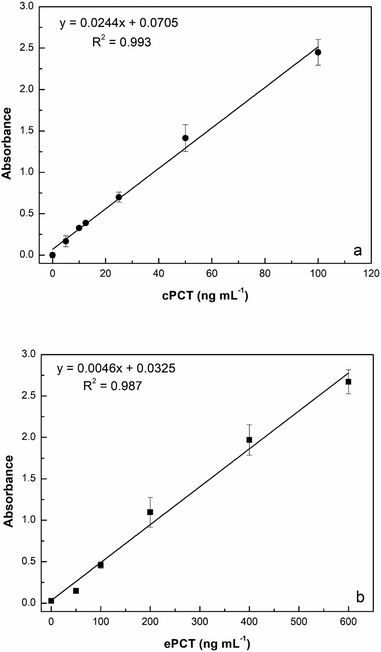
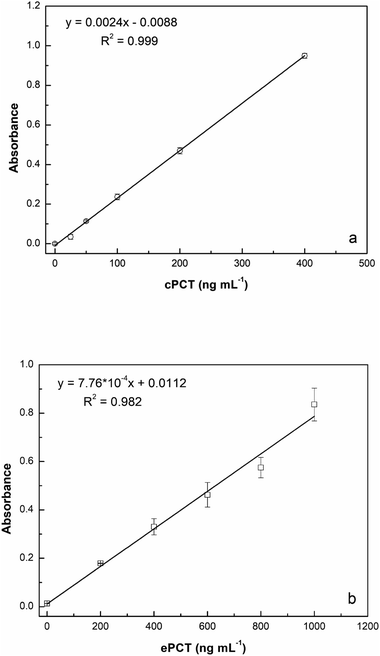
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 with buffer) for a final concentration of 25.0, 50.0, 100, 200 and 400 ng mL−1 for testing. As reported in Fig. 5a, the assay displays a linear response with cPCT concentrations, showing an excellent linear correlation (R2 = 0.999). The calibration range of cPCT in plasma resulted in 25–400 ng mL−1, showing a lower slope than in the buffer. The resulting LOD and LOQ were, respectively, 5.87 and 19.6 ng mL−1, with a CVav% of 10.0% vs. 6.57% obtained in buffer, but still in line with the EMA guidelines.78
10 with buffer) for a final concentration of 25.0, 50.0, 100, 200 and 400 ng mL−1 for testing. As reported in Fig. 5a, the assay displays a linear response with cPCT concentrations, showing an excellent linear correlation (R2 = 0.999). The calibration range of cPCT in plasma resulted in 25–400 ng mL−1, showing a lower slope than in the buffer. The resulting LOD and LOQ were, respectively, 5.87 and 19.6 ng mL−1, with a CVav% of 10.0% vs. 6.57% obtained in buffer, but still in line with the EMA guidelines.78
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5 and 1
5 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10). Once again, the 1
10). Once again, the 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution has been demonstrated to improve the output signal (data not shown). Analysis of dilutions of the recombinant equine protein in 1
10 dilution has been demonstrated to improve the output signal (data not shown). Analysis of dilutions of the recombinant equine protein in 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 diluted plasma (200, 400, 600, 800 and 1000 ng mL−1) generated a calibration curve with a R2 of 0.982 (Fig. 5b). The observed trend in plasma is very similar to that obtained in buffer; in contrast, the slope is significantly lower in the matrix than in the buffer and thus worse LOD and LOQ were estimated respectively at 4.46 ng mL−1 and 14.9 ng mL−1. On the other hand, the reproducibility, expressed as an average of coefficient of variation, was confirmed to be optimal also in plasma samples with values of 5.43% (intra-assay) and 7.61% (inter-assay).
10 diluted plasma (200, 400, 600, 800 and 1000 ng mL−1) generated a calibration curve with a R2 of 0.982 (Fig. 5b). The observed trend in plasma is very similar to that obtained in buffer; in contrast, the slope is significantly lower in the matrix than in the buffer and thus worse LOD and LOQ were estimated respectively at 4.46 ng mL−1 and 14.9 ng mL−1. On the other hand, the reproducibility, expressed as an average of coefficient of variation, was confirmed to be optimal also in plasma samples with values of 5.43% (intra-assay) and 7.61% (inter-assay).
In this study, for the first time, a PNE-based mimetic receptor was produced onto disposable 96-well microplates for detection of ePCT and cPCT. To the best of our knowledge, for PCT detection in animal species, there are only sandwich ELISA tests, completely based on antibodies, available on the market and not applicable in the clinical field. None of the well-conceived studies in the literature succeeded in the development of innovative analytical methods for the determination of these biomarkers in veterinary medicine. Moreover, in the context of biomimetic receptors, the only example available in the literature reports an optical biosensor based on a molecularly imprinted polymer for the real-time detection of human procalcitonin.80 Therefore, in this work, we aimed to realize new MIP-based sandwich assays for equine and canine PCT detection, in which biomimetic receptors were employed as target-capturing probes. The assay was capable of measuring e/c PCT in buffer and in untreated plasma (except for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution), thus being particularly useful in terms of speed and simplicity. The sensitivity of the assay, for ePCT quantification, is in accordance with the clinical performance demonstrated by previous studies in which home-made ELISA assay, based on human anti-PCT antibody, were used. Furthermore, Battaglia et al. (2020) have validated a commercially available ELISA assay based on human anti-PCT antibodies, by using an external standard, but showing a LOD of 56.0 ng mL−1 in plasma, significantly higher than the ePCT pseudo-ELISA assay here reported (4.46 ng mL−1). Anyway, it is difficult to establish an optimal LOD value for PCT detection assay in veterinary medicine since little information is still available on the reference ranges as well as cut-off values for the lack of validated analytical methods.
10 dilution), thus being particularly useful in terms of speed and simplicity. The sensitivity of the assay, for ePCT quantification, is in accordance with the clinical performance demonstrated by previous studies in which home-made ELISA assay, based on human anti-PCT antibody, were used. Furthermore, Battaglia et al. (2020) have validated a commercially available ELISA assay based on human anti-PCT antibodies, by using an external standard, but showing a LOD of 56.0 ng mL−1 in plasma, significantly higher than the ePCT pseudo-ELISA assay here reported (4.46 ng mL−1). Anyway, it is difficult to establish an optimal LOD value for PCT detection assay in veterinary medicine since little information is still available on the reference ranges as well as cut-off values for the lack of validated analytical methods.
Conclusions
A sandwich pseudo-ELISA method that exploits molecularly imprinted nanofilms as artificial antibodies for the detection of equine and canine PCT is presented. This method exhibited good performance both in buffer and plasma, suggesting a possible application to detect PCT in real dog and horse samples. The binding capacity, as well as the synthesis conditions of MIPs for both biomarkers were previously tested via a SPR platform by developing the polymer on the surface of gold chips. Here, the innovative assay has been developed allowing detection of the targets by using a portable, low-cost, and common platform, the ELISA microplate reader. Based on commercially available kits in the novel assay, working in a sandwich format, the capture antibody was replaced with a PNE-based MIP and secondary biotinylated Ab was maintained to bind S-HRP and to give a colorimetric signal. This ELISA method was able to detect both the biomarkers (canine and equine PCT) in buffer and in plasma samples (diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10). Under optimal conditions, the results obtained in plasma samples showed a limit of detection and a reproducibility of 5.87 ng mL−1 and 10.0% respectively for the canine sample and 4.46 ng mL−1 and 7.61% for the equine sample. Future studies could be dedicated to further improve the detection limit, by using more sensitive detection strategies such as fluorescence, and to accelerate the transition to a completely antibody-free assay. In this context, catecholamine-based MIPs, norepinephrine in this case, have shown to be a performing and very promising material for new generation MIPs, suitable for the effective detection of peptides and proteins in disposable microplates. The aim here was to maintain the routine protocol of commercial “ready to use” ELISA assay while introducing new and attractive elements as MIPs, in order to enhance the final assay in terms of stability, cost-effectiveness and reusability and open up new possibilities in the scenario of point of care (POC) tests. Moreover, to get closer to the real applicability of the method, in the future, the purpose of an advanced stage of this work could be the collection, over time, of “suspected septic patients” to be tested and the subsequent correlation analysis between the experimental results obtained by this method and the other traditional clinical evidence of sepsis.
10). Under optimal conditions, the results obtained in plasma samples showed a limit of detection and a reproducibility of 5.87 ng mL−1 and 10.0% respectively for the canine sample and 4.46 ng mL−1 and 7.61% for the equine sample. Future studies could be dedicated to further improve the detection limit, by using more sensitive detection strategies such as fluorescence, and to accelerate the transition to a completely antibody-free assay. In this context, catecholamine-based MIPs, norepinephrine in this case, have shown to be a performing and very promising material for new generation MIPs, suitable for the effective detection of peptides and proteins in disposable microplates. The aim here was to maintain the routine protocol of commercial “ready to use” ELISA assay while introducing new and attractive elements as MIPs, in order to enhance the final assay in terms of stability, cost-effectiveness and reusability and open up new possibilities in the scenario of point of care (POC) tests. Moreover, to get closer to the real applicability of the method, in the future, the purpose of an advanced stage of this work could be the collection, over time, of “suspected septic patients” to be tested and the subsequent correlation analysis between the experimental results obtained by this method and the other traditional clinical evidence of sepsis.
Ethics declarations
An owner's written consent was obtained for the collection of plasma for the horses and dogs included in this study.Author contributions
Federica Battaglia: conceptualization, methodology, investigation, writing—original draft preparation, writing—review & editing. Francesca Bonelli: visualization, project administration, funding acquisition. Micaela Sgorbini: visualization, project administration, funding acquisition. Luigi Intorre: writing—review & editing, supervision, project administration, funding acquisition. Maria Minunni: writing—review & editing, supervision. Simona Scarano: conceptualization, methodology, investigation, data curation, review & editing, supervision. Valentina Meucci: conceptualization, methodology, investigation, data curation, writing—review & editing, supervision, project administration, funding acquisition.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research did not receive any specific grant from funding agencies, commercial, or not-for-profit sectors.Notes and references
- X. Fu, Y. Liu, R. Qiu, M. F. Foda, Y. Zhang, T. Wang and J. Li, Biochem. Biophys. Rep., 2018, 13, 73–77 Search PubMed.
- S. Wang, Y. Liu, S. Jiao, Y. Zhao, Y. Guo, M. Wang and G. Zhu, J. Agric. Food Chem., 2017, 65(46), 10107–10114 CrossRef PubMed.
- K. Xu, H. Long, R. Xing, Y. Yin, S. A. Eremin, M. Meng and R. Xi, Talanta, 2017, 164, 341–347 CrossRef PubMed.
- Z. Liu, B. Zhang, J. Sun, Y. Yi, M. Li, D. Du, F. Zhu and J. Luan, Sci. Total Environ., 2018, 613–614, 861–865 CrossRef PubMed.
- A. Padoan, F. Bonfante, M. Pagliari, A. Bortolami, D. Negrini, S. Zuin, D. Bozzato, C. Cosma, L. Sciacovelli and M. Plebani, EBioMedicine, 2020, 62, 103101 CrossRef CAS PubMed.
- T. Peng, Y. Zong, M. Di Johnson, S. V. Menghani, M. L. Lewis and J. N. Galgiani, Diagn. Microbiol. Infect. Dis., 2021, 99(1), 115198 CrossRef CAS PubMed.
- R. M. Lequin, Clin. Chem., 2005, 51(12), 2415–2418 CrossRef CAS PubMed.
- G. N. Konstantinou, Methods Mol. Biol., 2017, 1592, 79–94 CrossRef CAS PubMed.
- G. Gonzalez-Sapienza, M. A. Rossotti and S. Tabares-da Rosa, Front. Immunol., 2017, 8, 997 CrossRef PubMed.
- A. Zhang, W. Long, Z. Guo, G. Liu, Z. Hu, Y. Huang, Y. Li, T. M. Grabinski, J. Yang, P. X. Zhao, A. D. Everett, Y. Zhang and B. B. Cao, J. Immunol. Methods, 2010, 355(1–2), 61–67 CrossRef CAS PubMed.
- P. Rebelo, E. Cosa-Rama, I. Seguro, J. G. Pacheco, H. P. A. Nouws, M. N. D. S. Cordeiro and C. Delerue-Matos, Biosens. Bioelectron., 2021, 172, 112719 CrossRef CAS PubMed.
- F. Cui, Z. Zhou and H. S. Zhou, Sensors, 2020, 20(4), 996 CrossRef CAS.
- J. Liu, Y. Wang, X. Liu, Q. Yuan, Y. Zhang and Y. Li, Talanta, 2019, 199, 573e580 Search PubMed.
- P. Palladino, M. Minunni and S. Scarano, Biosens. Bioelectron., 2018, 106, 93–98 CrossRef CAS PubMed.
- A. Turco, S. Corvaglia, E. Mazzotta, P. Pompa and C. Malitesta, Sens. Actuators, B, 2018, 255, 3374–3383 CrossRef CAS.
- W. Zhao, H. Yang, S. Xu, J. Cai, J. Luo, W. Wei, X. Liu and Y. Zhu, Colloids Surf., A, 2018, 555, 95e102 CrossRef.
- T. Vaneckova, J. Bezdekova, G. Han, V. Adam and M. Vaculovicova, Acta Biomater., 2020, 101, 444–458 CrossRef CAS PubMed.
- K. Haupt, P. X. Medina Rangel and B. Tse Sum Bui, Chem. Rev., 2020, 120, 9554–9582 CrossRef CAS.
- J. J. BelBruno, Chem. Rev., 2019, 119, 94–119 CrossRef PubMed.
- M. J. Whitcombe, I. Chianella, L. Larcombe, S. A. Piletsky, J. Noble, R. Porter and A. Horgan, Chem. Soc. Rev., 2011, 40, 1547–1571 RSC.
- R. Thoelen, R. Vansweevelt, J. Duchateau, F. Horemans, J. D'Haen, L. Lutsen, D. Vanderzande, M. Ameloot, M. vandeVen, T. J. Cleij and P. Wagner, Biosens. Bioelectron., 2008, 23(6), 913–918 CrossRef PubMed.
- L. Fang, M. Jia, H. Zhao, L. Kang, L. Shi, L. Zhou and W. Kong, Trends Food Sci. Technol., 2021, 116, 387–404 CrossRef.
- Y. Bow, E. Sutriyono, S. Nasir and I. Iskandar, Int. J. Adv. Sci. Eng. Inf. Technol., 2017, 7(2), 662–668 CrossRef.
- A. E. Radi, T. Wahdan and A. El-Basiony, Electrochemical Sensors Based on Molecularly Imprinted Polymers for Pharmaceuticals Analysis, Curr. Anal. Chem., 2019, 14(3), 219–239 CrossRef.
- B. Hatamluyi, A. Hashemzadeh and M. Darroudi, Sens. Actuators, B, 2020, 307, 127614 CrossRef CAS.
- M. R. Ali, M. S. Bacchu, M. R. Al-Mamun, M. M. Rahman, M. S. Ahommed, M. A. Saad Aly and M. Z. H. Khan, J. Mater. Sci., 2021, 56, 12803–12813 CrossRef CAS.
- Z. Mazouz, M. Mokni, N. Fourati, C. Zerrouki, F. Barbault, M. Seydou, R. Kalfat, N. Yaakoubi, A. Omezzine, A. Bouslema and A. Othmane, Biosens. Bioelectron., 2020, 151, 111978 CrossRef CAS PubMed.
- M. Pirzada, E. Sehit and Z. Altintas, Biosens. Bioelectron., 2020, 166, 112464 CrossRef CAS PubMed.
- Z. X. Xu, H. J. Gao, L. M. Zhang, X. Q. Chen and X. G. Qiao, Food Sci., 2011, 76(2), 69–75 CrossRef PubMed.
- C. Chen, J. Luo, C. Li, M. Ma, W. Yu, J. Shen and Z. Wang, J. Agric. Food Chem., 2018, 66, 2561–2571 CrossRef CAS PubMed.
- C. Zhang, H. Cui, Y. Han, F. Yu and X. Shi, Anal. Methods, 2018, 240, 893–897 CAS.
- L. Li, Z. Z. Lin, A. H. Peng, H. P. Zhong, X. M. Chen and Z. Y. Huang, J. Chromatogr., Biomed. Appl., 2016, 1035, 25–30 CrossRef CAS PubMed.
- C. Guoning, G. Pengqi, W. Yan, W. Lu, S. Hua, L. Yunzhe, J. Wanghui, C. Chun and F. Qiang, Talanta, 2019, 198, 55–62 CrossRef PubMed.
- S. P. Tang, F. Canfarotta, K. Smolinska-Kempisty, E. Piletska, A. Guerreiro and S. Piletsky, Anal. Methods, 2017, 9, 2853–2858 RSC.
- F. Torrini, L. Caponi, A. Bertolini, P. Palladino, F. Cipolli, A. Saba, A. Paolicchi, S. Scarano and M. Minunni, Anal. Bioanal. Chem., 2022, 414, 5423–5434 CrossRef CAS.
- J. Liu, Q. Deng, D. Tao, K. Yang, L. Zahng, Z. Liang and Y. Zhang, Sci. Rep., 2014, 4, 5487 CrossRef CAS PubMed.
- P. Palladino, F. Bettazzi and S. Scarano, Anal. Bioanal. Chem., 2019, 411, 4327–4338 CrossRef CAS PubMed.
- C. Xie, X. Wang, H. He, Y. Ding and X. Lu, Adv. Funct. Mater., 2020, 30(25), 1909954 CrossRef CAS.
- J. H. Ryu, P. B. Messersmith and H. Lee, ACS Appl. Mater. Interfaces, 2018, 10(9), 7523–7540 CrossRef CAS PubMed.
- V. Baldoneschi, P. Palladino, S. Scarano and M. Minunni, Anal. Bioanal. Chem., 2020, 412(24), 5945–5954 CrossRef CAS PubMed.
- A. Lamaoui, J. M. Palacios-Santander, A. Amine and L. Cubillana-Aguilera, Microchem. J., 2021, 164, 106043 CrossRef.
- G. H. Yao, R. P. Liang, C. F. Huang, Y. Wang and J. D. Qiu, Anal. Chem., 2013, 85(24), 11944–11951 CrossRef PubMed.
- J. Miao, A. Liu, L. Wu, M. Yu, W. Wei and S. Liu, Anal. Chim. Acta, 2020, 1095, 82–92 CrossRef PubMed.
- J. Yin, Z. Meng, M. Du, C. Liu, M. Song and H. Wang, J. Chromatogr. A, 2010, 1217, 5420e5426 Search PubMed.
- A. Tretjakov, V. Syritski, J. Reut, R. Boroznjak, O. Volobujeva and A. Opik, Microchim. Acta, 2013, 180, 1433–1442 CrossRef.
- B. Yang, S. Lv, F. Chen, C. Liu, C. Cai, C. Chen and X. Chen, Anal. Chim. Acta, 2016, 912, 125–132 CrossRef CAS PubMed.
- Y. Sun and S. Zhong, Colloids Surf., B, 2017, 159, 131–138 CrossRef CAS PubMed.
- X. Xu, R. Liu, P. Guo, Z. Luo, X. Cai, H. Shu, Y. Ge, C. Chang and Q. Fu, Food Chem., 2018, 256, 91–97 CrossRef CAS PubMed.
- W. Chen, M. Fu, X. Zhu and Q. Liu, Biosens. Bioelectron., 2019, 142, 111492 CrossRef CAS PubMed.
- X. Liu, W. Lin, P. Xiao, M. Yang, L.-P. Sun, Y. Zhang, W. Xue and B.-O. Guan, Chem. Eng. J., 2020, 387, 124074 CrossRef CAS.
- A. H. Nadim, M. A. Abd El-Aal, M. A. Al-Ghobashy and Y. S. El-Saharty, Microchem. J., 2021, 167, 106333 CrossRef CAS.
- S. Hong, J. Kim, Y. S. Na, J. Park, S. Kim, K. Singha, G. Im, D. K. Han, W. J. Kim and H. Lee, Angew. Chem., 2013, 52(35), 9187–9191 CrossRef CAS PubMed.
- V. Baldoneschi, P. Palladino, M. Banchini, M. Minunni and S. Scarano, Biosens. Bioelectron., 2020, 157, 112161 CrossRef CAS PubMed.
- F. Battaglia, V. Baldoneschi, V. Meucci, L. Intorre, M. Minunni and S. Scarano, Talanta, 2021, 230, 122347 CrossRef CAS.
- M. Jin and A. I. Khan, Lab. Med., 2010, 41(3), 173–177 CrossRef.
- J. Davies, J. Clin. Pathol., 2015, 68(9), 675–679 CrossRef CAS PubMed.
- M. Meisner, Procalcitonin: Biochemistry and Clinical Diagnosis, UNI-MED, Verlag AG, 1st edn, 2010 Search PubMed.
- A. L. Vijayan, S. Ravindran, R. Saikant, S. Lakshmi, R. Kartik and G. Manoj, J. Intensive Care, 2017, 5, 51 CrossRef PubMed.
- E. Matur, E. Eraslan and Ü. Çötelioğlu, J. Istanbul Vet. Sci., 2017, 2(1), 16–27 CrossRef.
- M. J. López-Martínez, L. Franco-Martínez, S. Martínez-Subiela and J. J. Cerón, Anim. Health Res. Rev., 2021, 23, 82–99 CrossRef PubMed.
- I. Nocera, F. Bonelli, V. Vitale, V. Meucci, G. Conte, E. Jose-Cunilleras, L. A. Gracia-Calvo and M. Sgorbini, Animals, 2021, 11, 2015 CrossRef.
- W. El-Deeb, M. Fayez, I. Elsohaby, H. V. Mkrtchyan and A. Alhaider, Comp. Immunol., Microbiol. Infect. Dis., 2020, 73, 101525 CrossRef.
- F. Bonelli, V. Meucci, T. J. Divers, E. Jose-Cunilleras, M. Corazza, R. Tognetti, G. Guidi, L. Intorre and M. Sgorbini, J. Vet. Intern. Med., 2015, 29, 1689–1691 CrossRef CAS.
- I. Kilcoyne, J. E. Nieto and J. E. Dechant, J. Am. Vet. Med. Assoc., 2020, 256, 927–933 CrossRef CAS PubMed.
- M. C. Costa, G. Silva, R. V. Ramos, H. R. Staempfli, L. G. Arroyo, P. Kim and J. S. Weese, Vet. J., 2016, 205, 74–80 CrossRef PubMed.
- Z. Yilmaz, Y. O. Ilcol and I. H. Ulus, Crit. Care Med., 2008, 85(24), 11944–11951 Search PubMed.
- R. Troia, M. Giunti and R. Goggs, BMC Vet. Res., 2018, 14, 111 CrossRef PubMed.
- R. Goggs, M. Milloway, R. Troia and M. Giunti, Vet. Rec. Open, 2018, 5, e000255 Search PubMed.
- F. Easley, M. K. Holowaychuk, E. W. Lashnits, S. K. Nordone, H. Marr and A. J. Birkenheuer, J. Vet. Intern. Med., 2020, 34(2), 653–658 CrossRef PubMed.
- F. Bonelli, V. Meucci, T. J. Divers, A. Boccardo, D. Pravettoni, M. Meylan, A. G. Belloli and M. Sgorbini, Vet. J., 2018, 234, 61–65 CrossRef PubMed.
- N. Ercan, N. Tuzcu, O. Başbug, M. Tuzcu and A. Alim, J. Vet. Diagn. Invest., 2016, 28, 180–183 CrossRef PubMed.
- F. Battaglia, V. Meucci, R. Tognetti, F. Bonelli, M. Sgorbini, G. Lubas, C. Pretti and L. Intorre, Animals, 2020, 10(9), 1511 CrossRef PubMed.
- M. Rieger, C. Kochleus, D. Teschner, D. Rascher, A. K. Barton, A. Geerlof, E. Kremmer, M. Schmid, A. Hartmann and H. Gehlen, Anal. Bioanal. Chem., 2014, 406(22), 5507–5512 CrossRef PubMed.
- D. Teschner, M. Rieger, C. Koopmann and H. Gehlen, PFERDEHEILKUNDE, 2015, 31(4), 371–377 CrossRef.
- A. K. Barton, A. Pelli, M. Rieger and H. Gehlen, BMC Vet. Res., 2016, 12, 5–7 CrossRef PubMed.
- A. N. K. Floras, M. K. Holowaychuk, D. C. Hodgins, H. S. Marr, A. Birkenheuer, S. Sharif, A. M. E. Bersenas and D. Bienzle, J. Vet. Intern. Med., 2014, 28(2), 599–602 CrossRef PubMed.
- A. M. Mostafa, S. J. Barton, S. P. Wren and J. Barker, Trends Anal. Chem., 2021, 144, 116431 CrossRef.
- EMA, Guideline on Bioanalytical Method Validation, 2011, https://www.ema.europa.eu/en/documents/scientific-guideline/guideline-bioanalytical-method-validation_en.pdf, accessed 15 April 2022 Search PubMed.
- R. E. Toribio, C. W. Kohn, G. W. Leone, C. C. Capen and T. J. Rosol, Mol. Cell. Endocrinol., 2003, 199(1–2), 119–128 CrossRef PubMed.
- G. Sener, E. Ozgur, A. Y. Rad, L. Uzun, R. Say and A. Denizli, Analyst, 2013, 138(21), 6422–6428 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ay01175a |
| This journal is © The Royal Society of Chemistry 2023 |