Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Label-free visualization of photosynthetic microbial biofilms using mid-infrared photothermal and autofluorescence imaging†
Ryo
Kato
 *abc,
Kaisei
Maeda
*d,
Taka-aki
Yano
abc,
Kan
Tanaka
d and
Takuo
Tanaka
*abc,
Kaisei
Maeda
*d,
Taka-aki
Yano
abc,
Kan
Tanaka
d and
Takuo
Tanaka
 abc
abc
aInstitute of Post-LED Photonics, Tokushima University, 2-1 Minamijosanjima-cho, Tokushima, Tokushima 770-0856, Japan. E-mail: kato.ryo@tokushima-u.ac.jp
bInnovative Photon Manipulation Research Team, RIKEN Center for Advanced Photonics, Wako, Saitama 351-0198, Japan
cMetamaterials Laboratory, RIKEN Cluster for Pioneering Research, Wako, Saitama 351-0198, Japan
dLaboratory for Chemistry and Life Science, Institute of Innovative Research, Tokyo Institute of Technology, Midori-Ku, Yokohama, Kanagawa 226-8503, Japan. E-mail: kmaeda@res.titech.ac.jp
First published on 10th November 2023
Abstract
The formation of photosynthetic microbial biofilms comprising multispecies biomolecules, such as extracellular polymeric substances (EPSs), and microbial cells play pivotal roles in maintaining or stimulating their biological functions. Although there are numerous studies on photosynthetic microbial biofilms, the spatial distribution of EPS components that are vital for microbial biofilm formation, such as exopolysaccharides and proteins, is not well understood. Visualization of photosynthetic microbial biofilms requires label-free methods, because labelling EPSs results in structural changes or aggregation. Raman spectroscopy is useful for label-free visualization of biofilm constituents based on chemical contrast. However, interference resulting from the bright autofluorescence of photosynthetic molecules and the low detection efficiency of Raman scattering make visualization a challenge. Herein, we visualized photosynthetic microbial biofilms in a label-free manner using a super-resolution optical infrared absorption imaging technique, called mid-infrared photothermal (MIP) microscopy. By leveraging the advantages of MIP microscopy, such as its sub-micrometer spatial resolution, autofluorescence-free features, and high detection sensitivity, the distribution of cyanobacteria and their extracellular polysaccharides in the biofilm matrix were successfully visualized. This showed that cyanobacterial cells were aligned along acidic/sulfated polysaccharides in the extracellular environment. Furthermore, spectroscopic analyses elucidated that during formation of biofilms, sulfated polysaccharides initially form linear structures followed by entrapment of cyanobacterial cells. The present study provides the foundation for further studies on the formation, structure, and biological functions of microbial biofilms.
Introduction
Generally, microorganisms live in biofilms consisting of extracellular polymeric substances (EPSs), which provide a suitable environment around the cell.1 Photosynthetic microorganisms, eucaryotic algae, and cyanobacteria, also form biofilms. The algal biofilms are also important for other species because they provide habitation, nutrient sources, and even control of the surrounding environment. Biological soil crusts (BSCs), which occupy approximately 12% of the land surface in arid and semi-arid regions, are biofilms formed with the EPSs of filamentous cyanobacterial cells as a core, and prevent land desertification.2 Transparent extracellular particles (TEPs), which are distributed on the surface of the ocean, are biofilms consisting of various microorganisms attached to EPSs produced by algae. They play a significant role in the carbon cycle of the ocean and provide a habitat for marine microorganisms.3 These EPSs mainly comprise exopolysaccharides of cyanobacteria or eukaryotic algae; additionally, it is assumed that their distribution is based on their roles in the biofilm. Proteins, nucleic acids, and lipids, which are components of EPSs, are known to cooperate to provide physiological functions such as mechanical stability and a three-dimensional polymer structure. Therefore, to develop an understanding of photosynthetic biofilms, it is important to assess the distribution of individual components, particularly exopolysaccharides—the primary constituents of EPSs. However, conventional methods for visualizing exopolysaccharides employ staining reagents, such as Alcian blue, which significantly alter the structure of the biofilms because of the interaction between the staining reagents and polysaccharides. Therefore, a substantial demand for a label-free method to visualize biochemical components in biofilms exists in the field of microbiology.Vibrational spectroscopies, such as Raman spectroscopy and Fourier transform infrared (FTIR) spectroscopy, are powerful label-free imaging approaches for visualizing the chemical compositions in a sample. These techniques directly measure the optical responses of molecules associated with molecular vibrations (molecular fingerprints). Raman spectroscopy has been used for the chemical characterization of a wide range of samples, including biofilms, cells, and polymers4,5 because its spatial resolution, which is at the sub-micrometer level, is high enough to resolve structures such as bacteria and organelles within cells. However, the autofluorescence signal of photosynthetic bacteria, such as cyanobacteria, and extracellular polysaccharides produced by the bacteria overwhelms the relatively weak Raman signal. The wavelength of the excitation light for Raman spectroscopy, which excites electronic transitions in photosynthetic molecules and autofluorescence, typically ranges from 488 to 660 nm. Because of this range, it is difficult to measure Raman signals from photosynthetic molecules using conventional Raman microscopy. Although the use of longer excitation wavelengths (785 or 1065 nm) for Raman measurements could prevent the effect of autofluorescence, the scattering efficiency in the near-infrared wavelength region is low. Therefore, high-resolution Raman imaging consumes time, e.g. it takes 25 h for acquiring a 300 × 300 pixelated hyperspectral image with 1.0 s exposure time. This results in a reduction of the measurement area or pixel resolution during Raman imaging of biofilms. In addition, a higher incident laser power is required to obtain clear Raman spectra because Raman scattering has a weak optical effect; this is usually accompanied by photodamage of the sample. This provides a case for photosynthetic bacteria and their biofilms, because they are fragile and photosensitive. In contrast, FTIR spectroscopy is an autofluorescence-free and non-destructive analytical technique that employs mid-IR light, which has low photon energies and does not excite the fluorescence of photosynthetic molecules. Hence, it has been extensively used for the chemical characterization of photosynthetic molecules and bacterial extracellular polysaccharides.6,7 However, the spatial resolution of FTIR spectroscopy is limited to several micrometers or more because of the diffraction limit of mid-IR light, which is not high enough to resolve single bacteria and their biofilm structures. Although AFM-based nanoscale vibrational imaging techniques are useful to study photosynthetic molecules,8–11 these techniques have a limited field-of-view, typically in a few to ten micrometers scale and require mechanical contact between a metallic tip and a sample. They also require a long acquisition time for imaging while the sample can be mechanically damaged by the tip when high-speed imaging is performed.
Herein, we present all-optical super-resolution mid-IR imaging of photosynthetic bacterial biofilms by means of mid-infrared photothermal (MIP) microscopy. MIP microscopy leverages both continuous visible and pulsed-IR beams, and monitors scattering signal in the visible region to measure photothermal responses of a sample originating from IR absorption. When the modulated pulsed mid-IR light is absorbed by a sample, local sample heating and expansion, known as the photothermal effect, occurs. The intensity of visible scattering or reflection modulated by the photothermal effect is detected to measure IR absorption of the sample. Since the spatial resolution of this technique can be determined by the wavelength of the continuous visible beam, MIP microscopy achieves both the sub-micrometer spatial resolution beyond the diffraction limit of mid-IR light and the high chemical specificity based on IR absorption.12–16 Owing to its super-resolution label-free imaging capability, MIP microscopy has shown versatile applications in life science including microbiology13,17,18 and material science.12,19 MIP microscopy also prevents the autofluorescence signals of photosynthetic bacteria from affecting vibrational imaging, which aids in the visualization of the extracellular polysaccharides of photosynthetic bacteria without signal deterioration. Moreover, MIP microscopy offers multi-modal optical imaging, in which both vibrational and autofluorescence imaging of the same sample can be performed. To our knowledge, the visualization of biofilms and bacterial cells using MIP has not been previously reported. In this work, the distribution of cyanobacteria and their extracellular polysaccharides in a biofilm matrix was successfully visualized in a label-free manner using multi-modal optical imaging. The formation of the cyanobacterial biofilms was investigated without inhibitions due to staining or labeling, and it was evidenced that cyanobacterial cells were distributed along acidic/sulfated polysaccharides in extracellular environment. Furthermore, our observation found the areas, where cyanobacterial cells were not accompanied with sulfated polysaccharides, which supports hypothesis that sulfated polysaccharides initially form the line structures followed by trapping cyanobacterial cells with EPSs. MIP microscopy also enabled large-area and high-resolution observations of biofilm components without causing photodamage to the samples. This study paves the way for a better understanding of the formation, structure, and functions of photosynthetic biofilms.
Experimental section
Sample preparation
Preparation of the biofilm samples followed the procedure described by Maeda et al.20 The motile substrain PCC-P of the unicellular cyanobacterium Synechocystis sp. PCC 6803 (here after S.6803), which exhibits phototaxis21 and forms bloom-like aggregates, were cultured in 50 mL of BG11 liquid medium22 under continuous illumination of fluorescent lamps from outside (30 μmol photons m−2 s−1) with bubbling of 2% CO2 in air at 31 °C, or on 1.5% agar plates. Cell density was monitored at 730 nm. Before the biofilm formation experiment, cells were precultured once in liquid after transfer from plates. In the first step, cells inoculated at OD730 = 0.2 were grown with vigorous aeration at 31 °C for 48 h. In the second step, the culture was shifted to the standing condition without bubbling under the same continuous light for another 48 h for cells to rise to the surface and form bloom-like viscous biofilm. The xanthan gum (SIGMA, G1253) was used as a control of non-sulfated acidic polysaccharides. The sulfated polysaccharide produced by S.6803, synechan, was purified by the protocol in the previous work20 and used as a positive control. The samples for both the MIP and FTIR measurements were casted onto a transparent CaF2 substrate.Fluorescent polymer beads with a diameter of 500 nm were purchased from Polysciences (Fluoresbrite, 529/547). The fluorescent beads were casted on a transparent CaF2 substrate for measurements.
Optical setup of MIP microscopy
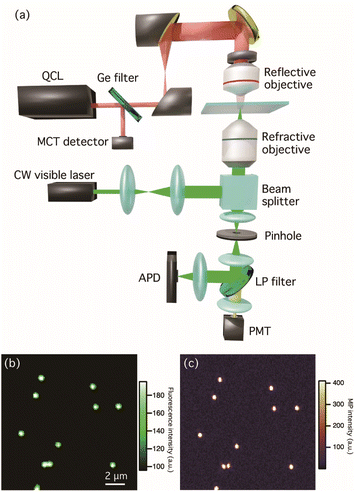
Fig. 1(a) shows the optical setup for multi-modal optical imaging. Our MIP imaging configuration employs a counter-propagation scheme for both mid-IR and visible light sources because the refractive objective lens with high numerical aperture (NA) can be used for visible light. This scheme affords both the high detection sensitivity of MIP signal and the high spatial resolution to visualize bacteria and their biofilms. A wavenumber-tunable quantum cascade laser (QCL, Daylight Solutions, MIRcat-QT-2100) ranging in wavenumber from 1000 to 1690 cm−1 was used as a pulsed mid-IR pump light source for MIP measurements. The power of the IR laser was set to less than 0.8 mW for all measurements to avoid photodamage. The operated repetition rate of the QCL was set to 80 kHz and a pulse width was set to 500 ns. A Ge window reflects 10% of the IR beam in the optical path guiding to monitor the power spectrum of the QCL by a mercury cadmium telluride (MCT) detector (Thorlabs, PDAVJ10). The IR beam was focused on the sample by a reflective objective (Thorlabs, 40×/NA 0.5). A continuous-wave (CW) visible laser with a wavelength of 532 nm (ELFORLIGHT, G4-200) was used for both MIP and autofluorescence imaging and the power density of the visible beam was less than 1.4 × 102 kW cm−2. The visible beam was guided to a high-NA water immersion objective (Olympus, 60×/NA 1.2) using a beam splitter (reflection![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) transmission = 10
transmission = 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90). The back-scattered beam was collected using the same objective. After passing through a pinhole with a diameter of 50 μm, the beam was split in two using a dichroic mirror: one beam containing a visible scattering signal was sent to an avalanche photodetector (APD, Thorlabs, APD430A2/M) to extract the photothermal responses of the samples, and the other beam originating from autofluorescence was detected using a photomultiplier tube (PMT, Thorlabs, PMT1002). The signal detected by the APD was sent to a lock-in amplifier (Zurich Instrument, MFLI) for the phase-sensitive detection, which was demodulated at the same frequency of the QCL repetition rate. Fluorescence and MIP images of fluorescent polymer beads with a diameter of 500 nm are shown in Fig. 1(b) and (c), respectively. The MIP image was constructed using the MIP signal intensity at 1494 cm−1, which corresponds to the aromatic vibrational mode of polystyrene. The contrast of fluorescence and MIP signals with sub-micrometer spatial resolution, which was beyond the diffraction limit of IR light, showed that the design enabled the visualization of polystyrene beads.
90). The back-scattered beam was collected using the same objective. After passing through a pinhole with a diameter of 50 μm, the beam was split in two using a dichroic mirror: one beam containing a visible scattering signal was sent to an avalanche photodetector (APD, Thorlabs, APD430A2/M) to extract the photothermal responses of the samples, and the other beam originating from autofluorescence was detected using a photomultiplier tube (PMT, Thorlabs, PMT1002). The signal detected by the APD was sent to a lock-in amplifier (Zurich Instrument, MFLI) for the phase-sensitive detection, which was demodulated at the same frequency of the QCL repetition rate. Fluorescence and MIP images of fluorescent polymer beads with a diameter of 500 nm are shown in Fig. 1(b) and (c), respectively. The MIP image was constructed using the MIP signal intensity at 1494 cm−1, which corresponds to the aromatic vibrational mode of polystyrene. The contrast of fluorescence and MIP signals with sub-micrometer spatial resolution, which was beyond the diffraction limit of IR light, showed that the design enabled the visualization of polystyrene beads.
For reference measurements, the IR absorption spectra of the samples were measured using a commercially available FTIR spectrometer in transmission mode (JASCO Corporation, FTIR-6800). The FTIR spectra ranged between 1000 and 3000 cm−1 with a spectral resolution of 2 cm−1. The reference FTIR spectra of the cyanobacterial cells and exopolysaccharides were measured to characterize the IR vibrational signatures of the biofilm components.
Results and discussion
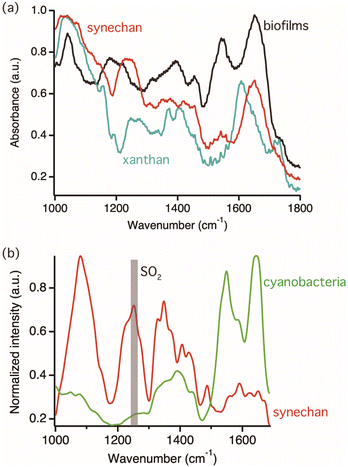
The target polysaccharides of this study are the sulfated polysaccharides, which are acidic polysaccharides modified by sulfate residue. In nature, various sulfated polysaccharides are distributed specifically in eukaryotic algae and cyanobacteria among microorganisms and found in their biofilms.23,24 In other words, sulfated polysaccharides are one of the important targets for photosynthetic biofilm research. A recent study showed that one model cyanobacterium, S. 6803, produces a novel sulfated polysaccharide, synechan, extracellularly and uses it to form a viscous biofilm like a bloom.20 This biofilm was selected for observation because it can be easily reproduced under laboratory conditions.First, reference FTIR spectra of cyanobacterial cells and exopolysaccharides were measured to characterize IR vibrational signatures of biofilm components. Fig. 2(a) shows FTIR spectra of the biofilm sample compose of cyanobacteria and all biofilm components, synechan, and xanthan as the example of non-sulfated acidic polysaccharides, shown by the black, red, and light blue colored spectra, respectively. In the spectrum of synechan, the characteristic vibrational mode of carboxyl group at 1643 cm−1 and the SO2 stretching mode at 1250 cm−1 deriving from sulfated polysaccharides were observed.25 On the other hand, the identical vibrational mode of the sulfated group was not observed in the spectrum of xanthan because of the absence of sulfated polysaccharides. The peaks emerging from 1350 to 1450 cm−1 in all spectra were assigned as vibrational modes of CH2 and COO− groups, respectively. Distinct peaks were observed in the MIP spectrum of the biofilm; for example, the amide I and II bands at 1640 and 1550 cm−1, respectively, the amide III band conjugated with the SO2 stretching mode at approximately 1200 cm−1, the PO4− stretching mode, and the vibrational mode of skeletal glucan at approximately 1050 cm−1. Because the FTIR spectrum of a biofilm reflects the vibrational signatures of all biofilm components, such as cyanobacteria, EPSs, and proteins, it is difficult to distinguish their characteristics. In order to identify vibrational signatures of cyanobacterial cells and sulfated exopolysaccharides, IR absorption of a single cyanobacterial cell and synechan were recorded by MIP microscopy, which has the high spatial resolution enough to measure a single bacterium. Fig. 2(b) shows MIP spectra of a single cyanobacterial cell and the synechan sample. Distinct MIP signal of the sulfated group at 1250 cm−1 could be obtained only from sulfated exopolysaccharides while the amide III band from proteins in cyanobacteria was weak but detectable in our MIP system. Hence, the SO2 stretching mode at 1250 cm−1 could be the marker band for sulfated exopolysaccharides. The slight variation between the FTIR and MIP spectra of synechan and cyanobacteria/biofilms could be due to the difference in molecular components within the detection volume of each technique and measurement principle or indicate variation of molecular state, namely exopolysaccharides and cyanobacteria were interacted with biofilm components in the biofilm matrix and vibrational energy might be perturbed.26,27 In synechan, its predominant constituent consists of sulfated polysaccharides, although contamination of trace amounts of protein and dye is possible due to the purification process. The MIP spectrum of synechan exhibits notably sharp peaks, in contrast to the relatively broad peaks observed in the FTIR spectrum. This discrepancy might arise from the fact that FTIR spectra show assemble information on all molecular components in synechan while MIP spectra show relatively local chemical information of samples. We would like to emphasize here that such vibrational signatures of cyanobacteria and synechan cannot be analyzed by conventional Raman spectroscopy measurements because of bright autofluorescence signal as shown in Fig. S1 in ESI.†
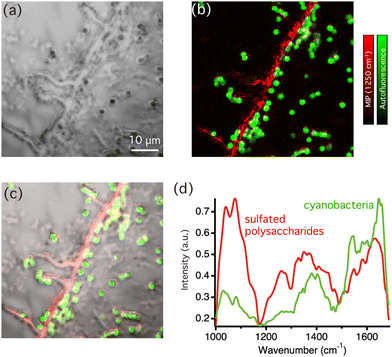
Next, we performed multi-modal optical imaging of cyanobacterial biofilms. Fig. 3(a) displays an optical image of biofilms casted on the CaF2 substrate. The optical image shows entire biofilm structures without chemical specificity, so that biofilm components could not be identified. Fig. 3(b) shows an overlaid image of autofluorescence intensity (green) and MIP signal intensity images at 1250 cm−1 (red) in the corresponding area. Because photosynthetic pigments, such as chlorophyll, in cyanobacteria yield bright fluorescence signals, the autofluorescence image specifically represents the distribution of cyanobacterial cells. The MIP image was constructed by the MIP signal intensity at 1250 cm−1, corresponding to the sulfated group to visualize sulfated exopolysaccharides. The line structures were visualized in Fig. 3(b), which indeed uncovers that the structures were formed by sulfated exopolysaccharides. The distribution of the molecular components of the biofilm matrix was further clarified in the bright-field image of the biofilm superimposed with the multi-modal optical imaging result as shown in Fig. 3(c). Notably, multi-modal optical imaging showed that most cyanobacterial cells were distributed along the line structures of the sulphated exopolysaccharides or in their vicinity. It is well known that acidic exopolysaccharides usually form line shape structures accompanied with cyanobacterial cells at macroscopic scale, but they were not observed in sub-micron scale.28 For example, in S.6803 under laboratory conditions, it was speculated that cyanobacterial cells and bubbles are first trapped in linearly distributed synechan chains and then rise to the surface due to the buoyancy of the bubbles to form bloom-like aggregates at the liquid surface.20 However, it was difficult to observe the original positional relationship between polysaccharides and the cells with microscale observations using basic dyes, such as Alcian blue, because the acidic polysaccharides were firmly aggregated by the staining agents. These results support the hypothesis that cyanobacterial cells are distributed along the extracellular acidic polysaccharides, even at the microscale level. Because the distribution is extracellular polysaccharide-dependent in the early stages of biofilm formation, the changes in the polysaccharide structure due to thixotropy or salinity changes may stimulate biofilm formation.
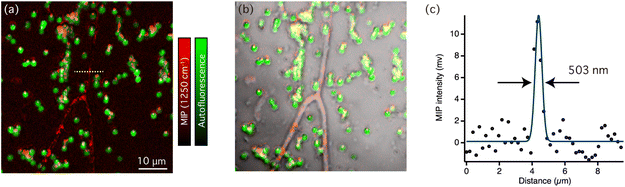
Fig. 3(d) shows comparison of typical MIP spectra recorded from the line structures of sulfated exopolysaccharides and a single cyanobacteria. Not only distinct peaks from sulfated exopolysaccharides but also weak amide II bands around 1550 cm−1 were also observed in the MIP spectra of sulfated exopolysaccharides. This suggested that the sulfated exopolysaccharides were associated with proteins. It has been shown that protein components in the biofilm matrix interacted with exopolysaccharides, which could lead to the formation of bundles in biofilms.29 Fig. S2(a) in ESI† shows the MIP image at 1550 cm−1, which corresponds to the amide II band. The contrast in Fig. S2(a)† is similar, but slightly varied compared with the MIP image at 1250 cm−1 because it predominantly reflects on the protein distribution. It is necessary to indicate that the chemical contrast in the MIP images displayed here should originate from vibrational signatures of biofilm components as the MIP image of the biofilms at 1180 cm−1 (see Fig. S2(b) in ESI†), which is off-resonant IR wavenumber of biofilm components, hardly showed any contrast. These insights into the molecular distributions in cyanobacterial biofilms were revealed because of the multi-modal imaging capability that was based on autofluorescence and the IR absorption of molecules. It should be noted that the slight difference of MIP spectra of cyanobacteria depicted in Fig. 2(b) and 3(d) is ascribed to variances of the sample conditions. The former shows the MIP spectrum of cyanobacterial cells isolated from biofilms, manifesting vibrational signatures of pure cyanobacteria while the latter shows the MIP spectra of a single cyanobacterial cell in the biofilm matrix. Consequently, extracellular substances within biofilms, such as sulfated exopolysaccharides and proteins, may interact with cyanobacteria and slightly contribute vibrational signatures. We further sought to find the area, where the distribution of sulfated exopolysaccharides and cyanobacterial cells varies. Fig. 4(a) shows an overlaid image of autofluorescence intensity (green) and MIP intensity images at 1250 cm−1 (red) in the biofilm sample. Fig. 4(b) displays the bright-field image of the corresponding area of the biofilm superimposed with the multi-modal optical imaging result. In this area, the sulfated exopolysaccharides form the line structures accompanied with several cyanobacterial cells while some cyanobacterial cells were not associated with the line structures in some area. The distribution of cyanobacterial cells apart from the sulfated exopolysaccharides suggests that the line structures of sulfated exopolysaccharides were self-formed. These cells were then surrounded by molecular interactions with sulfated exopolysaccharides. The label-free observation of biofilms at sub-micrometer scale allowed to gain spectroscopic evidence of formation of photosynthetic bacterial biofilms. Fig. 4(c) shows the intensity line-profile of the MIP image at 1250 cm−1 along the white dotted line in Fig. 4(a). According to the gaussian-fitted intensity profile in Fig. 4(c), the full-width half at maximum (FWHM) of the profile was estimated to be 503 nm, thereby we demonstrated that biofilm components can be visualized in our MIP system with at least the spatial resolution of 500 nm, which cannot be achieved by conventional FTIR imaging systems. The demonstrated performance of our MIP imaging and the fundamental performance of other vibrational imaging techniques were summarized in Table S1 in ESI.† Owing to its nonlinear signal-generating mechanism, MIP microscopy has three-dimensional sectioning capabilities that provide an advantage over conventional IR imaging techniques (linear IR absorption) when imaging microbial species and their biofilms.30 Hence, volumetric structures of biofilms formed in three dimensions, such as algal and cyanobacterial colonies, can be observed using MIP microscopy. Internal molecular structures of relatively large microorganisms spanning from 10 to 100 μm in size can also be observed, expanding the use of MIP imaging for microbiological applications.24 Because AFM-based nanoscale IR imaging is a surface sensitive technique that cannot resolve structures in three dimensions, the significance of the visualization of biofilms by MIP imaging is emphasized. In addition, the combination of wide-field imaging techniques with MIP microscopy achieved real-time MIP imaging as Bai et al., demonstrated.31 This would be profitable for visualizing the growth of biofilms in real-time based on the chemical contrast of molecules revealing the roles of each molecule in biofilm formation. Therefore, the visualization of biofilm components by MIP imaging in this study can be used to understand the fundamentals of biofilm formation and their biological functions.
Apart from the capability of MIP microscopy for label-free visualization of the distribution of biofilm components, we would like to emphasize here that MIP microscopy is a non-destructive analytical approach. Because photosynthetic bacteria (for example, cyanobacteria) are photosensitive, a low power density of incident light is required to better understand their biological functions in a non-destructive manner.32 The high power density (more than 1.0 × 104 kW cm−1) of incident light used for typical Raman spectroscopy measurement generally results in photodamage of samples and degradation of Raman spectra are seen.33 Although the visible incident beam was also employed in MIP and autofluorescence imaging, the power density of the incident beam was 2 orders lower than the case for Raman imaging. Fig. S3 in ESI† shows the MIP spectra of the same cyanobacterial cell continuously recorded on six occasions. No notable spectral degradation was observed in all spectra; therefore, MIP measurements did not result in the photodamage of the cyanobacteria. Optical measurements of photosynthetic bacteria and biofilms can be performed in a non-destructive manner. Given that bacterial communities are sensitive to light-induced stress, which may alter their biological functions, the implementation of a non-destructive chemical imaging approach would improve the understanding of the multifaceted roles of bacterial biofilms, such as their biological functions, formation, and motility.
Conclusion
We demonstrated label-free visualization of photosynthetic bacterial biofilms by utilizing MIP microscopy. Owing to its capability of multi-modal autofluorescence and super-resolution chemical imaging based on IR absorption, the distribution of sulfated exopolysaccharides and cyanobacterial cells were successfully visualized with the sub-micrometer spatial resolution, which is far beyond the diffraction limit of IR light. Additionally, imaging was not inhibited by staining or labelling and showed biofilm formation, where cyanobacterial cells were distributed along acidic/sulfated polysaccharides in the extracellular environment. Further, we found the area, where cyanobacterial cells were not associated with sulfated polysaccharides, which supports hypothesis that sulfated polysaccharides first form the line structures followed by trapping cyanobacterial cells with EPSs. Multi-modal optical imaging also showed that the protein components in the biofilm matrix may interact with sulphated exopolysaccharides, supporting the assumptions of previous studies. This study has demonstrated the capability of MIP microscopy to visualize the distribution of photosynthetic microorganisms and their biofilm components, surpassing the capabilities of conventional vibrational imaging methodologies. The high sensitivity with low incident input power, large field-of-view, and high-speed imaging demonstrated in this work, highlights the versatile potential of MIP microscopy in the study of microbiology. Our present work offers powerful ways to fetch important insights into chemical heterogeneities/homogeneities of microbial biofilms. These tools can be used to better understand the fundamentals of microbial communities, biofilm formation, and biosynthetic processes, which would accelerate the use of microorganisms and their EPSs for biotechnological and environmental applications.Author contributions
R. K. and K. M. conceptualized and supervised the present work and wrote the manuscript. K. M. carried out sample preparation and R. K. carried out spectroscopic experiments. All authors discussed the results and provided feedbacks on manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported in part by JST ACT-X Grant No. JPMJAX21B4 (R. K.) and JPMJAX20BG (K. M.), JST FOREST Grant No. JPMJFR202I (T. Y.), JST CREST Grant No. JPMJCR1904 (T. T.), JSPS KAKENHI Grant No. JP23H04139 (R. K.), Nakatani foundation, Shimadzu Science Foundation, and Research Foundation for Opto-Science and Technology. The authors also acknowledge the financial support from the project on the Promotion of Regional Industries and Universities by the Cabinet Office, research clusters of Tokushima University.References
- H.-C. Flemming and J. Wingender, Nat. Rev. Microbiol., 2010, 8, 623–633 CrossRef CAS PubMed.
- H.-F. Xu, H. Raanan, G.-Z. Dai, N. Oren, S. Berkowicz, O. Murik, A. Kaplan and B.-S. Qiu, FEMS Microbiol. Rev., 2021, 45, fuab036 CrossRef CAS PubMed.
- X. Mari, U. Passow, C. Migon, A. B. Burd and L. Legendre, Prog. Oceanogr., 2017, 151, 13–37 CrossRef.
- D. Mrđenović, D. Abbott, V. Mougel, W. Su, N. Kumar and R. Zenobi, ACS Appl. Mater. Interfaces, 2022, 14, 24938–24945 CrossRef PubMed.
- W. Zhang, Z. Dong, L. Zhu, Y. Hou and Y. Qiu, ACS Nano, 2020, 14, 7920–7926 CrossRef CAS PubMed.
- Y. Kato, S. Haniu, Y. Nakajima, F. Akita, J. R. Shen and T. Noguchi, J. Phys. Chem. B, 2020, 124, 121–127 CrossRef CAS PubMed.
- S. Abinandan, S. R. Subashchandrabose, K. Venkateswarlu and M. Megharaj, ACS Omega, 2020, 5, 6888–6894 CrossRef CAS PubMed.
- R. Kato, T. Umakoshi and P. Verma, J. Phys. Chem. C, 2021, 125, 20397–20404 CrossRef CAS.
- R. Kato, T. Moriyama, T. Umakoshi, T. Yano and P. Verma, Sci. Adv., 2022, 8, eabo4021 CrossRef CAS PubMed.
- T. Tanaka, T. Yano and R. Kato, Nanophotonics, 2022, 11, 2541–2561 CrossRef CAS.
- A. Dazzi and C. B. Prater, Chem. Rev., 2017, 117, 5146–5173 CrossRef CAS PubMed.
- R. Kato, T. Yano and T. Tanaka, Vib. Spectrosc., 2022, 118, 103333 CrossRef CAS.
- C. Li, D. Zhang, M. N. Slipchenko and J. X. Cheng, Anal. Chem., 2017, 89, 4863–4867 CrossRef CAS PubMed.
- K. Aleshire, I. M. Pavlovetc, R. Collette, X. T. Kong, P. D. Rack, S. Zhang, D. J. Masiello, J. P. Camden, G. V. Hartland and M. Kuno, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 2288–2293 CrossRef CAS PubMed.
- O. Klementieva, C. Sandt, I. Martinsson, M. Kansiz, G. K. Gouras and F. Borondics, Adv. Sci., 2020, 7, 1903004 CrossRef CAS PubMed.
- R. Kato, T. Yano and T. Tanaka, Analyst, 2023, 148, 1285–1290 RSC.
- J. Yin, L. Lan, Y. Zhang, H. Ni, Y. Tan, M. Zhang, Y. Bai and J. X. Cheng, Nat. Commun., 2021, 12, 1–11 CrossRef PubMed.
- X. Li, D. Zhang, Y. Bai, W. Wang, J. Liang and J. X. Cheng, Anal. Chem., 2019, 91, 10750–10756 CrossRef CAS PubMed.
- N. Baden, H. Kobayashi and N. Urayama, Int. J. Polym. Anal. Charact., 2020, 25, 1–7 CrossRef CAS.
- K. Maeda, Y. Okuda, G. Enomoto, S. Watanabe and M. Ikeuchi, eLife, 2021, 10, e66538 CrossRef CAS PubMed.
- S. Yoshihara, F. Suzuki, H. Fujita, X. X. Geng and M. Ikeuchi, Plant Cell Physiol., 2000, 41, 1299–1304 CrossRef CAS PubMed.
- R. Y. Stanier, R. Kunisawa, M. Mandel and G. Cohen-Bazire, Bacteriol. Rev., 1971, 35, 171–205 CrossRef CAS PubMed.
- S. Pereira, A. Zille, E. Micheletti, P. Moradas-Ferreira, R. De Philippis and P. Tamagnini, FEMS Microbiol. Rev., 2009, 33, 917–941 CrossRef CAS PubMed.
- F. Tsuneo, O. Takahira, M. Masayoshi, N. Kazuhiro, T. Yasunobu and H. Hideaki, Appl. Environ. Microbiol., 2004, 70, 3338–3345 CrossRef PubMed.
- M. Okajima-Kaneko, M. Ono, K. Kabata and T. Kaneko, Pure Appl. Chem., 2007, 79, 2039–2046 CrossRef CAS.
- E. J. Elzinga, J.-H. Huang, J. Chorover and R. Kretzschmar, Environ. Sci. Technol., 2012, 46, 12848–12855 CrossRef CAS PubMed.
- Z. Hong, W. Chen, X. Rong, P. Cai, K. Dai and Q. Huang, Chem. Geol., 2013, 360–361, 118–125 CrossRef CAS.
- D. C. Fabian, Z. Rui-Qian, O. Sabrina, S. Nils, W. Annegret and W. M. Conrad, J. Bacteriol., 2019, 201, e00344–e00319 Search PubMed.
- J. Böhning, M. Ghrayeb, C. Pedebos, D. K. Abbas, S. Khalid, L. Chai and T. A. M. Bharat, Nat. Commun., 2022, 13, 7082 CrossRef PubMed.
- D. Zhang, C. Li, M. Slipchenko, C. Zhang and J. X. Cheng, Opt. InfoBase Conf. Pap., 2017, Part F79-N, 1–8.
- Y. Bai, D. Zhang, L. Lan, Y. Huang, K. Maize, A. Shakouri and J.-X. Cheng, Sci. Adv., 2019, 5, eaav7127 CrossRef CAS PubMed.
- Y. Yawata, K. Toda, E. Setoyama, J. Fukuda, H. Suzuki, H. Uchiyama and N. Nomura, J. Biosci. Bioeng., 2010, 110, 377–380 CrossRef CAS PubMed.
- R. Kato, T. Yano, T. Minamikawa and T. Tanaka, Anal. Sci., 2022, 38, 1497–1503 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Raman spectra of cyanobacteria and biofilms, MIP images of biofilms at 1550 cm−1 and 1180 cm−1, comparison of imaging performance, and MIP spectra of cyanobacteria cells recorded 6 times. See DOI: https://doi.org/10.1039/d3an01453c |
| This journal is © The Royal Society of Chemistry 2023 |