Open Access Article
Open Access ArticleElucidation of N-/O-glycosylation and site-specific mapping of sialic acid linkage isomers of SARS-CoV-2 human receptor angiotensin-converting enzyme 2†
Liming
Wei‡
 a,
Yuning
Chen‡
bc,
Xiaoxiao
Feng
a,
Jun
Yao
a,
Lei
Zhang
a,
Xinwen
Zhou
a,
Guoquan
Yan
a,
Hong
Qiu
bc,
Chunhe
Wang
*bc and
Haojie
Lu
a,
Yuning
Chen‡
bc,
Xiaoxiao
Feng
a,
Jun
Yao
a,
Lei
Zhang
a,
Xinwen
Zhou
a,
Guoquan
Yan
a,
Hong
Qiu
bc,
Chunhe
Wang
*bc and
Haojie
Lu
 *a
*a
aInstitutes of Biomedical Sciences and Department of Chemistry, Fudan University, 131 Dongan Road, 20032 Shanghai, China. E-mail: luhaojie@fudan.edu.cn
bShanghai Institute of Materia Medica, Chinese Academy of Sciences Department, 555 Zuchongzhi Road, 201203 Shanghai, China. E-mail: wangc@simm.ac.cn
cSchool of Pharmacy, University of Chinese Academy of Sciences, No. 19A Yuquan Road, 100049 Beijing, China
First published on 5th September 2023
Abstract
Human angiotensin-converting enzyme 2 (hACE2) is the primary receptor for cellular entry of SARS-CoV-2 into human host cells. hACE2 is heavily glycosylated and glycans on the receptor may play a role in viral binding. Thus, comprehensive characterization of hACE2 glycosylation could aid our understanding of interactions between the receptor and SARS-CoV-2 spike (S) protein, as well as provide a basis for the development of therapeutic drugs targeting this crucial interaction. Herein, 138 N-glycan compositions were identified, most of which are complex-type N-glycans, from seven N-glycosites of hACE2. Among them, 67% contain at least one sialic acid residue. At the level of glycopeptides, the overall quantification of sialylated glycan isomers observed on the sites N322 and N546 have a higher degree of NeuAc (α2–3)Gal (over 80.3%) than that of other N-glycosites (35.6–71.0%). In terms of O-glycans, 69 glycan compositions from 12 O-glycosites were identified, and especially, the C-terminus of hACE2 is heavily O-glycosylated. The terminal sialic acid linkage type of H1N1S1 and H1N1S2 are covered highly with α2,3-sialic acid. These findings could aid the investigation of the interaction between SARS-CoV-2 and human host cells.
1. Introduction
Caused by severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2), coronavirus disease-2019 (COVID-19) is a severe respiratory disease that has spread throughout the world since emerging near the end of 2019.1 In the three and a half years since this virus emerged, seven SARS-CoV-2 mutants have evolved, each with high transmissibility and an increased risk for infection.2,3 To date, over 765.2 million people have been infected with SARS-CoV-2 and 6.9 million deaths due to COVID-19 have been recorded globally. Moreover, the epidemic has severely affected normal production and trade activities around the world, triggering a global economic recession. Even though the WHO announced that the ongoing COVID-19 has not been a public health emergency of international concern recently, continued research is still needed to better understand SARS-CoV-2 infection to resist the possible occurrence of other mutants.SARS-CoV-2 infects host cells via S protein, which interacts with hACE2 on the surface of host cells to initiate infection.4,5 hACE2 is a type I transmembrane protein expressed in human tissues such as lung, nasal, oral mucosa, heart, etc.6–9 Besides, the high expression of hACE2 is correlated with the high risk of SARS-CoV-2 infection.9 Notably, the function of favourable mutations in SARS-CoV-2 S protein exhibited higher binding affinity with hACE2.10 The key role of hACE2 may have contributed to multi-organ damage in COVID-19 patients to spark a pandemic by SARS-CoV-2 infection.11 Hence, hACE2 is critical for both in-depth understanding of the infectious disease and the development of therapeutics. Both SARS-CoV-2 S protein and hACE2 are heavily glycosylated, which affects the interaction between SARS-CoV-2 S protein and hACE2.12–15 hACE2 has been reported to have high glycan occupancy (73.2 to 100%) at seven N-glycosylation sites, and one novel O-glycosylation site was deduced to be involved in viral binding.15,16 The glycans at N53, N90, and N322 on hACE2 are critical for SARS-CoV-2 S protein binding with hACE2.17,18 Meanwhile, the glycan at N546 on hACE2 is reported to interact with the glycans at N74 and N165 on the SARS-CoV-2 S protein.15 Thus, glycans may play a role in the binding between hACE2 and both the protein and glycan components of SARS-CoV-2 S protein.14
As ubiquitous terminal residues of glycol conjugates (usually covering the cell surface), sialic acids are critical in viral tissue tropism and control the entry of viruses including influenza viruses, orthomyxoviruses, and coronaviruses.19–23 Previous studies showed that the S proteins from HCoV-OC43 and BCoV might mediate entry into host cells using 9-O-acetyl-sialic acid (9-O-Ac-Sia) as a receptor.21 Based on homology, in silico studies have suggested that the N-terminal domain of SARS-CoV-2 S protein is responsible for binding with terminal sialic acid attached to either proteins or lipids on the host cell surface.24 Assisted by a glycol nanoparticle decorated with N-acetylneuraminic acid (NeuAc), researchers discovered that sialic acid might bind with SARS-CoV-2 S protein.25 At the terminus of glycans on human glycoproteins, sialic acid is usually attached to galactose (Gal) residues through either α2,3- or α2,6-linkages. The specific configurations, mainly α2,3- or α2,6-sialic acid isomers, are preferentially recognized in virus infection.21,26,27 For example, human influenza viruses primarily bind to sialic acid terminated by α2,6-galactose, while avian influenza viruses bind preferentially to sialic acid, which is terminated by α2,3-galactose.19 Within the current pandemic of COVID-19, saturation transfer difference-1H,13C-HSQC NMR experiments have elegantly demonstrated how SARS-CoV-2 S protein effectively binds NeuAc in both α2,3- and α2,6-sialyl N-acetyllactosamine, especially for the α2,3-linked analogue.28 Through hACE2 glycosylation mutants and surface plasmon resonance, it was shown that the sialic acids on N90 of hACE2 may have hampered the interaction between SARS-CoV-2 S and hACE2.29 Yuen et al. found that removing sialic acids from hACE2 enhanced the binding between the receptor-binding domain (RBD) of SARS-CoV-2 S protein and hACE2.29 It was demonstrated that the sialic acid on hACE2 precluded a perfect interaction between SARS-CoV-2 S and hACE2, which is in line with the study that treating hACE2 with neuraminidase increased the binding with SARS-CoV-2 S protein.17,30 However, Klassen et al. showed that both RBD binding to and pseudo-virus infection of hACE2 expressing cells are decreased upon deletion of cell surface sialic acids in different ways.31 The discrepancy of these reports indicates that efficient SARS-CoV-2 infection may also require terminal sialic acid of hACE2 in a specific linkage type. A relatively higher level of α2,3-sialylated N-glycans on hACE2 has been revealed using the ESI-MSn method.16 However, the α2,3- or α2,6-linked sialic acid was analyzed without considering the glycosites; therefore, it might average the different ratios of the same glycan composition at different glycosites. To deeply understand the role of glycosylation in the SARS-CoV-2 S/hACE2 interaction, a thorough characterisation of the glycosylation of hACE2 is needed, including the detail of linkage types of terminal sialic acids associated with different glycosites and glycans.
In this study, we combined ion mobility (IM)-mass spectrometry (MS) and high-resolution MS to characterize N- and O-glycosylation on hACE2 with the pretreatment of glycopeptides using ZIC-HILIC media. The mapping of α2,3- or α2,6-sialic acid linkage isomers on hACE2 was first reported at the glycopeptide level. Meanwhile, the complex O-glycan compositions of novel O-glycosites were determined. The linkage types of terminal sialic acid associated with the highly abundant O-glycans at the C-terminus of hACE2 were elucidated as well.
2. Experimental section
2.1. Chemicals and materials
Sequencing-grade modified trypsin and Glu-C protease were obtained from Promega (Madison, WI, USA). Proteinase K was purchased from Takara (Beijing, China). Peptide-N-glycosidase F (PNGase F) was obtained from New England Biolabs (Ipswich, MA, USA). Other chemical reagents were obtained from Sigma-Aldrich (St Louis, MO, USA). Analytical grade solvents and zwitterionic chromatography-hydrophilic interaction liquid chromatography (ZIC-HILIC) medium (5 μm) were acquired from Merck (Darmstadt, Germany). A C18 solid-phase extraction cartridge was obtained from Waters Technology Co., Ltd (Shanghai, China).2.2. Expression and purification of hACE2
The codon-optimized DNA fragment encoding extracellular hACE2 (residues 18–740) was sub-cloned into the mammalian expression vectors pTT5 (Addgene) with a 6× His-tag at the C-terminus. Purified plasmids were transfected into HEK293F cells and cultured in suspension with FreeStyle 293 expression medium (Gibco, CA, USA). After 6 days of culture, the cell culture medium was collected and purified by Ni-NTA chromatography. Protein purity was measured using SDS-PAGE gel (Bio-Rad Laboratories, Inc., CA, USA).2.3. Sample preparation of hACE2 for glycosylation analysis
Purified hACE2 (200 μg in 25 mM NH4HCO3) was reduced and alkylated following the kit protocol (Applied Biosystems Inc., Foster City, CA). Subsequently, the protein was digested by incubating for 18 h at 37 °C first with trypsin and then with Glu-C.32 For O-glycosylation analysis of hACE2, N-linked glycans were removed from the denatured hACE2 sample by treating with PNGase F for 18 h at 37 °C. For the analysis of hACE2 C-terminal O-glycosylation, proteinase K was additionally applied at 37 °C overnight.For the specific enrichment of N- or O-glycopeptides of hACE2, the resulting peptides were purified using a C18 solid-phase extraction column, freeze-dried, and enriched using the ZIC-HILIC tips as previously described.33 The peptides were first dissolved in loading buffer consisting of 80% acetonitrile (ACN) and 1% trifluoroacetic acid (TFA) and then loaded onto a microtip containing 30 mg of ZIC-HILIC medium. After being washed with 200 μL of loading buffer 6 times, the enriched N- or O-glycopeptides were eluted with 200 μL of 0.1% TFA solution. The eluted N- or O-glycopeptides were then dried by vacuum centrifugation and stored at −80 °C.
2.4. Nano-liquid chromatography-mass spectrometry analysis
N- and O-glycopeptides of hACE2 were analyzed on a nano-HPLC (M-Class), equipped with a PST CSH130 reversed-phase analytical column (Waters, C18, 150 mm × 75 μm, 1.7 μm) maintained at 35 °C. Mobile phases (MPs) are A: 0.1% formic acid (FA) in water and B: 0.1% FA in water/ACN (2/98, v/v). The flow rate was 300 nL min−1, with a gradient from 5 to 40% MPs over 90 min. The separated glycopeptides were analyzed by hybrid ion trap-Orbitrap mass spectrometry (Orbitrap Exploris™ 480, Thermo Fisher Scientific, Sunnyvale, CA, USA). The parameters of mass spectrometry were set as followd: spray voltage, 2.2 kV; capillary temperature, 320 °C; full MS resolution, 120![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at m/z 200; mass range, 300–2000 (m/z); intensity threshold, 2E4; isolation width, 2.2 m/z; stepped collision energies, 20%, 30%, and 40%. The AGC target value for fragment spectra was set at 75% with a resolution of 15
000 at m/z 200; mass range, 300–2000 (m/z); intensity threshold, 2E4; isolation width, 2.2 m/z; stepped collision energies, 20%, 30%, and 40%. The AGC target value for fragment spectra was set at 75% with a resolution of 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 and an injection time of 50 ms, and the cycle time was set to 2.5 s.
000 and an injection time of 50 ms, and the cycle time was set to 2.5 s.
Meanwhile, the eluted N- or O-glycopeptides from the chromatographic column were also fed into the ESI-MS system for detection using quadrupole time-of-flight ion-mobility mass spectrometry (IM-MS) (Synapt G2-Si, Waters, Milford, Massachusetts, USA). The parameters of mass spectrometry were set as follows: spray voltage, 2.2 kV; cone voltages, 30 V; source temperature, 100 °C; desolvation temperature, 250 °C. To further obtain the terminal sialic acid linkage isomers, targeted glycopeptides were isolated in the quadrupole and fragmented in the trap region before ion mobility separations. The parameters of IM-MS were set as follows: collision energy, 30 V; IMS cell pressure, ∼2.66 mbar; N2 gas flow, 90 mL min−1; traveling wave height, 40 V; traveling wave velocity, 950 m s−1; for CCS calibration, the MajorMix (Waters) in 0.1% FA was infused at 5 μL min−1 with identical MS and IMS settings. Meanwhile, the signal intensity of precursor ions was limited at approximately 105 counts per second to improve the accuracy of relative quantification of the sialic acid linkage isomer.34 Three injection replications were carried out for all samples.
2.5. Data analysis
N- and O-glycopeptides of hACE2 were identified using Byonic software (Protein Metrics Inc., USA, V2.16.11) with a single database hACE2 protein (UniProt ID: Q9BYF1). The search parameters of the Orbitrap Exploris™ 480 were set as follows: enzyme, trypsin and Glu-C or proteinase K; missed cleavage, 1; precursor ion mass tolerance, 10 ppm; mass tolerance for the fragment ion, 20 ppm; static modification of Cys, carboxymethylation. Variable modifications were included: Met, oxidation; Asn and Gln, deamidation; possible common human, 182 N-glycans and 70 O-glycans. The identified N- or O-glycopeptides were filtered to 1% FDR and a manual score cut-off value of 150. To ensure the accuracy of identification, automated identifications were further examined manually including the pattern of glycan neutral loss, oxonium ions, and fragmentation of the peptide sequence. The relative proportions of glycans at the specific site were calculated by comparing the spectral counts assigned to the specific glycopeptides with all glycopeptides on identical glycosites.The proteinase K is a serine endopeptidase with wide cleavage activity toward the carboxy-terminated peptide bonds of aliphatic amino acids and aromatic amino acids. According to the ExPASy portal, the cleavage sites were set as follows: Ala(A), Ile(I), Leu(L), Phe(F), Tyr(Y), Trp(W) and Val(V). Semi-specific digestion was further added for interpreting O-glycosylation on the proline-rich C-terminus of hACE2. The identification of O-glycopeptides was examined manually and filtered using the reported standard criteria, including at least two glycan oxonium ions and at least 6 b/y ions in the tandem mass spectrum.35
The data from Synapt G2-Si were analyzed using the above parameters for the identification of N- and O-glycopeptides, except the mass tolerance for precursor and fragment ions was set as 20 ppm and 0.05 Da, respectively.
2.6. Site-specific mapping of sialic acid linkage isomers by IM-MS
MassLyn 4.1 and DriftScope v2.9 (Waters) were applied for the data analysis performed using IM-MS. Arrival time distributions (ATDs) were extracted from raw data using MassLynx. Collision cross-section (CCS) values were estimated as described previously.36 The area under the ATD of each isomer fragment was calculated using MassLyn 4.1 with the same spectral peak integral parameters and used for the mapping of α2,3- and α2,6-sialic acid linkage isomers on the specific glycopeptide.3. Results and discussion
3.1. N-Glycosylation of hACE2
Prior to analysis, the purity of the recombinant hACE2 was evaluated by SDS-PAGE (Bio-Rad Laboratories, Inc., CA, USA) (Fig. S1, ESI†). The single protein band shown in Fig. S1† proves the high purity of the recombinant hACE2. To allow for in-depth site-specific N-/O-glycosylation and sialic acid linkage isomer analysis of hACE2, a series of enzyme digestion, specific enrichment, separation, and advanced fragmentation strategies were utilized in this study (Scheme S1†). By employing the step-energy collision dissociation mode using the Orbitrap Exploris™ 480, 1166 oxonium ion-containing tandem mass spectra were assigned to 406 unique glycopeptides, which consist of 128 N-glycans (Table S1a, ESI†). Based on the analysis of N-glycopeptides with Synapt G2-Si in the ramp-energy collision dissociation mode, 1661 oxonium ion-containing tandem mass spectra were assigned to 434 unique glycopeptides, in which 107 N-glycans were identified (Table S1b†). The overlap of the N-glycans identified by using two different mass spectrometers was about 63% at 1% FDR (Fig. S2†). Lastly, a total of 138 N-glycans were determined from seven N-glycosites (N53, N90, N103, N322, N432, N546, and N690) on hACE2 (Table S2†). The spectral counts of N-glycopeptides and the detailed structural compositions of these glycans are available in Table S2.† The N-glycans on N90, N103, N546, and N690 (Fig. 1) were more heterogeneous than those on N53, N322, and N432. From the distribution and spectral counts of N-glycopeptides, we found that the N90, N103, N546, and N690 are also more heavily N-glycosylated than the other N-glycosites (Table S2 and Fig. S3†). Notably, the most dominant N-glycans on hACE2 were HexNAc4Hex5Fuc1NeuAc1 (H5N4F1S1) and HexNAc4Hex5Fuc1NeuAc2 (H5N4F1S2), which were located on N90 and N103, respectively. This phenomenon was also observed in the N-glycomics analysis of hACE2.15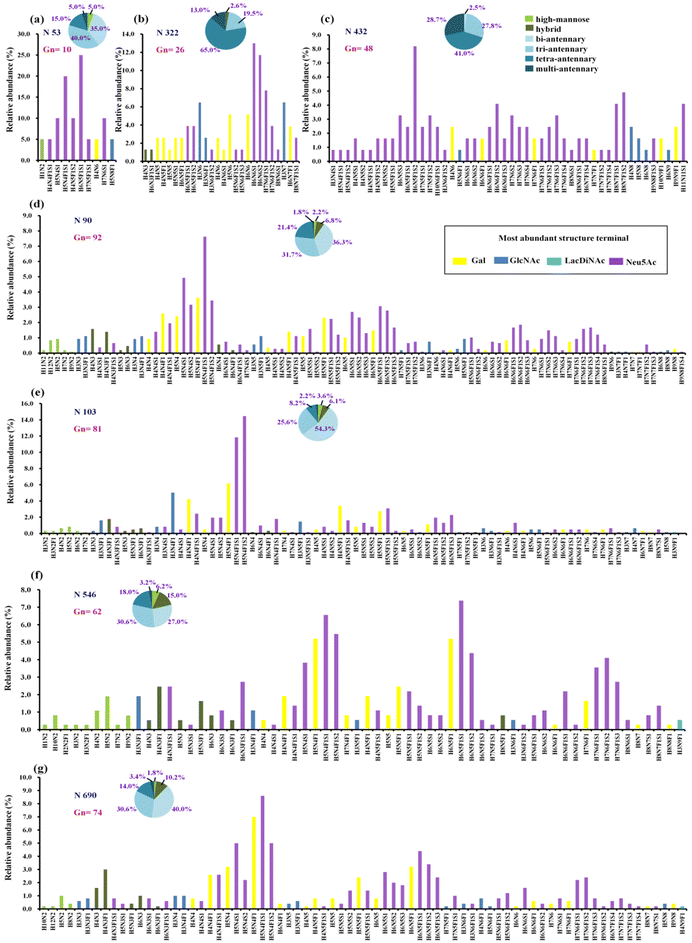 | ||
| Fig. 1 The quantitative profiles of glycan composition on hACE2. (a–g) Glycopeptides of hACE2 were analysed with Scheme S1(a).† All seven sites of N-glycosylation are presented here. Displayed in the bar graphs are the individual compositions observed in term of relative abundances. According to the most abundant terminal glycan structures, the graphs are categorized and colored with different colors shown in legend. The pie charts summarize the relative quantification of these glycans simplified into different categories on specific N-glycosites (more details in Table S3†). (a) N53, (b) N322, (c) N432, (d) N90, (e) N103, (f) N546 and (g) N690. Gn represents the number of glycan compsition assigned to the specific N-glycosites. N, HexNAc; H, Hex; F, fucose; S, sialic acid. | ||
To determine the relative proportion of site-specific N-glycans, the relative quantification was performed according to the spectral counts of each N-glycopeptide. Fig. 1 shows the quantitative glycosylation profiles of hACE2, which are summed into high-mannose, hybrid, and categories of complex-type glycosylation with different branches, as well as fucosylation and sialylation (Fig. 1 and Table S3†). In contrast to high-mannose and hybrid type glycans, complex-type glycans predominate across all N-glycosites. We discovered highly complex-type bi-antennary, tri-antennary, and tetra-antennary glycans were dominant on the N53, N90, N546, and N690 sites. Site N103 featured predominantly complex-type bi-antennary and tri-antennary glycans, whereas sites N322 and N432 carried abundant complex-type tri-antennary and tetra-antennary glycans (Table S3†). Meanwhile, high sialylation was observed across all N-glycosites of hACE2. The relative abundance of sialylated N-glycans on sites N53 and N432 was 85.0% and 82.5%, respectively. Over 60% of the glycans on sites N90, N546, and N690 were sialylated N-glycans (Table S3†), while the proportion of sialylated N-glycans on N103 and N322 was 57.3% and 57.1%, respectively. Except for site N322, other N-glycosites showed high levels of fucosylated glycans with the proportions ranging from 60.4% to 81.6%. Only 21.3–24.6% of the N-glycans on N53 and N432 contained terminal Gal, which was lower than that for other N-glycosites. Therefore, N-glycans assigned to sites N90, N103, N546, and N690 have more heterogeneity in terms of glycan structure than those on sites N53, N322, and N432.
As a common but complex post-translational modification, glycosylation on hACE2 has important implications for the SARS-CoV-2 S/hACE2 interaction.37 In previous studies, N53 glycan on ACE2 was expected to have a dual and possibly competing role in heterodimer (ACE2-RBD) and homodimer (intra-ACE2) interactions.38 Recently, N53 glycan has been proved to provide a positive contribution to hACE2-RBD binding and facilitate viral entry.18 It has also been shown that the glycans on N322 can strengthen S protein binding to hACE2.39,40 As a result of our findings, we hypothesize that fewer glycans on N53 may be advantageous in stabilizing the homodimer interface of hACE2 and interacting with the RBD heterodimer via glycan–glycan and glycan–protein binding. The low levels of glycosylation and heterogeneity on N322 may offer appropriate steric hindrance for the binding to SARS-CoV-2 S protein. In contrast, with their high heterogeneity and abundance, N-glycans on N90 may provide protection for host cells against viral infection, as also demonstrated by biophysical and mutation studies.7,41 The glycans on N90 and N103 in the above glycosylation elucidation of hACE2 are comparable in the great heterogeneity and distribution of each glycan type. It will benefit the synergic effect of glycans for the binding surface with the RBD of S protein.41 Meanwhile, the glycans on N546 may be involved in the interactions with the N74 and N165 glycans of S protein.15 The highly abundant glycosylation on N690 allows for glycan–protein interactions to maintain the hACE2 homodimer interface.38 With the medium heterogeneity of glycans, N432 is distant from hACE2-RBD interaction sites. In summary, the N-glycosylation of hACE2 plays important roles in the stabilization of two hACE2 dimers and interaction with SARS-CoV-2 S protein. All of these binding sites should be further investigated for exploring the detailed molecular mechanism of host–virus interactions.
3.2. Mapping sialic acid linkage isomers of N-glycopeptides by IM-MS
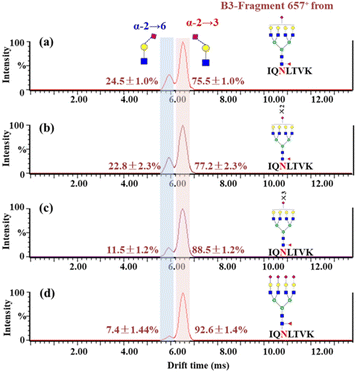
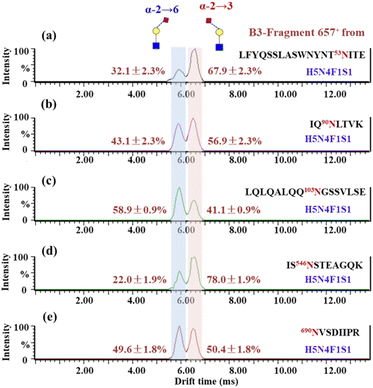
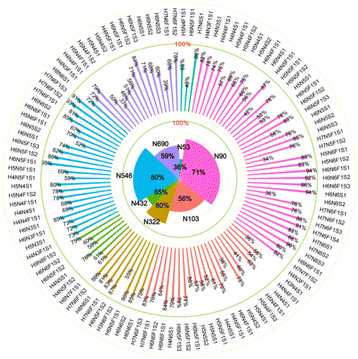
According to the list of identified N-glycopeptides, the sialic acid linkage isomers of N-glycopeptides were analyzed through our reported workflow (Scheme S1c†).34 The targeted glycopeptides were subsequently selected for tandem IM-MS analysis at a specific retention time, and the m/z 657 B3-type fragment, [NeuAcα2-6/α2,3Galβ1-4GlcNAc − H2O + H]+, was assigned to the sialylation of glycopeptides. For example, the amino acid sequence was IQ90N#LTVK, in which N was modified with the N-glycan HexNAc6Hex7Fuc1NueAc1 (H7N6F1S1), HexNAc6Hex7Fuc1NueAc2 (H7N6F1S2), HexNAc6Hex7Fuc1NueAc3 (H7N6F1S3) and HexNAc6Hex7Fuc1NueAc4 (H7N6F1S4), respectively. These glycopeptides was isolated and analyzed by IM-MS, respectively (Fig. S4†). In the IM-MS analysis, the ATD of the m/z 657 B3-type fragment showed remarkable separation of two trisaccharides in which the α2,6-fragment has a clear Gaussian peak at ∼5.95 ms and the α2,3-isomer shows a peak at ∼6.52 ms (Fig. 2). The travelling wave collision cross-section in nitrogen drift gas (TW CCS N2) is 235.8 Å2 for the α2,6-linked NeuAc fragment and 246.5 Å2 for the α2,3-linked NeuAc fragment. In the process of examining various glycoforms at the fixed N-glycosite, we can see a trend in which the higher degree of sialylation is correlated with an increase in the content of α2,3-sialylation, which increased from 75.5 ± 1.0% to 92.6 ± 1.4% (Fig. 2). Such correlations have also been formerly noticed during the IM-MS elucidation of sialic acid linkage isomers on α1-AGP and haptoglobin.36Presented on different glycosites of hACE2, we found that the expression of α2,3- and α2,6-sialylation on the same glycan composition was also different. Taking the most dominant bi-antennary glycoform H5N4F1S1 for example, the glycopeptide IQ90N#LTVK modified with H5N4F1S1 showed 56.9 ± 2.3% α2,3-sialylation (Fig. 3b), while the glycopeptide IS546N#STEAGQK modified with H5N4F1S1 showed 78.0 ± 1.9% α2,3-sialylation (Fig. 3d). Meanwhile, the α2,3-sialylation content values in response to other sites modified with H5N4F1S1, i.e. N53, N103, and N690 were, respectively, 67.9 ± 2.3%, 41.1 ± 0.9%, and 50.4 ± 1.8% (Fig. 3a, c and e). This implies that H5N4F1S1 modification on hACE2 bears, on average, 58.9 ± 1.8% α2,3-sialylation. These phenomena are also observed on the different glycosites modified with tri-antennary glycan H6N5F1S1, on which the content of α2,3-sialylation ranges from 42.6 ± 1.2% to 94.8 ± 2.0% (Fig. S5†). In our analysis, the sialylation patterns showed different proportions not only on the different glycan compositions but also on the same glycan composition assigned to different N-glycosites of hACE2. Therefore, it is necessary to map the sialic acid linkage isomers of N-glycopeptides for the detailed characterization of hACE2. A total of 115 identified N-glycopeptides of hACE2 with glycans containing at least one sialic acid residue were selected for IM-MS analysis (CV < 10%, n = 3) (Table S4†). With a proportion of 91.3%, 105 out of 115 sialylated glycopeptides represented higher α2,3-sialylation. To determine the content of α2,3- and α2,6-sialylation on specific N-glycosites, the actual abundance of α2,3- and α2,6-sialylation was corrected by considering the relative abundance of glycopeptides. The pattern and distribution of α2,3- or α2,6-sialylation on N-glycopeptides and specific N-glycosites are summarized in Fig. 4 and Table S4.† In Fig. 4, N322 and N546 show a higher degree of α2,3-sialylation, bearing 80%, than the other five N-glycosites bearing 36–71%. Except for N53, which is predominant with 67% α2,6-sialylation,the sialylation patterns on other N-glycosites are dominated by α2,3-sialylation at the levels of both N-glycans and N-glycopeptides. The mapping of sialic acid linkage isomers on hACE2 was first summarized at the level of glycopeptides, when we observed relatively higher α2,3-sialylation on hACE2 by IM-MS, which is the same as the result obtained by N-glycomics analysis on hACE2.16 It is worth mentioning that the proportion of sialic acid linkage isomers from the same glycan varied among hACE2 N-glycosites, which also shows the importance of site-specific N-glycans analysis.
Previous works reported that α2,6-sialylation on N-glycans was predominant in human serum compared to α2,3-sialylation, including haptoglobin.23,34,42 As previously reported, MERS-CoV S protein binds to α2,3-linked sialic acid preferentially over α2,6-linked sialic acid receptors, which are abundant in the major sites of replication in the upper and lower respiratory tracts of camels and humans, respectively.26 Meanwhile, the SARS-CoV-2 S protein presented higher binding affinity with α2,3-Sia-containing oligosaccharides than with α2,6-Sia ones.28 Therefore, we speculate that these oligosaccharides serve as entry points for SARS-CoV-2, where the special property of α2,3-sialylation on hACE2 endows it with positive binding ability to SARS-CoV-2 S protein. Glycans on N322 and N546 have proved to be involved in intermolecular glycan–glycan interaction with the S protein.15,40 In our detailed sialic acid isomer map of hACE2, the highest α2,3-sialylation was observed on N322 and N546 with 80% proportions. Thus, we speculate that the α2,3-sialylation on N322 and N546 may facilitate SARS-CoV-2 S protein binding. The detailed linkage types of terminal sialic acid on specific sites of hACE2 may be helpful for the elucidation of the binding mechanism between SARS-CoV-2 S protein and hACE2.
3.3. O-Glycosylation of hACE2
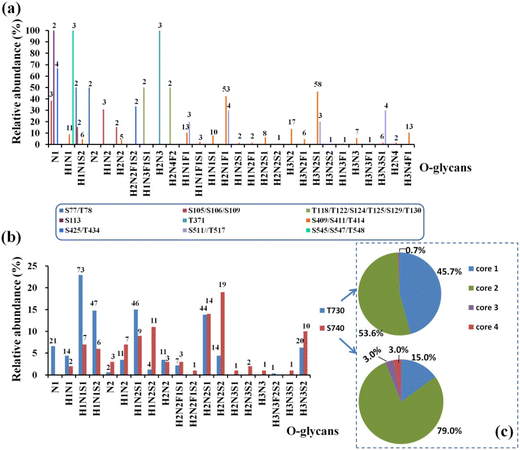
By using the step-energy collision dissociation mode in Orbitrap Exploris™ 480, 192 unique O-glycopeptides were identified from 958 oxonium ion-containing spectra, consisting of 42 O-glycan compositions (Table S5a†). Conversely, based on Synapt G2-Si in the ramp-energy collision dissociation mode, 38 O-glycan compositions of 160 unique O-glycopeptides from 928 oxonium ion-containing spectra were identified (Table S5b†). Based on these findings, a total of 12 O-glycosites were assigned to hACE2. Five O-glycosites, including S113, T371, T434, T730 and S740, were unambiguously identified. Meanwhile, the other seven O-glycosites, which included S77/T78, S105/S106/S109, T118/T122/S124/T125/S128/T129, S409/S411/T414, S425/T434, S511/T517 and S545/S547/T548, were ambiguously assigned to the corresponding glycopeptides with multi-O-glycosites. The representative mass spectra of O-glycopeptides are shown in the ESI (Fig. S6–Fig. S20†). After mapping these O-glycosites to the amino sequences, we found that the identified O-glycosites clustered in several positions, which are proximal to the known N-glycosites and the C-terminus of hACE2 (Fig. S21†). For example, S105/S106/S109 and S113 are adjacent to N103 and S425/T434 is adjacent to N432. Similarly, S545, S547 and T548 are located near N546. This phenomenon was also reported by Tian on SARS-CoV-2 S protein suggesting the possible “O-Follow-N” rule, where the O-glycosites are in close proximity to N-glycosylation.43In total, 69 O-glycan compositions, including 48 O-glycans attached to hACE2 were identified (Table S6†). Among the identified O-glycans, 22 and 25 O-glycans are fucosylated and sialylated, respectively. The spectral counts and distribution of O-glycans at specific sites are summarized in Fig. 5 and Table S6.† Except for S409/S411/T414, T730 and S740, other O-glycosites were modified with fewer than 4 O-glycans (Fig. S22†). For example, two O-glycans of H1N1S2 and N2 were identified on S77/T78, while H2N3 was only found on T371 (Fig. 5a). With 42 O-glycans with multi-types, the C-terminus of hACE2 showed the highest heterogeneity among all of the O-glycosites. Compared with the unmodified peptides, T730 and S740 showed highly abundant occupancy of O-glycan, at almost 97.5 ± 0.5%. In Fig. 5b, the O-glycans on T730 are mainly H1N1S1, H1N1S2, H1N2S1, and H2N2S1 with 66% occupancy. H2N2S1, H2N2S2, H1N2S1, H1N2S2, and H3N3S2 predominated on S740 accounting for 63% occupancy (Table S7†). The O-glycan profiles of T730 and S740 demonstrated the presence of core 1, core 2, core 3, and core 4 structures, capped with NeuAc, as shown in Fig. 5c. The presence of core 1 and core 2 structures accounts for 99.3% of O-glycans on T730. Meanwhile, the O-glycan composition assigned to S740 was mainly in core 2 structures accounting for 79% of O-glycans on S740.
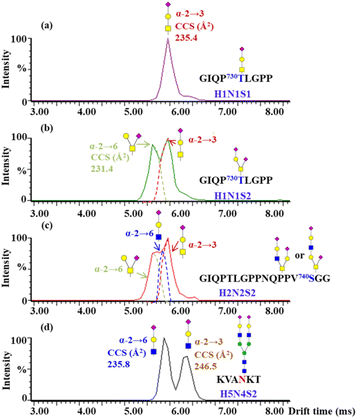
To further assign the α2,3- and α2,6-sialylation of O-glycans at T730 and S740, the O-glycopeptides with high abundances of glycoforms were examined by IM-MS. The glycopeptide GIQP730T#LGPP bearing a single O-glycosite, modified with H1N1S1 and H1N1S2, and the glycopeptide GIQPTLGPPNQPPV740S#GGGG modified with H2N2S2, were analyzed by IM-MS (Fig. S23†). Following IM-MS analysis, the m/z 657 B3-type fragment from the above O-glycopeptides was assigned the sialylation of the corresponding O-glycans, as shown in Fig. 6. The extracted ATD of m/z 657 B3 ions varies from distinct O-glycans. In Fig. 6a, the CCS of 235.4 Å2 of the only dominant peak from H1N1S1 at T730 is in agreement with the previously reported CCS of the α2,3 T-antigen sialylation.44 With the monitored ATDs of m/z 657 B3 ions from H1N1S2 at T730, two peaks were observed at 6.0 and 5.6 ms (Fig. 6b). The CCS of the first peak fits exactly the observed CCS of the α2,3 T-antigen sialylation, while the second peak with the CCS of 231.4 Å2 maps the CCS of α2,6-sialylation on NeuAc(α2–6)GalGalNAc. The ratio of the α2,3 T-antigen sialylation to the NeuAc(α2–6)GalGalNAc linkage is 1.2. There are three species with peaks at 6.0, 5.8 and 5.6 ms on S740 modified with H2N2S2 (Fig. 6c). The CCS of the first peak fits exactly that of α2,6-sialylation on NeuAc(α2–6)GalGalNAc, while the CCS of the third peak fits with that of α2,3 T-antigen sialylation. To characterize the second peak, the standard glycopeptide (KVAN#KT) modified with H5N4S2, where the terminal NeuAc was connected to Gal through α2–3 and α2–6 bonds, was analyzed using IM-MS (Fig. S24†). We determined that the CCS of the m/z 657 B3 ion fragment obtained by fragmentation of the standard glycopeptide with NeuAc(α2–6)GalGlcNAc (CCS of 235.8 Å2) is in agreement with the second peak at 5.85 ms, which is a shoulder peak of the α2,3 T-antigen sialylation shown in Fig. 6c. Through five-pass cyclic IM-MS analysis, the fragment of NeuAc(α2–6)GalGlcNAc and α2,3 T-antigen sialylation could attain obvious separation.44 Lastly, T730 not only has high occupancy of O-glycans but is also highly capped with α2,3-sialic acid of H1N1S1 and H1N1S2. In contrast to N-glycans, the O-glycan structures are more diverse, which greatly restricts the interpretation of glycan isomers. Thus, establishing the drift times of structures relevant to O-glycan in IM-MS will be essential for resolving different structures of O-glycans.
O-Glycans are associated with many critical biological functions, such as virus entry by binding onto its receptor, formation, and recognition by the cell immune system.45–48 Consistent with the O-glycomics, Zhao et al. reported that S155 and T730 at the C-terminus of hACE2 were O-glycosylated.15 Azadi et al. identified only one O-glycosite (T730) on hACE2 without pretreatment of O-glycopeptides.16 In our study, 12 O-glycosites were found in addition to the C-terminus of T730 and S740 using multi-enzymatic digestion and the HILIC enrichment method. However, the O-glycans present on hACE2 were of very low abundance at these identified O-glycosites, except for T730, S740, and S409/S411/T414. T730 and S740 are located in the collectrin-like domain close to the membrane. Adjacent to T730, L731 has been reported to be at the interface of ACE2 and B0AT1 (also called SLC6A19), which is a chaperone protein of ACE2. As an amino acid transporter, B0AT1 may block access of TMPRSS2s to the cutting site on ACE2. Further research indicates that the ACE2/B0AT1 dimer structure in the conformational binding of the SARS-CoV-2 S protein may affect the viral invasion of host cells.49 Thus, the heavily hydrophilic glycosylation at T730 and S740 may further affect the homodimer interaction and the ACE2/B0AT1 dimer structure on the cell surface. The simulation of RBD-bound and apo-ACE2 provides evidence that the flexibility in the ACE2 homodimer occurred not only at the linker connecting the transmembrane and head domains but also in the motions of the transmembrane helix in the membrane.38 Whether the heavy O-glycosylation at the C-terminus of hACE2 is attributed to the flexibility in the conformational variability of hACE2 remains unclear. Moreover, adjacent to the transmembrane (TM) domain of hACE2, S740 and its modified O-glycans may interact with the lipid rafts on the cell. The cellular and molecular mechanisms of the interaction still need to be clarified. Here, we illustrated the possible sialic acid linkage isomer at the heavy O-glycans located on the C-terminus of hACE2, including H1N1S1, H1N1S2, and H2N2S2. α2,3-Linked sialic acid is primarily capped on the terminus of H1N1S1and H1N1S2, especially the core 1 structure, H1N1S1, on which only α2,3-sialylation presented. Whether α2,3-sialylation, presented on the O-glycans of hACE2, is involved in the infection and transmission of SARS-CoV-2 needs more in-depth and in vivo studies, which could aid the development of inhibitors to block the entry of SARS-CoV-2 into host cells.
4. Conclusions
In this study, we comprehensively determined site-specific N- and O-glycosylation patterns of hACE2 using complementary enzymatic digestion and specific enrichment methods combined with high-resolution MS. A total of 138 N-glycan compositions were determined for N-glycosites (N53, N90, N103, N322, N432, N546, and N690) and 91% were complex-type N-glycans. Meanwhile, N322 and N546 showed a higher degree of α2,3-sialic acid linkage isomers of over 80.3% compared with the other N-glycosites (from 35.6 to 71.0%) using IM-MS analysis. For the first time, we identified 69 O-linked glycan compositions assigned to 12 O-glycosites, including T730 and S740 of hACE2. Furthermore, with high occupancy of O-glycans on T730, the terminal sialic acid linkage types of N1H1S1 and H1N1S2 were highly capped with α2,3-sialylation. Our study substantially expands the glycoforms on hACE2 and provides a basis for further exploration of the function of glycosylation on hACE2, as well as its interactions with SARS-CoV-2 S protein.Author contributions
All authors have accepted responsibility for the entire content of this manuscript and approved its submission. L. W., Y. C., H. Q., C. W. and H. L. designed the research; L. W., Y. C., X. F., J. Y., G. Y. and X. Z. performed the research; L. W., L. Z., H. Q., C. W. and H. L. analyzed data; and L. W., Y. C., X. F., J. Y., L. Z., G. Y., X. Z., H. Q., C. W., and H. L. wrote the paper.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The work was supported by the Shanghai Science and Technology Program (22DZ2291700 and 22142202400), the National Key Research and Development Program of China (2016YFA0501303 and 2020YFE0202200), the NSF of China (Grant 21974025), the Shanghai Pujiang Program (18PJD002), the project supported by the Shanghai Municipal Science and Technology Major Project (Grant No. 2017SHZDZX01), and NHC Key Laboratory of Glycoconjugates Research (Fudan University). We would like to thank Ziqing Lin and Zachery R. Gregorich for critical reading of this manuscript. We thank Synapt G2-Si (Waters Co. UK) for providing the IM-MS analysis.References
- L. Wang and Y. Xiang, Viruses, 2020, 12, 1289 CrossRef CAS PubMed.
- S. Kim, Y. Liu, Z. Lei, J. Dicker, Y. Cao, X. F. Zhang and W. Im, J. Chem. Theory Comput., 2021, 17, 7972–7979 CrossRef CAS PubMed.
- S. Pascarella, M. Ciccozzi, D. Zella, M. Bianchi, F. Benedetti, D. Benvenuto, F. Broccolo, R. Cauda, A. Caruso, S. Angeletti, M. Giovanetti and A. Cassone, J. Med. Virol., 2021, 93, 6551–6556 CrossRef CAS PubMed.
- A. C. Walls, Y. J. Park, M. A. Tortorici, A. Wall, A. T. McGuire and D. Veesler, Cell, 2020, 181, 281–292 CrossRef CAS PubMed.
- G. Lu, Q. Wang and G. F. Gao, Trends Microbiol., 2015, 23, 468–478 CrossRef CAS PubMed.
- D. Wrapp, N. Wang, K. Corbett, J. A. Goldsmith, C. Hsieh, O. Abiona, B. S. Graham and J. S. McLellan, Science, 2020, 367, 1260–1263 CrossRef CAS PubMed.
- W. Cao, C. Dong, S. Kim, D. Hou, W. Tai, L. Du, W. Im and X. F. Zhang, Biophys. J., 2021, 120, 1011–1019 CrossRef CAS PubMed.
- M. Y. Li, L. Li, Y. Zhang and X. S. Wang, Int. J. Med. Parasit. Dis., 2020, 9, 45 Search PubMed.
- H. Xu, L. Zhong, J. Deng, J. Peng, H. Dan, X. Zeng, T. Li and Q. Chen, Int. J. Oral Sci., 2020, 12, 8 CrossRef CAS PubMed.
- E. Socher, M. Conrad, L. Heger, F. Paulsen, H. Sticht, F. Zunke and P. Arnold, J. Cell. Biochem., 2021, 122, 1863–1872 CrossRef CAS PubMed.
- D. Santra, A. Banerjee, S. K. De, H. Thatoi and S. Maiti, Comp. Clin. Pathol., 2023, 32, 179–189 Search PubMed.
- J. Huang, D. Wang, R. D. Shipman, Z. Zhu, Y. Liu and L. Li, Anal. Bioanal. Chem., 2021, 413, 7295–7303 CrossRef CAS PubMed.
- D. Chang, J. A. Klein, M. R. Nalehua, W. E. Hackett and J. Zaia, Anal. Bioanal. Chem., 2021, 413, 7305–7318 CrossRef CAS PubMed.
- C. Huang, Z. Tan, K. Zhao, W. Zou, H. Wang, H. Gao, S. Sun, D. Bu, W. Chai and Y. Li, iScience, 2021, 24, 103272 CrossRef CAS PubMed.
- P. Zhao, J. L. Praissman, O. C. Grant, Y. Cai, T. Xiao, K. E. Rosenbalm, K. Aoki, B. P. Kellman, R. Bridger, D. H. Barouch, M. A. Brindley, N. E. Lewis, M. Tiemeyer, B. Chen, R. J. Woods and L. Wells, Cell Host Microbe, 2020, 28, 586–601 CrossRef CAS PubMed.
- A. Shajahan, S. Archer-Hartmann, N. T. Supekar, A. S. Gleinich, C. Heiss and P. Azadi, Glycobiology, 2020, 1–15 Search PubMed.
- J. D. Allen, Y. Watanabe, H. Chawla, M. L. Newby and M. Crispin, J. Mol. Biol., 2021, 433, 166762 CrossRef CAS PubMed.
- A. Isobe, Y. Arai, D. Kuroda, N. Okumura, T. Ono, S. Ushiba, S. I. Nakakita, T. Daidoji, Y. Suzuki, T. Nakaya, K. Matsumoto and Y. Watanabe, Commun. Biol., 2022, 5, 1188 CrossRef CAS PubMed.
- K. E. Shinya, M. Ebina, S. Yamada, M. Ono, N. Kasai and Y. Kawaoka, Nature, 2006, 440, 435–436 CrossRef CAS PubMed.
- E. Qing, M. Hantak, S. Perlman and T. Gallagher, mBio, 2020, 11, e02764 CrossRef CAS PubMed.
- M. A. Tortorici, A. C. Walls, Y. Lang, C. Wang, Z. Li, D. Koerhuis, G. J. Boons, B. J. Bosch, F. A. Rey, R. J. de Groot and D. Veesler, Nat. Struct. Mol. Biol., 2019, 26, 481–489 CrossRef PubMed.
- M. Matrosovich, G. Herrler and H. D. Klenk, Top. Curr. Chem., 2013, 367, 1–28 CrossRef PubMed.
- M. Cheng, H. Shu, M. Yang, G. Yan, L. Zhang, L. Wang, W. Wang and H. Lu, Anal. Chem., 2022, 94, 4666–4676 CrossRef CAS PubMed.
- M. Awasthi, S. Gulati, D. P. Sarkar, S. Tiwari, S. Kateriya, P. Ranjan and S. K. Verma, Viruses, 2020, 12, 909 CrossRef CAS PubMed.
- A. N. Baker, S. J. Richards, C. S. Guy, T. R. Congdon, M. Hasan, A. J. Zwetsloot, A. Gallo, J. R. Lewandowski, P. J. Stansfeld, A. Straube, M. Walker, S. Chessa, G. Pergolizzi, S. Dedola, R. A. Field and M. I. Gibson, ACS Cent. Sci., 2020, 6, 2046–2052 CrossRef CAS PubMed.
- W. Li, R. J. G. Hulswit, I. Widjaja, V. S. Raj, R. McBride, W. Peng, W. Widagdo, M. A. Tortorici, B. Dieren, Y. Lang, J. W. M. Lent, J. C. Paulson, C. A. M. Haan, R. J. Groot, F. J. M. Kuppeveld, B. L. Haagmans and B. Bosch, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, e8508–e8517 CAS.
- Y. J. Park, A. C. Walls, Z. Wang, M. M. Sauer, W. Li, M. A. Tortorici, B. J. Bosch, F. DiMaio and D. Veesler, Nat. Struct. Mol. Biol., 2019, 26, 1151–1157 CrossRef CAS PubMed.
- L. Unione, M. J. Moure, M. P. Lenza, I. Oyenarte, J. Ereno-Orbea, A. Arda and J. Jimenez-Barbero, Angew. Chem., Int. Ed., 2022, 61, e202201432 CrossRef CAS PubMed.
- H. Chu, B. Hu, X. Huang, Y. Chai, D. Zhou, Y. Wang, H. Shuai, D. Yang, Y. Hou, X. Zhang, T. T. Yuen, J. P. Cai, A. J. Zhang, J. Zhou, S. Yuan, K. K. To, I. H. Chan, K. Y. Sit, D. C. Foo, I. Y. Wong, A. T. Ng, T. T. Cheung, S. Y. Law, W. K. Au, M. A. Brindley, Z. Chen, K. H. Kok, J. F. Chan and K. Y. Yuen, Nat. Commun., 2021, 12, 134 CrossRef CAS PubMed.
- Q. Yang, T. A. Hughes, A. Kelkar, X. Yu, K. Cheng, S. Park, W. C. Huang, J. F. Lovell and S. Neelamegham, eLife, 2020, 9, e61552 CrossRef CAS PubMed.
- L. Nguyen, K. A. McCord, D. T. Bui, K. M. Bouwman, E. N. Kitova, M. Elaish, D. Kumawat, G. C. Daskhan, I. Tomris, L. Han, P. Chopra, T. J. Yang, S. D. Willows, A. L. Mason, L. K. Mahal, T. L. Lowary, L. J. West, S. D. Hsu, T. Hobman, S. M. Tompkins, G. J. Boons, R. P. de Vries, M. S. Macauley and J. S. Klassen, Nat. Chem. Biol., 2022, 18, 81–90 CrossRef CAS PubMed.
- H. Li, Y. Wang, L. Zhang, H. Lu, Z. Zhou, L. Wei and P. Yang, Analyst, 2015, 140, 7886–7895 RSC.
- P. Fang, J. Xie, S. Sang, L. Zhang, M. Liu, L. Yang, Y. Xu, G. Yan, J. Yao, X. Gao, W. Qian, Z. Wang, Y. Zhang, P. Yang and H. Shen, Anal. Chem., 2020, 92, 867–874 CrossRef CAS PubMed.
- X. Feng, H. Shu, S. Zhang, Y. Peng, L. Zhang, X. Cao, L. Wei and H. Lu, Anal. Chem., 2021, 93, 15617–15625 CrossRef CAS PubMed.
- Y. Zhang, W. Zhao, Y. Mao, Y. Chen, J. Zhu, L. Hu, M. Gong, J. Cheng and H. Yang, Front. Chem., 2021, 9, 689521 CrossRef CAS PubMed.
- M. Guttman and K. K. Lee, Anal. Chem., 2016, 88, 5212–5217 CrossRef CAS PubMed.
- J. Huang, S. Hou, J. An and C. Zhou, Anal. Bioanal. Chem., 2023, 415, 1455–1464 CrossRef CAS PubMed.
- E. P. Barros, L. Casalino, Z. Gaieb, A. C. Dommer, Y. Wang, L. Fallon, L. Raguette, K. Belfon, C. Simmerling and R. E. Amaro, Biophys. J., 2021, 120, 1072–1084 CrossRef CAS PubMed.
- W. Li, C. Zhang, J. Sui, J. H. Kuhn, M. Moore, S. Luo, S. Wong, I. Huang, K. Xu, Y. He, W. A. Marasco, Y. Guan, H. Choe and M. Farzan, EMBO J., 2005, 24, 1634–1643 CrossRef CAS PubMed.
- A. R. Mehdipour and G. Hummer, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2100425118 CrossRef CAS PubMed.
- B. Wang, J. Zhao, S. Liu, J. Feng, Y. Luo, X. He, Y. Wang, F. Ge, J. Wang, B. Ye, W. Huang, X. Bo, Y. Wang and J. J. Xi, Emerging Microbes Infect., 2022, 11, 1488–1499 CrossRef CAS PubMed.
- T. Nishikaze, H. Tsumoto, S. Sekiya, S. Iwamoto, Y. Miura and K. Tanaka, Anal. Chem., 2017, 89, 2353–2360 CrossRef CAS PubMed.
- W. Tian, D. Li, N. Zhang, G. Bai, K. Yuan, H. Xiao, F. Gao, Y. Chen, C. C. L. Wong and G. F. Gao, Cell Res., 2021, 31, 1123–1125 CrossRef CAS PubMed.
- M. Sanda, L. Morrison and R. Goldman, Anal. Chem., 2021, 93, 2003–2009 CrossRef CAS PubMed.
- I. Bagdonaite, R. Norden, H. J. Joshi, S. L. King, S. Y. Vakhrushev, S. Olofsson and H. H. Wandall, J. Biol. Chem., 2016, 291, 12014–12028 CrossRef CAS PubMed.
- E. J. Simon and A. D. Linstedt, J. Biol. Chem., 2018, 293, 19866–19873 CrossRef CAS PubMed.
- D. S. Roberts, M. Mann, B. H. Li, D. Kim, A. R. Braiser, S. Jin and Y. Ge, Chem. Sci., 2022, 13, 10944–10949 RSC.
- D. S. Roberts, M. Mann, J. A. Melby, E. J. Larson, Y. Zhu, A. R. Brasier, S. Jin and Y. Ge, J. Am. Chem. Soc., 2021, 143, 12014–12024 CrossRef CAS PubMed.
- R. Yan, Y. Zhang, Y. Li, L. Xia, Y. Guo and Q. Zhou, Science, 2020, 367, 1444–1448 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Supplementary Table. See DOI: https://doi.org/10.1039/d3an01079a |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2023 |