Open Access Article
Open Access ArticleA multiplatform metabolomics approach for comprehensive analysis of GIST xenografts with various KIT mutations†
Szymon
Macioszek
 a,
Danuta
Dudzik
a,
Margot
Biesemans
a,
Danuta
Dudzik
a,
Margot
Biesemans
 a,
Agnieszka
Wozniak
b,
Patrick
Schöffski
b and
Michal J.
Markuszewski
a,
Agnieszka
Wozniak
b,
Patrick
Schöffski
b and
Michal J.
Markuszewski
 *a
*a
aDepartment of Biopharmaceutics and Pharmacodynamics, Medical University of Gdańsk, Hallera 107, 80-416 Gdańsk, Poland. E-mail: michal.markuszewski@gumed.edu.pl; Tel: +48-58-349-1493
bLaboratory of Experimental Oncology, Department of Oncology, KU Leuven, and Department of General Medical Oncology, University Hospitals Leuven, Leuven Cancer Institute, Leuven, Belgium
First published on 14th July 2023
Abstract
Metabolites in biological matrices belong to diverse chemical groups, ranging from non-polar long-chain fatty acids to small polar molecules. The goal of untargeted metabolomic analysis is to measure the highest number of metabolites in the sample. Nevertheless, from an analytical point of view, no single technique can measure such a broad spectrum of analytes. Therefore, we selected a method based on GC-MS and LC-MS with two types of stationary phases for the untargeted profiling of gastrointestinal stromal tumours. The procedure was applied to GIST xenograft samples (n = 71) representing four different mutation models, half of which were treated with imatinib. We aimed to verify the method coverage and advantages of applying each technique. RP-LC-MS measured most metabolites due to a significant fraction of lipid components of the tumour tissue. What is unique and worth noting is that all applied techniques were able to distinguish between different mutation models. However, for detecting imatinib-induced alterations in the GIST metabolome, RP-LC-MS and GC-MS proved to be more relevant than HILIC-LC-MS, resulting in a higher number of significantly changed metabolites in four treated models. Undoubtedly, the inclusion of all mentioned techniques makes the method more comprehensive. Nonetheless, for green chemistry and time and labour saving, we assume that RP-LC-MS and GC-MS analyses are sufficient to cover the global GIST metabolome.
Introduction
A gastrointestinal stromal tumour (GIST) is a gastrointestinal tract sarcoma originating from the intestinal pacemaker Cajal cell precursors.1,2 GIST's best-defining common feature is the membranous expression of the receptor tyrosine kinase KIT; KIT and DOG1 (discovered on GIST 1) are the preferred diagnostic markers for this sarcoma subtype.3 Despite the common characteristic, the tumour's genetic background is diversified. Approximately 75–85% of GISTs have an activating mutation in KIT and 5–10% in the PDGFRA gene, while the remaining cases may exhibit a mutation in other genes.4 Furthermore, it was observed that a particular mutation type is related to the tumour location along the gastrointestinal tract or the response rate to an anticancer therapy with tyrosine kinase inhibitors (TKIs) such as imatinib.5–7Until now, no single comprehensive mass spectrometry-based study was performed to describe the metabolome of GIST. In our previous study, we selected the most suitable robust method for preparing GIST xenograft samples for analysis by liquid chromatography coupled with mass spectrometry (LC-MS).8 As the metabolite composition of GIST is not elucidated, the method prioritised the coverage of the broadest possible range of metabolites of various physicochemical properties with satisfactory reproducibility. Here, our goal is to verify the method applicability for the untargeted metabolomic analysis of GIST xenografts carrying four different KIT mutations defining their responsiveness to imatinib treatment. Metabolomics is a part of omics tools that provide a full global metabolic readout of the present function of a cell or another biological matrix. It has been successfully applied to identify drug-induced metabolic alterations,9,10 in treatment monitoring,11,12 personalised medicine,13–15 to explain the mechanism of action of a drug in use,16,17 or to propose new targets for novel therapies.18–20 Metabolomics has already been used to examine the effect of anticancer agents on tumour metabolomes.21–24 As regards research on imatinib, Ishida et al. analysed the metabolome of GIST cells from cell culture and compared it with the set of metabolites detected in generated drug-tolerant cells.25 In a study by Caocci et al., the plasma metabolome of leukaemia patients treated with imatinib and other TKIs was analysed to investigate the molecular causes of various degrees of cardiovascular adverse effects of the treatment.26 Also, employing metabolomics and proteomics, Contreras Mostazo et al. observed a strong metabolic rewiring of leukaemia cells acquiring resistance to imatinib.27
Maximising the metabolome coverage is essential to increase the probability of drawing biologically relevant conclusions from an untargeted metabolomic project.28,29 The most efficient and simple approach is to parallelly apply several analytical techniques suitable for measuring various groups of analytes. MS-based platforms are the gold standard for high throughput metabolomic profiling of biological samples mainly due to high sensitivity and their possible hyphenation to separation techniques with a wide range of available stationary phases.30,31 Although LC with a reversed phase is able to cover the majority of known metabolites, those with higher hydrophilicity and polarity are insufficiently retained.32 Therefore, hydrophilic interaction liquid chromatography (HILIC) columns have been additionally implemented in metabolomics to widen the metabolite coverage with carbohydrates, amino acids, or nucleosides.30,33–36 Another analytical technique that can cover a wider range of polar metabolites is gas chromatography coupled with mass spectrometry (GC-MS), which requires thermally stable and volatile analytes. Therefore, compounds with polar functional groups such as alcohols, carboxylic acids, carbohydrates, amides, and amines must be derivatized prior to analysis, most commonly through silylation and methoxyamination. Indeed, more analytical techniques such as capillary electrophoresis (CE)-MS or ion chromatography (IC)-MS covering polar metabolites are used in metabolomics.37,38 However, the most pronounced drawbacks of CE-MS include its deficient robustness as well as lower sensitivity due to sample dilution with sheath liquid.39,40 IC-MS is a suitable technique for analysing anionic metabolites while its inability to detect zwitterionic amino acids and uncharged molecules is of critical concern.41 Although IC-MS is considered a promising technique for targeted experiments, it is still not common for untargeted profiling.42 Moreover, for untargeted methods, the use of both anionic and cationic columns and two elute systems would be required. With the development of sheathless methods or in-capillary preconcentration in CE-MS,40 and further improvements in IC-MS, both techniques can match the popularity of GC-MS and HILIC-LC-MS in untargeted metabolomics. In this study, the previously developed untargeted metabolomics method was tested for application in finding the differences between KIT mutants and assessing the impact of imatinib treatment on GIST tissue. In addition, we verified the necessity to perform all analytical batches and examined which one yielded the most information relevant to the stated biological questions.
Materials and methods
Chemicals and reagents
MS-grade methanol, acetonitrile, and isopropanol were obtained from Thermo Fisher Scientific (Loughborough, UK). Ultrapure water was produced in-house using a Direct-Q® 3UV (Millipore, Vienna, Austria). Heptane was purchased from Witko (Lodz, Poland). Pyridine, methyl tert-butyl ether (MTBE), N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) with 1% chlorotrimethylsilane (TMCS), ammonium formate, methoxyamine hydrochloride, n-pentadecanoic acid, and the alkane standard mixture (C10–C40, all even) were from Sigma-Aldrich (Saint Louis, MO, USA). Ammonia solution in water (25%) was obtained from Merck KGaA (Darmstadt, Germany). MS additive formic acid (98%) was purchased from Chem-Lab NV (Zedelgem, Belgium). The tuning mixture for MS calibration was from Agilent Technologies (Santa Clara, CA, USA).Study design and sample collection
The current study was performed on leftover ex-mouse tumour materials from previously performed in vivo experiments including four patient-derived xenograft mouse models, established and maintained in the Laboratory of Experimental Oncology, KU Leuven, Belgium.43–45 Model establishment and sample collection were approved by the Ethics Committee, University Hospitals Leuven (S53483) and by the KU Leuven Ethics Committee for Animal Research (project number P175/2015). Each model was created by subcutaneously transplanting human GIST tissue from a consented donor patient into a nude mouse (nu/nu NMRI). When the tumour reached 1000 mm3, it was re-transplanted into the subsequent mouse passages. Such models remain genetically stable and represent the original patient-derived tumour. In each experiment, among mice from which the leftover material was used in this study,43–45 approximately half of them were treated orally with 50 mg kg−1 imatinib twice a day for three weeks, while the other half received water as control. A few characteristics of the GIST models are presented in Table 1 and the tumour volume evaluation over 21 days of treatment is presented the ESI (ESI Fig. S1†). Tumours were snap-frozen immediately after collection and were stored at −80 °C until the day of sample preparation.| Treatment group | ||||||
|---|---|---|---|---|---|---|
| Control | Imatinib | |||||
| Xenograft model | Mutation type | Imatinib responsiveness | n | Dose | n | Dose |
| GIST1 | KIT exon 11 p.V560D | Sensitive43 | 10 | Untreated | 10 | 50 mg kg−1 BID |
| GIST2 | KIT exon9 p.A502_Y503dup | Dose-dependent resistant43 | 10 | Untreated | 9 | 50 mg kg−1 BID |
| GIST4 | KIT exon11 p.K558_G565delinsR | Sensitive44 | 4 | Untreated | 9 | 50 mg kg−1 BID |
| GIST9 | KIT exon 11 p.P577del; W557LfsX5 | Resistant45 | 9 | Untreated | 8 | 50 mg kg−1 BID |
| KIT exon 17 p.D820G | ||||||
Sample processing and metabolite extraction
Frozen samples were cut into 50–70 mg fragments and placed in Eppendorf tubes containing 2 mm diameter zirconium oxide homogenisation beads. Then, for each mg of the tumour, 10 μL of a methanol–water mixture (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) was added. The homogenisation process was performed in a Bullet Blender tissue homogenizer (Next Advance, Averill Park, NY, USA) in two 5 min cycles with a speed set at 10 and samples were cooled on ice between the cycles.
1, v/v) was added. The homogenisation process was performed in a Bullet Blender tissue homogenizer (Next Advance, Averill Park, NY, USA) in two 5 min cycles with a speed set at 10 and samples were cooled on ice between the cycles.
To prepare samples for LC-MS analysis, the samples were treated according to the previously selected robust method.8 Therefore, the sample's final ratio of methanol, MTBE, and water was 1.3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1
1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1.2 (v/v/v). 300 μL of each homogenate was mixed with 90 μL of methanol containing an internal standard (1 μg mL−1 chenodeoxycholic acid). Samples were vortex-mixed for 5 min, and then 120 μL of MTBE was added, and samples were again vortex-mixed for 3 min. The next step involved the addition of another part of MTBE (190 μL) and water (140 μL) containing 1-(4-fluorobenzyl)-5-oxoproline as an internal standard (1 μg mL−1). The extraction resulted in the formation of a lipid layer at the top and a polar layer at the bottom of the tubes. Both fractions were collected in separate LC glass vials. Polar extracts required one additional step of sample evaporation and reconstitution in an acetonitrile
1.2 (v/v/v). 300 μL of each homogenate was mixed with 90 μL of methanol containing an internal standard (1 μg mL−1 chenodeoxycholic acid). Samples were vortex-mixed for 5 min, and then 120 μL of MTBE was added, and samples were again vortex-mixed for 3 min. The next step involved the addition of another part of MTBE (190 μL) and water (140 μL) containing 1-(4-fluorobenzyl)-5-oxoproline as an internal standard (1 μg mL−1). The extraction resulted in the formation of a lipid layer at the top and a polar layer at the bottom of the tubes. Both fractions were collected in separate LC glass vials. Polar extracts required one additional step of sample evaporation and reconstitution in an acetonitrile![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water mixture (9
water mixture (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v).
1, v/v).
For GC-MS analysis, 100 μL of the homogenates were vortex-mixed with 200 μL of cold methanol containing an internal standard (1 mg mL−1 pentadecanoic acid). Samples were centrifuged at 4000g and 4 °C for 15 min, and 200 μL of the resulting supernatant was evaporated in a MiVac DUO concentrator (GeneVac, UK). Next, metabolites present in the samples were methoximated and trimethylsilylated for their conversion to volatile derivatives. To the dried supernatant, 20 μL of N,O-bis(trimethylsilyl)trifluoroacetamide (BSTFA) with 1% trimethylchlorosilane (TMCS) was added, followed by sample incubation at 70 °C for 1 hour. Finally, 100 μL of heptane was added to each sample.
Quality controls (QCs) were prepared simultaneously with sample preparation. First, a pool of 15 μL of all homogenates was created. Next, the pool was processed according to the procedure for the analysed samples; for LC-MS, 300 μL of the pooled homogenate was processed and for GC-MS, it was 100 μL of the homogenate. Moreover, blank samples containing water instead of the homogenate were prepared.
Untargeted metabolomics by LC-MS
LC-MS analyses were performed using an Agilent 1200 HPLC coupled with a 6224 time-of-flight mass spectrometry (TOF/MS) system (Agilent Technologies, Germany). Two complementary analytical columns were used for the separated organic and polar phases. A reversed phase InfinityLab Poroshell 120 EC-C8 column (2.1 mm × 150 mm, 2.7 μm, Agilent Technologies, USA) was applied for lipid metabolites. Each run was 50 min long, the flow rate was 0.5 mL min−1, and the injection volume was 5 μL. For electrospray ionisation in the positive mode, solvent A was 10 mM ammonium formate in water, and solvent B was 10 mM ammonium formate in a mixture of methanol and isopropanol (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, v/v). For the negative mode, 0.1% formic acid in water was used as solvent A and 0.1% formic acid in a mixture of methanol and isopropanol (85
15, v/v). For the negative mode, 0.1% formic acid in water was used as solvent A and 0.1% formic acid in a mixture of methanol and isopropanol (85![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 15, v/v) was used as solvent B. The chromatographic gradient started at 82% mobile phase B and reached 100% phase B.
15, v/v) was used as solvent B. The chromatographic gradient started at 82% mobile phase B and reached 100% phase B.
Hydrophilic compounds were separated in a 2.1 mm × 100 mm, 2.7 μm Poroshell 120 HILIC (Agilent Technologies, USA) column. In both the ionisation modes, the aqueous mobile phase was 10 mM ammonium formate in water, and the organic phase was acetonitrile. The gradient started from 5% aqueous phase and ended at the maximum of 45% aqueous phase. The total run time with the re-equilibration step was 29 min. The flow rate was 0.4 mL min−1, and the injection volume was 2 μL. MS parameters are listed in the ESI.†
The analytical batches in both ionization modes were started from the injection of two blank samples to detect possible impurities. Then, 10 QCs were injected for system equilibration. Finally, GIST samples were analysed in blocks of eight, with a QC being injected after each block. For analyte fragmentation, QC samples were analysed in the MS/MS mode on an LC-Q-TOF/MS system 6550A (Agilent Technologies, Germany), using exactly the same protocols as those for the previous LC-TOF/MS analysis.
Untargeted metabolomics by GC-MS
A 7890B Agilent Technologies GC system with a 7000 triple quadrupole (Agilent Technologies, United States) was used for GC-MS analysis. Separation of volatile metabolites was carried out in a ZB-5MS column (30 m length, 0.25 mm i.d., 0.25 μm film 95% dimethyl/5% diphenylpolysiloxane) from Phenomenex (United States). The injection volume was 1 μL, and the splitless mode was used. The helium carrier gas flow rate was set at 1 mL min−1. The temperature gradient started at 60 °C in 1 min and was increased to 320 °C at an 8 °C min−1 rate. The duration of each run was 37.5 min. Additional parameters are presented in the ESI.†First, two heptane blank samples were analysed, followed by an extraction blank (the whole sample preparation procedure was the same but without the tissue homogenate), and an n-alkane standard solution (C10–C40). Then, the system was equilibrated with 10 QCs. Finally, GIST samples were injected in a randomised order, with a QC sample being injected at constant intervals.
Data processing and statistical analysis
Raw data obtained during LC-MS analysis were uploaded to MassHunter Profinder B.08.00 (Agilent Technologies) software for peak deconvolution, alignment, and peak integration improvement. Metabolic features detected in blank samples were filtered out from the datasets. Missing values were replaced using the k-Nearest Neighbors algorithm. Finally, signals with a coefficient of variation in QCs higher than 20% were filtered out. The reproducibility of the analytical runs was also verified with principal component analysis (PCA) score plots generated in SIMCA 16 (version 16.0.2.10561, Umetrics AB, Sweden). HCA (hierarchical cluster analysis) was also performed using the same software.For GC-MS data, Agilent Unknown Analysis software (Ver. B.09.00, Agilent Technologies) with the National Institute of Standards and Technology (NIST) mass spectra library (Ver. 2014) was used for spectral deconvolution. Unlike in LC-MS data, the whole set of detected metabolites was identified before statistical analysis, by comparing fragmentation patterns and retention indices with the library. The retention times of the analysed n-alkanes were used for retention index calculation. Due to possible retention time shifts during the batch, metabolic signals generated from the same analytes were aligned in Mass Profiler Professional ver. B.02.1 (Agilent Technologies) software. The next data processing steps, including verification of qualifier and quantifier ions and correction of integration errors, were performed in Agilent MassHunter Quantitative Analysis (Ver. B.09.00, Agilent Technologies). The analytical reproducibility was assessed based on the signal from the internal standard and signal variation in QC samples. Only metabolites with a coefficient of variation of their signals in QCs lower than 30% were retained in the final dataset.
Both LC-MS and GC-MS data were subjected to univariate and multivariate statistical analyses to compare imatinib-treated and non-treated samples. Univariate Student's t-test for normally distributed variables or the U Mann-Whitney test in the absence of normality were performed in MATLAB 2016b software (MathWorks, Natick, MA, USA). As regards multivariate analysis, variable importance into projection (VIP) and p(corr) parameters were calculated in SIMCA 16 (version 16.0.2.10561, Umetrics AB, Sweden). Variables that were significant in both univariate (p > 0.05) and multivariate (p(corr) > 0.5 or <−0.5, VIP > 1) analyses were selected as biologically relevant. At this stage, LC-MS signals were finally annotated by searching against publicly available databases (CEU Mass Mediator, LIPIDMAPS, Human Metabolome Database (HMDB)), considering the measured mass, formed adducts, and MS/MS fragmentation patterns. According to the identification confidence levels proposed by Schymanski et al., the level 2 was obtained for detected metabolites.46 To compare metabolite abundance levels in control samples from four GIST models, heatmaps were generated in the Metaboanalyst 5.0 platform (https://www.metaboanalyst.ca). The Euclidean distance and Ward method were selected as the distance and clustering method. Both ANOVA and multivariate (VIP) analyses were performed to detect specific differences between KIT models.
Results and discussion
Principal component analysis
Raw data were prepared as described in the Materials and methods section and used to build PCA models (Fig. S2 in the ESI†). It is clearly visible that QC samples are clustered tightly together in the plots, which means that they are highly similar to each other thanks to stable conditions during analytical batches. Therefore, it is assumed that differences between the compared groups of samples are biologically relevant and not due to experimental variability. In each of the five PCA score plots, the treated samples belonging to the GIST4 model are separated from the remaining samples. In this xenograft model, the tumour reaction to the treatment seems to be the most marked also because of the highest tumour size reduction. As they are outliers in PCA analysis, the whole GIST4 model should be removed from the analysis and considered separately. Identical observations can be made based on the HCA (Fig. S3 in the ESI†).Results of untargeted analysis by analytical techniques
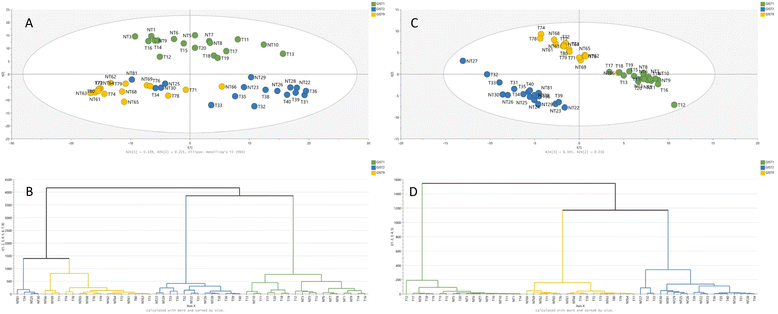
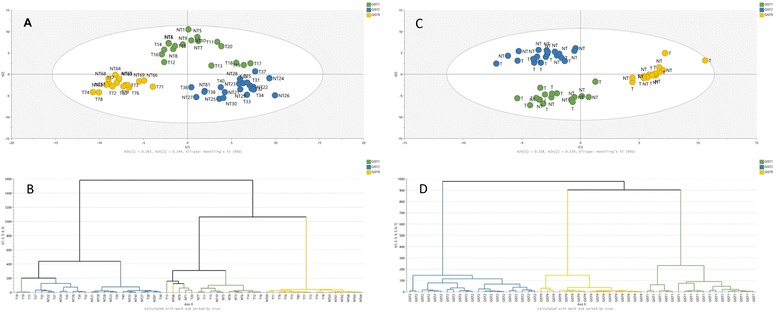
RP(+)-derived data can serve to distinguish between mutation models, which is confirmed in the PCA plot shown in Fig. 1. The samples spontaneously grouped according to their mutational status; however, non-treated samples from imatinib-resistant and dose-dependent models exhibit some similarities. On the other hand, the treatment effect is not emphasised in the part of the metabolome determined by the RP-LC-MS(+) analysis. Metabolites detected during RP-LC-MS(−) also contribute mostly to the separation of samples based on their KIT mutation type (Fig. 1).
More detailed information on the metabolic patterns in each GIST model can be obtained from the generated heatmaps. For GC-MS data, the heatmap is presented in Fig. 3, while the remaining heatmaps can be found in the ESI.† The most significant metabolites based on the univariate and multivariate comparison between the four models are also provided in the ESI.†
Impact of imatinib treatment on the GIST metabolome
Although the unsupervised statistical methods showed a substantial disparity in the metabolome composition between xenograft models, our question of interest was whether the applied untargeted metabolomic method could effectively detect alterations in the GIST metabolome due to imatinib treatment. We emphasise this clinically relevant problem as the results may help better understand the diversified response in GIST patients and improve therapy effectiveness. GIST resistance to imatinib, which is still the first-line therapy, remains one of the most severe challenges in the clinical management of GIST.3 To help find a solution, it is important to understand molecular mechanisms underlying the tumour response during the therapy. In this study, we compared the metabolome of imatinib-treated and non-treated GIST xenograft samples from two imatinib-sensitive models (GIST1, GIST4), one resistant model (GIST9), and one model characterised by dose-dependent resistance (GIST2). Unsurprisingly, the imatinib-resistant model produced the lowest number of statistically significant metabolites (Table 2). As the tumour did not respond to the introduced treatment, the biochemical processes inside the tumour were not affected considerably. The only altered metabolites included xanthosine, a few amino acid derivatives, acylcarnitines, and phosphatidylcholines.In contrast, the most pronounced differences were found in the drug-sensitive GIST4 model; however, this model has some limitations. The tumours reduced their size vastly after the treatment, presumably changing the tissue structure. To validate the findings, more data normalisation methods should be tested to correlate with the number of cancer cells inside the tumour tissue.
In general, the RP-LC-MS data gave the highest number of significant metabolites between treated and non-treated samples across the drug-responding models. Indeed, this is also because lipids are found in abundance in tissues, including the sarcomas.47,48 Lipids play essential roles in tumour cells as they are building blocks of cell membranes (phosphatidylcholine, phosphatidylethanolamine, phosphatidylserine, and phosphatidylinositol), constitute a source of energy (fatty acids), and take part in cell signalling. All of these functions are required for cell proliferation and tumour growth. In addition, metabolic reprogramming, which is the hallmark of cancers, is firmly represented in lipid metabolism alterations promoting tumorigenesis and cancer progression.49 Therefore, RP-LC-MS is the most essential analytical technique necessary for assessing changes in the GIST metabolome. However, regarding the HILIC-LC-MS analysis, this technique proved to be deficient in detecting alterations in the metabolome of the GIST1 model. In this respect, GC-MS is a more universal technique, suitable for all studied xenograft models. Moreover, as regards the techniques complementary to RP-LC-MS, it can be observed that a single GC-MS batch resulted in the generation of a larger amount of information than a single HILIC-LC-MS run in either positive or negative polarity, revealed from the number of statistically significant metabolites. The list of all significant compounds according to both univariate and multivariate statistical methods is presented in the ESI.†
Metabolite coverage overlap
Based on the statistical comparisons between imatinib-treated and non-treated GIST samples, the coverage overlap was examined between the applied analytical techniques. While lipids are usually detected in the RP-LC-MS analysis, single compounds can also be separated on a HILIC column. A few phosphatidylethanolamines, phosphatidylcholines, phosphatidic acids, and sphingomyelins were detected during the HILIC-LC-MS analysis. Purines and pyrimidines, which may be important for imatinib anticancer activity according to our previous research, can also be determined using GC-MS and HILIC-LC-MS. Some compounds such as adenine, adenosine, and xanthosine were observed in GC-MS, HILIC-LC-MS(+), and HILIC-LC-MS(−). Other metabolites like inosine and guanosine are detectable in GC-MS and HILIC-LC-MS(+), while hypoxanthine and uridine are detectable in GC-MS and HILIC-LC-MS(−). Other group of compounds that can be separated by both liquid and gas chromatography are fatty acids and organic acids. On the other hand, acylcarnitines, which were found to be affected by the imatinib treatment and are responsible for fatty acid transportation inside cells, were typically detected in the HILIC-LC-MS analysis.Overall, although RP-LC-MS assures the most comprehensive metabolite coverage of the GIST metabolome, it is necessary to include another technique to complement it for polar compounds. The choice between GC and HILIC mode LC should be made if one aims to avoid excessive use of solvents and save analysis time. As stated above, purines, pyrimidines, amino acids, organic acids, or carbohydrates can be detected by both techniques. In contrast to the HILIC-LC-MS analysis, the GC-MS analysis enabled the indication of statistically significant metabolites that are affected by the imatinib treatment in all studied mutation models (Table 1). However, the essential issue related to limiting only to GC-MS is the loss of acylcarnitines in the metabolite coverage.
Their general pros and cons should also drive the choice between both techniques. GC-MS offers higher reproducibility in retention times and fragmentation patterns in electron impact MS sources, making analyte identification more reliable with commercial libraries. However, the need for metabolite derivatization makes the sample preparation process more complicated.30 Moreover, this step requires the use of harmful reagents such as pyridine. On the other hand, HILIC is a more complex chromatographic system involving multiple retention mechanisms; the measurements are less reproducible than RP, and the peak shape is often unsatisfactory.50 It should be noted that among HILIC columns, various sorbents are used (zwitterionic, neutral diol, neutral amide, and aminoalkyl-based), and each of them is related with a slightly changed retention mechanism and exhibits different behaviours towards particular metabolites.51,52 In this study, only the bare silica HILIC column was considered; hence, another column type might ensure extra metabolite coverage.
Describing the GIST metabolome thoroughly and continuing the research on its response to TKIs can support the ongoing research on treatment effectiveness problems in GIST patients. Although the genetic causes of primary or secondary tumour resistance are described in the literature,53–55 metabolomic studies are necessary to find biochemical processes influenced by imatinib. By comparing a few mutational models, including imatinib-sensitive and resistant GISTs, specific metabolic pathways common for effective treatment may be recognised. It is possible that targeting the treatment via these essential pathways could constitute an approach to resensitise the tumour and improve treatment response.
Conclusions
To achieve the overriding aim of comprehensively elucidating the metabolome of GIST and relating it to KIT mutations and variable patient response to imatinib, the first step was to verify the untargeted metabolomic method for sample analysis. Tumour specimens were analysed using three techniques: RP-LC-MS and HILIC-LC-MS in two ionisation modes, and GC-MS. We found that each technique enables us to show differences in the GIST metabolome between KIT mutants. However, HILIC-LC-MS-derived data did not provide sufficient information enabling us to distinguish imatinib-treated samples from non-treated controls in all xenograft models. Therefore, GC-MS is a more appropriate technique to complement the GIST metabolome with non-lipid more polar compounds in this case. Overall, the application of parallel RP-LC-MS and GC-MS allows for capturing a comprehensive global picture of GIST tissue metabolism.Author contributions
Szymon Macioszek: conceptualization, investigation, formal analysis, and writing – original draft preparation. Danuta Dudzik: formal analysis, data curation, and visualisation. Margot Biesemans: formal analysis and data curation. Agnieszka Wozniak: conceptualization, resources and writing – reviewing and editing. Patrick Schöffski: resources and supervision. Michał J. Markuszewski: conceptualization, supervision, and writing – reviewing and editing.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Science Centre Poland [grant Preludium no 2018/29/N/NZ7/02908] and by the project POWR.03.02.00-00-I026/17-00 co-financed by the European Union through the European Social Fund under the Operational Programme Knowledge Education Development 2014–2020.References
- L. Thuneberg, Adv. Anat., Embryol. Cell Biol., 1982, 71, 1–130 CrossRef CAS PubMed.
- K. W. Min and M. Leabu, J. Cell. Mol. Med., 2006, 10, 995–1013 CrossRef CAS.
- P. G. Casali, J. Y. Blay, N. Abecassis, J. Bajpai, S. Bauer, R. Biagini, S. Bielack, S. Bonvalot, I. Boukovinas, J. V. M. G. Bovee, K. Boye, T. Brodowicz, A. Buonadonna, E. De Álava, A. P. Dei Tos, X. G. Del Muro, A. Dufresne, M. Eriksson, A. Fedenko, V. Ferraresi, A. Ferrari, A. M. Frezza, S. Gasperoni, H. Gelderblom, F. Gouin, G. Grignani, R. Haas, A. B. Hassan, N. Hindi, P. Hohenberger, H. Joensuu, R. L. Jones, C. Jungels, P. Jutte, B. Kasper, A. Kawai, K. Kopeckova, D. A. Krákorová, A. Le Cesne, F. Le Grange, E. Legius, A. Leithner, A. Lopez-Pousa, J. Martin-Broto, O. Merimsky, C. Messiou, A. B. Miah, O. Mir, M. Montemurro, C. Morosi, E. Palmerini, M. A. Pantaleo, R. Piana, S. Piperno-Neumann, P. Reichardt, P. Rutkowski, A. A. Safwat, C. Sangalli, M. Sbaraglia, S. Scheipl, P. Schöffski, S. Sleijfer, D. Strauss, S. J. Strauss, K. S. Hall, A. Trama, M. Unk, M. A. J. van de Sande, W. T. A. van der Graaf, W. J. van Houdt, T. Frebourg, A. Gronchi and S. Stacchiotti, Ann. Oncol., 2022, 33, 20–33 CrossRef CAS PubMed.
- A. Wozniak, G. Floris, M. Debiec-Rychter, R. Sciot and P. Schöffski, Cancer Invest., 2010, 28, 839–848 CrossRef CAS PubMed.
- J. Lasota and M. Miettinen, Histopathology, 2008, 53, 245–266 CrossRef CAS PubMed.
- M. Debiec-Rychter, R. Sciot, A. Le Cesne, M. Schlemmer, P. Hohenberger, A. T. van Oosterom, J. Y. Blay, S. Leyvraz, M. Stul, P. G. Casali, J. Zalcberg, J. Verweij, M. Van Glabbeke, A. Hagemeijer and I. Judson, Eur. J. Cancer, 2006, 42, 1093–1103 CrossRef CAS PubMed.
- H. Zhang and Q. Liu, Transl. Oncol., 2020, 13, 100812 CrossRef PubMed.
- S. Macioszek, D. Dudzik, J. Jacyna, A. Wozniak, P. Schöffski and M. J. Markuszewski, Metabolites, 2021, 11, 554 CrossRef CAS PubMed.
- P. Bhargava, K. C. Fitzgerald, S. L. V. Venkata, M. D. Smith, M. D. Kornberg, E. M. Mowry, N. J. Haughey and P. A. Calabresi, Ann. Clin. Transl. Neurol., 2019, 6, 33–45 CrossRef CAS PubMed.
- R. Kaddurah-Daouk, M. B. Bogdanov, W. R. Wikoff, H. Zhu, S. H. Boyle, E. Churchill, Z. Wang, A. J. Rush, R. R. Krishnan, E. Pickering, M. Delnomdedieu and O. Fiehn, Transl. Psychiatry, 2013, 3, e223–e223 CrossRef CAS PubMed.
- J. Wild, M. Shanmuganathan, M. Hayashi, M. Potter and P. Britz-Mckibbin, Analyst, 2019, 144, 6595–6608 RSC.
- C. Kamrath, M. F. Hartmann and S. A. Wudy, Steroids, 2019, 150, 108426 CrossRef CAS PubMed.
- S. Wei, L. Liu, J. Zhang, J. Bowers, G. A. N. Gowda, H. Seeger, T. Fehm, H. J. Neubauer, U. Vogel, S. E. Clare and D. Raftery, Mol. Oncol., 2013, 7, 297–307 CrossRef CAS PubMed.
- P. B. Phapale, S. D. Kim, H. W. Lee, M. Lim, D. D. Kale, Y. L. Kim, J. H. Cho, D. Hwang and Y. R. Yoon, Clin. Pharmacol. Ther., 2010, 87, 426–436 CrossRef CAS PubMed.
- Q. Huang, J. Aa, H. Jia, X. Xin, C. Tao, L. Liu, B. Zou, Q. Song, J. Shi, B. Cao, Y. Yong, G. Wang and G. Zhou, J. Proteome Res., 2015, 14, 3970–3981 CrossRef CAS PubMed.
- S. Halouska, O. Chacon, R. J. Fenton, D. K. Zinniel, R. G. Barletta and R. Powers, J. Proteome Res., 2007, 6, 4608–4614 CrossRef CAS PubMed.
- X. Bao, J. Wu, S. Kim, P. LoRusso and J. Li, J. Clin. Pharmacol., 2019, 59, 20–34 CrossRef CAS PubMed.
- P. Bhargava, M. D. Smith, L. Mische, E. Harrington, K. C. Fitzgerald, K. Martin, S. Kim, A. A. Reyes, J. Gonzalez-Cardona, C. Volsko, A. Tripathi, S. Singh, K. Varanasi, H. N. Lord, K. Meyers, M. Taylor, M. Gharagozloo, E. S. Sotirchos, B. Nourbakhsh, R. Dutta, E. M. Mowry, E. Waubant and P. A. Calabresi, J. Clin. Invest., 2020, 130, 3467–3482 CrossRef CAS PubMed.
- P. S. Ward, J. Patel, D. R. Wise, O. Abdel-Wahab, B. D. Bennett, H. A. Coller, J. R. Cross, V. R. Fantin, C. V. Hedvat, A. E. Perl, J. D. Rabinowitz, M. Carroll, S. M. Su, K. A. Sharp, R. L. Levine and C. B. Thompson, Cancer Cell, 2010, 17, 225–234 CrossRef CAS PubMed.
- L. Dang, D. W. White, S. Gross, B. D. Bennett, M. A. Bittinger, E. M. Driggers, V. R. Fantin, H. G. Jang, S. Jin, M. C. Keenan, K. M. Marks, R. M. Prins, P. S. Ward, K. E. Yen, L. M. Liau, J. D. Rabinowitz, L. C. Cantley, C. B. Thompson, M. G. Vander Heiden and S. M. Su, Nature, 2009, 462, 739–744 CrossRef CAS PubMed.
- L. R. Jensen, E. M. Huuse, T. F. Bathen, P. E. Goa, A. M. Bofin, T. B. Pedersen, S. Lundgren and I. S. Gribbestad, NMR Biomed., 2010, 23, 56–65 CrossRef CAS PubMed.
- J. Debik, L. R. Euceda, S. Lundgren, H. V. D. L. Gythfeldt, Ø. Garred, E. Borgen, O. Engebraaten, T. F. Bathen and G. F. Giskeødegård, J. Proteome Res., 2019, 18, 3649–3660 CrossRef CAS PubMed.
- N. Arias-Ramos, L. Ferrer-Font, S. Lope-Piedrafita, V. Mocioiu, M. Julià-Sapé, M. Pumarola, C. Arús and A. Candiota, Metabolites, 2017, 7, 20 CrossRef PubMed.
- K. M. Schuler, B. S. Rambally, M. J. Difurio, B. P. Sampey, P. A. Gehrig, L. Makowski and V. L. Bae-Jump, Cancer Med., 2015, 4, 161–173 CrossRef CAS PubMed.
- T. Ishida, T. Takahashi, Y. Kurokawa, T. Nishida, S. Hirota, S. Serada, M. Fujimoto, T. Naka, R. Teranishi, T. Saito, K. Yamashita, K. Tanaka, K. Yamamoto, T. Makino, M. Yamasaki, K. Nakajima, H. Eguchi and Y. Doki, Br. J. Cancer, 2021, 125, 1511–1522 CrossRef CAS PubMed.
- G. Caocci, M. Deidda, A. Noto, M. Greco, M. P. Simula, O. Mulas, D. Cocco, C. Fattuoni, G. Mercuro, G. La Nasa and C. Cadeddu Dessalvi, J. Clin. Med., 2020, 9, 1180 CrossRef CAS PubMed.
- M. G. Contreras Mostazo, N. Kurrle, M. Casado, D. Fuhrmann, I. Alshamleh, B. Häupl, P. Martín-Sanz, B. Brüne, H. Serve, H. Schwalbe, F. Schnütgen, S. Marin and M. Cascante, Cancers, 2020, 12, 1–26 Search PubMed.
- Y. Gagnebin, B. Julien, P. Belén and R. Serge, J. Pharm. Biomed. Anal., 2018, 161, 313–325 CrossRef PubMed.
- Ö. C. Zeki, C. C. Eylem, T. Reçber, S. Kır and E. Nemutlu, J. Pharm. Biomed. Anal., 2020, 190, 113509 CrossRef PubMed.
- D.-Q. Tang, L. Zou, X.-X. Yin and C. N. Ong, Mass Spectrom. Rev., 2016, 35, 574–600 CrossRef CAS PubMed.
- G. Theodoridis, H. G. Gika and I. D. Wilson, TrAC, Trends Anal. Chem., 2008, 27, 251–260 CrossRef CAS.
- S. Wernisch and S. Pennathur, Anal. Bioanal. Chem., 2016, 408, 6079–6091 CrossRef CAS PubMed.
- N. Sillner, A. Walker, E. M. Harrieder, P. Schmitt-Kopplin and M. Witting, J. Chromatogr. B: Anal. Technol. Biomed. Life Sci., 2019, 1109, 142–148 CrossRef CAS PubMed.
- H. Gallart-Ayala, I. Konz, F. Mehl, T. Teav, A. Oikonomidi, G. Peyratout, V. van der Velpen, J. Popp and J. Ivanisevic, Anal. Chim. Acta, 2018, 1037, 327–337 CrossRef CAS PubMed.
- A. M. King, L. G. Mullin, I. D. Wilson, M. Coen, P. D. Rainville, R. S. Plumb, L. A. Gethings, G. Maker and R. Trengove, Metabolomics, 2019, 15, 17 CrossRef PubMed.
- J. Guo and T. Huan, Anal. Chem., 2020, 92, 8072–8080 CrossRef CAS PubMed.
- Q. Hu, H. Tang and Y. Wang, J. Anal. Test., 2020, 4, 140–162 CrossRef.
- A. N. Macedo, A. T. Faccio, T. S. Fukuji, G. A. B. Canuto and M. F. M. Tavares, Adv. Exp. Med. Biol., 2021, 1336, 215–242 CrossRef CAS PubMed.
- C. Barbas, E. P. Moraes and A. Villaseñor, J. Pharm. Biomed. Anal., 2011, 55, 823–831 CrossRef CAS PubMed.
- T. Soga, TrAC, Trends Anal. Chem., 2023, 158, 116883 CrossRef CAS.
- J. Walsby-Tickle, J. Gannon, I. Hvinden, C. Bardella, M. I. Abboud, A. Nazeer, D. Hauton, E. Pires, T. Cadoux-Hudson, C. J. Schofield and J. S. O. McCullagh, Commun. Biol., 2020, 3, 247 CrossRef CAS PubMed.
- Y. Sun, K. Saito, R. Iiji and Y. Saito, SLAS Discovery, 2019, 24, 778–786 CrossRef CAS PubMed.
- T. Van Looy, A. Wozniak, G. Floris, H. Li, J. Wellens, U. Vanleeuw, R. Sciot, M. Debiec-Rychter and P. Schöffski, Transl. Oncol., 2015, 8, 112–118 CrossRef PubMed.
- T. Smyth, T. Van Looy, J. E. Curry, A. M. Rodriguez-Lopez, A. Wozniak, M. Zhu, R. Donsky, J. G. Morgan, M. Mayeda, J. A. Fletcher, P. Schöffski, J. Lyons, N. T. Thompson and N. G. Wallis, Mol. Cancer Ther., 2012, 11, 1799–1808 CrossRef CAS PubMed.
- P. Schöffski, Y. Gebreyohannes, T. Van Looy, P. Manley, J. D. Growney, M. Squires and A. Wozniak, Biomedicines, 2022, 10, 1135 CrossRef PubMed.
- E. L. Schymanski, J. Jeon, R. Gulde, K. Fenner, M. Ruff, H. P. Singer and J. Hollender, Environ. Sci. Technol., 2014, 48 Search PubMed.
- S. Vasseur and F. Guillaumond, Oncogenesis, 2022, 11, 46 CrossRef CAS PubMed.
- L. A. Broadfield, A. A. Pane, A. Talebi, J. V. Swinnen and S.-M. Fendt, Dev. Cell, 2021, 56, 1363–1393 CrossRef CAS PubMed.
- S. Beloribi-Djefaflia, S. Vasseur and F. Guillaumond, Oncogenesis, 2016, 5, e189–e189 CrossRef CAS PubMed.
- I. Kohler, A. Verhoeven, R. J. Derks and M. Giera, Bioanalysis, 2016, 8, 1509–1532 CrossRef CAS PubMed.
- D. V. McCalley, J. Chromatogr. A, 2010, 1217, 3408–3417 CrossRef CAS PubMed.
- S. Wernisch and S. Pennathur, Anal. Bioanal. Chem., 2016, 408, 6079–6091 CrossRef CAS PubMed.
- W.-C. Cho, Y.-K. Shin, Y.-S. Na, M.-H. Ryu, J.-L. Ku and Y.-K. Kang, Biochem. Biophys. Res. Commun., 2020, 529, 699–706 CrossRef CAS PubMed.
- M. Debiec-Rychter, J. Cools, H. Dumez, R. Sciot, M. Stul, N. Mentens, H. Vranckx, B. Wasag, H. Prenen, J. Roesel, A. Hagemeijer, A. Van Oosterom and P. Marynen, Gastroenterology, 2005, 128, 270–279 CrossRef CAS PubMed.
- X. Hu, Z. Wang, P. Su, Q. Zhang and Y. Kou, Front. Oncol., 2022, 12, 1–11 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d3an00599b |
| This journal is © The Royal Society of Chemistry 2023 |