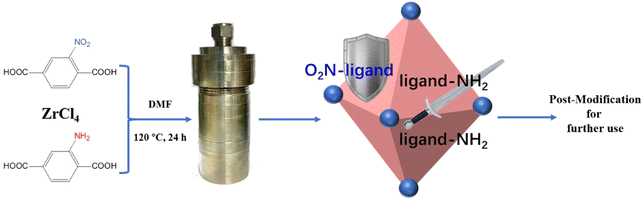
An alkali-resistant zirconium-biligand organic framework with dual-metal centers for highly selective capture of phosphopeptides†
Xiaoyu
Zhou
 abc,
Hongyan
Zhang
ab,
Li
Wang
ab and
Ren'an
Wu
abc,
Hongyan
Zhang
ab,
Li
Wang
ab and
Ren'an
Wu
 *ab
*ab
aLaboratory of High-Resolution Mass Spectrometry Technologies, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian, 116023, China. E-mail: wurenan@dicp.ac.cn; Tel: +86-411-84379828
bCAS Key Laboratory of Separation Science for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian, 116023, China
cUniversity of Chinese Academy of Sciences, Beijing 100049, China
First published on 24th November 2022
Abstract
The stability of MOFs plays one of the most important roles in material applications, while the delicate structure of MOFs suffers from the limitation of poor alkali tolerance. A new biligand Zr-MOF (biUIO-66–NH2NO2) with alkali-resistance performance and active functional groups has been synthesized in this study. The biUIO-66–NH2NO2 demonstrated a much better stability in 1% NH3·H2O solution than its parent material, UIO-66–NH2. Following further immobilization of Zr4+ ions, the biDZMOF consisting of dual-zirconium centers was prepared and was further applied in global enrichment of phosphopeptides by avoiding the instability of enrichment materials in the essential alkali elution procedure for the phosphopeptide enrichment workflow. The alkali-resistant elution of phosphopeptides from the biDZMOF can be directly coupled to a tandem mass spectrometry system for peptide analysis without desalting treatment. 425 phosphopeptides in total in 3 independent samples were identified from 10 μL human saliva after enrichment with biDZMOF. The improvement in alkali resistance and successful post-modification of biUIO-66–NH2NO2 suggest an efficient strategy to develop new types of MOF materials for application.
Introduction
As one of the most prevalent and significant post-translational modifications (PTMs), protein phosphorylation plays vital regulatory roles in numerous cellular behaviors and is closely related to diseases.1,2 Protein phosphorylation is related to almost all aspects of the cellular and molecular regulation mechanisms.3 Mass spectrometry (MS) based phosphoproteomics analysis has become one of the most attractive strategies due to its merits of fast speed, high resolution, and high throughput.4,5 Nevertheless, due to the low levels of phosphopeptides and the signal suppression caused by the high abundance of nonphosphopeptides, directly analyzing phosphopeptides by MS remains a significant challenge.6 The enrichment procedures before MS analysis are the crucial steps to harvest the reliable peaks of phosphoproteomes in complex biological samples.7,8Because of the unusual physical and chemical properties of phosphoryl groups, the specific interaction between the phosphoryl groups of phosphopeptides and enrichment materials was utilized for phosphopeptide enrichment.9 To date, metal oxide affinity chromatography (MOAC)10 and immobilized metal affinity chromatography (IMAC)8 have been the most commonly used strategies for the enrichment of phosphopeptides. Metal–organic frameworks (MOFs), which are well-organized architectures with bridging ligands and metal ions, have been developed as versatile platforms for a variety of applications, including phosphopeptide enrichment.11 The UIO series MOFs have been widely used in the peptide enrichment field due to their large surface areas, highly ordered open pores, amphoteric surface of metal oxides, and high stability in acidic solutions, which could provide the necessary specific interaction for phosphopeptide enrichment. The reversible affinity interaction between phosphate groups and the naturally exposed Zr–O clusters of the UIO series MOFs has been utilized in MOAC for the pre-separation of phosphopeptides in biological samples, and has been reported to be more selective for monophosphopeptides.12 At the same time, metal ions such as Fe3+, Ti4+, Zr4+, and so on have been immobilized on different chelation sites of the UIO series MOFs for phosphopeptide enrichment based on the strategy of IMAC, which preferred to enrich multi-phosphopeptides.13,14 The delicate structure and the open functional sites made the MOF-based material an ideal platform for combining the utilization of MOAC and IMAC.13–15
However, the instability of MOF materials in strongly alkaline solutions could definitely limit further MS analysis. The specific interaction force based on IMAC and MOAC mechanisms between phosphate groups and Zr-MOF materials needs to be destroyed in strongly basic elution buffer to release the anchored phosphopeptides for further MS analysis. Although the well-organized architectures, pore size, hydrophilicity, open functional sites, and other related properties of the MOF material are important for specific enrichment of peptides, the instability of MOFs in the basic NH3·H2O elution buffer makes the metal ions and organic ligands elute along with the phosphopeptides which is an unavoidable problem for MOF materials in phosphopeptide enrichment. Some of these MOF based enrichment methods require desalting treatment of the phosphopeptide eluate using reversed phase or other commercial materials before LC-MS analysis for obtaining detailed phosphorylation information.16–18 Otherwise, the salts in the eluate would suppress the MS signal of phosphopeptides and damage the hardware of the nano-LC system. Although the enrichment of phosphopeptides could harvest the reliable peaks in MS-based phosphoproteomics analysis after desalting, the additional desalting operation using a reversed phase material results in the potential loss of phosphopeptides with high hydrophilicity, low mass, or extremely low abundance. Meanwhile, the advantages of high selectivity, low limits of detection, and high anti-interference ability of well-designed enrichment materials that were detected by MALDI MS could not be fully embodied in further LC-MS analysis. Thus, an alkali-resistant MOF probe for the phosphopeptide enrichment workflow is expected.
Efforts have been devoted to the design of alkali-resistant Zr-MOFs. For alkali stability enhancement of UIO materials, various factors such as chemically stable ligands,19 the inclusion of thioether groups,20 and post-synthetic modification21 have been considered. But the changes in chemical sites and pore structures of newly synthesized MOFs were too profound to apply in further phosphopeptides enrichment. A series of UiO-66 MOFs were reported to be synthesized successfully with different functional groups on BDC ligands, which provide attractive methods to control chemical composition, functionality, and porosity while maintaining good stability and achieving the desired chemical and physical properties. A noticeable improvement in chemical resistance toward strongly basic media was noticed as a consequence of the nitro group on the BDC ligand. In a NaOH solution with pH = 14, UIO-66–NO2 could be stable.22 On the other hand, the amino group of UIO-66–NH2 is accessible as well as chemically reactive, which makes UIO-66–NH2 very attractive for further modification and application. However, UIO-66–NH2 does collapse in alkali solution, in contrast to alkali-resistant UIO-66–NO2.
Herein, inspired by combining the advantages of alkali-resistant UIO-66–NO2 with the advantages of the functional sites for metal immobilization provided by the –NH2 group, an alkali-resistant UIO-66 series MOF with double ligands (biUIO-66–NH2NO2) was synthesized. Furthermore, the biUIO-66–NH2NO2 was post-modified with Zr4+ ions to form the biDZMOF for global phosphopeptide enrichment. The mono- and multi-phosphopeptides were globally enriched using biDZMOF, which combines the MOAC and IMAC properties in dual zirconium centers. The improvement of the alkali resistance of the biUIO-66–NH2NO2 matrix greatly reduced the concentration of the MOF material that disintegrates in NH3·H2O solution during the elution step of the phosphopeptide enrichment workflow, so that the eluate of the enrichment MOF material could be directly compatible with LC-MS/MS system tandem analysis, which greatly improved the identification coverage of phosphopeptides in complex real samples. Alkali-resistant biUIO materials have great potential for further application.
Experimental
Synthesis of biUIO-66–NH2NO2
Biligand biUIO-66–NH2NO2 samples were synthesized by a modified solvothermal reaction with different molar ratios of the two organic linkers.23,24 Typically, 0.24 g of ZrCl4, 0.22 g of BDC–NH2 and different amounts of BDC–NO2 were dissolved in 20 mL of DMF. The mixture was transferred into a Teflon-lined stainless-steel autoclave. Then, the autoclave was heated at 120 °C for 24 h and cooled to room temperature. The product was collected for further characterization after extensive washing with DMF and methanol.Preparation of the biligand dual-metal centered zirconium–organic framework (biDZMOF)
The biDZMOF was prepared by post-synthetic functionalization of biUIO-66–NH2NO2. In brief, 50 mg of as-synthesized biUIO-66–NH2NO2 was incubated in an acetonitrile (ACN) solution containing 0.2 mmol of phosphorus oxychloride and 0.2 mmol of 2,4,6-collidine overnight at room temperature. After washing with ACN and water several times, the product was incubated in a 50 mM ZrOCl2 solution with gentle shaking overnight for the loading of Zr4+. Finally, the obtained biDZMOF was rinsed with water several times and dried under vacuum at 60 °C overnight for further use.Enrichment of global phosphopeptides
The digests of β-casein and BSA were utilized as the model proteins to test the enrichment performances of the biDZMOF. Typically, 50 μg of materials were incubated with the peptide mixture in 150 μL of loading buffer (85% ACN (v/v) and 6% trifluoroacetic acid (TFA) (v/v)) for 30 min at room temperature. The materials were then collected by centrifugation and stepwise rinsed with 150 μL of washing buffer 1 (50% ACN (v/v), 6% TFA (v/v), and 200 mmol L−1 NaCl), 100 μL of washing buffer 2 (50% ACN (v/v) and 0.1% TFA (v/v)) and 50 μL of washing buffer 3 (0.05% (w/w) NH3·H2O). Thereafter, the target peptides were eluted from materials using 10 μL of 10% (w/w) NH3·H2O solution. Then, the eluate was identified by matrix assisted laser desorption ionization-time of flight mass spectrometry (MALDI-TOF MS).Fresh human saliva was chosen as the complex biological sample for global phosphopeptide enrichment. Briefly, 10 μL of the human saliva sample collected from a healthy volunteer was mixed with 140 μL of loading buffer to form a homogeneous solution. Subsequently, the prepared saliva sample was treated with the biDZMOF following the above-mentioned model protein enrichment procedure. Finally, the eluate was identified by MALDI MS/MS and then acidized with FA for further nano reversed phase liquid chromatography-electrospray ionization tandem mass spectrometry (nano-RPLC-ESI-MS/MS) analysis.
Database searching
Raw data were processed with MaxQuant and its built-in Andromeda search engine for peptide identification. MS/MS spectra were searched against the Uniprot human reference proteome FASTA file (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 397 entries) using MaxQuant 1.4.1.2. Serine, threonine and tyrosine phosphorylation (+79.966330 Da) and methionine oxidation (Met, +15.994915 Da) were treated as variable modifications. Peptide precursor ions were searched with a maximum mass deviation of 4.5 ppm and fragment ions with a maximum mass deviation of 20 ppm. Peptide, protein and site identifications were filtered at an FDR of 1% using the decoy database strategy. The minimal peptide length was 7 amino acids and the minimum Andromeda score for modified peptides was 40 and the corresponding minimum delta score was 6 (default MaxQuant settings).
397 entries) using MaxQuant 1.4.1.2. Serine, threonine and tyrosine phosphorylation (+79.966330 Da) and methionine oxidation (Met, +15.994915 Da) were treated as variable modifications. Peptide precursor ions were searched with a maximum mass deviation of 4.5 ppm and fragment ions with a maximum mass deviation of 20 ppm. Peptide, protein and site identifications were filtered at an FDR of 1% using the decoy database strategy. The minimal peptide length was 7 amino acids and the minimum Andromeda score for modified peptides was 40 and the corresponding minimum delta score was 6 (default MaxQuant settings).
Results and discussion
Synthesis and characterization of alkali-resistant biUIO-66–NH2NO2
In general, even MOFs formed by metal clusters and ligands with high connections would hardly maintain their intact structure under extreme conditions. The framework decomposition occurs with the destruction of the coordination bond in a strongly basic solution. A series of newly synthesized ligands have been reported for obtaining a stable structure of MOFs with high alkali-resistance. However, the new ligand molecules changed the physical and chemical properties of the newly synthesized MOFs, limiting the application of alkali-resistant MOFs. The investigation of tagged UIO-66 Zr-MOFs obtained from different linker ligands, namely BDC–NO2 and BDC–NH2, respectively, has demonstrated that the functional groups present at the linker units could observably change the physicochemical and chemical properties of tagged UIO-66 MOFs.22 The BDC–NO2 ligands provided the alkali-resistance property to UIO-66–NO2 which could be stable in a NaOH solution of pH = 14. The BDC–NH2 ligands provided the ideal functional sites for further modification which could be used for further application. On the basis of the above considerations, herein, the strategy of biligand was adopted for the synthesis of alkali-resistant UIO-66 MOFs, which could result in better tolerance for basic solution before lattice collapse occurs. Thus, the ideal biligand UIO-66 MOF could maintain the functional sites for further modification with high alkali-resistance under basic conditions, similar to holding a sword and shield at the same time (Scheme 1).To investigate the resistance of the biUIO-66 MOF in basic solution, biUIO-66–NH2NO2 samples with extreme molar ratios of the two linkers of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 1
1 and 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 were first synthesized. All 2θ peaks in the wide-angle X-ray diffraction (XRD) profiles of the as-synthesized biUIO-66–NH2NO2 samples were consistent with those of the parent material UIO-66–NH2 as reported in the previous studies (Fig. 1a),22 which demonstrated that these biligand Zr-MOFs were topologically equivalent to UIO-66 MOFs without producing other structured materials. These as-synthesized MOFs were dispersed in NH3·H2O solution for 30 minutes, respectively, and then collected to test alkali resistance, which is shown by the XRD patterns of the recovered framework from NH3·H2O dispersions in Fig. 1b. The peaks of UIO-66–NH2 totally disappeared which proved the collapse of UIO-66–NH2 in basic solution. The vanishing of the fine structure of the biUIO-66–NH2NO2 sample with a molar ratio of 1
10 were first synthesized. All 2θ peaks in the wide-angle X-ray diffraction (XRD) profiles of the as-synthesized biUIO-66–NH2NO2 samples were consistent with those of the parent material UIO-66–NH2 as reported in the previous studies (Fig. 1a),22 which demonstrated that these biligand Zr-MOFs were topologically equivalent to UIO-66 MOFs without producing other structured materials. These as-synthesized MOFs were dispersed in NH3·H2O solution for 30 minutes, respectively, and then collected to test alkali resistance, which is shown by the XRD patterns of the recovered framework from NH3·H2O dispersions in Fig. 1b. The peaks of UIO-66–NH2 totally disappeared which proved the collapse of UIO-66–NH2 in basic solution. The vanishing of the fine structure of the biUIO-66–NH2NO2 sample with a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 indicated some disruption of the crystalline structure. Only the XRD patterns of recovered biUIO-66–NH2NO2 with a molar ratio of 10
10 indicated some disruption of the crystalline structure. Only the XRD patterns of recovered biUIO-66–NH2NO2 with a molar ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 showed the peaks of the structure which revealed the good tolerance to alkali solution. biUIO-66–NH2NO2 with ligands BDC–NH2 and BDC–NO2 at a molar ratio of 10
1 showed the peaks of the structure which revealed the good tolerance to alkali solution. biUIO-66–NH2NO2 with ligands BDC–NH2 and BDC–NO2 at a molar ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 is more resistant to basic solution than biUIO-66–NH2NO2 with ligands at a molar ratio of 1
1 is more resistant to basic solution than biUIO-66–NH2NO2 with ligands at a molar ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10. The preliminary result demonstrated that the minor addition of the BDC–NO2 ligand to UIO-66–NH2 could observably improve the performance of alkali resistance.
10. The preliminary result demonstrated that the minor addition of the BDC–NO2 ligand to UIO-66–NH2 could observably improve the performance of alkali resistance.
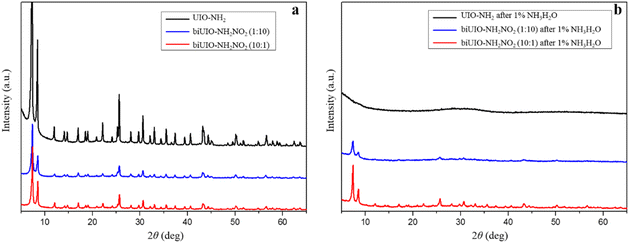 | ||
Fig. 1 XRD patterns of as-synthesized UIO-66–NH2, biUIO-66–NH2NO2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10), and biUIO-66–NH2NO2 (10 10), and biUIO-66–NH2NO2 (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) before (a) and after (b) soaking in NH3·H2O solution for 30 minutes, respectively. 1) before (a) and after (b) soaking in NH3·H2O solution for 30 minutes, respectively. | ||
To achieve the most alkali-resistant biUIO-66–NH2NO2 for further application, biUIO-66–NH2NO2 materials with different ratios of the two ligands were synthesized. The two ligands BDC–NH2 and BDC–NO2 with molar ratios from 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 to 12
2 to 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 were, respectively, utilized in the synthesis of biUIO-66. The as-synthesized biUIO-66–NH2NO2 samples were dispersed in NH3·H2O solution with higher concentration. The supernatants were collected, respectively, in which the Zr element was measured by coupled plasma-optical emission spectroscopy (ICP-OES) to calculate the concentration of Zr ions that disintegrated from biUIO-66–NH2NO2. ICP-OES results revealed that biUIO-66–NH2NO2 with the BDC–NO2 ligand introduced could obviously reduce the concentration of Zr that collapsed in basic solution (Table 1). Among them, the biUIO-66–NH2NO2 sample with a molar ratio of 12
6 were, respectively, utilized in the synthesis of biUIO-66. The as-synthesized biUIO-66–NH2NO2 samples were dispersed in NH3·H2O solution with higher concentration. The supernatants were collected, respectively, in which the Zr element was measured by coupled plasma-optical emission spectroscopy (ICP-OES) to calculate the concentration of Zr ions that disintegrated from biUIO-66–NH2NO2. ICP-OES results revealed that biUIO-66–NH2NO2 with the BDC–NO2 ligand introduced could obviously reduce the concentration of Zr that collapsed in basic solution (Table 1). Among them, the biUIO-66–NH2NO2 sample with a molar ratio of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 showed the best alkali resistance. Only 18.6% Zr4+ was present in the basic supernatant of biUIO-66–NH2NO2 with a molar ratio of 12
4 showed the best alkali resistance. Only 18.6% Zr4+ was present in the basic supernatant of biUIO-66–NH2NO2 with a molar ratio of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 compared with the case of UIO-66–NH2. All subsequent biUIO-66–NH2NO2 materials below represent biUIO-66–NH2NO2 with the BDC–NH2 and BDC–NO2 molar ratio of 12
4 compared with the case of UIO-66–NH2. All subsequent biUIO-66–NH2NO2 materials below represent biUIO-66–NH2NO2 with the BDC–NH2 and BDC–NO2 molar ratio of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4.
4.
| MOF materials | ppm of Zr |
|---|---|
| Conditions of the experiment: 2 mg of MOF in 2 mL NH3·H2O solution, the supernatant was diluted from 1.5 mL to 6 mL using 10% HNO3 after centrifugation. | |
| UIO66–NH2 | 3.692 |
biUIO-66–NH2NO2 with molar ratio of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 2 |
1.161 |
biUIO-66–NH2NO2 with molar ratio of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
0.875 |
biUIO-66–NH2NO2 with molar ratio of 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3 3 |
0.889 |
biUIO-66–NH2NO2 with molar ratio of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4 4 |
0.687 |
biUIO-66–NH2NO2 with molar ratio of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 6 |
1.342 |
The TEM images of biUIO-66–NH2NO2 showed its octahedral morphology structure which was consistent with the structure of the UIO-66 series Zr-MOFs (Fig. 2a). Nitrogen sorption/desorption isotherms were obtained to characterize the surface area and porous nature of biUIO-66–NH2NO2 (Fig. 2b). The calculated Brunauer–Emmett–Teller (BET) surface area was ca. 952.143 m2 g−1, which is in between the values observed for pristine UIO-66–NH2 and UIO-66–NO2.22 The estimated pore size by the DFT method was ca. 1.368 nm. Fourier transform infrared (FT-IR) spectroscopy of biUIO-66–NH2NO2 shown in Fig. 2c was further performed to confirm the existence of dual ligands. A coordinating metal center with linkers (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O in carboxylates) occurs in the region of 1540–1600 cm−1 through the deprotonating process. The C–O bond of carboxylic acid can also be recognized at around 1400 cm−1. The double peaks appearing at 3367 and 3475 cm−1 present the asymmetric and symmetric stretching vibrations of –NH2 functional groups on biUIO-66–NH2NO2.25,26 The asymmetric N–O vibration peaks at 1545 cm−1 appeared on biligand samples of biUIO-66–NH2NO2, which were not detected in the FT-IR spectra of UIO-66–NH2 (Fig. S1†).27 The regular morphology, high specific surface area, controllable core structure, active functional sites, and the most important property of alkali-resistance all indicated that biUIO-66–NH2NO2 is a promising application platform.
O in carboxylates) occurs in the region of 1540–1600 cm−1 through the deprotonating process. The C–O bond of carboxylic acid can also be recognized at around 1400 cm−1. The double peaks appearing at 3367 and 3475 cm−1 present the asymmetric and symmetric stretching vibrations of –NH2 functional groups on biUIO-66–NH2NO2.25,26 The asymmetric N–O vibration peaks at 1545 cm−1 appeared on biligand samples of biUIO-66–NH2NO2, which were not detected in the FT-IR spectra of UIO-66–NH2 (Fig. S1†).27 The regular morphology, high specific surface area, controllable core structure, active functional sites, and the most important property of alkali-resistance all indicated that biUIO-66–NH2NO2 is a promising application platform.
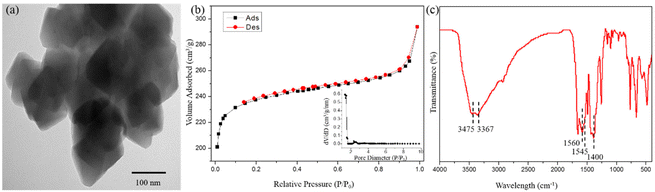 | ||
| Fig. 2 (a) The TEM image, (b) nitrogen sorption/desorption isotherms, (c) FT-IR spectra of as-synthesized biUIO-66–NH2NO2. | ||
Immobilization of Zr4+ and characterization of biDZMOF
Thanks to the inherent and naturally exposed Zr–O clusters and the improved alkali-resistance of the biUIO-66–NH2NO2 material, the demand for good tolerance to both acidic and basic solutions in the phosphopeptide enrichment workflow was fulfilled by biUIO-66–NH2NO2. The active functional groups–NH2 could be modified as chelation sites for immobilized Zr4+ ions, as shown in Fig. 3a. The global enrichment of mono- and multi-phosphopeptides was performed using biDZMOF with dual zirconium centers that combined the utilization of MOAC and IMAC strategies (Fig. 3b). The disintegration of biUIO-66–NH2NO2 in NH3·H2O elution buffer could be reduced greatly with the improvement of alkali resistance. The enriched phosphopeptides in the eluate could be directly injected into a LC-MS/MS system after simple acidification which maximized the enrichment performance and greatly improved the identification coverage of phosphopeptides.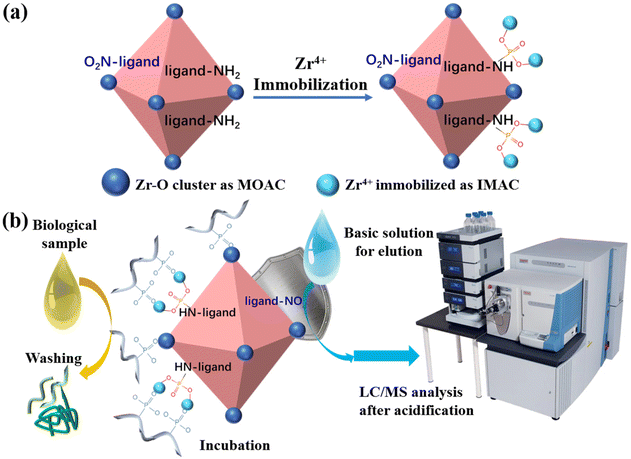 | ||
| Fig. 3 (a) Synthesis of biDZMOF and (b) the procedure of phosphopeptide enrichment and analysis using biDZMOF in a biological sample. | ||
The characterization of biDZMOF proved the success of post-modification. The TEM of biUIO-66–NH2NO2 after modifying the composite with the phosphonate group and Zr4+ showed that its structure and morphology have undergone no significant change (Fig. 4a). The XRD patterns confirmed that the characteristic cubic close-packed structure of the UIO-66 series MOFs was stable during the immobilization (Fig. 4b). FT-IR spectroscopy was performed to confirm the functionalization process (Fig. 4c). The absorption band that appeared at 1070 cm−1 is attributed to the P–O stretching vibration of phosphonate groups, while the double peaks of the primary aromatic amine disappeared, indicating the modification of the amine groups by phosphonate groups (Fig. 4c ii).14 The changes of zeta potential from a slightly negative (−12.00 ± 0.95 mV) to a positive value of +24.00 ± 1.68 mV with the modification proved the introduction of the BDC–NO2 ligand and the successful immobilization of the Zr4+ ions (Fig. 4d).
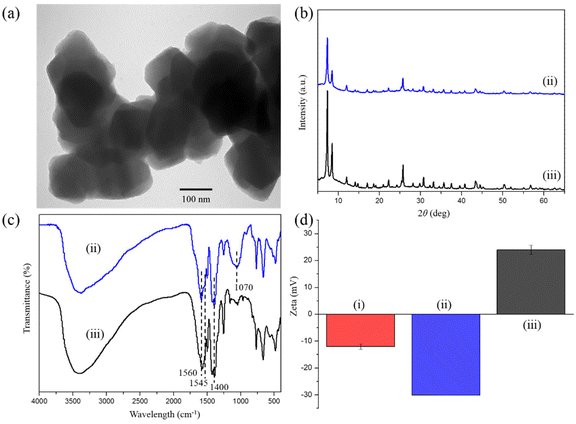 | ||
| Fig. 4 (a) The TEM image of biDZMOF, (b) XRD patterns, (c) FT-IR spectra, (d) zeta potential of as-synthesized (i) biUIO-66–NH2NO2, (ii) biUIO-66–NH2NO2–PO3 and (iii) biDZMOF. | ||
The zirconium content in biDZMOF was estimated to be ca. 25.4% including ca. 20.7% intrinsic Zr–O cluster and ca. 4.7% immobilized Zr4+via measuring the zirconium content of biUIO-66–NH2NO2, biUIO-66–NH2NO2–PO3, and biDZMOF, respectively, by ICP-OES. Considering the BDC–NH2 and BDC–NO2 molar ratio of 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4, it suggested that 30.7% of BDC–NH2 ligands of biUIO-66–NH2NO2 were modified by Zr4+. The observed contact angle of biUIO-66–NH2NO2 was 24.3°, which was far smaller than the 35.7° of UIO-66–NH2 owing to the electron-withdrawing NO2-groups instead of the electron-donating NH2-groups (Table S1†).7 The addition of BDC–NO2 ligands made the matrix materials of the affinity probe more hydrophilic to capture the target phosphopeptides selectively and specifically. The contact angle was found to be further decreased to 21.0° in the final product biDZMOF indicating that the modification of biUIO-66–NH2NO2 could improve the hydrophilicity of this MOF material and minimize non-specific interactions in the isolation of phosphopeptides from biological samples. The XPS survey spectrum further confirmed the existence of C, O, N, P, and Zr elements in biDZMOF (Fig. 5a).28 The curves of the Zr 3d region (Fig. 5b) could be distinguished into two peaks for Zr 3d5/2 and Zr 3d3/2 located at around 182.79 and 185.41 eV, respectively. Compared with the XPS spectrum of biUIO-66–NH2NO2 (Fig. S2†), the shift of the Zr peak in the direction of high energy was beneficial to capture negatively charged groups such as the phosphoryl groups from phosphopeptides. The hydrophilic biUIO-66–NH2NO2 which is modified with Zr4+ could act as a specific enrichment material for phosphopeptides.
4, it suggested that 30.7% of BDC–NH2 ligands of biUIO-66–NH2NO2 were modified by Zr4+. The observed contact angle of biUIO-66–NH2NO2 was 24.3°, which was far smaller than the 35.7° of UIO-66–NH2 owing to the electron-withdrawing NO2-groups instead of the electron-donating NH2-groups (Table S1†).7 The addition of BDC–NO2 ligands made the matrix materials of the affinity probe more hydrophilic to capture the target phosphopeptides selectively and specifically. The contact angle was found to be further decreased to 21.0° in the final product biDZMOF indicating that the modification of biUIO-66–NH2NO2 could improve the hydrophilicity of this MOF material and minimize non-specific interactions in the isolation of phosphopeptides from biological samples. The XPS survey spectrum further confirmed the existence of C, O, N, P, and Zr elements in biDZMOF (Fig. 5a).28 The curves of the Zr 3d region (Fig. 5b) could be distinguished into two peaks for Zr 3d5/2 and Zr 3d3/2 located at around 182.79 and 185.41 eV, respectively. Compared with the XPS spectrum of biUIO-66–NH2NO2 (Fig. S2†), the shift of the Zr peak in the direction of high energy was beneficial to capture negatively charged groups such as the phosphoryl groups from phosphopeptides. The hydrophilic biUIO-66–NH2NO2 which is modified with Zr4+ could act as a specific enrichment material for phosphopeptides.
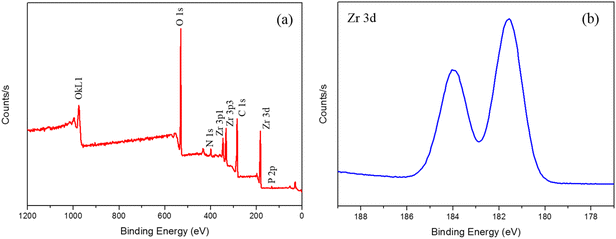 | ||
| Fig. 5 XPS patterns of biDZMOF. (a) Wide scan XPS spectrum and (b) high-resolution XPS spectrum of Zr 3d. | ||
The enrichment of standard phosphopeptides by biDZMOF
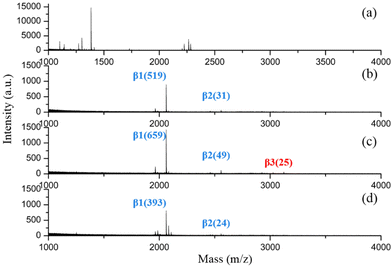
It is reported that MOAC materials favour a stronger interaction with mono-phosphorylated peptides, while IMAC materials prefer to enrich multiply phosphorylated peptides.29,30 It has been an attractive strategy to develop MOAC and IMAC hybrid materials for global phosphopeptide enrichment.13,31,32 However, the application of most of the materials reported for combining the utilization of MOAC and IMAC was limited to MALDI MS analysis due to the instability of the structure. The strongly acidic and basic buffers used in the phosphopeptide enrichment workflow still harmed the delicate structure of the MOAC and IMAC hybrid materials, making the eluate unsuitable with direct injection into a nano-LC-MS/MS system due to the coexistence of phosphopeptides and the disintegration of unstable MOF materials in alkali elution buffer.Fortunately, as the alkali-resistant metal-affinity probe with dual-zirconium centers, biDZMOF, could specifically enrich both mono- and multi-phosphopeptides, the eluted phosphopeptides could be analyzed by nano-LC-MS/MS without desalting. However, the addition of BDC–NO2 ligands improved alkali resistance as well as changed the hydrophilicity of biDZMOF. Special conditions in loading buffers should be optimized for biDZMOF. β-Casein digests were utilized as model phosphopeptides to evaluate the performance of biDZMOF toward phosphopeptide enrichment via MALDI-TOF analysis. As shown in Fig. 6, almost no signal of phosphopeptides was identified in the MALDI-TOF spectra of original β-casein digests, while two mono-phosphopeptides and one multi-phosphopeptide of tryptic digests of β-casein were captured using biDZMOF in the loading buffer with 85% ACN. The conventional concentration of 80% ACN was too low to prevent non-specific peptides.
The sensitivity and selectivity of biDZMOF toward phosphopeptides remain to be investigated since phosphopeptides have low abundance and are often interfered by other peptides in real biological samples. When the concentration of β-casein digests was as low as 0.05 fmol μL−1, two mono-phosphorylated peptides and one multi-phosphopeptide were still identified with a S/N ratio over 3 (Fig. S4†), which suggests the highly efficient enrichment of phosphopeptides by biDZMOF. The tryptic digests of BSA were utilized as interfering peptides for selectivity evaluation. As depicted in Fig. S5,† the main phosphorylated fragments were identified with high S/N ratio even when the molar ratio of β-casein digests to BSA digests was up to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000. These data demonstrate that the biDZMOF has a low limit of detection and excellent anti-interference ability toward capturing phosphopeptides.
5000. These data demonstrate that the biDZMOF has a low limit of detection and excellent anti-interference ability toward capturing phosphopeptides.
Highly specific enrichment of phosphopeptides from human saliva
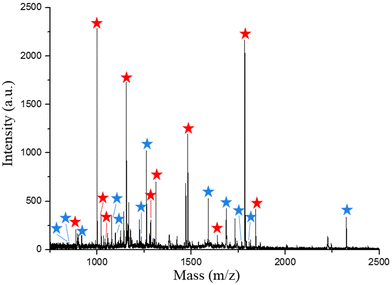
Encouraged by the emerging advantages of biDZMOF, the capture of endogenous phosphopeptides from a human saliva sample which is considered an ideal clinical specimen on account of noninvasive collection was carried out to evaluate the extraction performance in a complex biological sample. The endogenous phosphopeptides in human saliva are at low levels, but could be potential markers in diagnosis and therapy.33 After treating with biDZMOF, the extracted peptides from 10 μL human saliva sample were analyzed by MALDI first. Almost each of the observed major peaks in the mass spectrum (Fig. 7) could be identified to be the phosphopeptides by MALDI-TOF MS/MS (Fig. S6†). As a result, 23 phosphopeptides including 12 mono-phosphorylated peptides and 11 multi-phosphorylated peptides could be extracted and identified from a tiny volume of saliva sample by MALDI-TOF.Compared with other metal-affinity probes which need desalting of the eluate before LC-MS analysis,34 the eluate of biDZMOF could be injected into a nano-HPLC system directly after being acidized with FA and the alkali-resistant biDZMOF successfully prevented the yellow ligands from precipitation at the tip of the capillary column like in the direct injection of the eluate of UIO-66–NH2 (Fig. S7†). Three parallel identifications were implemented and the MS data obtained were analyzed by database search. The MaxQuant software suite was used to analyze the MS data in this protocol. For phosphopeptide identification, a localization probability cutoff was applied. This filter is used to select phosphopeptides with a high confidence level (greater than 0.75) in phosphoresidue identification.35–37 An average number of 148 (133, 158 and 155) unique phosphopeptides could be identified from 10 μL saliva by biDZMOF enrichment in three independent LC-MS/MS analyses (Table S3†). The mean numbers of peptides obtained from three parallel analyses of washing buffer 3 (0.05% (w/w) NH3·H2O) were 22 phosphopeptides and 93 non-phosphopeptides, which indicated the necessity of a weakly alkaline solution to wash the non-phosphopeptides when using biDZMOF. The traditional UIO-66–NH2 would disintegrate in a basic solution with a pH over 9, and even in 0.05% (w/w) NH3·H2O the instability of enrichment materials was still incompatible with a nano LC-MS system. The stability of biDZMOF in basic solution made the majority of phosphopeptides anchored for further analysis.
Notably, compared with the average number of 63 (65, 62 and 64) phosphopeptides obtained from desalting the biDZMOF eluate using the commercial Ti-IMAC material and the average number of 67 (57, 73 and 73) phosphopeptides obtained after enrichment with Ti-IMAC, the increased number of the identified mono- and multi-phosphopeptides in the eluate of biDZMOF which is shown in Fig. 8 demonstrates that the loss of identification coverage of phosphopeptides is avoided and the combined features of MOAC and IMAC are more fully embodied by the enrichment with the alkali-resistant MOF material with dual zirconium centers. In total, 425 phosphopeptides were detected from saliva in all three independent samples after enrichment with biDZMOF, showing its practicality in enriching global phosphopeptides.
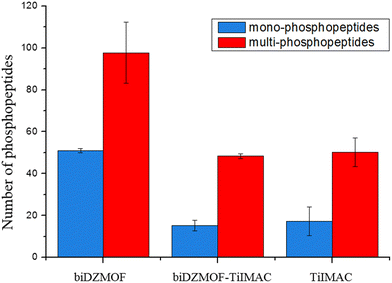 | ||
| Fig. 8 Mono-phosphorylated and multi-phosphorylated peptides identified from saliva after enrichment with biDZMOF, biDZMOF–TiIMAC and TiIMAC. | ||
In addition, a comparison of the reported MOF-based enrichment materials is listed in Table 2. Human saliva is almost always chosen as the real sample for MALDI MS analysis. Even though some hybrid MOF-based materials have shown more phosphopeptides in MALDI spectra than biDZMOF, the spectra of LC-MS analysis with direct injection of biDZMOF eluate could show much more phosphopeptides in a tiny volume of saliva when compared to the limited number of peptides identified in MALDI-MS. Moreover, multi-phosphopeptides made up 62.4% of all identified phosphopeptides from the saliva sample, demonstrating the combined MOAC and IMAC characteristics without the risk of desalting loss, as opposed to the multi-phosphopeptide proportion of the reported MOF-based materials. The above results further demonstrated that the improvement of alkali-resistance of MOF-based phosphopeptide enrichment materials has resulted in excellent performance for global phosphoproteomics analysis.
| MOF enrichment materials | Specificity | LODs (fmol μL−1) | Phosphopeptides from real samples detected by MALDI MS | Pretreatment before nano-LC-MS | Phosphopeptides from real samples detected by nano-LC-MS | Ref. |
|---|---|---|---|---|---|---|
| The symbol “—” means not mentioned in the article. The percentage number means multi-phosphopeptide percentage in the identified phosphopeptides from real samples. | ||||||
| Fe3O4@Hf/Ti-MOF-Gua | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2000 2000 |
0.2 | Human saliva (15) | — | — | 38 |
| mMOF-FBP-Ti4+ | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 500 |
0.2 | Human saliva (14) | — | — | 39 |
| Fe3O4@SiO2@Ce-Zr-MOF@PA | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 800 800 |
0.1 | Human saliva (15) | — | — | 40 |
| Ti4+@Zr-MOF | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
0.1 | Human saliva (25) | Lyophilized | Human saliva (105, 20%) | 41 |
| SiO2@PDA@Zr-MOF | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 1000 |
0.2 | Human saliva (25) | Lyophilized | Human saliva (240, 35%) | 42 |
| Fe3O4@ZIF-8@Zn–Ga LDH | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 5000 |
0.1 | Human saliva (17) | Freeze-dried | Human saliva digest (324, 32.5%) | 43 |
| biDZMOF | 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) : :![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 5000 |
0.05 | Human saliva (23) | Acidified with FA and injected directly | Human saliva (425, 62.4%) | This work |
Conclusions
In summary, a new biligand Zr-MOF biUIO–NH2NO2 has been successfully synthesized, and the addition of BDC–NO2 ligands could obviously enhance the alkali-resistance of biUIO–NH2NO2. The chemical activity of the –NH2 groups on biUIO–NH2NO2 was retained and a further immobilization of Zr4+ was carried out to prepare the biligand dual-metal centered zirconium–organic framework (biDZMOF) for the global enrichment of phosphopeptides. The improved alkali-resistance of the biDZMOF reduced material disintegration in NH3·H2O solution that is an essential elution buffer during the phosphopeptide enrichment workflow. The high sensitivity (0.05 fmol μL−1) and excellent anti-interference ability (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5000 of β-casein
5000 of β-casein![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) BSA) of biDZMOF were achieved in enrichment of global phosphopeptides from biological samples. 425 phosphopeptides in total were detected from 10 μL human saliva after directly coupling the basic eluate with LC-ESI-MS/MS analysis, by avoiding the sample loss of desalting treatment and consequently increasing the identification coverage of both mono- and multi-phosphopeptides. The further application of the biUIO-66–NH2NO2 is believed to be available in sample pretreatments due to its alkali-resistant property.
BSA) of biDZMOF were achieved in enrichment of global phosphopeptides from biological samples. 425 phosphopeptides in total were detected from 10 μL human saliva after directly coupling the basic eluate with LC-ESI-MS/MS analysis, by avoiding the sample loss of desalting treatment and consequently increasing the identification coverage of both mono- and multi-phosphopeptides. The further application of the biUIO-66–NH2NO2 is believed to be available in sample pretreatments due to its alkali-resistant property.
Author contributions
All authors have given approval to the final version of the manuscript.Conflicts of interest
There are no conflicts to declare. Informed consent was obtained from all human subjects.Acknowledgements
The financial support from the National Natural Science Foundation of China (NSFC) (Grant No. 21974138) is greatly acknowledged.References
- J. M. Irish, R. Hovland, P. O. Krutzik, O. D. Perez, O. Bruserud, B. T. Gjertsen and G. P. Nolan, Cell, 2004, 118, 217–228 CrossRef PubMed.
- J. G. Abelin, P. D. Trantham, S. A. Penny, A. M. Patterson, S. T. Ward, W. H. Hildebrand, M. Cobbold, D. L. Bai, J. Shabanowitz and D. F. Hunt, Nat. Protoc., 2015, 10, 1308–1318 CrossRef PubMed.
- R. F. Dreier, E. Ahrne, P. Broz and A. Schmidt, J. Proteome Res., 2019, 18, 493–507 Search PubMed.
- L. P. Li, L. N. Xu, Z. Li, Y. Bai and H. W. Liu, Anal. Bioanal. Chem., 2014, 406, 35–47 CrossRef PubMed.
- J. V. Olsen and M. Mann, Mol. Cell. Proteomics, 2013, 12, 3444–3452 CrossRef PubMed.
- T. E. Thingholm, T. J. D. Jorgensen, O. N. Jensen and M. R. Larsen, Nat. Protoc., 2006, 1, 1929–1935 CrossRef CAS PubMed.
- J. X. Peng, H. Niu, H. Y. Zhang, Y. T. Yao, X. Y. Zhao, X. Y. Zhou, L. H. Wan, X. H. Kang and R. A. Wu, ACS Appl. Mater. Interfaces, 2018, 10, 32613–32621 CrossRef CAS PubMed.
- H. J. Zhou, M. L. Ye, J. Dong, E. Corradini, A. Cristobal, A. J. R. Heck, H. F. Zou and S. Mohammed, Nat. Protoc., 2013, 8, 461–480 CrossRef CAS PubMed.
- J. F. Huang, F. J. Wang, M. L. Ye and H. F. Zou, J. Chromatogr. A, 2014, 1372, 1–17 CrossRef CAS PubMed.
- J. Yao, N. Sun, C. Deng and X. Zhang, Talanta, 2016, 150, 296–301 CrossRef CAS.
- G. Maurin, C. Serre, A. Cooper and G. Fereyd, Chem. Soc. Rev., 2017, 46, 3104–3107 RSC.
- X. Y. Zhu, J. L. Gu, J. Yang, Z. Wang, Y. S. Li, L. M. Zhao, W. R. Zhao and J. L. Shi, J. Mater. Chem. B, 2015, 3, 4242–4248 RSC.
- J. Q. Zhou, Y. L. Liang, X. W. He, L. X. Chen and Y. K. Zhang, ACS Sustainable Chem. Eng., 2017, 5, 11413–11421 CrossRef CAS.
- J. X. Peng, H. Y. Zhang, X. Li, S. J. Liu, X. Y. Zhao, J. Wu, X. H. Kang, H. Q. Qin, Z. F. Pan and R. A. Wu, ACS Appl. Mater. Interfaces, 2016, 8, 35012–35020 CrossRef PubMed.
- N. Zhang, T. Huang, P. S. Xie, Z. Yang, L. Zhang, X. P. Wu and Z. W. Cai, ACS Appl. Mater. Interfaces, 2022, 14, 39364–39374 CrossRef.
- T. E. Thingholm and M. R. Larsen, in Phospho-Proteomics: Methods and Protocols, ed. M. D. Graauw, Humana Press, Totowa, NJ, 2009, pp. 57–66, DOI:10.1007/978-1-60327-834-8_5.
- N. Zhang, N. R. Sun and C. H. Deng, Chem. Commun., 2020, 56, 13999–14002 RSC.
- R. Liu, W. Q. Gao, J. Q. Yang, S. Zhang, C. L. Wang, J. Lin, S. J. Zhang, J. C. Yu and K. Q. Tang, Anal. Bioanal. Chem., 2022, 414, 2251–2263 CrossRef.
- E. X. Chen, M. Qiu, Y. F. Zhang, Y. S. Zhu, L. Y. Liu, Y. Y. Sun, X. H. Bu, J. Zhang and Q. P. Lin, Adv. Mater., 2018, 30, 1704388 CrossRef PubMed.
- C. D. Fast, J. Woods, J. Lentchner and T. A. Makal, Dalton Trans., 2019, 48, 14696–14704 RSC.
- J. Aguilera-Sigalat and D. Bradshaw, Chem. Commun., 2014, 50, 4711–4713 RSC.
- M. Kandiah, M. H. Nilsen, S. Usseglio, S. Jakobsen, U. Olsbye, M. Tilset, C. Larabi, E. A. Quadrelli, F. Bonino and K. P. Lillerud, Chem. Mater., 2010, 22, 6632–6640 CrossRef CAS.
- Z. H. Rada, H. R. Abid, H. Sun and S. Wang, J. Chem. Eng. Data, 2015, 60, 2152–2161 CrossRef CAS.
- Z. H. Rada, H. R. Abid, H. Q. Sun, J. Shang, J. Y. Li, Y. D. He, S. M. Liu and S. B. Wang, Prog. Nat. Sci.: Mater. Int., 2018, 28, 160–167 CrossRef CAS.
- J. Clarkson and W. Ewen Smith, J. Mol. Struct., 2003, 655, 413–422 CrossRef CAS.
- S. Subudhi, S. Mansingh, S. P. Tripathy, A. Mohanty, P. Mohapatra, D. Rath and K. Panda, Catal. Sci. Technol., 2019, 9, 6585–6597 RSC.
- W. Q. Zhang, J. Chen, Y. C. Li, W. X. Yang, Y. D. Zhang and Y. P. Zhang, RSC Adv., 2017, 7, 5628–5635 RSC.
- Y. R. Du, X. Q. Li, X. J. Lv and Q. Jia, ACS Appl. Mater. Interfaces, 2017, 9, 30925–30932 CrossRef PubMed.
- Y. Yao, J. Dong, M. Dong, F. Liu, Y. Wang, J. Mao, M. Ye and H. Zou, J. Chromatogr. A, 2017, 1498, 22–28 CrossRef.
- C. Gao, J. Bai, Y. He, Q. Zheng, W. Ma, Z. Lei, M. Zhang, J. Wu, F. Fu and Z. Lin, ACS Appl. Mater. Interfaces, 2019, 11, 13735–13741 CrossRef.
- D. S. Yang, X. Y. Ding, H. P. Min, B. Li, M. X. Su, M. M. Niu, B. Di and F. Yan, J. Chromatogr. A, 2017, 1505, 56–62 CrossRef CAS PubMed.
- J. W. Wang, Z. D. Wang, N. R. Sun and C. H. Deng, Microchim. Acta, 2019, 186, 1–9 CrossRef.
- H. J. Zheng, J. C. Zhang, J. T. Ma and Q. Jia, ACS Appl. Mater. Interfaces, 2020, 12, 57468–57476 CrossRef CAS.
- A. L. Capriotti, C. Cavaliere, F. Ferraris, V. Gianotti, M. Laus, S. Piovesana, K. Sparnacci, R. Zenezini Chiozzi and A. Laganà, Talanta, 2018, 178, 274–281 CrossRef CAS.
- J. Cox and M. Mann, Nat. Biotechnol., 2008, 26, 1367–1372 CrossRef CAS PubMed.
- C. M. Potel, M. H. Lin, A. J. R. Heck and S. Lemeer, Nat. Methods, 2018, 15, 187–190 CrossRef CAS PubMed.
- A. Hogrebe, L. von Stechow, D. B. Bekker-Jensen, B. T. Weinert, C. D. Kelstrup and J. V. Olsen, Nat. Commun., 2018, 9, 1045 CrossRef PubMed.
- J. Y. Li, S. Zhang, W. Gao, Y. Hua and H. Z. Lian, ACS Sustainable Chem. Eng., 2020, 8, 16422–16429 CrossRef CAS.
- W. Gao, F. Zhang, S. Zhang, J.-Y. Li and H.-Z. Lian, Sep. Purif. Technol., 2023, 305, 122426 CrossRef CAS.
- S. Yan, B. Luo, J. He, F. Lan and Y. Wu, J. Mater. Chem. B, 2021, 9, 1811–1820 RSC.
- H. Y. Zheng, J. X. Wang, M. X. Gao and X. M. Zhang, Microchim. Acta, 2019, 186, 1–11 CrossRef PubMed.
- H. Lin, H. Chen, X. Shao and C. Deng, Microchim. Acta, 2018, 185, 562 CrossRef PubMed.
- H. Qi, G. Chen and Q. Jia, Talanta, 2022, 247, 123563 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an01604d |
| This journal is © The Royal Society of Chemistry 2023 |