Combining multidimensional chromatography-mass spectrometry and feature-based molecular networking methods for the systematic characterization of compounds in the supercritical fluid extract of Tripterygium wilfordii Hook F†
Boquan
Qu
ab,
Yanfang
Liu
 *ac,
Aijin
Shen
*ac,
Zhimou
Guo
*ac,
Aijin
Shen
*ac,
Zhimou
Guo
 ac,
Long
Yu
ac,
Dian
Liu
a,
Feifei
Huang
c,
Ting
Peng
c and
Xinmiao
Liang
ac,
Long
Yu
ac,
Dian
Liu
a,
Feifei
Huang
c,
Ting
Peng
c and
Xinmiao
Liang
 ac
ac
aKey Laboratory of Separation Science for Analytical Chemistry, Dalian Institute of Chemical Physics, Chinese Academy of Sciences, Dalian 116023, China. E-mail: liuyanfang@dicp.ac.cn; ajshen@dicp.ac.cn
bUniversity of Chinese Academy of Sciences, Beijing 100049, China
cJiangxi Provincial Key Laboratory for Pharmacodynamic Material Basis of Traditional Chinese Medicine, Ganjiang Chinese Medicine Innovation Center, Nanchang 330000, China
First published on 28th November 2022
Abstract
Tripterygium wilfordii Hook F from the family Celastraceae is a traditional Chinese medicine (TCM) whose principal chemical constituents are terpenoids, including sesquiterpene alkaloids and diterpenoids, which have unique and diverse structures and remarkable biological activities. In order to advance pharmacological research and guide the preparation of monomer compounds derived from T. wilfordii, a systematic approach to efficiently discover new compounds or their derivatives is needed. Herein, compound separation and identification were performed by offline reversed-phase × supercritical fluid chromatography coupled mass spectrometry (RP × SFC-Q-TOF-MS/MS) and Global Natural Product Social (GNPS) molecular networking. The 2D chromatography system exhibited a high degree of orthogonality and significant peak capacity, and SFC has an advantage during the separation of sesquiterpene alkaloid isomers. Feature-based molecular networking offers the great advantage of quickly detecting and clustering unknown compounds, which greatly assists in intuitively judging the type of compound, and this networking technique has the potential to dramatically accelerate the identification and characterization of compounds from natural sources. A total of 324 compounds were identified and quantitated, including 284 alkaloids, 22 diterpenoids and 18 triterpenoids, which means that there are numerous potential new compounds with novel structures to be further explored. Overall, feature-based molecular networking provides an effective method for discovering and characterizing novel compounds and guides the separation and preparation of targeted natural products.
Introduction
Natural products have been a significant source of new compounds for a long time, and many novel drugs have been derived from natural sources in recent years.1,2 In comparison to synthetic compounds, natural products display remarkably diverse structures and a broad range of bioactivities.3 Among all approved drugs by the end of 2019, approximately 60% were related to natural products.4 Consequently, exploring natural products is a very effective method to find new compounds, especially those from traditional Chinese medicine (TCM), which has the characteristics of a complex material basis and structural diversity.5 The traditional method finds new compounds through step-by-step preparation in an untargeted manner, which greatly limits the efficiency of new compound discovery.In addition to its high sensitivity, wide application range, and high separation efficiency, LC-MS/MS is broadly used for the separation and characterization of complex multicomponent systems.6–9 This strategy is effective for discovering new compounds in TCMs, especially those present in low abundance, as TCMs typically contain a large number of new compounds. Currently, the applied technologies include one-dimensional liquid chromatography-mass spectrometry (1DLC-MS) and multidimensional liquid chromatography-mass spectrometry (MDLC-MS). Conventional one-dimensional separation methods have significant methodological limitations in chromatographic separation and peak capacity.10 Two-dimensional liquid chromatography is an effective method based on integrating different separation mechanisms to improve peak capacity and resolution.11 Compared with online 2D chromatography, the advantages of offline 2D chromatography systems include their compatibility with solvents and ease of use, which makes them a powerful tool for research into natural products.12 It is worth mentioning that SFC has the significant advantages of low viscosity, high separation efficiency, and environmental friendliness because of the use of supercritical CO2 as the mobile phase.13 More importantly, SFC techniques have been applied to separate natural products and isomers14 with unique advantages for the separation of isomers due to its great selectivity difference from the traditional reversed-phase chromatography, leading to a high degree of orthogonality by the RP × SFC two-dimensional system.15
Molecular networking (MN) has become a novel strategy based on GNPS, which consists of visualization and annotation of the chemical space information from the nontargeted mass spectrometry data.16 The GNPS infrastructure includes a feature-based molecular network (FBMN) that uses chromatographic feature detection and alignment tools, the representative MS/MS spectra are obtained by averaging the MS/MS spectra from the MS features extracted in FBMN. The method clusters molecular ions which have similar fragmentation patterns to create molecular networks, which, combining artificial fragment analysis with natural product discovery is helpful in finding new natural products.17 The detailed information of components can be quickly obtained according to the clustering, which is widely applied in the broad areas of biologics, pharmaceutical analysis, metabolomics, and drug discovery research.18 FBMN has played an important role in targeting the separation of natural products in recent years, particularly for isolating new compounds and new chemical skeletons with distinct and important biological activities.19–21 This allows recommendations to be made to help guide and prioritize research,22 and unknown compound types can be used as preparation targets.
Tripterygium wilfordii Hook F (T. wilfordii) belongs to the family Celastraceae, which is widely distributed in southeast China and East Asia,23 has been reported to exhibit a variety of bioactivities and pharmacological effects. For example, triptolide exhibits high anti-inflammatory and antitumor properties,24,25 and the sesquiterpene alkaloids triptonines A and B have been suggested to have high inhibitory activity against HIV transcription and replication.26 Celastrol has significant functional effects on modulating inflammation, autoimmune disease, and the antitumor immune response and reducing obesity.27,28 Importantly, celastrol is one of five traditional natural medicines with the potential for modern molecular drug development.29 Despite the importance of its extensive pharmacological activities, there remains a paucity of evidence on its toxic mechanisms of action, which limits its clinical applications to a great extent;30 consequently, it is essential to understand the chemical composition of celastrol and to explore its new compounds. Using mass spectrometry, more than 100 compounds can be simultaneously identified at present.31 For revealing the mechanism of drug action and discovering new bioactive molecules, a new strategy for discovering and characterizing components from T. wilfordii is needed.
Herein, an offline RP × SFC tandem mass spectrometry strategy was developed. The supercritical fluid extract of T. wilfordii was systematically characterized and identified through feature-based molecular networks. This study contributes to the discovery of novel compounds from a TCM and clarifies the composition of its substance matrix, while providing strong and efficient technical support for the development of targeted natural product separation.
Experimental
Chemicals and apparatus
T. wilfordii was purchased from a local market in Hubei, China. HPLC grade methanol and acetonitrile were purchased from Fisher Company (Thermo Fisher Scientific (China) Co., Ltd, Shanghai, China), and LC-MS grade acetonitrile (ACN), methanol, and formic acid were also purchased from Fisher Company. Anhydrous ethanol and methanol were obtained from Shanghai Titan Scientific Co., Ltd, Shanghai, China. N-Hexane, isopropanol, and dichloromethane were purchased from Tianjin Kemiou Chemical Reagent Co. Ltd, China. Ultra-pure water was prepared with a Milli-Q ultrapure water system (Merck Millipore, Milford, MA, USA). High-purity carbon dioxide was purchased from East China Specialty Gases Co., Ltd, Jiangxi Province, China. Standards of Wilforgine (CAS: 37239-47-7, 99.0% purity), Triptophenolide (CAS: 74285-86-2, >99.0% purity) and Corydaline (CAS: 518-69-4, >99.0% purity) were purchased from Chengdu Pusi Biotechnology Co., Ltd (Sichuan, China).For chromatography, we utilized the following columns: C18-HD (4.6 mm × 250 mm, 10 μm, Acchrom, China); C18-HD (2.1 mm × 100 mm, 3.5 μm, Acchrom, China); Silica gel (4.6 mm × 250 mm, 10 μm, Acchrom, China); X5H (4.6 mm × 250 mm, 10 μm, Acchrom, China); NH2 (4.6 mm × 250 mm, 10 μm, Acchrom, China); XAmide (4.6 mm × 250 mm, 10 μm, Acchrom, China); Torus 2-PIC (4.6 mm × 250 mm, 5 μm, Waters, USA) and 2-EP (4.6 mm × 250 mm, 5 μm, Waters, USA).
Sample preparation
Supercritical carbon dioxide extraction of T. wilfordii (pulverized and screened through 50-mesh) was performed with a SFE220–50–6L supercritical CO2 extraction apparatus (Jiangsu Gaoke Pharmaceutical Equipment Co. Ltd, Jiangsu, China). The extraction parameters were as follows: the extraction pressure was set to 30 MPa, the extraction temperature was maintained at 50 °C, 10% anhydrous ethanol was chosen as a modifier, and the extraction time was 20 h. The obtained extracts were pretreated by normal-phase chromatography LC6000 (Jiangsu Hanbang Science and Technology Co., Ltd, Jiangsu, China) with a silica gel column (50 mm × 310 mm, 10 μm) to remove the nonpolar components. The mobile phase for elution was solvent A (dichloromethane) and B (methanol) with the following gradient: 0–18.6 min 100% (A), 18.7–43.4 min 98% (A), 43.5–55.8 min, 95% (A), 55.9–68.2 min, 90% (A), and 68.3–80.6 min, 10% (A) with a flow rate of 82.7 mL min−1. The detection wavelengths were 230 nm and 268 nm. To further remove the nonpolar components and improve solubility, reversed-phase C18YE packing (synthesized and saved by our laboratory) was used to treat samples in a proportion of 9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 followed by transfer to an SPE column. The mixed samples were eluted with 70% acetonitrile and the eluate was evaporated to dryness. The residue was redissolved in 60% acetonitrile and used for subsequent follow-up experiments.
1 followed by transfer to an SPE column. The mixed samples were eluted with 70% acetonitrile and the eluate was evaporated to dryness. The residue was redissolved in 60% acetonitrile and used for subsequent follow-up experiments.
Offline 2D by RP × SFC-Q-TOF-MS/MS conditions
For this work, we established an offline 2D chromatography system for the separation of T. wilfordii extracts. Specifically, first dimensional separation was performed using a C18HD column with a flow rate of 0.7 mL min−1, and HPLC analyses were performed on a Waters 2695 HPLC system (Waters, Milford, MA, USA) equipped with a Waters 2998 photodiode array detector (PDA) and a Waters e2695 separation module. The mobile phase consisted of A (acetonitrile/0.1% formic acid) and B (water/0.1% formic acid), and gradient elution was performed as follows: 0.0–8.0 min, 60% (A), 8.1–18.0 min, 63% (A), 18.1–25.0 min, 80% (A), 25.1–40.0 min, and 95% (A). A total of ten fractions were collected and analyzed in the following experiments.We used supercritical fluid chromatography (SFC), which is based on different separation modes for second dimensional separation. Supercritical fluid chromatography was performed using a 1260 Infinity analytical system (Agilent Technologies Co., Ltd, USA) coupled with a PDA detector. A 1260 Infinity SFC control module was used to separate the components of the sample, and the BPR temperature and pressure were set to 60 °C and 150 bar, respectively. The column was used a Torus 2-PIC, and the mobile phases included mobile phases A (Supercritical Fluid CO2) and B (methanol) at a flow rate of 2.0 mL min−1. Gradient elution was performed as follows: 0.0 min–6.0 min, 98%–80% (A), 6.0 min–8.0 min, 80%–70% (A), 8.0 min–11.0 min, 70%–60% (A), and 11.0 min–13.0 min, 60% (A). The postcolumn compensation solution was pure methanol pumped by an Agilent 1260 isocratic pump at a flow rate of 0.2 mL min−1. For the above separations, the column temperature was maintained at 30 °C, and the UV detector was set to 230 nm and 268 nm.
In the second dimension, the SFC system was coupled to an Agilent 6540 Q-TOF spectrometry system (Agilent Technologies Co., Ltd, USA) equipped with an electrospray ionization interface. For MS detection, the ESI† source was operated in positive ion mode to acquire MS/MS spectra because it showed a good response. The ESI-Q-TOF-MS/MS detection conditions were optimized as follows. High-purity nitrogen was used as both the nebulizer and drying gas, and the drying gas temperature was 320 °C at a flow rate of 8 L min−1 and a pressure of 35 psi. Additionally, the capillary voltage was 3500 V, the collision energy was 40 eV and 20 eV, the fragmentor voltage was 175 V, the skimmer voltage was 65 V, and the octopole 1 RF voltage was 750 V. The MS and MS/MS ranges of data acquisition were 100–1200 Da. MassHunter 10.0 software (Agilent) was used for data acquisition and to control the Q-TOF mass spectrometry system.
Peak capacity and orthogonality evaluation
The orthogonality evaluation was carried out using a geometric approach. Briefly, orthogonality is defined as the normalized area occupied by data points in the two-dimensional separation space.32 The retention times of the peaks within the two-dimensional plot were first transformed into normalized values according to eqn (1), and then a division of the rectangle in the planar two-dimensional drawing was determined according to eqn (2). Finally, the orthogonality (O) and practical peak capacity (NP) were calculated from eqn (3) and (4).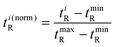 | (1) |
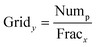 | (2) |
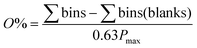 | (3) |
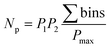 | (4) |
Feature-based molecular networking design
A feature-based molecular network (FBMN) for T. wilfordii was generated using the online platform at Global Natural Products Social Molecular Networking (GNPS) (https://gnps.ucsd.edu). The raw mass spectrometry data were converted to *.abf format with ABF (Analysis Base File) converter 1.3 (https://www.reifycs.com/AbfConverter/) and then converted from *.abf format to *.mgf format with MS-DIAL Software 4.70 (https://prime.psc.riken.jp/Metabolomics_Software/MS-DIAL/). The main MS-DIAL parameters were as follows: positive ionization mode was used, the data type was selected centroid data, the MS1 and MS2 tolerances were set as 0.01 Da and 0.025 Da, respectively, the mass slice width was set to 0.1, the identification score cutoff was set to 85%, the retention time tolerance was set to 0.1 min, and the accurate mass tolerance was set to 0.01 Da. For ions and adducts, we retained the [M]+, [M + H]+, and [M + Na]+ peaks. Finally, the *.mgf files were uploaded to the GNPS, and the network parameters were set as follows: minimum pairs cosine of 0.70, fragment ion mass tolerance of 0.02, network TopK of 10, minimum matched fragment ions of 6, and minimum cluster size of 2. The output from feature-based molecular networking was visualized and analyzed with Cytoscape software 3.8.2 (https://cytoscape.org/).Method validation
Methanol (LC-MS grade) was used for the preparation of the standard compound, the concentrations of Wilforgine, Triptophenolide and Corydaline were 1.66 mg mL−1, 1.98 mg mL−1, and 1.02 mg mL−1, respectively. A series of calibration curves were constructed by diluting the standard solution with methanol to yield a series of standard solutions at appropriate concentrations. An analytical curve and linear correlation coefficient (R2) have been determined from linear regression. A signal-to-noise method was used to determine the limit of detection (LOD) and limit of quantitation (LOQ) sensitivity, and these were expressed by the measurements at certain signal-to-noise ratios (S/N = 3 and 10). To calculate intra- and inter-day precisions, repeated analyses were conducted on the same day (intra-day precision) and three days apart (inter-day precision). Through the use of the standard addition method, recovery experiments at three different levels were conducted to validate the method's accuracy.Results and discussion
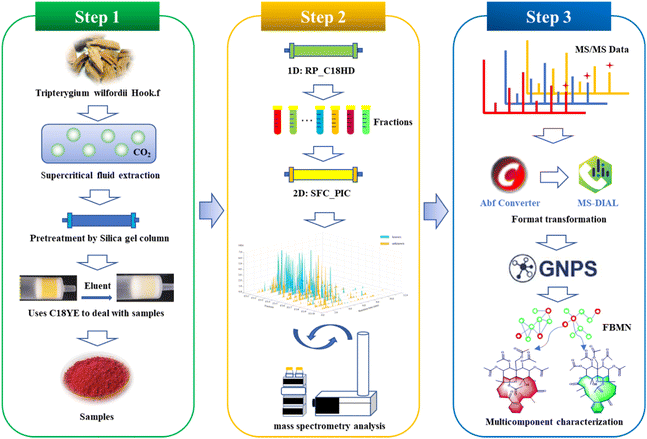
T. wilfordii contains mainly sesquiterpene alkaloids, diterpenes, and triterpenoids with medium and weak polarity, structural diversity and additional isomers. To better determine and systematically characterize the main components compounds in this vine, we developed the following workflow (Fig. 1). First, the samples were extracted with supercritical fluid and pretreated with silica gel. Second, the offline RP × SFC two-dimensional orthogonal system with tandem mass spectrometry was constructed to obtain sample information. Finally, identification of the compounds was completed by combining feature-based molecular networking and fragment information from mass spectrometry.Sample extraction and pretreatment
Supercritical fluid extraction (SFE) is environmentally friendly and highly efficient, making it suitable for extracting weakly or moderately polar ingredients after adding modifiers.33T. wilfordii was extracted with supercritical carbon dioxide mixed with 10% anhydrous ethanol. To remove the nonpolar components from the extract, normal-phase chromatography was used for pretreatment, and four different types of stationary phases were evaluated for column screening. The results were shown in Fig. S1.† Based on the comparison, a superior separation effect was observed with silica gel, and irreversible adsorption behaviors between the gel and sample were not observed during SPE experiment. The sample was divided into a total of 4 fractions from the silica gel column (Fig. S2†), and the RP-UHPLC analysis result was shown in Fig. S3.† These findings show that the main, moderately polar components are concentrated in Fr. 3, so Fr. 3 was selected for subsequent experiments.Offline two-dimensional system
Retention behaviors are significantly different between reversed-phase liquid chromatography and supercritical fluid chromatography; therefore, the combination 2D RPLC and SFC system was considered to achieve high orthogonality and improve peak capacity and separation selectivity.12In comparison to SFC, RPLC has the clear advantage of wide sample applicability and high stability during the preparation of natural products, making it easier to operate and scale up; thus, RPLC was selected as the first dimensional separation mode to better guide the separation of natural products. In order to improve the retention of samples with medium polarity and weak polarity, the high-density bonded stationary phase C18HD was selected during the development of the method in the first dimension. A total of 10 fractions (Fig. S4†) were collected to establish the second dimensional method. Separating isomers and chiral compounds with SFC is superior to high-pressure liquid chromatography.34,35 To achieve better separation of the isomers in T. wilfordii, we used SFC in the second dimension, and the effects of 2-aminomethylpyridine (2-PIC) and 2-ethylpyridine (2-EP) columns were investigated. The results showed that the 2-PIC column provided better separation (Fig. S5†). The separation of T. wilfordii was shown in Fig. 2a.
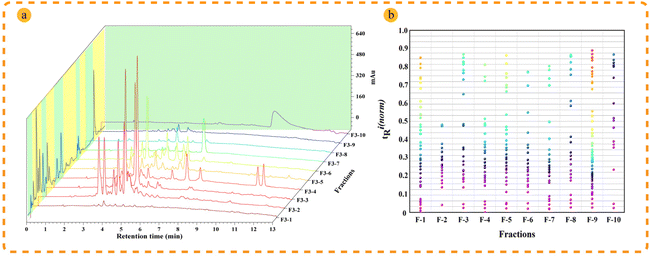 | ||
| Fig. 2 Orthogonality evaluation of the established 2D chromatography system. (a) Separation effect of T. wilfordii extract in 2D RP × SFC system; (b) 2D separation orthogonality plan of RP × SFC. | ||
Orthogonality evaluation
Orthogonality and peak capacity are important parameters that affect the separation ability in multidimensional chromatography. According to the procedure in the section “Peak capacity and orthogonality evaluation”, the following results were obtained and were shown in Fig. 2b. The orthogonality was 80.95%, and the practical peak capacity was 1941. In addition, SFC also has clear advantages in the separation of isomers. The sesquiterpene macrocyclic lactone alkaloids peritassine A and euonymine with the same molecular weight were successfully separated and identified from the same fraction (Fr. 3-2) as shown in Fig. 6a and b. These results demonstrated that the established method had great separation ability and selectivity.Annotation of the compounds in the feature-based molecular network
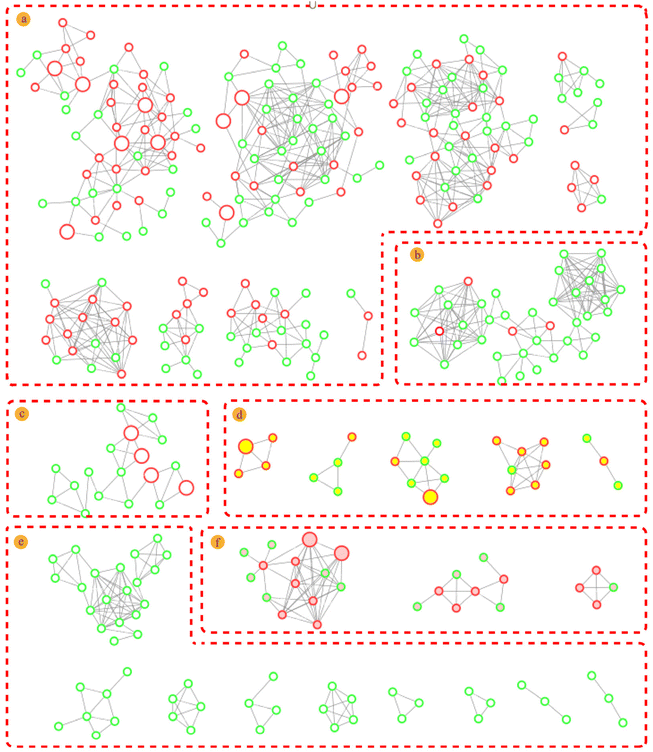
Molecular networks are constructed based on similarities in MS/MS fragmentation data. The same types of compounds can be clustered together and resolved by annotating the known compounds.36The mass spectrometry data from 10 fractions were collected by SFC-Q-TOF-MS/MS and visualized as feature-based molecular networks. The results were shown in Fig. 3. The nodes in the FBMN can be analyzed and commented on to obtain information on retention times and mass spectra of compounds within each node, and match the molecular weight of the compound reported in the literature. Compounds corresponding to this node will be identified as known compounds when the matching error is less than 20 ppm. Additionally, the FBMN has a representative MS/MS spectrum that can be used to identify isomers, by extracting the MS/MS fragment information of the node and combining the mass spectrum fragmentation rules of the compounds reported in the literature, it is possible to deduce the fragmentation process of identified compounds. This molecular network contains 28 clusters (nodes ≥ 3). The nodes with red edges represent known compounds and nodes with green edges represent unknown compounds. The white-filled nodes are alkaloids, yellow-filled nodes are diterpenes and pink-filled nodes are triterpenoids. Then, the mass spectrometry data of each cluster were analyzed, and there was a certain similarity between the MS/MS fragmentation patterns and cleavage rules. For example, these compounds all had the same basic skeleton and similar neutral loss and fragmentation pathways. The detailed process is discussed in the next section. In Part a, we identified these clusters as sesquiterpene macrolide alkaloids, including those with new molecular weights. It can be intuitively concluded that these alkaloids are among the most abundant in T. wilfordii, showing an advantage of the application of molecular networks in natural product research. Part b was identified as a non-macrolide sesquiterpene alkaloid, which is another type of highly abundant alkaloid contained within T. wilfordii. The diversity of these compounds depends on the positions of the connected groups and the complexity of the substituents. There were also distinct differences in the basic frameworks between the two types of alkaloids. Other types of alkaloids are shown in Part c. They have low molecular weights and relatively simple structures compared with the sesquiterpene alkaloids. According to the same identification process, Part d and Part f were identified as diterpenes and triterpenoids, respectively. Some nodes were not matched to known compounds in the GNPS database and related literature, and these compounds had low abundances. We divided these compounds into Part e. The information for the main cluster was shown in Fig. S7.† Compiling these new molecular weights and different MS/MS fragmentation data, we speculate that Part e may be a class of compounds with a new skeleton structure.
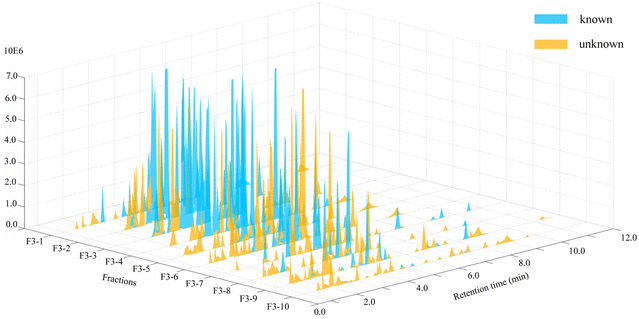
To more intuitively show the information for the compounds in each fraction, the processed raw EIC data were visualized and exported as a four-dimensional (4D) data plot (the fractions in 1-D as the x-axis, 2D retention time as the y-axis, peak intensity as the z-axis, and known and unknown of compounds denoted by peak color) using MATLAB 6.5, and the results were shown in Fig. 4. A total of 421 peaks with molecular weight distributions in the range of m/z 100–1200 are listed and showed good separation. From the 4D diagram, it can be seen that the compounds with the new molecular weights were mainly present in Fr. 3-8–Fr. 3-10, and there is a high probability of finding new compounds in these three fractions. Herein, the targeted separation priority for finding new molecular weight compounds was established, which has important guiding significance for the separation and purification of novel compounds of T. wilfordii. Overall, we were able to unequivocally determine the types of compounds, and 324 compounds were tentatively identified, including 284 alkaloids, 22 diterpenoids and 18 triterpenoids. The information on these compounds was listed in Table S1.† A large number (202) of new molecular weights were found among the various types of compounds from the molecular networks as shown in Fig. 3. These data show that there are still many unknown compounds in T. wilfordii to be explored.
The solubilities of the components in a sample differ widely depending on the extraction method. Most of the substances in T. wilfordii have polarities that range from medium to weak. Supercritical fluid extraction technology uses nonpolar carbon dioxide as the extraction solvent and anhydrous ethanol as the modifier. The extraction capacity of components with medium to weak polarity can be greatly improved by this method. Through silica gel column pretreatment, the main components can be separated, and the trace components can be further enriched. In this work, we found a large number of new compounds, some of which may potentially have novel structures. The results showed that supercritical fluid extraction technology had a very significant advantage for the extraction of terpenoids from T. wilfordii.
Compound identification by MS/MS
Eighteen compounds were tentatively identified and confirmed by MS/MS fragment ions and the GNPS database, including alkaloids and terpenoids, which were shown in Fig. 5. Detailed information on these compounds was listed in Table 1. Additionally, the fragmentation patterns of these compounds were discussed, which is helpful for their identification.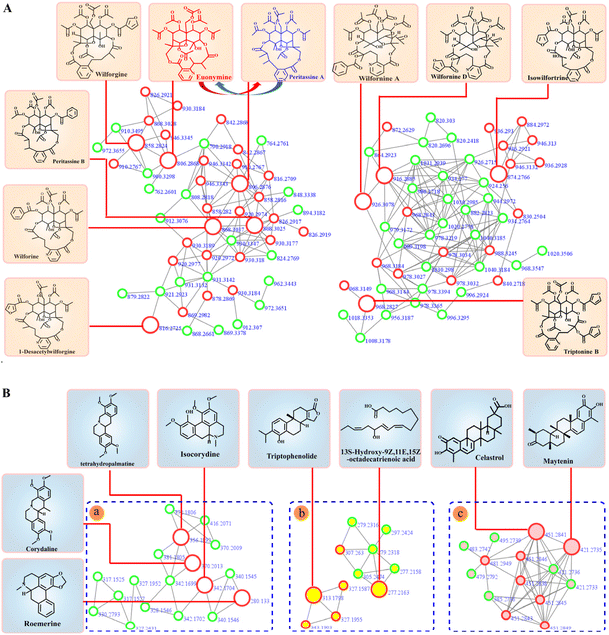 | ||
| Fig. 5 Identification of T. wilfordii compounds by FBMN annotation. (A) Sesquiterpene alkaloids; (B) other types of compounds: (a) other types of alkaloids; (b) diterpenes; (c) triterpenoids. | ||
| No. | Compounds | Molecular formula | Molecular mass | Observed m/z | Error (ppm) | MS/MS fragments | Identification |
|---|---|---|---|---|---|---|---|
| a The compound 17 parent ion matched in GNPS database is 295.2228, the actual parent ion obtained is 277.2163, we speculate that it formed [M + H − H2O]+. | |||||||
| 1 | Wilforgine | C41H47NO19 | 858.2815 | 858.2880 | 7.57 | 840.2772, 798.2663, 746.2179, 738.2447, 686.2445,626.2195, 206.0819, 188.0705, 178.0872, 160.0779, 132.0810, | 37 |
| 2 | 1-Desacetylwilforgine | C39H45NO18 | 816.2709 | 816.2711 | 2.45 | 798.2600, 756.2487, 724.2540, 686.2435, 644.2328, 584.2112, 206.0806, 178.0859 | 37 |
| 3 | Euontmine | C38H47NO18 | 806.2865 | 806.2867 | 2.48 | 788.2759, 746.2649, 686.2440, 626.2225, 566.2004, 206.0806, 188.0703, 178.0859 | 37 |
| 4 | Peritassine A | C38H47NO18 | 806.2865 | 806.2865 | 0.00 | 788.2759, 778.2915, 764.2748, 686.2440, 626.2225, 421.1491, 361.1275, 259.0955, 206.0808, 160.0755 | 37 |
| 5 | Wilfornine D | C43H49NO21 | 916.2869 | 916.2880 | 1.20 | 888.2904, 874.2754, 804.2710, 684.2280, 624.2046, 316.0805, 204.0652, 176.0707 | 37 |
| 6 | Wilfornine A | C45H51NO20 | 926.3077 | 926.3069 | −8.64 | 898.3123, 880.3108, 804.2705, 744.2486, 684.2283, 642.2177, 326.1017, 204.0652 | 37 |
| 7 | Celastrol | C29H38O4 | 451.2842 | 451.2839 | −6.65 | 405.2781, 257.1532, 201.0907, | 38 |
| 8 | Isowilfortrine | C41H47NO20 | 874.2764 | 874.2751 | −1.49 | 856.2652, 846.2817, 814.2550, 754.2333, 694.2120, 674.2439, 204.0653, 176.0705 | 37 |
| 9 | Peritassine B | C43H49NO18 | 868.3022 | 868.3024 | 2.30 | 850.2929, 840.3066, 808.2809, 766.2701, 748.2603, 688.2385, 644.2358, 421.1481, 361.1265, 213.0901 | 37 |
| 10 | Wilforine | C43H49NO18 | 868.3022 | 868.3067 | 5.18 | 850.2954, 840.3118, 822.7811, 746.2671, 718.2776, 686.2505, 218.0432, 176.0717 | 37 |
| 11 | Triptonine B | C46H49NO22 | 968.2818 | 968.2818 | 0.00 | 940.2865, 922.2763, 856.2658, 833.2553, 814.2554, 754.2341, 694.2120, 796.2447, 676.2026, 316.0815, 260.0924, 204.0655, 186.0547, 158.0598 | 37 |
| 12 | Isocorydine | C20H23NO4 | 342.1667 | 342.1695 | 8.18 | 327.1727, 311.1265, 296.1043, 279.1011, 264.0783, 249.1052, 178.0856, 165.1015, | GNPS |
| 13 | Roemerine | C18H17NO2 | 280.1293 | 280.1320 | 9.64 | 265.0842, 249.0909, 237.0886, 217.0795, 191.0847, 147.0913, 121.0758 | GNPS |
| 14 | Corydaline | C22H27NO4 | 370.1974 | 370.2003 | 7.83 | 354.1692, 340.1538, 322.1443, 218.1174, 192.1218, 177.0781, 165.1518, 150.0609 | GNPS |
| 15 | Tetrahydropalmatine | C21H25NO4 | 356.1807 | 356.1867 | 16.85 | 341.1615, 192.1019, 165.0908, 150.0671 | GNPS |
| 16 | Triptophenolide | C20H24O3 | 313.1759 | 313.1798 | 12.45 | 295.1684, 271.1322, 253.1220, 235.1116, 225.1270, 213.1267, 183.0798 | GNPS |
| 17a | 13S-Hydroxy-9Z,11E,15Z-octadecatrienoic acid | C18H30O3 | 295.2228 | 277.2150 | 10.11 | 262.1398, 259.2039, 249.1053, 234.0418, 223.0695, 214.0906, 201.0452 | GNPS |
| 18 | Maytenin | C28H36O3 | 421.2698 | 421.2736 | 9.02 | 243.1437, 215.1060, 201.0904, 189.0620 | GNPS |
Sesquiterpene alkaloids
The cleavage pathways of five different subtypes of sesquiterpene alkaloids were deduced in this study, including wilfordate, iso-wilfordate, evoninate, iso-evoninate and hydroxy-wilfordate. Sesquiterpene macrolide alkaloids exist mainly in two forms: neutral molecule losses and ring-opening bond scission. Different subtypes of sesquiterpene alkaloids create different fragment ions, and the major fragment ions investigated in this study were primarily above m/z 550 and below m/z 300.37 The iso-evoninate-type alkaloid Peritassine A and evoninate-type alkaloid Euonymine are isomers in which the nitrogen atoms are in the ortho and meta positions of pyridine dicarboxylic acid, respectively; these compounds were isolated from the same fraction (Fig. 6a and b). In positive ion mode, the mass spectrum of Peritassine A revealed a molecular ion peak at m/z 806.2865 ([M + H]+). The MS/MS fragmentation ions were predominantly of neutral losses from the parent ion, and the parent ion could also be dehydrated to form m/z 788.2759 ([M + H − H2O]+) or decarboxylated to form m/z 778.2915 ([M + H − CO]+) during collision cleavage. Acetyl groups can also be continuously lost to yield the fragments m/z 688.2440 ([M + H − 2AcOH]+) and m/z 626.2225 ([M + H − 3AcOH]+). Euonymine easily lost acetyl groups from the [M + H]+ quasi-molecular ion m/z 806.2867 in a stepwise manner to form the fragments m/z 746.2649, m/z 686.2400, m/z 626.2225, and m/z 566.2004. Ring fracture produces fragments of these two compounds that differ from each other. The fragment ion of peritassine A at m/z 481.1695 yielded the fragment m/z 421.1491 (B+) by losing the acetyl group and then forming the fragment ions [B+ − AcOH]+ and [B+ − 2AcOH − C2H2O]+ at m/z 361.1275 and m/z 259.0955. The fragment ion at m/z 206.0808 (A+) from pyridine dicarboxylic acid also produced the fragment ion m/z 160.0755 ([M + H − HCOOH]+) by losing the formyl group. The fragment ion of Euonymine between m/z 200 and m/z 500 was too low to be detected. However, the peak at m/z 206.0806 (A+) was dehydrated to produce the fragment ion m/z 188.0703 ([M + H − H2O]+) and also lost a formyl group to form a peak at m/z 178.0859 ([M + H − HCOOH]+). These fragmentation pathways were shown in Fig. 6a and b. These results fully illustrate that the position of the nitrogen atom affects the detected mass spectrometry fragments; therefore, the position of the nitrogen atom on the parent ring can be determined according to the response intensity of the fragment ions.37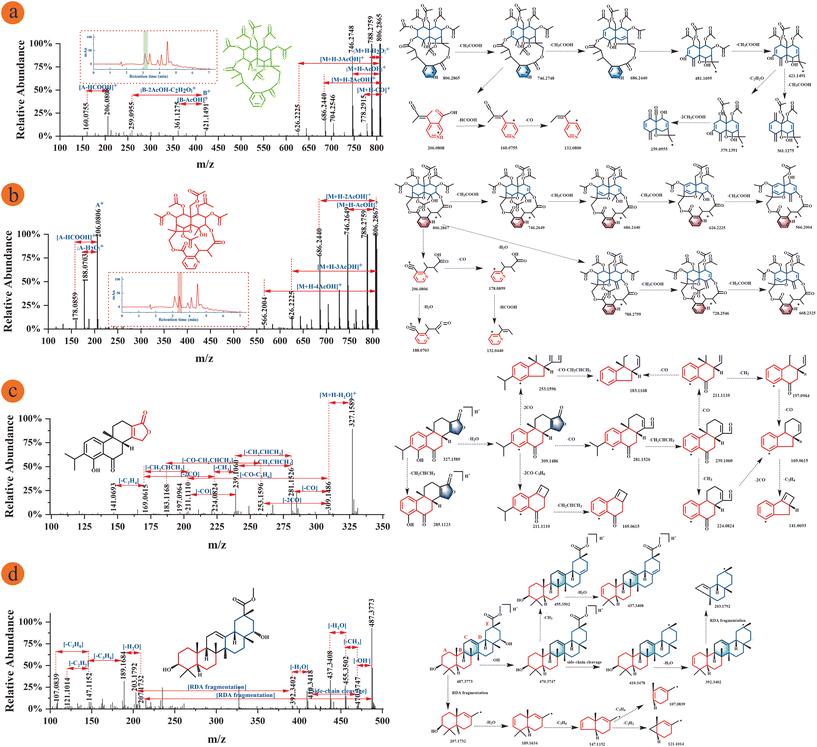 | ||
| Fig. 6 MS/MS fragments information and fragmentation pathway of T. wilfordii compounds. (a) Peritassine A, tR1 = 3.108 min; (b) euonymine, tR2 = 3.408 min; (c) triptonolide; (d) regelindiol B. | ||
Wilfornine A and Wilfornine D are hydroxy-wilfordate-type alkaloids with similar chemical structures that will produce the same fragments in their mass spectra (Fig. S8a and b†). The Wilfornine A molecular ion peak at m/z 926.3069 [M + H]+ produced a fragment ion at m/z 804.2705 ([M + H − BzOH]+) by losing the benzoyl group, the fragment ion at m/z 804.2705 yielded fragments at m/z 744.2486 ([M + H − AcOH]+), m/z 702.2383 ([M + H − AcOH − C2H2O]+), m/z 684.2283 ([M + H − 2AcOH]+), m/z 642.2177 ([M + H − 2AcOH − C2H2O]+), and m/z 582.1970 ([M + H − 3AcOH − C2H2O]+) by sequentially losing acetyl groups and C2H2O. Macrocycle cleavage also occurred to form fragment ions at m/z 326.1017 (A+) and m/z 204.0652 ([A+ − BzOH]+). Both compounds have the same substituent on the sesquiterpenoid skeleton, which may account for their almost identical mass fragments. The fragmentation pathways were shown in Fig. S7a.† The Wilfornine D parent ion at m/z 916.2880 ([M + H]+) lost a furanoyl group to yield fragment m/z 804.2710 ([M + H − FuOH]+), and then the acetyl group was sequentially lost from m/z 804.2710 to form m/z 744.2490 ([M + H − AcOH]+), m/z 684.2280 ([M + H − 2AcOH]+), and 624.2048 ([M + H − 3AcOH]+). Alternatively, the parent ion also yielded a fragment at m/z 874.2755 ([M + H − C2H2O]+) by losing C2H2O. Wilfornine D also underwent macrocycle cleavage to form a fragment ion at m/z 316.0805 (A+) and then gave the fragment m/z 204.0605 ([M + H − FuOH]+) by losing the furanoyl group and formed the peak at m/z 176.0707 ([M + H − CO]+) by decarbonylation. The fragmentation pathways were shown in Fig. S8b.† In summary, macrolide sesquiterpene alkaloids show a degree of structural similarity, and different alkaloid subtypes exhibit certain regularity during their cleavage which is beneficial for accurate and comprehensive guide to structurally identify new compounds.
Diterpenoids
Diterpenes are composed of a lactone ring, a hydroxyl group, and an isopropyl group, and these compounds typically undergo neutral loss, resulting in characteristic fragments, such as H2O (18 Da), CO2 (44 Da), and CO (28 Da). Among them, CH2CHCH3 (42 Da) is characteristic of the loss of isopropyl groups from C-13 sites.31The Triptolide parent ion at m/z 327.1589 ([M + H]+) formed fragment ion 285.1123 by losing the isopropyl group and produced a fragment ion at m/z 309.1486 after dehydration. Fragment ions at m/z 281.1526 and m/z 239.1060 were formed by removing carbonyl and isopropyl groups, respectively. The fragment ion at m/z 239.1060 produced the fragment ion m/z 169.0615 by decarbonylation and the loss of a methylene, and these same fragment ions can also be formed by the loss of methyl and carbonyl groups. Then, the fragment ion at m/z 141.0693 was formed by the loss of a vinyl group. The fragment ion at m/z 309.1486 formed other fragment ions by removing carbonyl and isopropyl groups. The fragmentation pathways were shown in Fig. 6c. The Neotriptophenolide molecular ion peak at m/z 343.1898 ([M + H]+) produced fragment ions at m/z 281.1307 and m/z 239.1420 by losing the –H2O–CO2 and isopropyl groups or formed the fragment ions at m/z 327.1735 and m/z 313.1433 by the losing CH4 and CH2. In this case, the loss of the methoxy group results in the formation of a fragment ion at m/z 283.1317. Next, the fragment ion at m/z 283.1317 formed fragment ions at m/z 237.1263, m/z 169.1012 and m/z 141.0536 through decarbonylation, dehydration and the loss of isopropyl. These fragmentation pathways were shown in Fig. S8c.†
Triterpenoids
Triterpenoids are mainly composed of carboxyl, hydroxyl and carbonyl groups. In positive ion mode, neutral loss occurs by the loss of CO (28 Da), CH3 (15 Da), CH2 (14 Da), and CH4 (16 Da). If the C ring contains double bonds, retro Diels–Alder (RDA) cleavage occurs. If the C ring lacks double bonds, the main method of cleavage is C ring fracture. Since most structures contain carboxyl groups, they tend to lose HCOOH (46 Da) in positive ion mode and produce characteristic fragment ions.31The molecular structure of Regelindiol B contains an unsaturated double bond in the C ring, causing its molecular ion peak m/z 487.3773 to be converted into the fragment ion at m/z 207.1732 from RDA cleavage, and dehydration and the loss of C3H6 produced fragment ions m/z 189.1634 and m/z 147.1152. In addition, the Regelindiol B molecular ion peak at m/z 487.3773 lost a hydroxyl radical to produce the fragment ion at m/z 470.3747. Fragment ions at m/z 410.3478 and m/z 392.3402 were formed sequentially by means of side-chain cleavage and dehydration, and the fragment ion at m/z 203.1792 was formed by RDA cleavage. The detailed fragmentation pathways were shown in Fig. 6d. Celastrol is a representative triterpenoid in T. wilfordii that contains a carboxyl group in its molecular structure. Therefore, the parent ion at m/z 451.2837 formed the fragment ion at m/z 405.2781 by losing the carboxyl group, which also generated other fragment ions. There is no double bond in the C ring of the celastrol structure, and thus C ring cleavage occurred to produce a fragment ion at m/z 215.1065. These fragmentation pathways were shown in Fig. S8d.† In conclusion, the terpenoids in T. wilfordii are mainly fractured by the loss of neutral molecules and ring-opening cleavage.
Quantitative analysis of compounds
The applicability and performance of the method in the determination of T. wilfordii compounds were validated, including linearity, intra-day precision, inter-day precision, LOD, LOQ and recovery (Table 2).| Analyst | Regression equations | R 2 | Linear range (ng mL−1) | Precision (%) | LOD (ng mL−1) | LOQ (ng mL−1) | Recovery | Contents in fractions | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Inter-day | Intra-day | original (ng mL−1) | Spiked (ng mL−1) | Found (ng mL−1) | Recovery (%) | F3-1 (mg kg−1) | F3-3 (mg kg−1) | F3-4 (mg kg−1) | ||||||
| N.D. = not detected. | ||||||||||||||
| Wilforgine | y = 204.57x + 1657.7 | 0.9974 | 41.50–830.00 | 2.41 | 1.73 | 5.19 | 10.37 | 290.00 | 280.00 | 582.14 | 104.34 | N.D. | 223.61 ± 12.79 | N.D. |
| 560.00 | 905.05 | 109.83 | ||||||||||||
| 1120.00 | 1547.62 | 112.29 | ||||||||||||
| Triptophenolide | y = 118.55x + 9676.5 | 0.9944 | 49.5–990.00 | 2.96 | 2.35 | 6.18 | 19.8 | 501.36 | 403.50 | 844.07 | 94.85 | N.D. | N.D. | 25.84 ± 3.26 |
| 807.00 | 1200.93 | 86.69 | ||||||||||||
| 1614.00 | 2386.88 | 116.82 | ||||||||||||
| Corydaline |
y = 21189x − 82![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 747 747 |
0.9986 | 3.19–1020.00 | 2.07 | 2.02 | 0.32 | 2.05 | 95.56 | 73.5 | 166.07 | 95.53 | 0.03 ± 0.001 | N.D. | N.D. |
| 147.00 | 228.80 | 90.64 | ||||||||||||
| 294.00 | 372.58 | 91.62 | ||||||||||||
The results indicated good linearity between peak areas and concentrations within the test concentration range (R2 > 0.99). The measured LOD range was 0.32 to 6.18 ng mL−1, LOQ range was 2.05 to 19.80 ng mL−1. And superior intra-day precision (ranged from 1.73% to 2.35%, n = 6) and inter-day precision (ranged from 2.07% to 2.96%, n = 3) were achieved. Besides, the recovery experiment, which was performed by adding an accurately known concentration of the analytes, was also carried out at three levels to evaluate the accuracy of the method. Acceptable recovery ranged from 86.69% to 116.82% were acquired, which indicated that the established method is accurate for the quantitative determination of the main compounds in T. wilfordii. The absolute quantitative concentrations of Wilforgine, Triptophenolide and Corydaline are 223.61 ± 12.79 mg kg−1, 25.84 ± 3.26 mg kg−1 and 0.03 ± 0.001 mg kg−1, shown in Table 2. For the other identified 321 compounds without standards, a relative quantitative analysis was calibrated based on the calibration curve in the same compound type. The corresponding results were shown in Table S1.† These results demonstrated the feasibility and reliability of this analytical method for the quantitative detection of complex natural products.
Conclusions
In this study, an offline two-dimensional chromatography system was established, which was combined with a mass spectrometry molecular network to quickly discover and characterize the compounds in T. wilfordii. The molecular weight distribution and types of compounds in this vine were determined. The RP × SFC two-dimensional methodology provided a high degree of orthogonality and significantly higher peak capacity; in addition, a powerful separation ability was exhibited for more efficiently characterized isomers. The structures of eighteen compounds were identified by the secondary mass spectrometry fragments and GNPS database. The fragmentation patterns of the different compounds in T. wilfordii were summarized, which is helpful for their characterization. Overall, 202 new molecular weights were found from feature-based molecular networking (FBMN) of T. wilfordii, including some new molecular weights of unknown classification, which suggests the presence of compounds with a high number of potentially novel structures, and the identified compounds were quantitatively analyzed. According to the four-dimensional diagram, by prioritizing compound preparation in light of their new molecular weights, it is possible to further improve the efficiency of compound preparation. This research can serve as a strong foundation and provide a feasible direction to deeply investigate T. wilfordii in future work.Author contributions
Boquan Qu, formal analysis, methodology, data curation, writing – original draft; Yanfang Liu, conceptualization, funding acquisition, investigation, writing – review & editing; Aijin Shen, investigation, methodology, writing – review & editing; Zhimou Guo, methodology, writing – review & editing; Long Yu, writing – review & editing; Dian Liu, investigation; Feifei Huang, investigation; Ting Peng, investigation; Xinmiao Liang, conceptualization, supervision, project administration.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
This work was supported by the Central Asian Drug Discovery and Development Center of Chinese Academy of Sciences (No. CAM202104), the Shandong Provincial Key Research and Development Program (Major Technological Innovation Project) (2021CXGC010508), the LiaoNing Revitalization Talents Program (XLYC1908035) and the Jiangxi “Double Thousand Plan”.References
- F. Torres, S. M. Robledo, W. Quinones, G. Escobar, R. Archbold, E. Correa, J. F. Gil, N. Arbelaez, J. Murillo and F. Echeverri, Front. Pharmacol., 2020, 11, 584668 CrossRef PubMed.
- B. Qiang, J. Y. Lai, H. W. Jin, L. R. Zhang and Z. M. Liu, Int. J. Mol. Sci., 2021, 22, 4632 CrossRef PubMed.
- F. Abdallah, G. Lecellier, P. Raharivelomanana and C. Pichon, Sci. Rep., 2019, 9, 4132 CrossRef PubMed.
- D. J. Newman and G. M. Cragg, J. Nat. Prod., 2020, 83, 770–803 CrossRef PubMed.
- H. P. Hao, N. Cui, G. J. Wang, B. R. Xiang, Y. Liang, X. Y. Xu, H. Zhang, J. Yang, C. N. Zheng, L. Wu, P. Gong and W. Wang, Anal. Chem., 2008, 80, 8187–8194 CrossRef PubMed.
- R. Wei, G. D. Li and A. B. Seymour, Anal. Chem., 2010, 82, 5527–5533 CrossRef CAS PubMed.
- L. J. Chen, Y. Liu, D. C. Jia, J. Yang, J. H. Zhao, C. L. Chen, H. S. Liu and X. Liang, J. Agric. Food Chem., 2016, 64, 3445–3455 CrossRef CAS PubMed.
- Y. X. Qian, W. W. Li, H. M. Wang, W. D. Hu, H. D. Wang, D. X. Zhao, Y. Hu, X. Li, X. M. Gao and W. Z. Yang, Arabian J. Chem., 2021, 14, 102957 CrossRef CAS.
- W. J. Zhou, Z. M. Guo, L. Yu, H. Zhou, A. J. Shen, Y. Jin, G. W. Jin, J. Y. Yan, F. Yang, Y. M. Liu, C. R. Wang, J. T. Feng, Y. F. Liu and X. M. Liang, Talanta, 2018, 186, 73–79 CrossRef CAS PubMed.
- B. J. Dong, C. S. Peng, P. Ma and X. B. Li, Anal. Bioanal. Chem., 2021, 413, 3511–3527 CrossRef CAS PubMed.
- D. R. Stoll, D. C. Harmes, G. O. Staples, O. G. Potter, C. T. Dammann, D. Guillarme and A. Beck, Anal. Chem., 2018, 90, 5923–5929 CrossRef CAS PubMed.
- A. S. Kaplitz, M. E. Mostafa, S. A. Calvez, J. L. Edwards and J. P. Grinias, J. Sep. Sci., 2020, 1–12 Search PubMed.
- L. Baldino, G. D. Porta, L. S. Osseo, E. Reverchon and R. Adami, J. Supercrit. Fluids, 2018, 133, 65–69 CrossRef CAS.
- W. L. Wei, J. J. Hou, C. L. Yao, Q. R. Bi, X. Wang, Z. W. Li, Q. H. Jin, M. Lei, Z. J. Feng, W. Y. Wu and D. A. Guo, J. Chromatogr. A, 2019, 1603, 179–189 CrossRef CAS.
- W. J. Lv, X. Z. Shi, S. Y. Wang and G. W. Xu, TrAC, Trends Anal. Chem., 2019, 120, 115302 CrossRef.
- L. F. Nothias, D. Petras, R. Schmid, K. Duhrkop, J. Rainer, A. Sarvepalli, I. Protsyuk, M. Ernst, H. gawa, M. Fleischauer, F. Aicheler, A. A. Aksenov, O. Alka, P. M. Allard, A. Barsch, X. Cachet, A. M. aballo-Rodriguez, R. R. Da Silva, T. Dang, N. Garg, J. M. Gauglitz, A. Gurevich, G. Isaac, A. K. musch, Z. Kamenik, K. B. Kang, N. Kessler, I. Koester, A. Korf, A. Le Gouellec, M. Ludwig, H. C. rtin, L. I. McCall, J. McSayles, S. W. Meyer, H. Mohimani, M. Morsy, O. Moyne, S. Neumann, H. uweger, N. H. Nguyen, M. Nothias-Esposito, J. Paolini, V. V. Phelan, T. Pluskal, R. A. Quinn, S. gers, B. Shrestha, A. Tripathi, J. J. J. van der Hooft, F. Vargas, K. C. Weldon, M. Witting, H. Yang, Z. ng, F. Zubeil, O. Kohlbacher, S. Bocker, T. Alexandrov, N. Bandeira, M. K. Wang and P. C. Dorrestein, Nat. Methods, 2020, 17, 905–908 CrossRef CAS.
- S. S. Chen, G. X. Huang, W. L. Liao, S. L. Gong, J. B. Xiao, J. Bai, W. L. W. Hsiao, N. Li and J. L. Wu, Food Chem., 2021, 347, 129008 CrossRef CAS PubMed.
- R. A. Quinn, L. F. Nothias, O. Vining, M. Meehan, E. Esquenazi and P. C. Dorrestein, Trends Pharmacol. Sci., 2017, 38, 143–154 CrossRef CAS PubMed.
- A. E. F. Ramos, L. Evanno, E. Poupon, P. Champy and M. A. Beniddir, Nat. Prod. Rep., 2019, 36, 980 Search PubMed.
- Q. F. He, Z. L. Wu, L. Li, W. Y. Sun, G. Y. Wang, R. W. Jiang, L. J. Hu, L. Shi, R. R. He, Y. Wang and W. C. Ye, Angew. Chem., Int. Ed., 2021, 60, 19609–19613 CrossRef.
- P. Zhao, Z. Y. Li, S. Y. Qin, B. S. Xin, Y. Y. Liu, B. Lin, G. D. Yao, X. X. Huang and S. J. Song, J. Org. Chem., 2021, 86, 15298–15306 CrossRef.
- D. P. Demarque, R. G. Dusi, F. D. M. de Sousa, S. M. Grossi, M. R. S. Silverio, N. P. Lopes and L. spindola, Sci. Rep., 2020, 10, 1051 CrossRef.
- A. M. Brinker, J. Ma, P. E. Lipsky and I. Raskin, Phytochemistry, 2007, 68, 1819 CrossRef.
- R. Li, Z. Zhang, J. Wang, Y. Huang, W. Sun, R. Xie, F. Hu and T. Lei, Biomed. Phrmacother., 2017, 95, 771–779 CrossRef.
- Q. Y. Liu, Int. Immunopharmacol., 2011, 11, 377–383 CrossRef CAS.
- X. D. Wang, W. Jia, W. Y. Gao, R. Zhang, Y. W. Zhang, J. Zhang, Y. Takaishi and H. Q. Duan, J. Asian Nat. Prod. Res., 2005, 7, 755–759 CrossRef CAS PubMed.
- J. Liu, J. Lee, M. A. S. Hernandez, R. Mazitschek and U. Ozcan, Cell, 2015, 161, 999–1011 CrossRef CAS PubMed.
- Y. X. Liu, N. Y. Xiao, H. K. Du, M. Kou, L. L. Lin, M. Huang, S. T. Zhang, S. Xu, D. L. Li and Q. Chen, Biochem. Pharmacol., 2020, 178, 114090 CrossRef CAS PubMed.
- T. W. Corson and C. M. Crews, Cell, 2007, 130, 769–774 CrossRef CAS.
- M. Cameron, J. J. Gagnier and S. Chrubasik, Cochrane Database Syst. Rev., 2011, 1–56, DOI:10.1002/14651858.CD002948.pub2.
- G. Xun, Y. H. Tian, Y. H. Gao, J. Zhang, X. Liu, S. Sun, Q. Qian, F. F. Liu, Q. Wang and X. Wang, J. Pharm. Biomed. Anal., 2022, 216, 114811 CrossRef.
- Y. M. Liu, X. Y. Xue, Z. M. Guo, Q. Xu, F. F. Zhang and X. M. Liang, J. Chromatogr. A, 2008, 1208, 133–140 CrossRef PubMed.
- A. David, F. Wang, X. M. Sun, H. N. Li, J. R. Lin, P. L. Li and G. Deng, Molecules, 2019, 24, 1897 CrossRef.
- K. Zawatzky, M. Biba, E. L. Regalado and C. J. Welch, J. Chromatogr. A, 2016, 1429, 374–379 CrossRef.
- C. Hamman, D. E. Schmidt Jr., M. Wong and M. Hayes, J. Chromatogr. A, 2011, 1218, 7886–7894 CrossRef CAS.
- A. T. Aron, E. C. Gentry, K. L. McPhail, L. F. Nothias, M. Nothias-Esposito, A. Bouslimani, D. Petras, J. M. Gauglitz, N. Sikora, F. Vargas, J. J. J. van der Hooft, M. Ernst, K. B. Kang, C. M. Aceves, M. Caraballo-Rodriguez, I. Koester, K. C. Weldon, S. Bertrand, C. Roullier, K. Y. Sun, R. M. Tehan, P. Boya, M. H. Christian, M. Gutierrez, A. M. Ulloa, J. A. T. Mora, R. Mojica-Flores, J. Lakey-tia, V. Vasquez-Chaves, Y. L. Zhang, A. I. Calderon, N. Tayler, R. A. Keyzers, F. Tugizimana, N. Ndlovu, A. Aksenov, A. K. Jarmusch, R. Schmid, A. W. Truman, N. Bandeira, M. X. Wang and P. C. Dorrestein, Nat. Protoc., 2020, 15, 1954–1991 CrossRef CAS.
- Q. Fu, Z. Y. Li, C. C. Sun, H. X. Xin, Y. X. Ke, Y. Jin and X. M. Liang, J. Supercrit. Fluids, 2015, 104, 85–93 CrossRef CAS.
- L. M. Zhou, J. Du and C. M. Wu, Chin. Chem. Lett., 2010, 21, 600–602 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2an01471h |
| This journal is © The Royal Society of Chemistry 2023 |