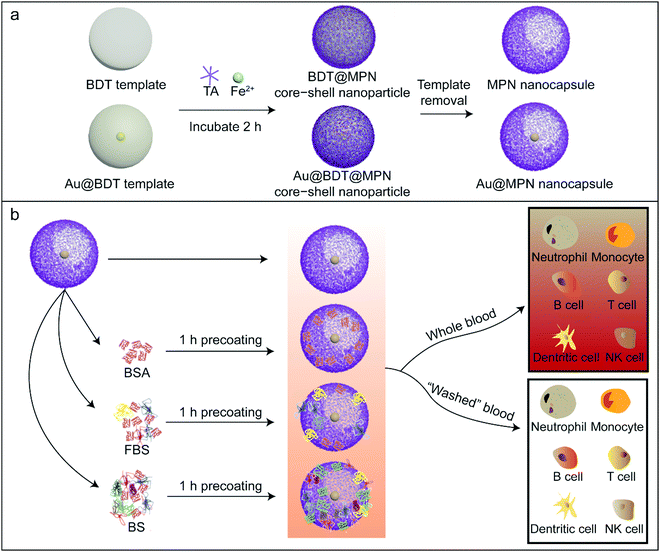
Protein precoating modulates biomolecular coronas and nanocapsule–immune cell interactions in human blood†
Shiyao
Li
 a,
Yi
Ju
a,
Yi
Ju
 *ab,
Jiajing
Zhou‡
*ab,
Jiajing
Zhou‡
 a,
Matthew
Faria
c,
Ching-Seng
Ang
a,
Matthew
Faria
c,
Ching-Seng
Ang
 d,
Andrew J.
Mitchell
e,
Qi-Zhi
Zhong
a,
Tian
Zheng
e,
Stephen J.
Kent
*f and
Frank
Caruso
d,
Andrew J.
Mitchell
e,
Qi-Zhi
Zhong
a,
Tian
Zheng
e,
Stephen J.
Kent
*f and
Frank
Caruso
 *a
*a
aDepartment of Chemical Engineering, The University of Melbourne, Parkville, Victoria 3010, Australia. E-mail: y.ju@unimelb.edu.au; fcaruso@unimelb.edu.au
bSchool of Health and Biomedical Sciences, RMIT University, Bundoora, Victoria 3083, Australia. E-mail: david.ju@rmit.edu.au
cSystems Biology Laboratory, School of Mathematics and Statistics, and the Department of Biomedical Engineering, The University of Melbourne, Parkville, Victoria 3010, Australia
dBio21 Molecular Science and Biotechnology Institute, The University of Melbourne, Parkville, Victoria 3010, Australia
eDepartment of Chemical Engineering, Materials Characterisation and Fabrication Platform, The University of Melbourne, Parkville, Victoria 3010, Australia
fDepartment of Microbiology and Immunology, Peter Doherty Institute for Infection and Immunity, The University of Melbourne, Parkville, Victoria 3010, Australia. E-mail: skent@unimelb.edu.au
First published on 9th June 2022
Abstract
The biomolecular corona that forms on particles upon contact with blood plays a key role in the fate and utility of nanomedicines. Recent studies have shown that precoating nanoparticles with serum proteins can improve the biocompatibility and stealth properties of nanoparticles. However, it is not fully clear how precoating influences biomolecular corona formation and downstream biological responses. Herein, we systematically examine three precoating strategies by coating bovine serum albumin (single protein), fetal bovine serum (FBS, mixed proteins without immunoglobulins), or bovine serum (mixed proteins) on three nanoparticle systems, namely supramolecular template nanoparticles, metal–phenolic network (MPN)-coated template (core–shell) nanoparticles, and MPN nanocapsules (obtained after template removal). The effect of protein precoating on biomolecular corona compositions and particle–immune cell interactions in human blood was characterized. In the absence of a pre-coating, the MPN nanocapsules displayed lower leukocyte association, which correlated to the lower amount (by 2–3 fold) of adsorbed proteins and substantially fewer immunoglobulins (more than 100 times) in the biomolecular corona relative to the template and core–shell nanoparticles. Among the three coating strategies, FBS precoating demonstrated the most significant reduction in leukocyte association (up to 97% of all three nanoparticles). A correlation analysis highlights that immunoglobulins and apolipoproteins may regulate leukocyte recognition. This study demonstrates the impact of different precoating strategies on nanoparticle–immune cell association and the role of immunoglobulins in bio–nano interactions.
Introduction
Nanoparticle-based drug delivery systems have emerged as promising platforms for overcoming the pharmaceutical limitations of traditional drug formulations, due in part to their potential to prolong the circulation time and release encapsulated therapeutics.1,2 An important biological process that accounts for undesirable in vivo outcomes of nanoparticle-based drugs is nanoparticle sequestration by circulating immune cells and tissue phagocytes.3 Intravenously administrated nanoparticles adsorb biomolecules in the blood and form a biomolecular corona.4 The opsonin proteins in the biomolecular corona, which include immunoglobulins and complement proteins, facilitate phagocytic recognition, resulting in sequestration and reduced bioavailability of the nanoparticles.5Several strategies have been developed to limit the opsonization of nanoparticles, including the surface functionalization of nanoparticles with hydrophilic or zwitterionic polymers, such as poly(ethylene glycol) (PEG),6–10 dextran,6,11,12 poly(ethylenimine),13–15 and chitosan.16–18 Among the different surface functionalization strategies, surface functionalization of nanoparticles with PEG (known as PEGylation)19–24 is the most commonly used strategy to enhance nanoparticle circulation time. The high hydrophilicity of PEG enables the formation of a hydrating layer around the nanoparticles, thus reducing the adsorption of plasma proteins. However, studies have shown that healthy humans have pre-existing anti-PEG antibodies, which can affect the fate and utility of PEGylated nanomedicines.25–27 Therefore, alternative strategies are needed to endow nanoparticles with stealth properties. Owing to the intrinsic biocompatibility and low immunogenicity of biomolecules, precoating nanoparticles with biomolecules (prior to their administration in the body/contact with blood) is emerging as a promising strategy for providing a protective coating on nanoparticles. Many strategies, including cell-membrane coating28–34 and CD47 peptide functionalization,35 have been shown to effectively endow nanoparticles with stealth properties. Previous studies have also demonstrated that precoating nanoparticles with plasma proteins, such as albumin,36–39 serum,40 or plasma,41–43 reduces cytotoxicity and enhances the stability and stealth properties of the nanoparticles, which eventually lowers their recognition by immune cells. However, the effects of precoating nanoparticles with serum proteins may vary depending on the nanoparticles.44 To date, few studies have directly compared the effectiveness of different protein precoating strategies.43,44 It is not fully clear how protein precoating modulates biomolecular corona formation and downstream immune responses.
Metal–phenolic networks (MPNs), which are formed largely through the coordination of polyphenols and metal ions, have emerged as promising candidates for biomedical applications because of their high biocompatibility, pH sensitivity, and drug-loading capabilities.45–51 MPN-based (nano)particles have been widely applied in advanced therapies, including checkpoint blockade immunotherapy, adoptive cell therapy, chemodynamic therapy, photodynamic therapy, and photothermal therapy.49,52 MPN capsules are engineered through the deposition of MPN coatings onto sacrificial templates and subsequent template removal. They can serve as drug delivery vehicles by encapsulating drugs during capsule synthesis.45 Previous studies have reported the use of different phenolic ligands and metal ions in the synthesis of MPNs to control their drug release profiles,45 to tailor their stealth and targeting properties,53–55 and to enable bioimaging.47 Our recent studies showed that PEG-based MPN nanocapsules displayed significantly less association with human blood immune cells compared with their counterpart core–shell particles of the same size and that the sequestration of MPN nanocapsules by leukocytes strongly influenced their in vivo circulation time.56 However, the composition of the biomolecular corona on MPN nanocapsules in human plasma and how this influences nanocapsule–immune cell associations remain unexplored.
Herein, a systematic study investigating the effect of protein precoating on biomolecular corona formation and nanoparticle–immune cell interactions was conducted by precoating nanoparticles with bovine proteins (single type protein vs protein mixtures with or without immunoglobulins), followed by incubation with human plasma or whole blood for biomolecular corona composition and nanoparticle–leukocyte association analysis (Scheme 1). Bovine proteins were used for protein precoating to be distinguished from human plasma proteins in the biomolecular corona composition analysis. Three nanoparticle systems were synthesized and compared: supramolecular core nanoparticle templates (∼100 nm); MPN-coated template (denoted core–shell) nanoparticles; and MPN nanocapsules (obtained after template/core removal). To quantitatively track the interactions of the nanoparticles with diverse biological systems, a gold nanoparticle (AuNP) of 14 nm was encapsulated into each core nanoparticle, which enabled highly accurate quantification on a per-cell basis using mass cytometry. The three nanoparticle systems were precoated with different bovine serum proteins—bovine serum albumin (BSA, a single type bovine protein), fetal bovine serum (FBS, bovine protein mixture without immunoglobulins), or bovine serum (BS, bovine protein mixture with immunoglobulins)—and the interactions between the nanoparticles and human blood leukocytes were studied in human blood assays that we previously developed.56,57 The compositions of the biomolecular coronas on the three nanoparticle systems were characterized by mass spectrometry-based proteomics and the key proteins were identified by correlating proteomics data with the blood assays. In the absence of a bovine protein precoating, we found that the MPN nanocapsules had lower association with leukocytes compared with the template and core–shell nanoparticles due to their lower adsorption of human plasma proteins and smaller enrichment of immunoglobulins. In contrast, when precoated with bovine proteins (BSA, FBS or BS), all three nanoparticle systems had lower association with most types of leukocytes compared with the uncoated nanoparticle counterparts. In particular, precoating with FBS was the most effective strategy to reduce leukocyte recognition of the nanoparticles among the three precoating strategies examined. A correlation analysis highlighted that nanoparticle–leukocyte association was correlated to key proteins, including immunoglobulins and apolipoproteins.
Experimental
Materials
Iron(II) chloride tetrahydrate (FeCl2·4H2O), tannic acid (TA), anhydrous dimethylformamide (DMF), BSA, FBS, BS, CH3CN, trifluoroacetic acid (TFA), formic acid and ethylenediaminetetraacetic acid (EDTA) were purchased from Sigma-Aldrich (USA). Dulbecco's phosphate-buffered saline (DPBS) was purchased from Life Technologies (USA). Paraformaldehyde (16% w/v) (PFA) and RPMI 1640 medium were purchased from Thermo Fisher Scientific (USA). Lysing buffer (10×) was purchased from BD Biosciences (USA). For human blood cell phenotyping, purified mouse anti-human CD3 (Clone OKT3), HLA-DR (Clone G46-6), CD45 (Clone HI30), CD56 (Clone NCAM16.2), CD66 (Clone B1.1), CD16 (Clone 3G8), CD11b (Clone ICRF44), CD19 (Clone HIB19), and CD20 (Clone 2H7) antibodies were purchased from BD Biosciences (USA), and purified anti-human CD14 (Clone M5E2) antibody was purchased from BioLegend (USA). Cell-ID™ Intercalator-Ir 500, Cell-ID™ Cisplatin, 100 μL and Maxpar® X8 Antibody Labeling Kits were purchased from Fluidigm. High-purity water (Milli-Q water) with a resistivity greater than 18.2 MΩ cm was obtained from a three-stage Millipore Milli-Q plus 185 purification system (Millipore Corporation, USA).Synthesis of 100 nm BDT@MPN, Au@BDT@MPN core–shell nanoparticles, and MPN and Au@MPN nanocapsules
The 100 nm diameter polymerized benzene-1,4-dithiol (BDT) and AuNP-encapsulated BDT (Au@BDT) nanoparticles were synthesized according to a previously reported method.58 To prepare MPN-coated BDT core–shell (BDT@MPN) or MPN-coated Au@BDT (Au@BDT@MPN) core–shell nanoparticles, approximately 1010 BDT or Au@BDT nanoparticles were suspended in Milli-Q water (300 μL). Then, TA aqueous solution (80 μL, 10 mg mL−1) and FeCl2·4H2O aqueous solution (40 μL, 10 mg mL−1) were added to the nanoparticle suspension. The mixture was then incubated for 2 h for oxidation of Fe2+ to Fe3+ and MPN deposition to proceed. The nanoparticles were then washed with Milli-Q water three times. To prepare MPN or Au@MPN nanocapsules, DMF (800 μL) was added to the BDT@MPN or Au@BDT@MPN nanoparticle suspension, and the resulting suspension was incubated for 2 h at room temperature. Then, the nanocapsules were spun down at 14![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min and washed with Milli-Q water three times to remove the dissolved BDT.
000g for 10 min and washed with Milli-Q water three times to remove the dissolved BDT.
Formation of biomolecular corona on nanoparticles
To precoat the nanoparticles with a bovine protein corona, a suspension of nanoparticles (Au@MPN nanocapsules, Au@BDT@MPN core–shell nanoparticles, or Au@BDT templates, 50 μL, ∼5 × 109) was incubated with BSA (300 μL, 25 mg mL−1 in DPBS), neat FBS, or neat BS at 37 °C with constant shaking (600 rpm) on an Eppendorf Thermomixer Comfort (Eppendorf, Germany) for 1 h, followed by washing two times with DPBS. To form a human protein corona on the nanoparticles, the precoated or nonprecoated nanoparticles were incubated with human plasma at 37 °C with constant shaking (600 rpm) on the thermomixer for 1 h. The nanoparticles were subsequently washed five times with DPBS at 4 °C for the proteomics analysis. The obtained corona-coated nanoparticles were stored at −80 °C before proteomics analysis.Whole blood and washed blood assays
Whole blood from healthy volunteers was collected into sodium heparin vacuette tubes (Greiner Bio-One) after obtaining their informed consent in accordance with The University of Melbourne Human Ethics Committee Approval #1443420 and the Australian National Health and Medical Research Council Statement on Ethical Conduct in Human Research. Washed blood was obtained by washing the blood cells with RPMI medium at 900g for 15 min to remove the plasma proteins. The blood cells were then resuspended in the RPMI medium to a concentration of 5 × 106 leukocytes mL−1. The nanoparticles were incubated with whole blood (200 μL) or washed blood (200 μL) at 37 °C for 1 h in 5 mL polystyrene tubes (BD Biosciences, San Jose, CA, USA). The leukocyte-to-particle ratio was around 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 or 1
200 or 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600. After incubation, 4 mL 1× lysing buffer was added twice to each blood sample to lyse red blood cells. Then, the samples were washed once with flow cytometry staining (FACS) buffer (0.1% BSA, 2 mM EDTA in DPBS). Leukocytes were stained for the mass cytometry analysis using metal-labeled antibodies by incubating with the antibody panel on ice for 40 min, based on a reported methodology.56 Following antibody incubation, each sample was suspended in cisplatin (500 μL, 1/4000 in DPBS) for 5 min to label nonviable leukocytes. The samples were then washed twice with FACS buffer to remove excess cisplatin. Finally, the cell pellets were dispersed in 4% PFA containing 0.025% Ir intercalator for at least 2 h to fix cells and to label cells with Ir. Prior to performing the CyTOF analysis, the samples were washed twice with Milli-Q water.
600. After incubation, 4 mL 1× lysing buffer was added twice to each blood sample to lyse red blood cells. Then, the samples were washed once with flow cytometry staining (FACS) buffer (0.1% BSA, 2 mM EDTA in DPBS). Leukocytes were stained for the mass cytometry analysis using metal-labeled antibodies by incubating with the antibody panel on ice for 40 min, based on a reported methodology.56 Following antibody incubation, each sample was suspended in cisplatin (500 μL, 1/4000 in DPBS) for 5 min to label nonviable leukocytes. The samples were then washed twice with FACS buffer to remove excess cisplatin. Finally, the cell pellets were dispersed in 4% PFA containing 0.025% Ir intercalator for at least 2 h to fix cells and to label cells with Ir. Prior to performing the CyTOF analysis, the samples were washed twice with Milli-Q water.
Liquid chromatography–tandem mass spectrometry (LC-MS/MS) and protein identification
All proteins from the biomolecular coronas were first digested with trypsin and the resultant peptides were analyzed on the mass spectrometer. A QExactive Plus Orbitrap mass spectrometer (Thermo Fisher Scientific) with a nanoESI interface was used in conjunction with an Ultimate 3000 RSLC nanoHPLC (Dionex Ultimate 3000) to perform LC-MS/MS analyses. An Acclaim PepMap nano-trap column (Dionex-C18, 100 Å, 75 μm × 2 cm) and an Acclaim PepMap RSLC analytical column (Dionex-C18, 100 Å, 75 μm × 50 cm) were used in the LC system. Before the enrichment column was switched in-line with the analytical column, the tryptic peptides were injected into the enrichment column at an isocratic flow of 5 L min−1 of 2% v/v CH3CN containing 0.05% v/v TFA for 5 min; 0.1% v/v formic acid (solvent A) and 100% v/v CH3CN in 0.1% v/v formic acid (solvent B) were used as eluents. The flow gradient in the column was (a) 3% B for 0–6 min, (b) 3–25% B (6–40 min), (c) 25–45% B (40–48 min), (d) 40–80% B (48–50 min), (e) 85–85% B (50–53 min), and (f) 85–3% B (53–54 min). Before the next sample injection, the flow was equilibrated at 3% B for 10 min. Full MS1 spectra were collected in the positive mode with an automatic gain control (AGC) target of 3e6, 70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 resolution, and a maximum injection time (IT) of 50 ms on the QExactive Plus mass spectrometer in data-dependent mode. MSMS was then applied to 15 of the most intense peptide ions with charge states ≥ 2 and intensity thresholds of 1.7e4. The isolation window was set at 1.2 m/z, and precursors were fragmented with a normalized collision energy of 30, 17
000 resolution, and a maximum injection time (IT) of 50 ms on the QExactive Plus mass spectrometer in data-dependent mode. MSMS was then applied to 15 of the most intense peptide ions with charge states ≥ 2 and intensity thresholds of 1.7e4. The isolation window was set at 1.2 m/z, and precursors were fragmented with a normalized collision energy of 30, 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 500 resolution, 1e5 AGC target, and 100 ms maximum IT time. The dynamic exclusion time was set at 30 s.
500 resolution, 1e5 AGC target, and 100 ms maximum IT time. The dynamic exclusion time was set at 30 s.
Raw files were processed on MaxQuant platform (version 1.6.17.0) and searched against the UniProt human and bovine database (20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 394 reviewed January 2021) using the default label-free quantification (LFQ) search parameters: LFQ min. ratio count = 2 with unique + razor peptides were used for quantitation, and match between runs feature was activated. Trypsin/P cleavage specificity was applied with 2 maximum missed cleavages. Protein N-Term acetylation and methionine oxidation were defined as variable modifications, and carbamidomethylation of cysteine was specified as the fixed modification. For both peptides and proteins, false discovery rate (FDR) values were calculated using a target-decoy method with a 1% FDR. Log2 transformation was then applied to the LFQ intensities imported from the proteinGroups.txt results.
394 reviewed January 2021) using the default label-free quantification (LFQ) search parameters: LFQ min. ratio count = 2 with unique + razor peptides were used for quantitation, and match between runs feature was activated. Trypsin/P cleavage specificity was applied with 2 maximum missed cleavages. Protein N-Term acetylation and methionine oxidation were defined as variable modifications, and carbamidomethylation of cysteine was specified as the fixed modification. For both peptides and proteins, false discovery rate (FDR) values were calculated using a target-decoy method with a 1% FDR. Log2 transformation was then applied to the LFQ intensities imported from the proteinGroups.txt results.
Quantitation of identified proteins
A minimum of two peptides with at least one razor or unique peptide were needed by LFQ and the protein groups identified in MaxQuant were imported into Perseus for further analysis. Three replicates of each sample were analyzed, and only proteins that were identified in at least two replicates were evaluated for abundance. The protein contents (RPA (%) of total proteins) for semiquantitative assessment of protein abundance were calculated using eqn (1):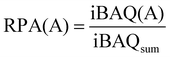 | (1) |
As the iBAQ value is well correlated with the peptide moles,59 as follows:
| iBAQ(A) = c × Mol(A) | (2) |
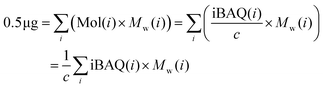 | (3) |
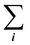 is the sum of all the proteins. As the iBAQ(i) and Mw(i) of protein i are known values, c could be calculated.
is the sum of all the proteins. As the iBAQ(i) and Mw(i) of protein i are known values, c could be calculated.
For the biomolecular corona sample s, the protein mass Mass(s) equals to:
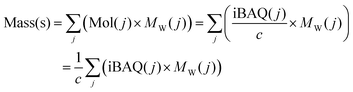 | (4) |
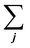 is the sum of all the proteins. As the iBAQ(j) and Mw(j) of protein j and c are known values, Mass(s) could be calculated. As the number of nanoparticles that was used for the preparation of the biomolecular corona samples was measured, the averaged protein mass per particle could be obtained.
is the sum of all the proteins. As the iBAQ(j) and Mw(j) of protein j and c are known values, Mass(s) could be calculated. As the number of nanoparticles that was used for the preparation of the biomolecular corona samples was measured, the averaged protein mass per particle could be obtained.
Calculation of coronal differential enrichment
Protein localization was determined from the Uniprot database, and proteins without the “secreted” location were excluded. Proteins that are nonsecreted are internal to the cells and thus do not influence the corona (which forms prior to cellular uptake).Undetected proteins in any sample (including control) but detected in any other sample (including control) were assumed to be present in at half the minimum detected abundance. This replacement procedure allows us to estimate and compare differential enrichment for rare proteins as opposed to excluding them or assuming that the enrichment index of the detected protein is infinite.
RPA was renormalized to sum to one in each sample (including control).
Differential protein enrichment was calculated by dividing the relative abundance in each sample by the abundance in the control (whole blood).
Proteins were grouped according to their biological function that is acute phase, apolipoproteins, coagulation, complement, immunoglobulins, and tissue leakage.
Heatmap graphs were generated with custom-written python code, which is available at https://bitbucket.org/mwfcomp/corona_comparison.
Correlation analysis between corona protein abundance and nanoparticle–leukocyte association
The nonparametric Spearman correlation between corona protein abundance (RPA) and nanoparticle–leukocyte association (particle number per cell) was determined across triplicate values from 12 types of nanoparticles (Au@MPN nanocapsules, Au@BDT@MPN core–shell nanoparticles, or Au@BDT templates without or with BSA, FBS, or BS precoating). The correlation coefficients (r) and p-values were calculated using GraphPad Prism 8.3. The FDR was controlled by adjusting the p-values using the Benjamini–Hochberg adjustment. The volcano plot was then generated by plotting significance (−log10(p-value)) versus correlation coefficient (r) in GraphPad Prism 8.3. The proteins with p-values of ≤0.0001 and |correlation coefficient (r)| > 0.5 were considered as significant correlating proteins.Characterization methods
Transmission electron microscopy (TEM) images were acquired on a Tecnai Spirit (Biosciences) at an operation voltage of 120 kV. The TEM samples were prepared by dropping the nanoparticle suspension (5 μL) onto a plasma-treated copper grid and subsequent air-drying. Atomic force microscopy (AFM) images were acquired on a Cypher ES AFM instrument (Asylum Research, USA) equipped with a Tap 300 cantilever (BudgetSensors) with a spring constant of 40 N m−1. The measurements were performed in tapping mode in air. The nanocapsule aqueous suspension was deposited on piranha-cleaned silicon wafers and allowed to dry in air. Caution: Piranha solution is strongly oxidizing and corrosive! Extreme care should be taken during preparation and use! UV-vis absorption spectra of the nanoparticle suspensions recorded within a wavelength range of 400–800 nm were obtained using a Varian Cary 4000 UV-vis spectrophotometer (Varian, USA). X-ray photoelectron spectroscopy (XPS) analysis was performed on a Kratos Axis ULTRA X-ray photoelectron spectrometer with a 165 mm hemispherical electron energy analyzer. The XPS spectra were processed on CasaXPS. ζ-Potential measurements were carried out on a Malvern Zetasizer Nano ZS fitted with a 4 mW He–Ne laser (633 nm). Particle size and particle concentration (particles mL−1) measurements were performed via nanoparticle tracking analysis (NTA) on a Malvern NanoSight NS300 instrument fitted with a 405 nm laser (65 mW output).Minimum information reporting in bio–nano experimental literature (MIRIBEL)
The studies conducted herein, including material characterization, biological characterization, and experimental details, conform to the MIRIBEL reporting standard for bio–nano research,60 and we include a companion checklist of these components in the ESI.†Results and discussion
MPN and Au@MPN nanocapsule synthesis, characterization and precoating strategies
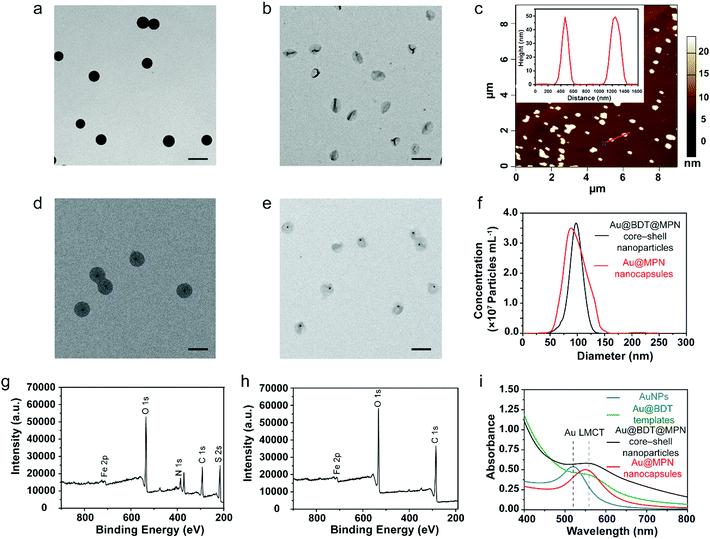
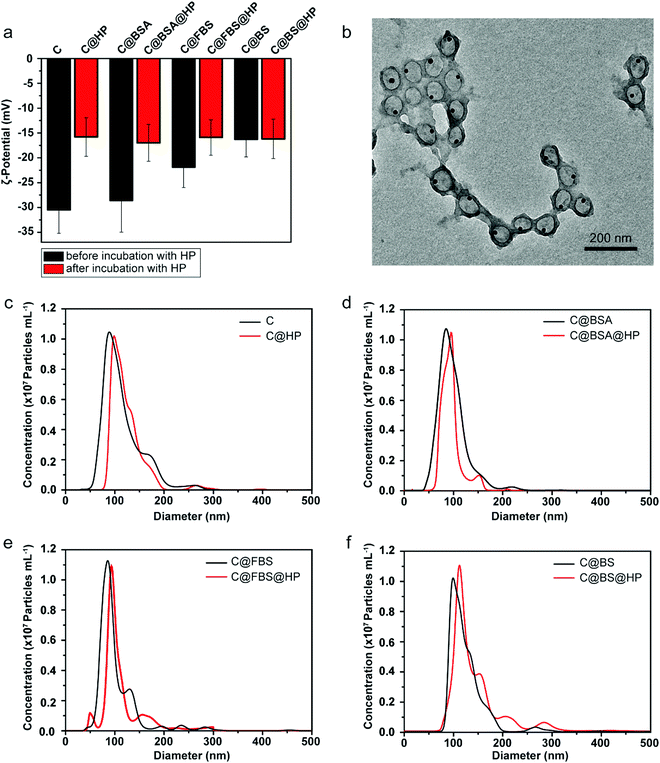
MPN nanocapsules were assembled via a template-assisted assembly process using BDT particles as templates, which allowed for template removal in organic solvent.58 The 100 nm BDT and AuNP-encapsulated BDT (Au@BDT) nanoparticles were first synthesized as templates for subsequent assembly of the MPN-coated template core–shell nanoparticles and MPN nanocapsules. A 14 nm diameter AuNP was encapsulated into each BDT nanoparticle during synthesis as a labeling agent for mass cytometry analysis. TEM images of the BDT and Au@BDT nanoparticles in Fig. S1 (ESI†) show the spherical morphology and uniform size of the templates, as well as the successful encapsulation of individual AuNPs. MPN-coated BDT core–shell (BDT@MPN) nanoparticles and MPN-coated Au@BDT (Au@BDT@MPN) nanoparticles were obtained through oxidation-mediated MPN deposition via the addition of TA and FeCl2 aqueous solution into BDT and Au@BDT nanoparticle suspensions, respectively, followed by incubation for 2 h.61 MPN and Au@MPN nanocapsules were obtained following dissolution of BDT using DMF. The morphology of the MPN and Au@MPN nanocapsules was characterized by TEM (Fig. 1b and e). Compared with the BDT@MPN and Au@BDT@MPN core–shell nanoparticles (Fig. 1a and d), the MPN and Au@MPN nanocapsules had lower contrast and a nonspherical shape because of template removal. AFM measurements performed on MPN nanocapsules in the “air-dried” state revealed that the thickness of the capsules was ∼50 nm (Fig. 1c, Table S1, ESI†), indicating that the single-layer thickness of the MPN capsules was ∼25 nm. Both Au@BDT@MPN core–shell nanoparticles and Au@MPN nanocapsules were monodisperse and ∼100 nm in size, as determined from NTA (Fig. 1f). Comparison of the XPS patterns of the BDT@MPN core–shell nanoparticles and MPN nanocapsules (Fig. 1g and h) confirmed the successful removal of BDT, as indicated by the absence of the S 2s signal peak at ∼226 eV and N 1s signal peak at 399 eV in the XPS pattern of the MPN nanocapsules. The ultraviolet-visible (UV-vis) absorption spectrum of the 100 nm Au@BDT@MPN core–shell nanoparticles and Au@MPN nanocapsules showed a ligand-to-metal charge transfer (LMCT) band at ∼565 nm,61 suggesting the successful formation of bis-complexes between TA and FeIII (Fig. 1i).To investigate the impact of protein precoating on the stealth properties of the Au@MPN nanocapsules (hereafter referred to as “C” for simplicity), three different types of bovine serum proteins, i.e., BSA, FBS, and BS, were used. Bovine proteins were selected to form protein precoating on the nanocapsules in order to distinguish from human corona proteins formed in human plasma for the subsequent proteomics study. BSA is the most abundant protein in FBS and BS. Compared to BS that comes from adult bovine, FBS comes from bovine fetus and commonly lacks immunoglobulins.62,63 Previous studies have shown that preincubation of inorganic nanoparticles (metallic, magnetic, and silica nanoparticles), polymeric nanoparticles, or lipid nanoparticles with BSA or FBS can enhance the stability and stealth properties and reduce the toxicities of the nanoparticles.36–38,40,42 However, the impact of precoating proteins on the biomolecular corona formation and nanoparticle–immune cell interactions in blood is poorly understood. The Au@MPN nanocapsules were incubated with BSA, FBS, or BS at 37 °C for 1 h to form nanocapsules precoated with a bovine protein corona. After precoating with BSA, FBS, or BS, there is a negligible change in particle size (Table S1, ESI†). The ζ-potential of the nanocapsules increased from around −31 mV (C) to −22 mV (C@FBS) or −16 mV (C@BS) after coating with FBS or BS, respectively (Fig. 2a, Table S1, ESI†), whereas there is a negligible change in zeta potential after BSA coating likely due to the negative charge of BSA at pH 7.4. The difference in zeta potential for the three precoated nanocapsules likely resulted from the different corona composition. The uncoated and precoated nanocapsules were then incubated with HP (C@HP, C@BSA@HP, C@FBS@HP, C@BS@HP, respectively) to mimic the condition when the nanocapsules are administered into blood. After incubating with HP, the ζ-potential of all of the examined nanocapsule types increased to a similar value of approximately −16 mV. TEM analysis of the Au@MPN nanocapsules incubated in HP (C@HP) (Fig. 2b) and the size distribution of the different types of nanocapsules, as determined by NTA, (Fig. 2c–f) showed that after coating with HP biomolecular corona, the nanocapsules remained monodisperse with a minimal change in size (<6%).
Ex vivo assessment of nanoparticle and nanocapsule interactions with human blood
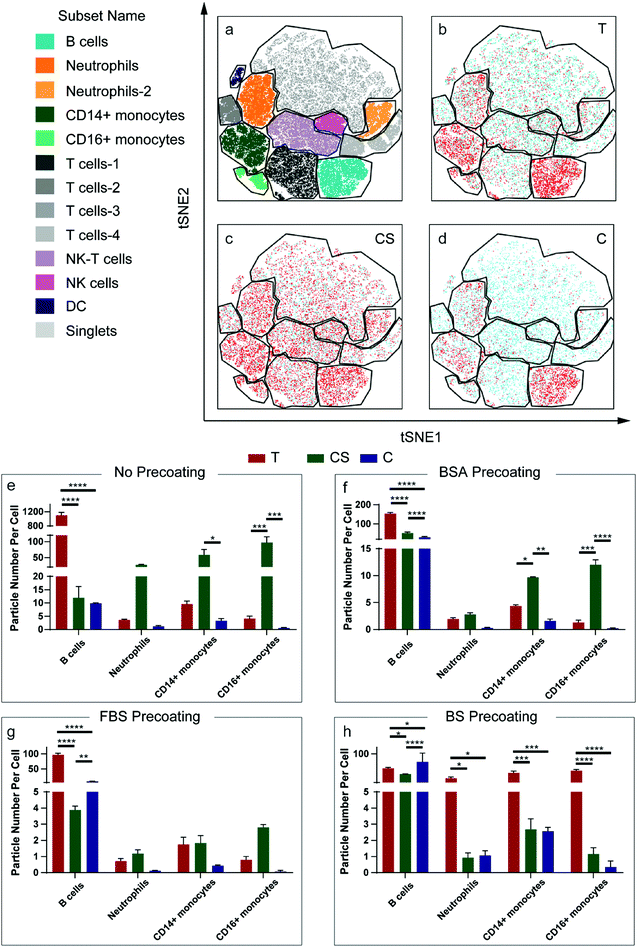
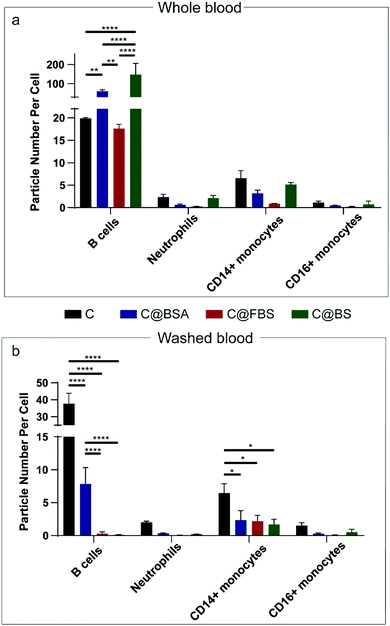
Ex vivo human blood assays were conducted with the three nanoparticle systems, i.e., Au@BDT template nanoparticles, Au@BDT@MPN core–shell nanoparticles, and Au@MPN nanocapsules—hereafter referred to as T, CS, and C, respectively, for simplicity—to evaluate their interactions with leukocytes in the human blood environment. Seven separate white blood cell populations, that is B lymphocytes (B cells), T lymphocytes (T cells), neutrophils, CD14 positive (CD14+) monocytes, CD16 positive (CD16+) monocytes, dendritic cells (DC), and natural killer (NK) cells, were identified through antibody labeling based on the gating strategy reported therein (Fig. S2 and S3, ESI†).56 The association of the leukocyte with nanoparticles without any protein precoating (uncoated T, CS, and C) and nanoparticles coated with BSA, FBS, or BS (T/CS/C@BSA, T/CS/C@FBS, T/CS/C@BS) after incubation of the nanoparticles in whole blood or “washed blood” (plasma protein-free blood) for 1 h at a leukocyte-to-particle ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 was investigated by cytometry by time-of-flight (CyTOF). The association patterns of the uncoated nanoparticles were first shown as t-distributed stochastic neighbor embedding (t-SNE) plots (Fig. 3a–d). The Au signal of the associated nanoparticles was overlaid with the population of leukocytes and shown as red dots (Fig. 3b–d). For all the uncoated and precoated three nanoparticle systems, the majority of the Au signal appeared within the B cell, neutrophil, and monocyte populations, indicating that most nanoparticles associated with those types of leukocytes. The high association of the nanoparticles with B cells is likely attributed to the formation of a biomolecular corona, which mediates nanoparticle binding with B cells through complement receptors, as consistent with our previous studies.44,57 Monocytes and neutrophils are also expected to have high nanoparticle association due to their main function of phagocytosing microbes and particulates. As the B cells, neutrophils, and monocytes are the populations that showed high nanoparticle association, they are referred to as the main leukocyte types; nanoparticle association of all leukocyte types is presented in Fig. S4–S13 (ESI†). The cell association results were characterized by the average particle number per cell (PPC) (Fig. 3e), calculated according to a standard methodology.56 Compared to the uncoated Au@BDT templates (T) and Au@BDT@MPN core–shell nanoparticles (CS), the uncoated Au@MPN nanocapsules (C) showed reduced association with neutrophils and monocytes. The average nanoparticle number per neutrophil values of the Au@BDT templates (T) and Au@BDT@MPN core–shell nanoparticles (CS) were 3.6 and 27.1, respectively, whereas only an average of 1.2 C associated with neutrophils. To study the effect of protein precoating on leukocyte association, the nanoparticles were precoated with BSA, FBS, or BS, and then incubated with human whole blood (Fig. 3f–h). Consistent with the trend observed for the uncoated nanoparticles, the bovine protein-precoated Au@MPN nanocapsules displayed reduced (up to 99%) association with neutrophils and monocytes compared with the bovine protein-precoated nanoparticle templates and Au@BDT@MPN core–shell nanoparticles. Furthermore, compared with the uncoated Au@MPN nanocapsules (C), the protein-precoated Au@MPN nanocapsules (C@BSA, C@FBS, C@BS) showed reduced association (by 8–90%) with neutrophils and monocytes. For instance, C@FBS had only 0.12 PPC with neutrophils, which is ∼1/10 of the PPC of the uncoated nanocapsules with neutrophils. Whole blood assays with Au@MPN nanocapsules were further performed at a higher leukocyte-to-capsule ratio (1
200 was investigated by cytometry by time-of-flight (CyTOF). The association patterns of the uncoated nanoparticles were first shown as t-distributed stochastic neighbor embedding (t-SNE) plots (Fig. 3a–d). The Au signal of the associated nanoparticles was overlaid with the population of leukocytes and shown as red dots (Fig. 3b–d). For all the uncoated and precoated three nanoparticle systems, the majority of the Au signal appeared within the B cell, neutrophil, and monocyte populations, indicating that most nanoparticles associated with those types of leukocytes. The high association of the nanoparticles with B cells is likely attributed to the formation of a biomolecular corona, which mediates nanoparticle binding with B cells through complement receptors, as consistent with our previous studies.44,57 Monocytes and neutrophils are also expected to have high nanoparticle association due to their main function of phagocytosing microbes and particulates. As the B cells, neutrophils, and monocytes are the populations that showed high nanoparticle association, they are referred to as the main leukocyte types; nanoparticle association of all leukocyte types is presented in Fig. S4–S13 (ESI†). The cell association results were characterized by the average particle number per cell (PPC) (Fig. 3e), calculated according to a standard methodology.56 Compared to the uncoated Au@BDT templates (T) and Au@BDT@MPN core–shell nanoparticles (CS), the uncoated Au@MPN nanocapsules (C) showed reduced association with neutrophils and monocytes. The average nanoparticle number per neutrophil values of the Au@BDT templates (T) and Au@BDT@MPN core–shell nanoparticles (CS) were 3.6 and 27.1, respectively, whereas only an average of 1.2 C associated with neutrophils. To study the effect of protein precoating on leukocyte association, the nanoparticles were precoated with BSA, FBS, or BS, and then incubated with human whole blood (Fig. 3f–h). Consistent with the trend observed for the uncoated nanoparticles, the bovine protein-precoated Au@MPN nanocapsules displayed reduced (up to 99%) association with neutrophils and monocytes compared with the bovine protein-precoated nanoparticle templates and Au@BDT@MPN core–shell nanoparticles. Furthermore, compared with the uncoated Au@MPN nanocapsules (C), the protein-precoated Au@MPN nanocapsules (C@BSA, C@FBS, C@BS) showed reduced association (by 8–90%) with neutrophils and monocytes. For instance, C@FBS had only 0.12 PPC with neutrophils, which is ∼1/10 of the PPC of the uncoated nanocapsules with neutrophils. Whole blood assays with Au@MPN nanocapsules were further performed at a higher leukocyte-to-capsule ratio (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600) (Fig. S6 and S9, ESI†) and a longer incubation time (4 h) (Fig. S8 and S9, ESI†) to assess the stealth properties of the bovine protein-precoated nanocapsules. Increasing the leukocyte-to-capsule ratio and incubation time led to higher possibility of nanocapsules interacting with the blood cells, which resulted in higher association results, when compared with the results obtained for the whole blood assays that were conducted at a leukocyte-to-capsule ratio of 1
600) (Fig. S6 and S9, ESI†) and a longer incubation time (4 h) (Fig. S8 and S9, ESI†) to assess the stealth properties of the bovine protein-precoated nanocapsules. Increasing the leukocyte-to-capsule ratio and incubation time led to higher possibility of nanocapsules interacting with the blood cells, which resulted in higher association results, when compared with the results obtained for the whole blood assays that were conducted at a leukocyte-to-capsule ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 and incubation time of 1 h (Fig. 4a). Consistent with the results of the whole blood assay performed at a leukocyte-to-capsule ratio of 1
200 and incubation time of 1 h (Fig. 4a). Consistent with the results of the whole blood assay performed at a leukocyte-to-capsule ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 and an incubation time of 1 h, as discussed above, precoating nanocapsules with bovine proteins reduced the association of the nanocapsules with most types of leukocytes. In particular, precoating the nanocapsules with FBS resulted in the lowest association of the precoated nanocapsules with leukocytes among the three precoating strategies examined.
200 and an incubation time of 1 h, as discussed above, precoating nanocapsules with bovine proteins reduced the association of the nanocapsules with most types of leukocytes. In particular, precoating the nanocapsules with FBS resulted in the lowest association of the precoated nanocapsules with leukocytes among the three precoating strategies examined.
Washed blood (blood with HP removed) assays with the nanoparticles were conducted to investigate the role of HP on leukocyte recognition (Fig. 4b, Fig. S5, S7, S11, S13, ESI†). As shown in Fig. 4b, when the HP proteins were removed from the blood, the association between the precoated capsules and B cells decreased considerably (from ∼60 to ∼7 PPC for C@BSA, from ∼17 to ∼0.3 PPC for C@FBS, and from 147 to 0.11 PPC for C@BS). C@FBS and C@BS showed negligible association with B cells, neutrophils, or monocytes. This result indicates that the formation of a biomolecular corona from HP is essential for leukocyte recognition of the nanocapsules in blood. The effects of HP on leukocyte association with Au@MPN nanocapsules were further confirmed by changing the leukocyte-to-capsule ratio from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 200 to 1
200 to 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 600 (Fig. S7, ESI†). In addition, similar results were found in the washed blood assays with the Au@BDT templates and Au@BDT@MPN core–shell nanoparticles (Fig. S11 and S13, ESI†).
600 (Fig. S7, ESI†). In addition, similar results were found in the washed blood assays with the Au@BDT templates and Au@BDT@MPN core–shell nanoparticles (Fig. S11 and S13, ESI†).
Proteomics analysis of bovine protein precoating
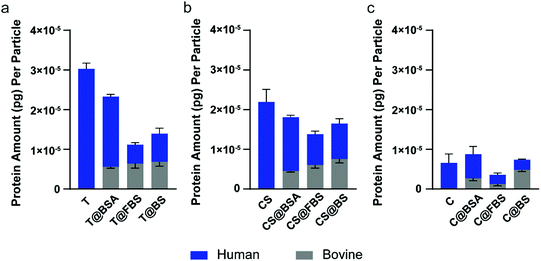
Proteomics analysis was performed on the uncoated nanoparticles, and the BSA-, FBS-, and BS-precoated nanoparticles to examine the protein corona composition of the nanoparticles using mass spectrometry-based proteomics. The precoated bovine proteins were separated from human proteins in the corona based on the organism of origin of the proteins. The average protein amount (pg) per particle was calculated based on the iBAQ value and the molecular weight of each protein, with 0.5 μg HP used as the reference sample (Fig. 5). Upon preincubation with different bovine preparations, the Au@BDT templates (T) and Au@BDT@MPN core–shell nanoparticles (CS) adsorbed a similar level of bovine proteins of ∼0.6 × 10−5 pg per particle, whereas the Au@MPN nanocapsules adsorbed various amounts of bovine proteins from 0.1–0.5 × 10−5 pg per particle. Although no distinct trend was observed for the amount of adsorbed bovine proteins, the findings showed that BSA was not the most abundant bovine corona protein in either of the FBS- and BS-precoated nanoparticles, which displayed a relative protein abundance (RPA) of 0.63–15% (Table S2, ESI†). This range of values is much lower than the BSA abundance in the original FBS or BS solution (33–39%, Table S4, ESI†). The composition of the precoated bovine proteins resulted in the different nanoparticle–leukocyte associations observed in the washed blood assays. Although BSA precoating reduced the nanoparticle–leukocyte association to a large extent, FBS and BS precoating led to negligible nanoparticle association with most types of leukocytes (Fig. S5, S7, S11, S13, ESI†), which illustrates the important role of some commonly enriched bovine proteins in the FBS- and BS-precoated nanoparticles in downregulating leukocyte recognition. Comparison of the top 20 abundant bovine proteins in the FBS- and BS-precoated nanoparticles revealed that apolipoproteins (APOA1, APOA2, APOE, and AOPH) made up over 5% of the corona proteins in all the FBS- and BS-precoated nanoparticles (30% for T@FBS, 23% for T@BS, 19% for CS@FBS, 35% for CS@BS, 6% for C@FBS, and 6% for C@BS) (Table S2, ESI†), which may be related to the lower leukocyte association of the FBS- and BS-precoated nanoparticles, compared with that of the BSA-precoated nanoparticles. This finding is consistent with previous studies that have shown apolipoproteins function as dysopsonins, which limit phagocytosis.57,64Proteomics analysis of human plasma coronas
The quantification of the adsorbed HP proteins on different types of nanoparticles without or with bovine protein precoating are shown in Fig. 5. Without precoating, Au@MPN nanocapsules (C; 0.7 × 10−5 pg per particle) adsorbed 3–4 times less proteins than the uncoated Au@BDT templates (T; 3.0 × 10−5 pg per particle) or the Au@BDT@MPN core–shell nanoparticles (CS; 2.2 × 10−5 pg per particle). The higher amount of corona proteins adsorbed onto Au@BDT templates was likely due to their highly hydrophobic surface, which has been previously observed for other nanoparticle systems with a hydrophobic surface, such as polystyrene nanoparticles.41,43 The coating of MPNs onto the Au@BDT templates changed the surface chemistry of the nanoparticles, thereby reducing the plasma protein adsorption. The difference in protein adsorption of the templated particles before and after core removal has also been observed on PEG@mesoporous silica and PEG particles.65 The decreased protein adsorption onto the nanocapsules may lead to their lower association with leukocytes in human blood. This is consistent with previous reports wherein the reduction in the adsorption of plasma proteins was correlated to the stealth properties of nanoparticles, leading to reduced immune cell recognition.42,57,66,67Compared to the uncoated nanoparticles, the nanoparticles precoated with bovine proteins adsorbed a lower total amount of HP proteins after interacting with HP (Fig. 5). Among the nanoparticles subjected to the different precoating strategies and regardless of the nanoparticle type (T, CS, or C), the nanoparticles with FBS precoating demonstrated the highest resistance to HP adsorption. Specifically, the BSA-, FBS-, and BS-precoated Au@BDT templates adsorbed 1.8 × 10−5, 4.8 × 10−6, and 7.1 × 10−6 pg HP proteins per particle, respectively. The BSA-, FBS-, and BS-precoated Au@BDT@MPN core–shell nanoparticles adsorbed 1.4 × 10−5, 7.8 × 10−6, and 9.0 × 10−6 pg HP proteins per particle, respectively. The BSA-, FBS-, and BS- precoated Au@MPN nanocapsules adsorbed 6.1 × 10−6, 2.4 × 10−6, and 2.6 × 10−6 pg HP proteins per particle, respectively. The decreased HP protein adsorption obtained from the FBS and BS precoating strategies may result from the diversity of the proteins in the precoated bovine protein corona, in which some types of proteins, such as apolipoproteins that are highly enriched on all the FBS- and BS-precoated nanoparticles, compete for surface binding with opsonins, leading to the overall lower HP protein adsorption.
In addition to the overall amount of plasma corona, the corona composition may play a more important role in nanoparticle–leukocyte association. Among the top 20 most abundant HP proteins detected in the biomolecular corona of the nanoparticles, fibrinogen (FGA, FGB, and FGG) made up a larger part in all the uncoated nanoparticles (49% for Au@BDT templates, 64% for Au@BDT@MPN core–shell nanoparticles, and 59% for Au@MPN nanocapsules) compared with the composition observed in the bovine protein-precoated nanoparticles (Table S3, ESI†); this larger proportion of fibrinogen observed is expected to result from hydrogen bonding and hydrophobic interactions between TA and fibrinogen.68 Although fibrinogen has commonly been identified as a blood coagulation protein, there have been some studies showing that it can regulate leukocyte function and promote auto-immunity.69–71
Differential enrichment of the HP corona proteins
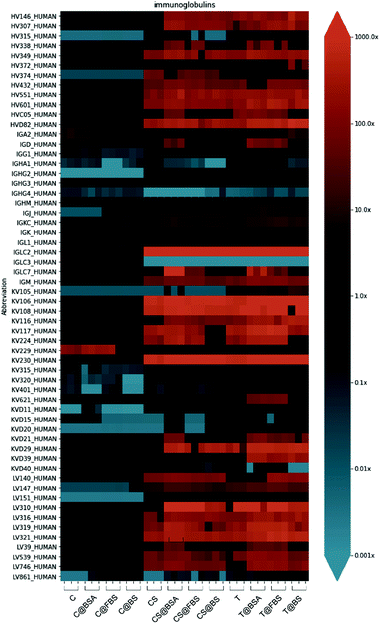
To determine the composition of the biomolecular corona, the enrichment index of each corona protein was quantified. The enrichment index was calculated based on the ratio between the RPA in each sample and the protein abundance in the control sample (see Experimental section for details). This method is an improvement on our previous coronal quantification differential enrichment analysis introduced therein.44 A key goal of understanding the biomolecular corona is to capture how it differs from the background/baseline (i.e., the amount of proteins enriched in whole blood).Differential enrichment of immunoglobulins and other protein groups, classified according to their biological function (acute phase, apolipoproteins, coagulation, complement, tissue leakage), is shown in the form of a heatmap to obtain an overall visualization of the biomolecular corona enrichment in the different types of nanoparticles examined (Fig. 6, Table S5 and Fig. S14–S19, ESI†). Immunoglobulins in the biomolecular corona of nanoparticles play important roles in activating the complement cascade and triggering immune responses.72 Although no significant difference in the enrichment of complement components on the nanoparticles with different precoatings was observed (Fig. S17, ESI†), a distinct trend was observed when comparing the differential enrichment of immunoglobulins on the capsules and the other two nanoparticle systems (Fig. 6). Immunoglobulins were enriched to a considerably lesser extent on the Au@MPN nanocapsules compared with the enrichment observed on the Au@BDT templates and Au@BDT@MPN core–shell nanoparticles. Nine types of immunoglobulins (IGLC2, IGKV1-8, IGKV2D-30, IGKV1-6, IGKV1-17, IGLV3-21, IGHV4-61, IGHV5-51, and IGLV3-19) were over 100 times (enrichment index > 100) enriched in the coronas of the uncoated and BSA-, FBS-, and BS-precoated Au@BDT templates and Au@BDT@MPN core–shell nanoparticles, whereas their enrichment level in the uncoated and BSA-, FBS-, and BS-precoated Au@MPN nanocapsules was similar to baseline human plasma (enrichment index ∼1) (Table S5, ESI†). Combining the results of the total protein adsorption presented in Fig. 5, we conclude that the nanocapsules adsorbed fewer of these immunoglobulins compared with the templates and core–shell nanoparticles, which may account for the reduced leukocyte recognition and association in blood observed for the nanocapsules.
Correlation of specific HP corona protein to nanoparticle association with leukocytes
To further confirm the role of immunoglobulins in immune cell recognition, a correlation analysis was performed, which allowed identification of key proteins that regulate leukocyte recognition. The RPA of each protein (with RPA larger than 0.1%) identified in the HP biomolecular corona of the nanoparticles was correlated to the leukocyte association in whole blood assays using a Spearman correlation analysis (Fig. 7, Table S6, ESI†). Human proteins with p-value ≤ 0.0001 and |correlation coefficient (r)| > 0.5 were identified as significant correlating proteins. It is worth noting that although being identified as key correlating proteins, not all of them played essential roles in regulating leukocyte association. The abundance of proteins is another important parameter that should be considered together with the correlation coefficient. Among the 10 key proteins that are positively correlated to the leukocyte association, 6 of them are immunoglobulins (IGJ, IGHA1, IGHG2, IGHM, immunoglobulin lambda-1 light chain, and immunoglobulin kappa light chain). The relative abundance of those immunoglobulins ranged from 10–28% in the uncoated and precoated Au@BDT templates and Au@BDT@MPN core–shell nanoparticles and 2–3% in the uncoated and precoated Au@MPN nanocapsules. The results further confirm the crucial role of immunoglobulins in stimulating leukocyte recognition of the nanoparticles. For the key proteins that showed a negative correlation to leukocyte association, kininogen-1 (KNG1) and apolipoprotein E (APOE) were the only two proteins that were present in a relatively high abundance (>1%) in the nanoparticles and showed a negative correlation to association with most types of leukocytes (neutrophils, CD14+, and CD16+ monocytes). KNG1 is a blood coagulation protein, and its role in regulating immune responses is unclear.73 APOE has been shown to contribute to some non-lipid functions, including suppressing immune and inflammatory responses.74,75 The enrichment of apolipoproteins and KNG1 may compete with surface binding with opsonins, leading to reduced immune cell recognition.54,76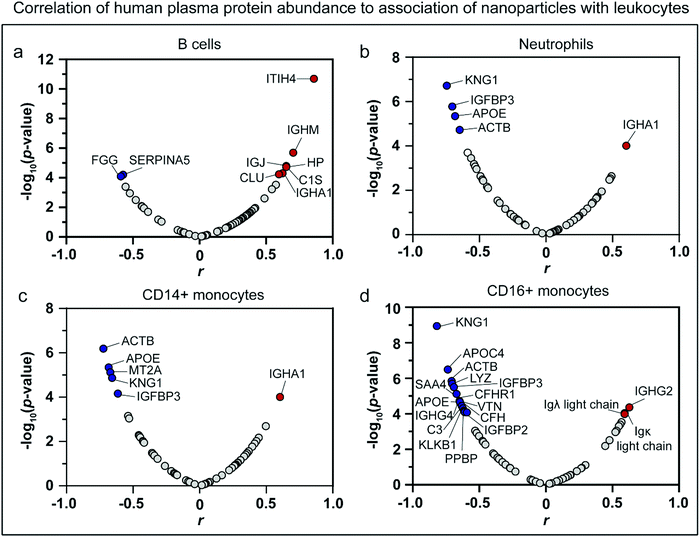 | ||
| Fig. 7 Spearman correlation analysis of HP protein abundance on biomolecular coronas to nanoparticle association with leukocytes. (a–d) Volcano plots of significance (−log10(p-value)) versus correlation coefficient (r) identifying the key proteins within the biomolecular coronas that are highly correlated to nanoparticle association (12 types of nanoparticles that is three nanoparticle systems with and without precoating) with (a) B cells, (b) neutrophils, (c) CD14+ monocytes, and (d) CD16+ monocytes. The top proteins that have a significant correlation (p-value of ≤0.0001 and |correlation coefficient (r)| > 0.5) are presented as red or blue dots (positive or negative correlation, respectively). The key identified proteins are listed in Table S6 (ESI†). | ||
Conclusions
Three protein precoating strategies on three different nanoparticle systems were examined and directly compared to assess the efficacy of the different strategies in regulating biomolecular corona compositions and immune cell responses. The 100 nm TA-Fe-based MPN core–shell nanoparticles and nanocapsules were prepared through a one-step assembly on polymerized BDT templates, with AuNPs encapsulated into the templates as a tracking agent for mass cytometry analysis. Bovine serum proteins (BSA, FBS, or BS) were introduced to form a precoating layer onto the Au@BDT templates, Au@BDT@MPN core–shell nanoparticles, and Au@MPN nanocapsules. Ex vivo whole blood assays with the precoated nanoparticles revealed that FBS precoating effectively reduced nanoparticle association with leukocytes. Proteomics studies were performed to analyze the composition of the biomolecular coronas formed, and the results were correlated to the leukocyte association data. The nanocapsules adsorbed smaller amounts of proteins onto their surface when compared with the template and core–shell nanoparticles. Furthermore, among the different types of nanoparticles without and with precoating examined, the FBS-precoated nanoparticles adsorbed the least amount of proteins. Heatmap analysis of the proteomics data further revealed that the immunoglobulins in the biomolecular corona of the MPN nanocapsules were significantly less enriched compared with the enrichment observed in the biomolecular coronas of the template and core–shell nanoparticles. Several key corona proteins from the human plasma that are highly correlated to the nanoparticle–leukocyte association, including immunoglobulins and apolipoproteins, were identified through the correlation analysis. Overall, our study shows that FBS precoating can effectively alter the biomolecular corona composition in human plasma, leading to a lower overall protein abundance and immunoglobulin enrichment, which is correlated to reduced nanoparticle–leukocyte association in human blood. Therefore, precoating nanoparticles with FBS is demonstrated as a promising strategy to evade immune cell sequestration, highlighting the role of immunoglobin enrichment in biomolecular coronas on modulating nanoparticle–leukocyte interactions.Author contributions
S. L. synthesized and characterized the nanoparticles and performed the full blood assay and the correlation analysis, and drafted the manuscript. Y. J. conceived, designed, and co-supervised the studies. J. Z. synthesized the BDT particles and assisted with capsule preparation. M. F. performed the biomolecular corona enrichment analysis. C.-S. A. carried out the LC-MS/MS experiments of the biomolecular corona samples. A. J. M. assisted with the full blood assay and associated analysis. Q.-Z. Z. assisted with the nanocapsule preparation. T. Z. performed the AFM experiments. S. J. K. and F. C. contributed to the design of the project and data analysis, and supervised the studies. All authors edited the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This research was supported by the Australian Research Council Discovery Project (DP210103114). F. C. acknowledges the award of a National Health and Medical Research Council Senior Principal Research Fellowship (GNT1135806). Y. J. acknowledges the award of an Early Career Researcher Grant from The University of Melbourne (ECR1032020) and an RMIT Vice-Chancellor's Postdoctoral Fellowship from RMIT University. This work was performed in part at the Materials Characterisation and Fabrication Platform at The University of Melbourne and the Victorian Node of the Australian National Fabrication Facility. The authors acknowledge Dr Hannah Gabrielle Kelly for helpful discussions and assistance with the whole blood assays.Notes and references
- M. J. Hawkins, P. Soon-Shiong and N. Desai, Adv. Drug Delivery Rev., 2008, 60, 876–885 CrossRef CAS PubMed.
- Y. Barenholz, J. Controlled Release, 2012, 160, 117–134 CrossRef CAS PubMed.
- E. Blanco, H. Shen and M. Ferrari, Nat. Biotechnol., 2015, 33, 941–951 CrossRef CAS PubMed.
- D. Docter, D. Westmeier, M. Markiewicz, S. Stolte, S. K. Knauer and R. H. Stauber, Chem. Soc. Rev., 2015, 44, 6094–6121 RSC.
- D. E. Owens, 3rd and N. A. Peppas, Int. J. Pharm., 2006, 307, 93–102 CrossRef.
- D. Bamberger, D. Hobernik, M. Konhauser, M. Bros and P. R. Wich, Mol. Pharmaceutics, 2017, 14, 4403–4416 CrossRef CAS.
- J. Lipka, M. Semmler-Behnke, R. A. Sperling, A. Wenk, S. Takenaka, C. Schleh, T. Kissel, W. J. Parak and W. G. Kreyling, Biomaterials, 2010, 31, 6574–6581 CrossRef CAS.
- A. S. Karakoti, S. Das, S. Thevuthasan and S. Seal, Angew. Chem., Int. Ed., 2011, 50, 1980–1994 CrossRef CAS PubMed.
- Z. Amoozgar and Y. Yeo, Wiley Interdiscip. Rev.: Nanomed. Nanobiotechnol., 2012, 4, 219–233 CAS.
- Q. Feng, Y. Liu, J. Huang, K. Chen, J. Huang and K. Xiao, Sci. Rep., 2018, 8, 1–13 CAS.
- Z. Shaterabadi, G. Nabiyouni and M. Soleymani, Mater. Sci. Eng., C, 2017, 75, 947–956 CrossRef CAS PubMed.
- M. P. A. Ferreira, V. Talman, G. Torrieri, D. Liu, G. Marques, K. Moslova, Z. Liu, J. F. Pinto, J. Hirvonen, H. Ruskoaho and H. A. Santos, Adv. Funct. Mater., 2018, 28, 1705134 CrossRef.
- H. N. Yang, J. S. Park, S. Y. Jeon, W. Park, K. Na and K. H. Park, Biomaterials, 2014, 35, 8439–8449 CrossRef PubMed.
- Y. Kapilov-Buchman, E. Lellouche, S. Michaeli and J. P. Lellouche, Bioconjugate Chem., 2015, 26, 880–889 CrossRef CAS PubMed.
- T. Patino, J. Soriano, L. Barrios, E. Ibanez and C. Nogues, Sci. Rep., 2015, 5, 1–12 Search PubMed.
- J. Zhang, W. Xia, P. Liu, Q. Cheng, T. Tahirou, W. Gu and B. Li, Mar. Drugs, 2010, 8, 1962–1987 CrossRef CAS PubMed.
- S. Cui, H. Chen, H. Zhu, J. Tian, X. Chi, Z. Qian, S. Achilefu and Y. Gu, J. Mater. Chem., 2012, 22, 4861–4873 RSC.
- K. V. Kozhikhova, M. N. Ivantsova, M. I. Tokareva, I. D. Shulepov, A. V. Tretiyakov, L. V. Shaidarov, V. L. Rusinov and M. A. Mironov, Pharm. Dev. Technol., 2018, 23, 334–342 CrossRef CAS.
- F. M. Veronese and G. Pasut, Drug Discovery Today, 2005, 10, 1451–1458 CrossRef CAS.
- T. H. Chow, Y. Y. Lin, J. J. Hwang, H. E. Wang, Y. L. Tseng, S. J. Wang, R. S. Liu, W. J. Lin, C. S. Yang and G. Ting, Anticancer Res., 2009, 29, 2111–2120 CAS.
- J. Cui, Y. Ju, Z. H. Houston, J. J. Glass, N. L. Fletcher, S. Alcantara, Q. Dai, C. B. Howard, S. M. Mahler and A. K. Wheatley, Adv. Healthcare Mater., 2019, 8, 1801607 CrossRef.
- R. I. El-Gogary, N. Rubio, J. T.-W. Wang, W. T. Al-Jamal, M. Bourgognon, H. Kafa, M. Naeem, R. Klippstein, V. Abbate and F. Leroux, ACS Nano, 2014, 8, 1384–1401 CrossRef CAS PubMed.
- J. Cui, K. Alt, Y. Ju, S. T. Gunawan, J. A. Braunger, T. Y. Wang, Y. Dai, Q. Dai, J. J. Richardson, J. Guo, M. Bjornmalm, C. E. Hagemeyer and F. Caruso, Biomacromolecules, 2019, 20, 3592–3600 CrossRef CAS.
- H. Zhou, Z. Fan, P. Y. Li, J. Deng, D. C. Arhontoulis, C. Y. Li, W. B. Bowne and H. Cheng, ACS Nano, 2018, 12, 10130–10141 CrossRef CAS PubMed.
- B. M. Chen, T. L. Cheng and S. R. Roffler, ACS Nano, 2021, 15, 14022–14048 CrossRef CAS PubMed.
- H. Freire Haddad, J. A. Burke, E. A. Scott and G. A. Ameer, Regener. Eng. Transl. Med., 2021, 8, 32–42 CrossRef.
- Y. Ju, W. S. Lee, E. H. Pilkington, H. G. Kelly, S. Li, K. J. Selva, K. M. Wragg, K. Subbarao, T. H. O. Nguyen, L. C. Rowntree, L. F. Allen, K. Bond, D. A. Williamson, N. P. Truong, M. Plebanski, K. Kedzierska, S. Mahanty, A. W. Chung, F. Caruso, A. K. Wheatley, J. A. Juno and S. J. Kent, medRxiv, 2022 DOI:10.1101/2022.01.08.22268953.
- W. Gao, C. M. Hu, R. H. Fang, B. T. Luk, J. Su and L. Zhang, Adv. Mater., 2013, 25, 3549–3553 CrossRef CAS PubMed.
- A. Parodi, N. Quattrocchi, A. L. van de Ven, C. Chiappini, M. Evangelopoulos, J. O. Martinez, B. S. Brown, S. Z. Khaled, I. K. Yazdi, M. V. Enzo, L. Isenhart, M. Ferrari and E. Tasciotti, Nat. Nanotechnol., 2013, 8, 61–68 CrossRef CAS.
- K. Hayashi, S. Yamada, W. Sakamoto, E. Usugi, M. Watanabe and T. Yogo, ACS Biomater. Sci. Eng., 2018, 4, 2729–2732 CrossRef CAS PubMed.
- M. Xuan, J. Shao, J. Zhao, Q. Li, L. Dai and J. Li, Angew. Chem., Int. Ed., 2018, 57, 6049–6053 CrossRef CAS.
- Y. Chen, Y. Zhang, J. Zhuang, J. H. Lee, L. Wang, R. H. Fang, W. Gao and L. Zhang, ACS Nano, 2019, 13, 7209–7215 CrossRef CAS PubMed.
- J.-G. Piao, L. Wang, F. Gao, Y.-Z. You, Y. Xiong and L. Yang, ACS Nano, 2014, 8, 10414–10425 CrossRef CAS.
- O. C. Farokhzad, Nature, 2015, 526, 47–48 CrossRef CAS PubMed.
- P. L. Rodriguez, T. Harada, D. A. Christian, D. A. Pantano, R. K. Tsai and D. E. Discher, Science, 2013, 339, 971–975 CrossRef CAS PubMed.
- A. Aires, S. M. Ocampo, D. Cabrera, L. de la Cueva, G. Salas, F. J. Teran and A. L. Cortajarena, J. Mater. Chem. B, 2015, 3, 6239–6247 RSC.
- J. Mariam, S. Sivakami and P. M. Dongre, Drug Delivery, 2016, 23, 2668–2676 CrossRef CAS.
- J. Wang and B. Zhang, Curr. Med. Chem., 2018, 25, 2938–2953 CrossRef CAS PubMed.
- A. O. Elzoghby, W. M. Samy and N. A. Elgindy, J. Controlled Release, 2012, 157, 168–182 CrossRef CAS PubMed.
- H. Yin, R. Chen, P. S. Casey, P. C. Ke, T. P. Davis and C. Chen, RSC Adv., 2015, 5, 73963–73973 RSC.
- J. Simon, L. K. Muller, M. Kokkinopoulou, I. Lieberwirth, S. Morsbach, K. Landfester and V. Mailander, Nanoscale, 2018, 10, 10731–10739 RSC.
- F. Giulimondi, L. Digiacomo, D. Pozzi, S. Palchetti, E. Vulpis, A. L. Capriotti, R. Z. Chiozzi, A. Lagana, H. Amenitsch, L. Masuelli, G. Peruzzi, M. Mahmoudi, I. Screpanti, A. Zingoni and G. Caracciolo, Nat. Commun., 2019, 10, 1–11 CrossRef PubMed.
- L. K. Müller, J. Simon, S. Schöttler, K. Landfester, V. Mailänder and K. Mohr, RSC Adv., 2016, 6, 96495–96509 RSC.
- A. C. G. Weiss, H. G. Kelly, M. Faria, Q. A. Besford, A. K. Wheatley, C. S. Ang, E. J. Crampin, F. Caruso and S. J. Kent, ACS Nano, 2019, 13, 4980–4991 CrossRef CAS PubMed.
- Y. Ping, J. Guo, H. Ejima, X. Chen, J. J. Richardson, H. Sun and F. Caruso, Small, 2015, 11, 2032–2036 CrossRef CAS PubMed.
- H. Ejima, J. J. Richardson, K. Liang, J. P. Best, M. P. van Koeverden, G. K. Such, J. Cui and F. Caruso, Science, 2013, 341, 154–157 CrossRef CAS PubMed.
- J. Guo, Y. Ping, H. Ejima, K. Alt, M. Meissner, J. J. Richardson, Y. Yan, K. Peter, D. von Elverfeldt, C. E. Hagemeyer and F. Caruso, Angew. Chem., Int. Ed., 2014, 53, 5546–5551 CrossRef CAS PubMed.
- H. Ejima, J. J. Richardson and F. Caruso, Nano Today, 2017, 12, 136–148 CrossRef CAS.
- D. Wu, J. Zhou, M. N. Creyer, W. Yim, Z. Chen, P. B. Messersmith and J. V. Jokerst, Chem. Soc. Rev., 2021, 50, 4432–4483 RSC.
- J. H. Park, K. Kim, J. Lee, J. Y. Choi, D. Hong, S. H. Yang, F. Caruso, Y. Lee and I. S. Choi, Angew. Chem., Int. Ed., 2014, 126, 12628–12633 CrossRef.
- Y. Ju, C. Cortez-Jugo, J. Chen, T. Y. Wang, A. J. Mitchell, E. Tsantikos, N. Bertleff-Zieschang, Y. W. Lin, J. Song, Y. Cheng, S. Mettu, M. A. Rahim, S. Pan, G. Yun, M. L. Hibbs, L. Y. Yeo, C. E. Hagemeyer and F. Caruso, Adv. Sci., 2020, 7, 1902650 CrossRef CAS PubMed.
- Y. Ju, H. Liao, J. J. Richardson, J. Guo and F. Caruso, Chem. Soc. Rev., 2022, 51, 4287–4336 RSC.
- Y. Ju, J. Cui, M. Müllner, T. Suma, M. Hu and F. Caruso, Biomacromolecules, 2015, 16, 807–814 CrossRef CAS PubMed.
- Y. Ju, J. Cui, H. Sun, M. Mullner, Y. Dai, J. Guo, N. Bertleff-Zieschang, T. Suma, J. J. Richardson and F. Caruso, Biomacromolecules, 2016, 17, 2268–2276 CrossRef CAS PubMed.
- Y. Ju, Q. Dai, J. Cui, Y. Dai, T. Suma, J. J. Richardson and F. Caruso, ACS Appl. Mater. Interfaces, 2016, 8, 22914–22922 CrossRef CAS PubMed.
- S. Li, Y. Ju, J. Zhou, K. F. Noi, A. J. Mitchell, T. Zheng, S. J. Kent, C. J. H. Porter and F. Caruso, ACS Appl. Mater. Interfaces, 2021, 13, 35494–35505 CrossRef CAS PubMed.
- Y. Ju, H. G. Kelly, L. F. Dagley, A. Reynaldi, T. E. Schlub, S. K. Spall, C. A. Bell, J. Cui, A. J. Mitchell, Z. Lin, A. K. Wheatley, K. J. Thurecht, M. P. Davenport, A. I. Webb, F. Caruso and S. J. Kent, ACS Nano, 2020, 14, 15723–15737 CrossRef PubMed.
- J. Zhou, Z. Lin, M. Penna, S. Pan, Y. Ju, S. Li, Y. Han, J. Chen, G. Lin, J. J. Richardson, I. Yarovsky and F. Caruso, Nat. Commun., 2020, 11, 1–8 CrossRef PubMed.
- B. Schwanhäusser, D. Busse, N. Li, G. Dittmar, J. Schuchhardt, J. Wolf, W. Chen and M. Selbach, Nature, 2011, 473, 337–342 CrossRef PubMed.
- M. Faria, M. Bjornmalm, K. J. Thurecht, S. J. Kent, R. G. Parton, M. Kavallaris, A. P. R. Johnston, J. J. Gooding, S. R. Corrie, B. J. Boyd, P. Thordarson, A. K. Whittaker, M. M. Stevens, C. A. Prestidge, C. J. H. Porter, W. J. Parak, T. P. Davis, E. J. Crampin and F. Caruso, Nat. Nanotechnol., 2018, 13, 777–785 CrossRef CAS PubMed.
- Q.-Z. Zhong, S. Li, J. Chen, K. Xie, S. Pan, J. J. Richardson and F. Caruso, Angew. Chem., Int. Ed., 2019, 58, 12563–12568 CrossRef CAS PubMed.
- J. R. Cann, R. A. Brown and J. G. Kirkwood, J. Am. Chem. Soc., 1949, 71, 1609–1614 CrossRef CAS.
- X. Zheng, H. Baker, W. S. Hancock, F. Fawaz, M. McCaman and E. Pungor Jr, Biotechnol. Prog., 2006, 22, 1294–1300 CrossRef CAS PubMed.
- N. Bertrand, P. Grenier, M. Mahmoudi, E. M. Lima, E. A. Appel, F. Dormont, J. M. Lim, R. Karnik, R. Langer and O. C. Farokhzad, Nat. Commun., 2017, 8, 1–8 CrossRef CAS PubMed.
- J. Song, Y. Ju, T. H. Amarasena, Z. Lin, S. Mettu, J. Zhou, M. A. Rahim, C. S. Ang, C. Cortez-Jugo, S. J. Kent and F. Caruso, ACS Nano, 2021, 15, 10025–10038 CrossRef PubMed.
- Q. He, Z. Zhang, F. Gao, Y. Li and J. Shi, Small, 2011, 7, 271–280 CrossRef CAS PubMed.
- R. Garcia-Alvarez, M. Hadjidemetriou, A. Sanchez-Iglesias, L. M. Liz-Marzan and K. Kostarelos, Nanoscale, 2018, 10, 1256–1264 RSC.
- L. Yang, L. Han, Q. Liu, Y. Xu and L. Jia, Acta Biomater., 2017, 64, 187–199 CrossRef CAS PubMed.
- T. K. Boehm and E. DeNardin, Hum. Antibodies, 2008, 17, 45–56 CAS.
- J. K. Ryu, M. A. Petersen, S. G. Murray, K. M. Baeten, A. Meyer-Franke, J. P. Chan, E. Vagena, C. Bedard, M. R. Machado, P. E. Rios Coronado, T. Prod'homme, I. F. Charo, H. Lassmann, J. L. Degen, S. S. Zamvil and K. Akassoglou, Nat. Commun., 2015, 6, 1–15 Search PubMed.
- V. Vitorino de Almeida, A. Silva-Herdade, A. Calado, H. Rosario and C. Saldanha, Clin. Hemorheol. Microcirc., 2015, 59, 97–106 CAS.
- V. P. Vu, G. B. Gifford, F. Chen, H. Benasutti, G. Wang, E. V. Groman, R. Scheinman, L. Saba, S. M. Moghimi and D. Simberg, Nat. Nanotechnol., 2019, 14, 260–268 CrossRef CAS PubMed.
- C. M. Liu, J. Chen, S. Yang, L. G. Mao, T. T. Jiang, H. H. Tu, Z. L. Chen, Y. T. Hu, L. Gan, Z. J. Li and J. C. Li, J. Ethnopharmacol., 2018, 225, 271–278 CrossRef PubMed.
- K. Ali, M. Middleton, E. Pure and D. J. Rader, Circ. Res., 2005, 97, 922–927 CrossRef CAS PubMed.
- G. S. Getz and C. A. Reardon, J. Lipid Res., 2009, S156–S161 CrossRef CAS PubMed.
- S. Ritz, S. Schottler, N. Kotman, G. Baier, A. Musyanovych, J. Kuharev, K. Landfester, H. Schild, O. Jahn, S. Tenzer and V. Mailander, Biomacromolecules, 2015, 16, 1311–1321 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Nanoparticle and nanocapsule characterization (size, ζ-potential, thickness); TEM images of the templates; gating strategy used in whole blood assays; cell association data of the whole blood assays with statistical analysis included; whole blood assay mass cytometry antibody panel; types and enrichment indexes of corona proteins; and MIRIBEL checklist for reporting research in bio–nano science. See DOI: https://doi.org/10.1039/d2tb00672c |
| ‡ Present address: College of Biomass Science and Engineering, National Engineering Research Center of Clean Technology in Leather Industry, Sichuan University, Chengdu 610065, China |
| This journal is © The Royal Society of Chemistry 2022 |