Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Direct imaging of polymer filaments pulled from rebounding drops†
Zi Qiang
Yang
 a,
Peng
Zhang
b,
Meng
Shi
a,
Peng
Zhang
b,
Meng
Shi
 a,
Ali
Al Julaih
a,
Ali
Al Julaih
 a,
Himanshu
Mishra
a,
Himanshu
Mishra
 b,
Enzo
Di Fabrizio
b,
Enzo
Di Fabrizio
 c and
Sigurdur T.
Thoroddsen
c and
Sigurdur T.
Thoroddsen
 *a
*a
aDivision of Physical Science and Engineering, King Abdullah University of Science and Technology (KAUST), Thuwal, 23955-6900, Saudi Arabia. E-mail: sigurdur.thoroddsen@kaust.edu.sa
bDivision of Biological and Environmental Science and Engineering, King Abdullah University of Science and Technology (KAUST), Thuwal, 23955-6900, Saudi Arabia
cDepartment of Applied Science and Technology, Politecnico di Torino, Torino, 10129, Italy
First published on 21st June 2022
Abstract
Polymer filaments form the foundation of biology from cell scaffolding to DNA. Their study and fabrication play an important role in a wide range of processes from tissue engineering to molecular machines. We present a simple method to deposit stretched polymer fibers between micro-pillars. This occurs when a polymeric drop impacts on and rebounds from an inclined superhydrophobic substrate. It wets the top of the pillars and pulls out liquid filaments which are stretched and can attach to adjacent pillars leaving minuscule threads, with the solvent evaporating to leave the exposed polymers. We use high-speed video at the microscale to characterize the most robust filament-forming configurations, by varying the impact velocity, substrate structure and inclination angle, as well as the PEO-polymer concentration. Impacts onto plant leaves or a randomized nano-structured surface leads to the formation of a branched structure, through filament mergers at the free surface of the drop. SEM shows the deposition of filament bundles which are thinner than those formed by evaporation or rolling drops. Raman spectroscopy identifies the native mode B stretched DNA filaments from aqueous-solution droplets.
1 Introduction
Methods to produce and collect polymer filaments are of primary importance in biology, tissue engineering, medicine, pharmacology and textiles.1–6 Several attempts have been reported to obtain single molecular-level polymer filaments like DNA suspended between micro-pillars, using methods like stamp peeling,7,8 evaporation of fakir drops9–14 and drop sliding.15–17Herein, we demonstrate a robust approach to produce and collect highly ordered arrays of polymer nanostrands with well-defined length and orientation. This involves impacting a drop containing polymers on a microfabricated superhydrophobic pillared surface, where the rebounding pulls out the filaments and then deposits them onto the pillars. To leave the filaments intact on the pillars, the droplet must bounce away from the original impact center, which we accomplish by tilting the substrate away from the horizontal. We show that both PEO and DNA filaments can become thinner by drop impact, than by the evaporative technique. We use time-resolved 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fps high-speed video imaging, on the micro-scale, to study the details of this process. Different filament structures can be pulled from the drop during the rebounding, depending on whether the substrate has random roughness or regular pillars, as shown in Fig. 1. This raises the prospect of targeted pesticide delivery, especially in dry climates, when molecular structures on these filaments could embed targeted molecules, protecting against the arrival of pests. The details of the filament pulling could also help design optimal spraying techniques to increase droplet retention on superhydrophobic leaves. Our preliminary experiments with lemongrass, show filament formation for repeated bounces of a micro-drop from the leaf surface, as shown in Fig. 1(c). For biomedical applications, our method, in conjunction with existing purification techniques, presents a platform to rapidly attach very thin polymer filaments between the micro-pillars to probe molecular structure, conformations or other properties for diagnostics.18
000 fps high-speed video imaging, on the micro-scale, to study the details of this process. Different filament structures can be pulled from the drop during the rebounding, depending on whether the substrate has random roughness or regular pillars, as shown in Fig. 1. This raises the prospect of targeted pesticide delivery, especially in dry climates, when molecular structures on these filaments could embed targeted molecules, protecting against the arrival of pests. The details of the filament pulling could also help design optimal spraying techniques to increase droplet retention on superhydrophobic leaves. Our preliminary experiments with lemongrass, show filament formation for repeated bounces of a micro-drop from the leaf surface, as shown in Fig. 1(c). For biomedical applications, our method, in conjunction with existing purification techniques, presents a platform to rapidly attach very thin polymer filaments between the micro-pillars to probe molecular structure, conformations or other properties for diagnostics.18
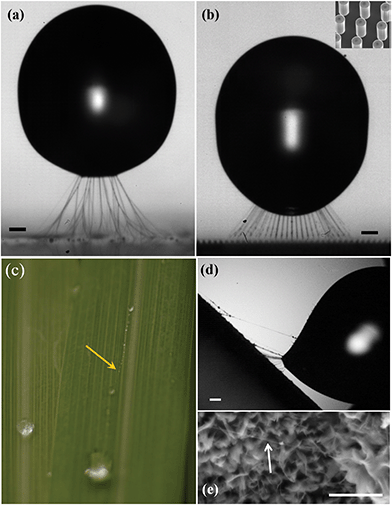 | ||
| Fig. 1 Polymer filaments pulled from rebounding drops after impact on a nano-particle-coated surface in (a) and on micro-pillars in (b). In (a) the glass substrate has four layers of “glaco” nanoparticle coating,19 with impact conditions D = 1.40 mm, U = 0.36 m s−1, Re = 463 & We = 2.64, while in (b) the micro-pillars are coated with FDTS, for D = 1.42 mm, U = 0.42 m s−1; Re = 552 & We = 3.67. The inset shows an SEM image of the pillars, which are 50 μm tall and spaced by 50 μm. For the random-roughness glaco-surface the filaments become branched, while for the pillars regularly-spaced linear filaments are pulled out. The scale bars are 150 μm long. For more details, see Supplementary Videos 1 & 2 (ESI†). (c) Similar polymer drops leave polymer filaments (arrow) when bouncing from superhydrophobic leaves of lemon-grass. (d) Video frame of the configuration in (c). (e) SEM image of filaments left behind on the leave. Scale bar is 5 μm. | ||
Droplets can bounce at low impact velocities due to the presence of an unbroken lubricating air film between the impinging droplet and the surface, as has been studied extensively using interferometry.10,20–24 Superhydrophobic surfaces with specific surface structures like various pillar arrangements also enable drop bouncing25–31 with reduced contact time.32,33 Chen et al.34 and Li et al.35 have previously observed these filaments and found them to delay the drop retraction. However, their 20 μm px−1 resolution could not capture the details of the finest filaments observed herein.
2 Experimental section
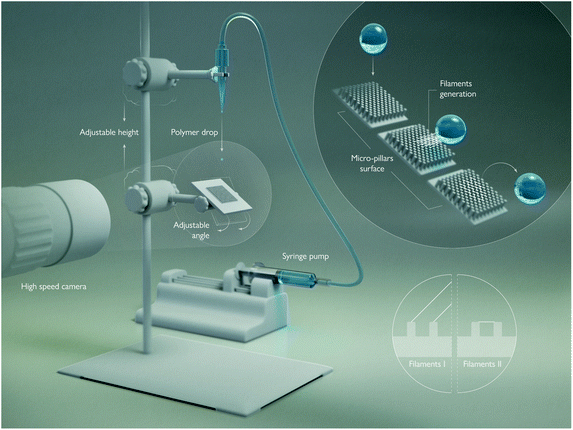
Fig. 2 shows a schematic of the experimental setup. We use Phantom V2511 ultra-fast CMOS video cameras at up to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 frame-per-second (fps), with a Leica Z16 APO long-distance microscope with adjustable magnification of up to 29.2, which gives pixel resolution up to about 1 μm px−1. In some experiments we used two synchronized cameras, for different viewing angles.
000 frame-per-second (fps), with a Leica Z16 APO long-distance microscope with adjustable magnification of up to 29.2, which gives pixel resolution up to about 1 μm px−1. In some experiments we used two synchronized cameras, for different viewing angles.
The drops are released by gravity from a glass nozzle with 100 μm inner diameter. Liquid was fed using a syringe pump at slow flow-rate of 2 μl min−1 to generate repeatable drops through static pinch-off.
Micro-pillar fabrication
Silicon wafers (4 inch diameter and 500 μm thickness) with a 2.4 μm thickness silica layer were used as substrates. The arrays of cylindrical pillars (diameter: 20 μm, height: 50 μm, and pitch: 50 μm) were fabricated by photolithography and dry etching. After fabrication, the wafers with features were cleaned by Piranha solution (H2SO4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) H2O2= 3
H2O2= 3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 by volume) for 15 min at 110 °C, and then flushed by de-ionized water and dried in a Spin Rinse Dryer. To achieve hydrophobicity, these wafers were then coated with perfluorodecyltrichlorosilane (FDTS) by molecular vapor deposition (ASMT 100E).
1 by volume) for 15 min at 110 °C, and then flushed by de-ionized water and dried in a Spin Rinse Dryer. To achieve hydrophobicity, these wafers were then coated with perfluorodecyltrichlorosilane (FDTS) by molecular vapor deposition (ASMT 100E).
PEO
The polyethylene-oxide (PEO) was purchased from Sigma-Aldrich (Mw = 4 × 106 g mol−1). It was in powder form, which was mixed with DI water (Milli-Q Plus system) using a magnetic mixer for at least 10 hours with low angular speeds before use.SEM
The PEO polymer filaments stretched between the pillars are plasma-coated with 2 nm iridium to increase the contrast and prevent it from breaking from the SEM electric beam, which was usually kept below 3 kV. 5 kV was used in Fig. 5(c) and a magnification of 3500. Twice the thickness of the iridium layer was subtracted from the diameter measurements.λ-DNA
λ-DNA was purchased from Thermo Fisher Scientific with molecular weight of 31.5 × 106 Dalton and concentration of 300 ppm from Thermo Fisher Scientific. The solvent is mostly water, but contains 10 mM tris–HCl (pH 7.6) and 1 mM EDTA. The 48.5 kb λ-DNA molecule has a B-form contour length of ∼16.5 μm (with an upper limit of 0.34 nm bp−1).9Raman spectroscopy
Laser-confocal Raman microscopy (WiTec Apyron) was used to deploy the Raman measurements. To avoid breaking the PEO nano-filaments, a low dose (5 mW) 532 nm laser was used as the excitation source. A 100× objective lens (NA = 0.9) was used to focus the laser spot on DNA nano-filaments and collect Raman scattering signal.3 Results and discussion
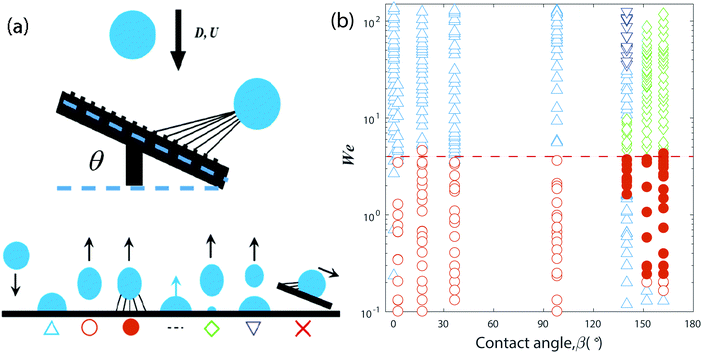
The experimental set-up used for drop impacts on inclined micro-pillared substrates, to collect PEO and DNA filament bundles, is sketched in Fig. 2 and 3. We first map the bouncing regimes of a polymer drop impacting on different solid substrates, while keeping the PEO concentration fixed at 100 ppm. The surfaces are characterized by the effective contact angle β, listed in the caption of Fig. 3(a). They include flat mica sheet, flat silica, silica surface treated with plasma, silica surface coated with FDTS, one & four times Glaco-nanoparticle-coated surfaces and surface with cylindrical micro-pillar array (height h = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm; diameter d = 20
μm; diameter d = 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm; spacing w = 50
μm; spacing w = 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm) coated with FDTS, see also Table S1 (ESI†).
μm) coated with FDTS, see also Table S1 (ESI†).
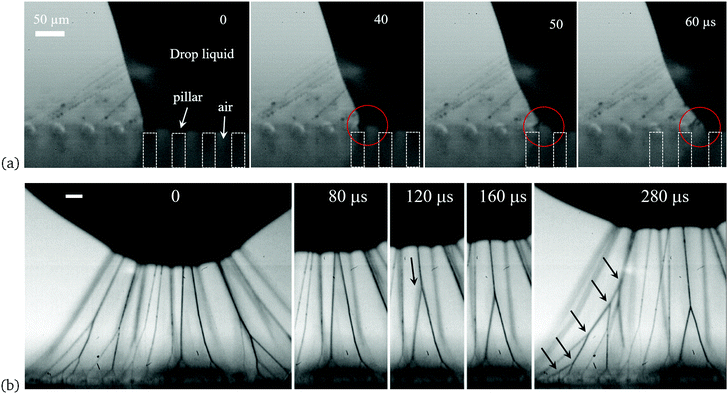 | ||
| Fig. 4 Filament formation for impact on horizontal substrates. (a) The process of liquid pinning on top of the pillars and pulling out of filaments, as the rim of liquid body moves to the next pillars during the retraction, marked by the red circles. The impact conditions are the same as in Fig. 1(b). (b) Branching of filaments during rebound from a random nano-particle coated glass surface, under the same impact conditions as in Fig. 1(a). The first frames are chosen as time reference and the scale bars are 50 μm long. See also Supplementary Movies 3 & 4 (ESI†). The first arrow indicates the merging motion of the two filaments. The arrows in the last panel identify the nodal points of the multiply-branched structure. | ||
The phase-diagram in Fig. 3(b) sums up the bouncing behavior of 100 ppm PEO droplets, of diameter D, for normal impact on these surfaces at different velocities U, characterized in terms of the Weber number We = ρDU2/σ, describing the ratio between inertial forces and surface tension, where ρ is the liquid density and σ the surface tension. Starting with the uncoated micropillar surface (β ≃ 0°), a complete deposition is observed over the whole range of We, because the liquid wets the whole superhydrophilic substrate immediately following contact with the pillars. On the other hand, for all of the smooth flat surfaces (β = 2° → 100°), the drop bounces within a very similar range starting at Wemin ∼ 0.1 to Wemax ∼ 4 (dashed red line in Fig. 3(b)), above this value the drop deposits on the surface. This is true even for the hydrophilic substrate and agrees well with earlier results,20,21 indicating the 100 ppm PEO drops are not in direct contact with the solid, with a stable lubricating air-layer between drop and substrate. This lack of contact prevents any filament formation.
For the rough or structured superhydrophobic surfaces (β ≥ 140°) the outcomes are more varied. The most prominent change, is the repeatable formation of polymer filaments during the rebounding. Furthermore, for the lowest impact velocities, i.e. the smallest We, direct deposition occurs. Above this we observe repeatable rebounding which always generates fiber filaments. For the FDTS-coated micropillars (β = 140°) this rebounding window is narrowest between We = 2.19–3.74, followed by partial rebounding and deposition at larger We.
It is therefore clear that in rebounding cases, there must be penetration through the air-layer and local wetting to pull liquid filaments from the drop. We conclude that the combined requirement of rebounding and local contact demands the use of substrates with surface structure for filament pull-out. Whether this liquid filament forms into a polymer filament is shown below to depend on the polymer concentration.
Similar progression of outcomes is observed for the random roughness of the nano-particle-coated Glaco surfaces. Both one and four coatings (β = 152° & 162°) show direct deposition for the lowest impact velocities. Furthermore, we find a narrow range of bouncing without filaments, which is followed, starting at We ≃ 0.2, by a large range of rebounding with polymer filaments. This filament-formation regime covers a much wider range of We than for the regular pillars. Again, for We > 4 one sees partial rebounding, with prominent liquid blob left behind (see inset sketch in Fig. 3(a)). We note that the critical We = 4 needed for the bottom of the drop to penetrate the air film is not changed for textured or flat surfaces, as marked by the dashed red line in Fig. 3(b). Finally, for even larger We, we see partial rebounding over a large range of We. Further increase in We above the region shown in Fig. 3(b) splashing begins from the rapidly expanding lamellae.36
These results show that the most regular filaments occur for impacts on the superhydrophobic micropillar array, which we now focus on. Fig. 5(a) shows how changing the concentration of PEO affects the impact dynamics on these pillars. We characterize this concentration, going from low to high at 0, 10, 50, 100, 200, 400, 1000 ppm, i.e. from left to right in the figure, in terms of the Deborah number,37 which is the ratio of time-scales De = τE/τR, where τE is the extensional relaxation time of the polymers, while Rayleigh capillary-inertial timescale 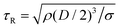 marks the hydrodynamic reaction time, from balancing inertia of the drop with the driving force of the surface tension. Starting with pure water we see a large rebounding region while deposition occurs for both lower and higher impact velocities. For low PEO concentrations of 10 & 50 ppm, while similar to water, the rebounding regime reduces, replaced by partial rebounding for We > 5 and then above We ≃ 10 larger wetting and deposition occurs with vertically ejected droplets. However, filaments are not observed during these rebounds, perhaps due to insufficient polymer concentrations to stabilize the liquid filaments emanating from the contacts with the pillar surfaces. Drops of both 100 and 200 ppm concentrations show clear We-ranges of rebounds with filaments. For 200 ppm the region moves to higher We, thereby requiring larger impact velocities. The pulling of more filaments reduces the rebounding height (see Fig. S2, ESI†) and partial rebound no longer occurs. The 200 ppm also generates thicker filaments with more droplets along them, which leads to a more irregular filament formation. Considering the importance of the rebounding height, for filament-collection scenarios, we therefore conclude that 100 ppm is the optimal concentration for our purposes. For even higher concentrations of PEO, of 400 ppm and 1000 ppm in Fig. 5(a), the rebounding regime disappears and deposition rules the entire range of We.
marks the hydrodynamic reaction time, from balancing inertia of the drop with the driving force of the surface tension. Starting with pure water we see a large rebounding region while deposition occurs for both lower and higher impact velocities. For low PEO concentrations of 10 & 50 ppm, while similar to water, the rebounding regime reduces, replaced by partial rebounding for We > 5 and then above We ≃ 10 larger wetting and deposition occurs with vertically ejected droplets. However, filaments are not observed during these rebounds, perhaps due to insufficient polymer concentrations to stabilize the liquid filaments emanating from the contacts with the pillar surfaces. Drops of both 100 and 200 ppm concentrations show clear We-ranges of rebounds with filaments. For 200 ppm the region moves to higher We, thereby requiring larger impact velocities. The pulling of more filaments reduces the rebounding height (see Fig. S2, ESI†) and partial rebound no longer occurs. The 200 ppm also generates thicker filaments with more droplets along them, which leads to a more irregular filament formation. Considering the importance of the rebounding height, for filament-collection scenarios, we therefore conclude that 100 ppm is the optimal concentration for our purposes. For even higher concentrations of PEO, of 400 ppm and 1000 ppm in Fig. 5(a), the rebounding regime disappears and deposition rules the entire range of We.
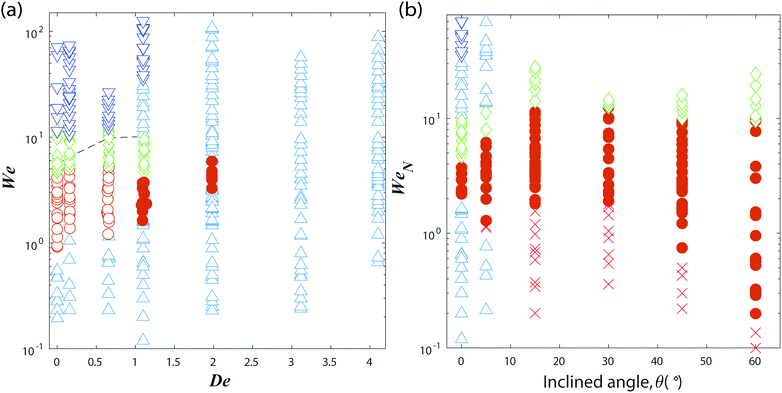 | ||
Fig. 5 Parameter regimes for the impact of a polymeric water drop: (a) effect of PEO concentration for a drop impacting on a horizontal surface with cylindrical micro-pillars coated with FDTS. The PEO concentration is characterized by the Deborah number De, going from low to high at 0, 10, 50, 100, 200, 400, 1000 ppm, i.e. from left to right in the figure. (b) Influence of the surface inclination angle θ. Here we use the Weber number based on the normal component of the impact velocity, UN = U![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ. Meaning of the symbols is the same as in Fig. 3. θ. Meaning of the symbols is the same as in Fig. 3. | ||
In the parameter space of Figure panels 3(b) & 5(a) the impacts were all perpendicular to the horizontal substrates. To deposit and study the filaments the drop cannot be allowed to return straight back down, so we instead incline the substrate and let the drop bounce downwards to leave the filaments attached to the pillars, as sketched in Fig. 3(a). Fig. 5(b) shows the results for the 100 ppm PEO drop impacting on the pillars, over a large range of inclination angles. Here we use the normal component of the impact velocity, UN = U![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) θ, in the Weber number WeN. The inclined substrates all show larger regions of filament formation than for first impact on horizontal pillars, θ = 0. For low impact velocities the drops tend to slide along the pillars before rebounding. For θ > 10° this sliding leaves filaments and occurs over a large range of low impact velocities, reminiscent of the pure sliding tested by.15–17 At slightly higher U, for WeN ∼ 1, the rebounding with filaments starts and persists until WeN ≃ 12, above which partial rebound occurs. While similar dynamics occur for θ between 15–60°, for consistency we limit the detailed study below primarily to the 45° angle.
θ, in the Weber number WeN. The inclined substrates all show larger regions of filament formation than for first impact on horizontal pillars, θ = 0. For low impact velocities the drops tend to slide along the pillars before rebounding. For θ > 10° this sliding leaves filaments and occurs over a large range of low impact velocities, reminiscent of the pure sliding tested by.15–17 At slightly higher U, for WeN ∼ 1, the rebounding with filaments starts and persists until WeN ≃ 12, above which partial rebound occurs. While similar dynamics occur for θ between 15–60°, for consistency we limit the detailed study below primarily to the 45° angle.
We now study the details of the filament formation using time-resolved high-speed video at micron-level spatial resolution, from different viewing angles (see ESI†). Fig. 1 showed overall snapshots after the drops have risen far above the surface, with long filaments reaching from drop to substrate. Fig. 4 and 6 show close-up video frames of this process for horizontal and inclined surfaces respectively. During the retraction of the edge of the drop leading to the rebound, in Fig. 4(a), we see the free surface of the drop attached to the top of a pillar, being pulled away leaving the filament. We also analyzed the location and moving velocity of the contact line between the micro-pillars and droplet in Fig. 4(a) and (b), which shows a clear stepwise evolution of the pinning dynamics and the process of filaments generation and stretch, corresponding to the images in Fig. 4(a). The average retraction velocity is around 0.2 m s−1 while the velocity jumps to a higher value of 1.24 m s−1 when the droplet liquid jumps from an outer pillars onto an inner one. The cylindrical pillars are marked by dashed white boxes and air is trapped between the pillars and the droplet. The liquid thread, spanning from the top of the pillar to the drop surface, is rapidly stretched reducing in diameter from about 50 μm to ∼5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm in only 60 μs. By assuming the conservation of liquid volume in the thread, its length must grow by the inverse square of its diameter, suggesting an effective stretching rate of the order of 106 s−1, thereby straightening the polymers.
μm in only 60 μs. By assuming the conservation of liquid volume in the thread, its length must grow by the inverse square of its diameter, suggesting an effective stretching rate of the order of 106 s−1, thereby straightening the polymers.
Fig. 6(a) shows a close-up sideview of filaments being pulled out of the drop's free surface, during rebounding from a 30° inclined substrate. The arrow in the second panel points out where a filament attaches to the liquid surface. One notices a small conical region around the filament where the two surface curvatures adjust. This matching region appears to reduce in size with time, as the filament thins. The size of the cones is much smaller than the spacing of the filaments, which corresponds directly to the distance between pillars, just like observed for the horizontal impacts in Fig. 1(b).
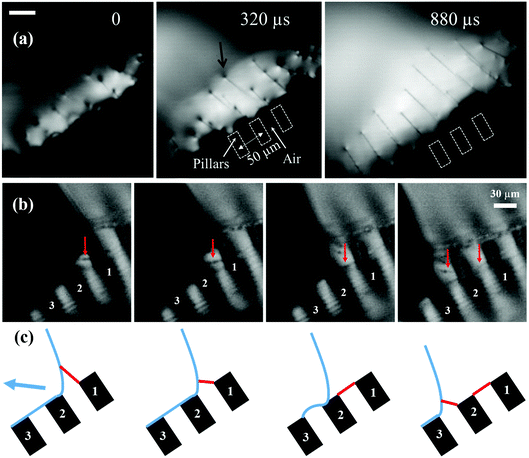 | ||
| Fig. 6 Close-up video frame of filaments generation attached on the top of two pillars from impinging droplets of aqueous PEO solution on hydrophobic surfaces for 100 ppm at angle 30°. The impact condition is D = 1.62 mm, U = 0.38 m s−1, Fr = 9, We = 3.The bright area in (a) is light reflected from the drop surface. A small conical region where the filament connects to the free surface is marked by the black arrow. The red arrows in (b) point out the generated filaments attached on the pillars. The blue arrow in (c) note the drop bouncing direction. The scale bars are 30 μm long. Frames from a video clip taken at 25 kf.p.s. See also Supplementary Movies 5 and 6 (ESI†). | ||
In contrast to the almost fixed-spaced filaments from impacts on the micro-pillars, the filaments generated by impacts on the randomly-spaced roughness-peaks of the Glaco-coated surfaces do not stay separate, as many of them are generated quite close to each other. The high-speed videos show that adjacent filaments often merge to form a unique branching structure, which was shown in Fig. 1(a). When filaments are too close to each other on the free surface, their conical menisci are pulled together by the surface tension to minimize surface energy, akin to the “Cheerios effect”38 experienced by adjacent objects floating on a liquid surface. This is shown in the sequence of frames in Fig. 4(b) and Supplemental Video 4 (ESI†). Once the cones merge, between 2nd and 3rd frame, two filaments meet at a cuspy corner. This is an unstable configuration, as is well-known in the evolution of foam, i.e., the stable angle between the filaments (Plateau borders) in a static 2-D configuration must be 120°. The initial angle between the two bottom branches are smaller than this value and the nodal point is rapidly pulled downwards to increase this angle. The downward zipping speed of the nodal point is as high as 3.29 m s−1, as marked by the arrow in Fig. 4(b). This merger can occur multiple times in sequence to form a distinctive branching structure. The last frame in the sequence points at five levels of nodes leading to the same filament attaching to the drop surface.
Our goal is to deposit polymer filaments between adjacent pillars to allow their study using molecular structure characterizations, such as determining chain conformation. Therefore after being pulled out they must attach to the next pillar and not be pulled out vertically as in Fig. 1(b) and (c). Fig. 6(b) shows how this is accomplished for impacts on an inclined substrate. The filament (marked by a red arrow) is pulled from pillar marked 1, but as the free surface is pulled towards pillar 2 it attaches to this pillar and is left behind, free-standing between these two pillars, while a new filament is pulled from pillar 2 and so forth. This attachment between adjacent pillars occurs during the retraction phase of the pancake, but not during the final rebound of the drop where it moves primarily up and away from the pillars. Regular deposition of these filaments between pillars is therefore limited to a sub-region of the area of the maximum impact footprint. This process is clearly seen in the Supplementary Video clip 6 (ESI†).
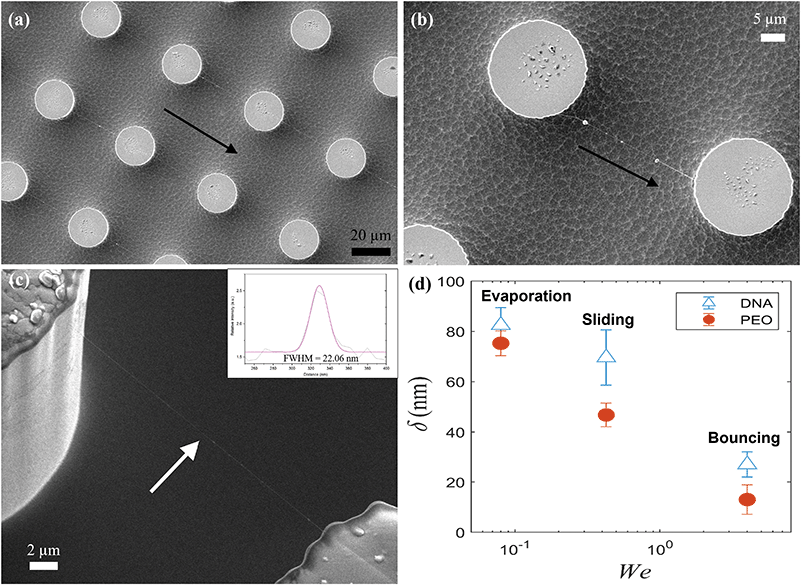
Soon after these impacts the substrates are removed and subjected to SEM imaging to characterize the size and structure of the deposited filaments. Fig. 7 shows typical SEM images of filaments attached between the tops of adjacent pillars. In panel (b) three tiny nodules indicate remnants of micro-droplets on the filament before drying. In (a) regular filaments are aligned towards bottom right between all pillars. This coincides with the inclination angle of the substrate, i.e. filaments are pulled out in the bouncing direction. Fig. S1 (ESI†) shows even larger number of regularly deposited filaments. Fig. S2 (ESI†) shows a more complicated filament structure, resulting from multiple drop impacts coming from different directions, which is accomplished by rotating the substrate between impacts. In the inset in panel (c) we fit the intensity profile across one filament to determine the Full–Width–Half–Max diameter, giving here a thickness value of δ = 22 nm. This thickness measurement was performed on numerous filaments and the results are shown in panel 3(d), where we contrast our drop-impact method to earlier work using drop-evaporation or rolling drops.11,12,14,39 This also includes results from drops containing λ-DNA. This shows that our impact method can generate thinner filaments than the other two approaches, which we verified with evaporating and rolling drops under the same experimental conditions as the impacts.
The filaments span approximately 30 μm distance between the pillars, which is much longer that the 16 μm λ-DNA segments. From Fig. 7(d), we therefore conclude that filaments are formed from bundles of overlapping λ-filaments. Due to the wide range of possible bouncing regimes, the attached location and thickness of the DNA filaments on the pillars could be optimized further by controlling the droplet impact condition and micro-pillar shapes.
Using Raman spectroscopy (see ESI†) we confirmed that the B-form DNA is the favoured conformation in nano-filaments at 45% humidty. During the drop bouncing, the DNA molecules are stretched by the hydrodynamic forces, which where described in the previous report. Below 80% humidity the stable structure is the B form, as verified in several experiments.11,14 Our impact technique therefore provides stretched λ-DNA in the B-from conformation, which is closer to the native structure in solution.
In summary, our experimental results present a simple but robust method to deposit stretched polymer fibers between micro-pillars, using inpacting drops. This deposition occurs much more rapidly than for the drop-evaporation technique. While the entire drop liquid evaporates on top of the pillars, leaving a heap of polymers at the center, the bouncing drop can be collected after the rebound, to bounce again on a new substrate, thereby using much smaller sample volume of liquid. Furthermore, the stronger stretching during the dynamic impact, appears to produce thinner filaments than the rolling technique, which can be beneficial for their detailed electron micro-scope imaging.
Author contributions
S. T. T. and Z. Q. Y. conceived the whole project, designed the research & participated in all phases of the work. Z. Q. Y. took the initial high-speed video data; S. T. T. and E. D. F. provided the original idea for DNA; Z. Q. Y. performed most experiments, participated in PEO, DNA preparation, high speed imaging and data analysis. A. A. J. carried out some of the experiments; H. M., M. S. and P. Z. designed and fabricated the micro pillar substrates; Z. Q. Y. and P. Z. performed the SEM imaging; P. Z. worked on Raman spectroscopy measurements and analysis; Z. Q. Y., S. T. T. and P. Z. wrote the manuscript and all authors discussed the results and edited the paper.Conflicts of interest
There are no conflicts of interest, but we have filed a provisional patent application.Acknowledgements
This work was supported by King Abdullah University of Science and Technology (KAUST), under grants URF/1/2126-01-01, URF/1/3727-01-01 & BAS/1/1352-01-01.Notes and references
- A. Goyanes, A. B. Buanz, A. W. Basit and S. Gaisford, Int. J. Pharm., 2014, 476, 88–92 CrossRef CAS PubMed.
- A. J. Van Brabant, R. Stan and N. A. Ellis, Annu. Rev. Genomics Hum. Genet., 2000, 1, 409 CrossRef CAS PubMed.
- D. Zhang, Advances in filament yarn spinning of textiles and polymers, Elsevier, 2014 Search PubMed.
- C. Soto and S. Pritzkow, Nat. Neurosci., 2018, 21, 1332–1340 CrossRef CAS.
- S. Stassi, M. Marini, M. Allione, S. Lopatin, D. Marson, E. Laurini, S. Pricl, C. F. Pirri, C. Ricciardi and E. Di Fabrizio, Nat. Neurosci., 2019, 10, 1–10 Search PubMed.
- D. H. Reneker and A. L. Yarin, Polymer, 2008, 49, 2387 CrossRef CAS.
- R. Lebofsky and A. Bensimon, Briefings Funct. Genomics, 2003, 1, 385–396 CrossRef CAS PubMed.
- J. Guan and L. J. Lee, Proc. Natl. Acad. Sci. U. S. A., 2005, 102, 18321–18325 CrossRef CAS PubMed.
- W. Wang, J. Lin and D. C. Schwartz, Biophys. J., 1998, 75, 513–520 CrossRef CAS PubMed.
- D. Quéré, Nat. Mater., 2002, 1, 14–15 CrossRef PubMed.
- F. Gentile, M. Moretti, T. Limongi, A. Falqui, G. Bertoni, A. Scarpellini, S. Santoriello, L. Maragliano, R. Proietti Zaccaria and E. Di Fabrizio, Nano Lett., 2012, 12, 6453–6458 CrossRef CAS PubMed.
- M. Marini, G. Das, R. La Rocca, F. Gentile, T. Limongi, S. Santoriello, A. Scarpellini and E. Di Fabrizio, Microelectron. Eng., 2014, 119, 151–154 CrossRef CAS.
- G. Ciasca, M. Papi, M. Chiarpotto, A. De Ninno, E. Giovine, G. Campi, A. Gerardino, M. De Spirito and L. Businaro, Micro Nano Lett., 2014, 6, 280–286 CrossRef.
- M. Marini, A. Falqui, M. Moretti, T. Limongi, M. Allione, A. Genovese, S. Lopatin, L. Tirinato, G. Das and B. Torre, et al. , Sci. Adv., 2015, 1, e1500734 CrossRef PubMed.
- B. Su, S. Wang, Y. Wu, X. Chen, Y. Song and L. Jiang, Adv. Mater., 2012, 24, 2780–2785 CrossRef CAS PubMed.
- B. Su, S. Wang, J. Ma, Y. Wu, X. Chen, Y. Song and L. Jiang, Adv. Mater., 2012, 24, 559–564 CrossRef CAS.
- G. Ciasca, L. Businaro, M. Papi, A. Notargiacomo, M. Chiarpotto, A. De Ninno, V. Palmieri, S. Carta, E. Giovine and A. Gerardino, et al. , Nanotechnology, 2013, 24, 495302 CrossRef CAS.
- E. Di Fabrizio, S. Schlücker, J. Wenger, R. Regmi, H. Rigneault, G. Calafiore, M. West, S. Cabrini, M. Fleischer and N. F. Van Hulst, et al. , J. Opt., 2016, 18, 063003 CrossRef.
- I. U. Vakarelski, N. A. Patankar, J. O. Marston, D. Y. Chan and S. T. Thoroddsen, Nature, 2012, 489, 274–277 CrossRef CAS PubMed.
- J. De Ruiter, R. Lagraauw, D. Van Den Ende and F. Mugele, Nat. Phys., 2015, 11, 48–53 Search PubMed.
- J. M. Kolinski, L. Mahadevan and S. Rubinstein, Europhys. Lett., 2014, 108, 24001 CrossRef.
- R. C. van der Veen, T. Tran, D. Lohse and C. Sun, Phys. Rev. E: Stat., Nonlinear, Soft Matter Phys., 2012, 85, 026315 CrossRef PubMed.
- R. C. van der Veen, M. H. Hendrix, T. Tran, C. Sun, P. A. Tsai and D. Lohse, Soft Matter, 2014, 10, 3703–3707 RSC.
- E. Li and S. T. Thoroddsen, J. Fluid Mech., 2015, 780, 636–648 CrossRef.
- W. Barthlott and C. Neinhuis, Planta, 1997, 202, 1–8 CrossRef CAS.
- D. Richard and D. Quéré, Europhys. Lett., 2000, 50, 769 CrossRef CAS.
- M. Reyssat, A. Pépin, F. Marty, Y. Chen and D. Quéré, Europhys. Lett., 2006, 74, 306 CrossRef CAS.
- D. Bartolo, F. Bouamrirene, E. Verneuil, A. Buguin, P. Silberzan and S. Moulinet, Europhys. Lett., 2006, 74, 299 CrossRef CAS.
- T. Deng, K. K. Varanasi, M. Hsu, N. Bhate, C. Keimel, J. Stein and M. Blohm, Appl. Phys. Lett., 2009, 94, 133109 CrossRef.
- D. H. Kwon, H. K. Huh and S. J. Lee, Exp. Fluids, 2013, 54, 1–11 CrossRef.
- H.-M. Kwon, A. T. Paxson, K. K. Varanasi and N. A. Patankar, Phys. Rev. Lett., 2011, 106, 036102 CrossRef.
- J. C. Bird, R. Dhiman, H.-M. Kwon and K. K. Varanasi, Nature, 2013, 503, 385–388 CrossRef CAS PubMed.
- Y. Liu, L. Moevius, X. Xu, T. Qian, J. M. Yeomans and Z. Wang, Nat. Phys., 2014, 10, 515–519 Search PubMed.
- L. Chen, Y. Wang, X. Peng, Q. Zhu and K. Zhang, Macromolecules, 2018, 51, 7817–7827 CrossRef CAS.
- B. Li, S. Lin, Y. Wang, Q. Yuan, S. Joo and L. Chen, New J. Phys., 2020, 22(12), 123001 CrossRef CAS.
- C. Josserand and S. T. Thoroddsen, Annu. Rev. Fluid Mech., 2016, 48, 365–391 CrossRef.
- M. Y. Pack, A. Yang, A. Perazzo, B. Qin and H. A. Stone, Phys. Rev. Fluids, 2019, 4, 123603 CrossRef.
- D. Vella and L. Mahadeva, Am. J. Phys., 2005, 73, 817–825 CrossRef.
- F. De Angelis, F. Gentile, F. Mecarini, G. Das, M. Moretti, P. Candeloro, M. Coluccio, G. Cojoc, A. Accardo and C. Liberale, et al. , Nat. Photonics, 2011, 5, 682–687 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available: Videos corresponding to Fig. 1, 4 and 6. See DOI: https://doi.org/10.1039/d2sm00599a |
| This journal is © The Royal Society of Chemistry 2022 |