 Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Correction: Guided run-and-tumble active particles: wall accumulation and preferential deposition
Chamkor
Singh
*ab
aDepartment of Physics, Central University of Punjab, Bathinda 151401, India. E-mail: iamchamkor@gmail.com
bDepartment of Chemical and Biological Engineering, Northwestern University, Evanston, IL 60208, USA
First published on 22nd December 2021
Abstract
Correction for ‘Guided run-and-tumble active particles: wall accumulation and preferential deposition’ by Chamkor Singh, Soft Matter, 2021, 17, 8858–8866, DOI: 10.1039/D1SM00775K.
The author regrets the following mistake in Section 2 of the original article, while presenting the computational procedure in the text. The procedure consistent with the computational program (which is public at https://gitlab.com/iamchamkor/bio3d) is described below.
The change in angle due to rotational noise and tumble events is first calculated in eqn (1) of the paper (page 8859, left column, line 40), as
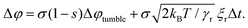 | (1) |
e* = σ[e![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Δφ + (n × e)sin Δφ + (n × e)sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Δφ] Δφ] | (2) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin
sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) φw (section 4, page 8861, right column, line 49) is also applied, then an additional change in angle due to this torque is calculated as Δφ2 = σ[−Γ
φw (section 4, page 8861, right column, line 49) is also applied, then an additional change in angle due to this torque is calculated as Δφ2 = σ[−Γ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) sin
sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) φ/γr]Δt. Here, φ is the angle between −k and e* (positive counterclockwise about the unit rotation axis, Fig. 4) and the unit rotation axis is w = −e* × k/|−e* × k|. In general, w is different from n. The bacteria is then reorientated using: e′ = σ[e*
φ/γr]Δt. Here, φ is the angle between −k and e* (positive counterclockwise about the unit rotation axis, Fig. 4) and the unit rotation axis is w = −e* × k/|−e* × k|. In general, w is different from n. The bacteria is then reorientated using: e′ = σ[e*![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) cos
cos![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Δφ2 + (w × e*)sin
Δφ2 + (w × e*)sin![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Δφ2]. If there is no guiding torque then obviously e′ = e*. Once reoriented due to rotational noise, tumbling, and the guiding torque, the bacteria is translated:
Δφ2]. If there is no guiding torque then obviously e′ = e*. Once reoriented due to rotational noise, tumbling, and the guiding torque, the bacteria is translated:  , and the program moves to the next time step. Here, ξt is a vector whose components are white Gaussian randoms. The rotational as well as translational diffusion remains active throughout runs as well as tumbles. The expression for torque in Section 4, page 8861, right column, line 46, has a typo and should be read as T = Γe × d.
, and the program moves to the next time step. Here, ξt is a vector whose components are white Gaussian randoms. The rotational as well as translational diffusion remains active throughout runs as well as tumbles. The expression for torque in Section 4, page 8861, right column, line 46, has a typo and should be read as T = Γe × d.
The Royal Society of Chemistry apologises for these errors and any consequent inconvenience to authors and readers.
| This journal is © The Royal Society of Chemistry 2022 |
