Open Access Article
Open Access ArticleMetal organic framework encapsulated tamavidin-Gluc reporter: application in COVID-19 spike antigen bioluminescent immunoassay†
Sherwin
Reyes
 ab,
Emily
Rizzo
a,
Albert
Ting
a,
Emre
Dikici
ab,
Emily
Rizzo
a,
Albert
Ting
a,
Emre
Dikici
 ab,
Sylvia
Daunert
ab,
Sylvia
Daunert
 abc and
Sapna K.
Deo
abc and
Sapna K.
Deo
 *ab
*ab
aDepartment of Biochemistry and Molecular Biology, University of Miami – Miller School of Medicine, Miami, FL 33136, USA. E-mail: SDeo@med.miami.edu
bThe Dr. John T. McDonald Foundation Bionanotechnology Institute of University of Miami, Miami, FL 33136, USA
cClinical and Translational Science Institute of University of Miami, FL 33136, USA
First published on 29th August 2022
Abstract
Enzyme linked immunosorbent assay (ELISA) is one of the most utilized serological methods to diagnose and identify etiologic agents of many infectious diseases and other physiologically important analytes. ELISA can be used either alone or adjunct to other diagnostic methods such as molecular arrays, and other serological techniques. Most ELISA assays utilize reagents that are proteinaceous in nature, which are not very stable and require cold-chain transport systems. Development of a desirable immunoassay requires stability of reagents used and its ability to be stored at room temperature without sacrificing the activity of the reagents or the protein of interest. Metal organic frameworks (MOFs) are a rapidly emerging and evolving class of porous polymeric materials used in a variety of biosensor applications. In this study, we introduce the use of MOFs to stabilize a universal reporter fusion protein, specifically, avidin-like protein (Tam-avidin2) and the small bioluminescent protein Gaussia luciferase (Gluc) forming the fusion reporter, tamavidin2-Gluc (TA2-Gluc). This fusion protein serves as a universal reporter for any assays that utilize biotin–avidin binding strategy. Using SARS-CoV2 S1 spike antigen as the model target antigen, we demonstrated that encapsulation of TA2-Gluc fusion protein using a nano-porous material, zeolitic imidazolate framework-8 (ZIF-8), allows us to store and preserve this reporter protein at room temperature for over 6 months and use it as a reporter for an ELISA assay. Our optimized assay was validated demonstrating a 0.26 μg mL−1 limit of detection, high reproducibility of assay over days, detection of spiked non-virulent SARS-COV2 pseudovirus in real sample matrix, and detection in real COVID-19 infected individuals. This result can lead to the utilization of our TA2-Gluc fusion protein reporter with other assays and potentially in diagnostic technologies in a point-of-care setting.
1. Introduction
Quantitative, accurate, specific, sensitive, rapid, and field deployable immunoassays are currently in-demand due to increasing cases of new and re-emerging infectious diseases. A variety of approaches including agglutination testing,1 immunofluorescent assay (IFA),2,3 radioimmunoassay (RIA),4 enzyme-linked immunosorbent assays (ELISA), and other immunoassay platforms have come a long way for the rapid detection of antigens or antibodies corresponding to such infectious agents.5 Amongst these methods, ELISA has been the go-to method in a clinical setting due to its simple procedure, specificity, short turn-around time (TAT), and high sensitivity.4,6 A typical ELISA utilizes an enzyme linked antigen or antibody conjugates. The formation of product of the enzymatic reaction provides for the generation of the signal that can be correlated with the target analyte/pathogen in a sample.6 Although RIA is one of the most sensitive immunoassay methods, owing to the use of radioactive labels, its application in diagnostic testing is now diminished. New ELISA and immunoassays using labels such as bioluminescent proteins termed bioluminescent immunoassay (BIA) and chemiluminescent substrates have achieved sensitivity comparable to RIA.4,7,8 Specifically, horseradish peroxidase label combined with chemiluminescent substrates and bioluminescent proteins such as Gaussia luciferase (Gluc), Renilla luciferase (Rluc), and aequorin have become popular labels for the design of new immunoassays. The use of bioluminescent proteins, which are small and can be expressed in bacteria, provides an additional advantage to create genetic fusions, allowing for better reproducibility of reagents and conjugate preparation. Our laboratory recently reported the construction and bacterial expression of a fusion protein composed of the biotin-binding protein tamavidin 2 (TA2) and bright luminescent reporter Gluc to generate the fusion protein TA2-Gluc.9 TA2 protein provides for binding to biotin and Gluc provides for signal generation with high signal/noise ratio upon binding of TA2 to biotin. We hypothesize that this fusion protein can serve as a universal reporter protein since it can be employed as a reporter in many assay types, including immunoassays, where avidin–biotin type binding is commonly utilized in an assay design. We have demonstrated that this fusion protein can be prepared in large quantities using bacterial expression and purification system and yields high bioluminescent signal.9 We also showed its utility as a reporter in DNA hybridization assays previously.9 Now we show that this reporter protein could be easily employed as a label in immunoassays for the development of highly sensitive, accurate, and rapid immunoassays for targets of interest such as pathogens. An additional characteristic that is desired in any immunoassay development is the stability of the reagents used and the ability to store them at room temperature or higher temperatures, to avoid the use of refrigeration. In that regard, lyophilization based approaches with different cryoprotectants are commonly used to impart stability to proteins for storage at room temperature. However, this method does not always yield desired stability without sacrificing the activity of the reagent/protein of interest. Therefore, additional methods such as encapsulation with polymers10,11 and metal–organic frameworks12–14 have been reported. In that regard, in this manuscript we have prepared room temperature stable TA2-Gluc based on encapsulation with metal organic frameworks and showed their application as a label in the design of an immunoassay using COVID-19 spike antigen as the model target analyte. We believe that the creation of a stable, extended shelf-life reporter that is based on universally used avidin–biotin binding and high signal generating bioluminescent protein will find application in a variety of assay designs.Metal–organic frameworks (MOFs) can be used to preserve and store labile biomolecules such as proteins at room temperature for longer periods.15–17 MOFs includes different composites of functional materials such as metal nanoparticles, oxides, quantum dots, polyoxometalates polymers, graphene, carbon nanotubes, biomolecules and many preparation methods.17–32 Amongst MOFs methods, encapsulation pave its way due to its distinct advantage as the MOF pore size is independent of the size of protein and its simplicity. The one step encapsulation method offers the highest level of protection and recyclability, as encapsulated enzymes generally retain better performance during reconstitution because the enzyme is embedded in the MOF particles. There are a multitude of possible combinations for the variety of metal ions, organic linkers, and structural motifs that can provide a great degree of structural diversity, wide range of chemical and physical tunability, low operational cost, good adsorption and desorption kinetics, ease of handling, and facile functionality.17,20 Among the many MOF related studies, more than half are related to zeolitic imidazolate framework-8 (ZIF-8) using imidazole as linker.33 As opposed to infiltration method that uses PVP or alcohol for synthesis, a study conducted by Liang et al.,22 showed that encapsulation using one-pot strategy known as biomimetic mineralization technique can be helpful in maintaining the viability and stability of biomacromolecules under various conditions. In this study, they have encapsulated two different enzymes, urease and horseradish peroxidase, within a ZIF-8 framework and demonstrated that the enzymes were shown to maintain their activity even after being exposed to denaturation conditions. They showed that the ZIF-8 framework was formed simultaneously in water by simply combining the enzymes, 2-methylImidazole (mIM) and zinc acetate. This system is preferable due to its high stability, straight forward production, and excellent performance.23,34 ZIF-8 is a highly porous type of MOF and an analogue of zeolites and is formed by tetrahedral N atom linkages of metal ions such as Zn2+ and Co2+.35 Thus, giving rise to a strong bond that provides enormous chemical and thermal stability compared to classic MOFs. Due to its structural flexibility, ZIF-8 possesses good functionality and a wide range of porosities that can be easily manipulated.32 MOFs ZIF-8, which contains zinc ions, can be used as a protective material for biomolecules to maintain the ability to recognize antibodies or viral proteins on the surface of biosensors stored at ambient and high temperatures.23,36–38
In this manuscript, we designed a bioluminescent immunoassay (BIA) using MOFs encapsulated TAM2-Gluc reporter for a model analyte SARS-CoV-2 antigen. Here we describe encapsulation of TA2-Gluc protein with ZiF-8 and demonstrate enhanced room temperature stability of TA-2 Gluc for an extended time period. Tamavidin-2 are a group of avidin-like biotin-binding proteins found in the tamogitake mushroom (Pleurotus comucopiae), commonly known as “oyster mushroom”. When compared to biotin-binding proteins avidin and streptavidin, TAM2 confers high affinity and is easily produced in soluble form in the bacterial expression system.39–41 Gluc is a small Renilla-like luciferase (19.9 kDa, emission at 470 nm) derived from copepods (Gaussia princeps) that has been cloned since 2002.42 Gluc, compared to other bioluminescent proteins, gives a high bioluminescent intensity. The encapsulated stable TA2-Gluc protein was employed as a label in the design of a highly sensitive bioluminescent immunoassay for the detection of COVID-19 spike antigen. Coronavirus disease (COVID-19) is an infectious respiratory disease caused by the novel coronavirus strain, SARS-CoV-2, that has spread throughout the world leading to a global pandemic.43,44 SARS-CoV-2 spreads primarily through droplets discharged from the nose or mouth when an infected person coughs or sneezes and can be detected using immunoassays.45–47 Therefore, SARS-COV2 antigen serves as a good example of antigen to demonstrate the use of stabilized universal bioluminescent reporter system.
2. Experimental section
2.1 Materials and methods
TA2-Gluc was expressed and purified using these reagents: Difco LB Broth Miller (Luria Bertani) (Cat. No. 244620) was purchased from VWR International, LLC (Radnor, PA). Ampicillin sodium (Cat. No. MBPC-1901) purchased from Molecular Biologicals International, Inc., (Irvine, CA). 500 mL 1× PBS (Cat. No. 10010049) and Ni–NTA resin (Cat. No. 88221) was purchased from ThermoFisher Scientific (Waltham, MA). Protein was visualized using mini-PROTEAN TGX Gels (Cat. No. 4561096) (BIORAD Life Sciences, Hercules, CA).
The COVID-19 Incubation buffer (CIB) was composed of 10 mM sodium phosphate dibasic, 2 mM potassium phosphate monobasic, 137 mM sodium chloride, 2.7 mM potassium chloride, 0.05% Tween 20, 1.0% bovine serum albumin, 2% poly (ethylene glycol) 6000 (PEG) at pH 7.4. The coating buffer was composed of 100 mM sodium bicarbonate at pH 9.6. Wash buffer was composed of phosphate buffered saline and Tween 20 (PBST) at pH 7.40. 5 μg mL−1 coelenterazine was used as bioluminescent substrate. Blocking buffer PBST (protein-free) (Cat. No. 786-665) was purchased at G-Biosciences (St. Louis, MO). The MOF-ZIF8 solubilizing solution was composed of 200 mM sodium phosphate, and 2 mM EDTA containing 0.01% Tween 20 at pH 5.6.
Cell culture and SARS-CoV-2 pseudovirus propagation was done using MA104 cell line (ATCC CRL-2378.1) purchased from ATCC (Manassas, VA) and medium 199 (1×) (Cat. No. 12340-030) purchased from GIBCO, Thermo Fisher Scientific, (Waltham, MA).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 MWCO Slide-A-lyzer dialysis cassettes G2 (Cat. No. 87730) purchased from ThermoFisher Scientific (Waltham, MA).
000 MWCO Slide-A-lyzer dialysis cassettes G2 (Cat. No. 87730) purchased from ThermoFisher Scientific (Waltham, MA).
Protein expression and purification was done using QSonica sonicator (Qsonica Llc, Newtown, CT), rotary mixer (IKA Trayster Digital) purchased from IKA Works, Inc., (Wilmington, NC) and visualized using SDS page electrophoresis machine purchased from BIORAD Life Sciences, (Hercules, CA).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g to collect the ZIF-8 encapsulated TA2-Gluc. The supernatant was removed, and the pellet was dried overnight. Representative samples were stored at various temperatures (25 °C, 4 °C and −20 °C) for future testing.
000 × g to collect the ZIF-8 encapsulated TA2-Gluc. The supernatant was removed, and the pellet was dried overnight. Representative samples were stored at various temperatures (25 °C, 4 °C and −20 °C) for future testing.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 × g.22 To release the encapsulated TA2-Gluc, 1 mL of solubilizing solution was added, vortexed thoroughly and incubated at room temperature for 45 minutes. After incubation, the tube was vortexed again and the luminescence signal of the released TA2-Gluc was determined by injecting 100 μL of 5 μg mL−1 coelenterazine and measured using a luminometer at 7.40 seconds interval.
000 × g.22 To release the encapsulated TA2-Gluc, 1 mL of solubilizing solution was added, vortexed thoroughly and incubated at room temperature for 45 minutes. After incubation, the tube was vortexed again and the luminescence signal of the released TA2-Gluc was determined by injecting 100 μL of 5 μg mL−1 coelenterazine and measured using a luminometer at 7.40 seconds interval.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution of pooled human saliva (PHS) sample containing live SARS-CoV-2 pseudovirus to simulate infection.
10 dilution of pooled human saliva (PHS) sample containing live SARS-CoV-2 pseudovirus to simulate infection.
3. Results and discussion
3.1 Tamavidin 2 – Gaussia luciferase
A novel genetic fusion protein was created in our laboratory combining a bioluminescent reporter (Gluc) and avidin-like protein, tamavidin, TA2 (TA2-Gluc). This novel genetic fusion protein promises a cost-effective universal reporter based on binding of TA2 to biotin and high luminescent output upon adding the substrate, coelenterazine for Gluc. This fusion can be readily produced using the bacterial expression system in a reproducible manner. This genetic fusion has 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 conjugation ratio and anything biotinylated can be detected using this system as compared to performing chemical conjugation between streptavidin/avidin and a reporter moiety where heterogeneous conjugate mixture is a pitfall.9
1 conjugation ratio and anything biotinylated can be detected using this system as compared to performing chemical conjugation between streptavidin/avidin and a reporter moiety where heterogeneous conjugate mixture is a pitfall.9
3.2 Room temperature stability of MOF-ZIF 8 encapsulated tamavidin 2 – Gaussia luciferase
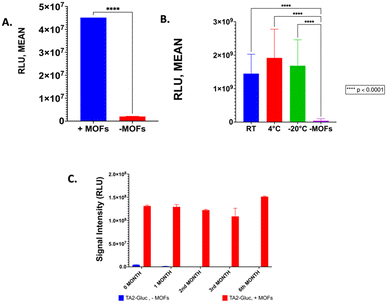
We have previously demonstrated that TA2-Gluc, was found to be stable for >6 months at 4 °C.9 Although this stability is desired, this requires cold chain infrastructure in order to use this fusion protein in assays at point of care or resource limited settings. To be able to store a protein-based reagent at room temperature is very desirable, however, our fusion protein loses its activity in 6 to 8 hours at room temperature. In order to address this problem, we decided to use metal organic framework encapsulation to stabilize TA2-Gluc. It has been shown in the literature that metal organic framework can be used to stabilize biological molecules very efficiently.14,20,23 For example, zinc ions coupled with methyl-imidazole molecules forms a cage-like encapsulation that traps the fusion protein (Fig. 1A), stabilizing the protein in the process. When needed, the encapsulated protein can be released from this cage like structure by solubilizing the MOFs-ZIF 8 using a slightly acidic buffer (pH ∼6) to shift the ionization of the imidazole moiety and thereby disrupting the Zn–imidazole interactions, and EDTA to chelate the released zinc ions (Fig. 1B). TA2-Gluc fusion protein, once released, can then react with its substrate, coelenterazine, to produce a bright luminescence (Fig. 1C). To demonstrate that this particular MOF encapsulated TA2-Gluc protein is more stable than the unencapsulated fusion protein, an equimolar solution of TA2-Gluc encapsulated with MOF-ZIF 8 was tested for stability and compared to native TA2-Gluc. The bioluminescence activity was used as the benchmark for stability, and we demonstrated that TA2-Gluc preserved by encapsulation exhibited higher luminescence intensity when compared to the native TA2-Gluc (Fig. 2A). Encapsulation or immobilization of various proteins on surfaces have been previously shown to improve protein activity and hence our observation of higher luminescence upon encapsulation is not surprising.18,53 The stability of the encapsulated fusion protein at various temperatures (room temperature (RT), 4 °C and −20 °C) was monitored and indicated that we can preserve this fusion protein with MOF-ZIF 8 (Fig. 2B) in any of the tested temperatures. We monitored the stability of this MOF-ZIF 8 fusion protein and demonstrated its stability for up to 6 months at ambient storage conditions that can be potentially utilized as part of an assay in a point-of- care setting (Fig. 1C).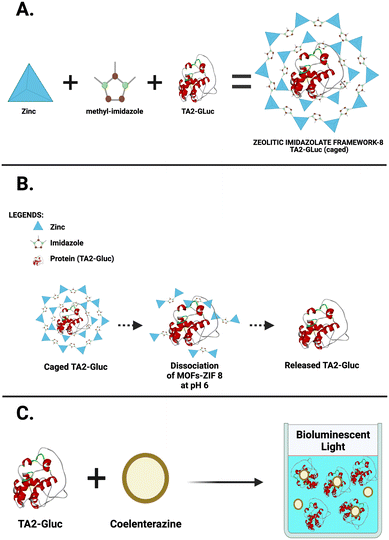 | ||
| Fig. 1 Metal organic frameworks (ZIF-8) A. Encapsulation of TA2-Gluc. B. TA2-Gluc release at pH 6. C. TA2-Gluc with its substrate coelenterazine producing bioluminescent signal. | ||
3.3 Optimization and evaluation of MOFs-ZIF 8 encapsulated TA2-Gluc in a bioluminescent immuno-assay (BIA)
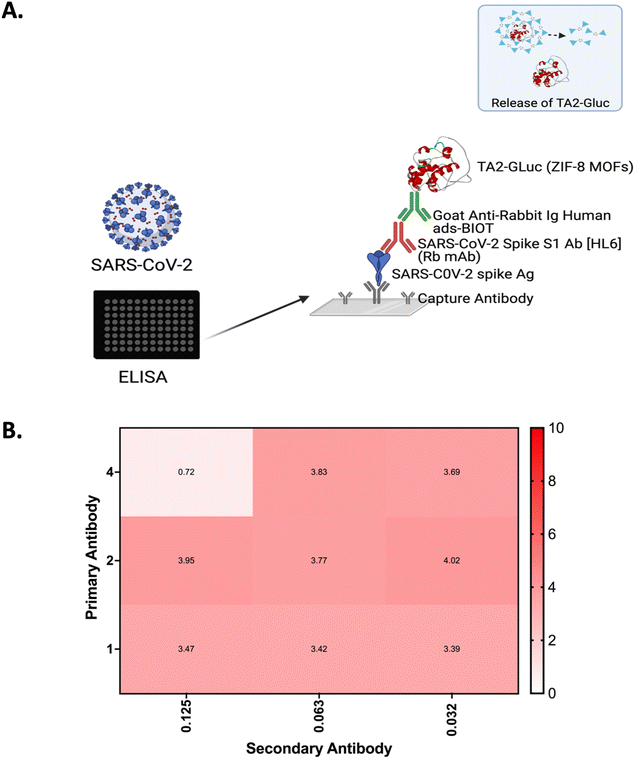
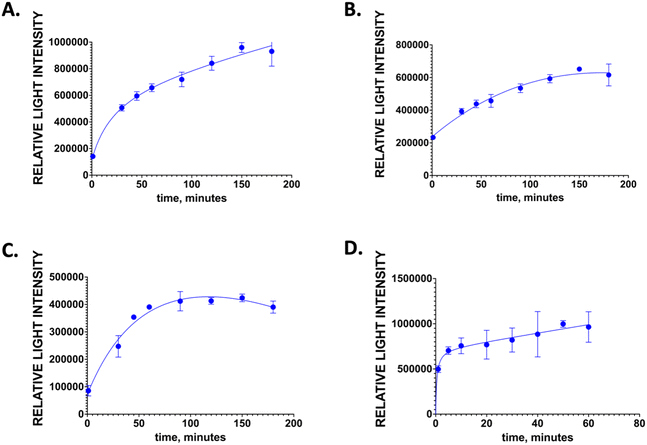
BIA are an excellent assay system for the detection of biomolecules54 as they afford high sensitivity and specificity. To further expand on their potential applications, we prepared room temperature stable universal bioluminescent reporter, MOF-ZIF 8 TA2-Gluc and showed its application in a BIA system. In this study, we used SARS-CoV-2 spike antigen as the model analyte for the design of BIA since this antigen is of importance in the detection of virus causing COVID-19 pandemic. The schematic for the BIA assay is depicted in Fig. 3A.For this assay, COVID-19 S1 spike antigen is first captured by SARS-CoV-2 spike antibody that is coated into the wells of the microtiter plate. When the antigen is captured, the primary antibody against the COVID-19 S1 spike antigen will bind to spike antigen forming an antibody sandwich. Once this antibody sandwich is formed, a biotinylated secondary antibody is added, which binds to the Fc receptor of the primary antibody. The reporter, MOF encapsulated TA2-Gluc binds to the biotin and a measurable signal is produced upon adding the substrate for the Gluc, coelenterazine. The signal produced is correlated to the amount of spike antigen and in turn the presence of SARS-CoV-2 viral copies present in the sample. The availability of this MOF-ZIF8 encapsulated fusion protein could be an avenue for a new and more stable reporters that can be used to detect target analytes of interest potentially in immunoassays applicable to a point-of-care setting. Moreover, since we have demonstrated its stability at room temperature for over six months, this MOF ZIF8 encapsulated protein can be shipped across the world without the need for a cold chain infrastructure. We believe this is a tremendous advantage for low resource countries. A checkerboard assay was done to optimize the concentration of the antibody pair (2 μg mL−1, primary antibody, and 0.32 μg mL−1 biotinylated secondary antibody) to improve the sensitivity of the assay. Signal to noise ratio was calculated by dividing the light intensity of those that have the COVID-19 S1 spike antigen (positive control) to the light intensity by those that do not have the COVID-19 s1 spike antigen (negative control) (Fig. 3B). The blocking conditions for this assay was optimized to reduce the non-specific binding. Several different blocking buffers and buffering conditions were tested, and we found that the blocking step is not necessary for this assay. The optimized incubation time for the antigen, primary antibody, biotinylated secondary antibody, and the reporter MOF-ZIF8 TA2-Gluc time are summarized in Fig. 4 and Table 1. We optimized the time of incubation to minimize overall assay time without compromising the sensitivity of the assay. The optimized incubation times and concentrations are listed in Table 1. Overall, the total assay time for the laboratory-based BIA was less than 6 hours. This is comparable to other laboratory-based immunoassays reported in the literature. The signal generation using MOF-ZIF 8 TA2-Gluc as a reporter is rapid within 3 s and provide a great addition to the existing reporters that are currently available on the market as a room temperature stable reporter.
| Components | Concentrations | Incubation time |
|---|---|---|
| Capture antibody | 3 μg mL−1 | Overnight |
| COVID-19 spike Ag | 2 μg mL−1 | 120 minutes |
| Primary antibody | 2 μg mL−1 | 120 minutes |
| Biotinylated secondary antibody | 0.032 μg mL−1 | 60 minutes |
| ZIF-8 encapsulated TA2-Gluc | 1 μM | 10 minutes |
3.4 Dose–response curve for SARS-CoV-2 S1 spike antigen and assay reproducibility
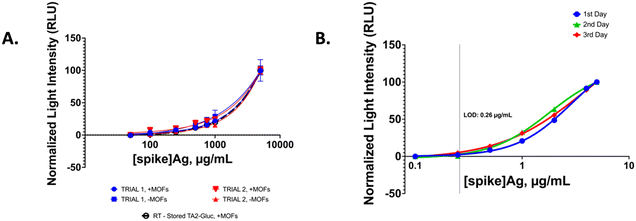
Using the optimized BIA conditions we generated a dose–response curve by varying SARS-CoV-2 spike antigen to obtain a detection limit. First, we compared the performance of the assay using TA2-Gluc without any MOFs encapsulation and compared the dose–response curve with the MOF encapsulated TA2-Gluc. We observed that the MOF encapsulated fusion protein gave similar results as compared to using the fusion protein alone (Fig. 5A). Similar dose–response curve was obtained while using MOFs encapsulated TA2-Gluc stored at room temperature over a longer period (Fig. 5A). This indicates that encapsulation of the fusion protein does not affect the assay performance while affording better stability to the reporter protein. Moreover, the room-temperature stored protein can be used as a reporter without affecting the performance of the assay. We also performed a reproducibility assay using the optimal conditions. For this purpose, three separate assays were performed on three separate days and the results were compared. Fig. 5B shows day to day reproducibility. There was no significant difference in the curves obtained over three days and the limit of detection of 0.26 μg mL−1 was obtained on all three days. The detection limit was obtained by interpolation from the GraphPad Prism software using the following formula LOD = SB + 3 × SDB where SB and SDB are the mean and the standard deviation of the blank signals, respectively.553.5 Detection of SARS-COV-2 pseudo virus in a physiological fluid and SARS-CoV-2 Ag in nasal swab samples of infected individuals
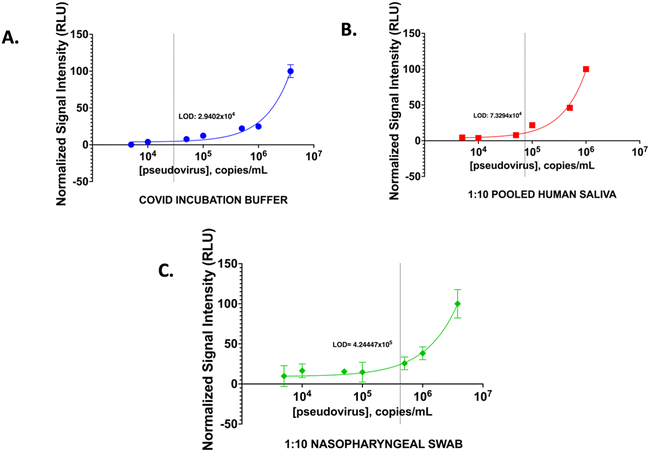
To demonstrate that we can use the BIA developed for the detection of virus if present in the physiological fluid we performed analysis using spiked pooled human saliva (PHS) and nasopharyngeal swab samples. We compared the performance in these samples with SARS-CoV-2 pseudovirus spiked into the COVID incubation buffer (CIB). Pseudovirus was used since it contains the spike protein of the virus but does not have the virulence and hence it can be safely handled. Different known copies per mL of pseudovirus were spiked into the CIB and PHS samples. BIA was performed following the optimized conditions and using these spiked samples (Fig. 6). A limit of detection of 2.94 × 104 copies per mL in CIB samples, 7.33 × 104 copies per mL in PHS samples and 4.24 × 105 for nasopharyngeal swab medium was obtained. For both physiological fluids 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
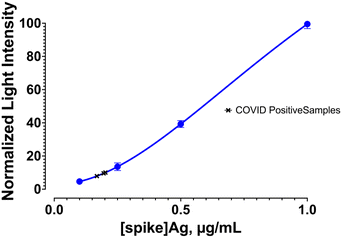
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10 dilution showed similar detection limit as the buffer. The levels of viral copies reported in saliva and nasopharyngeal swabs range from 104 to 107 copies per mL in infected individuals56,57 with symptoms. Although, RT-PCR-based analysis can yield much lower limit of detection due to the amplification process and allow early identification of COVID-19 infection, currently available immunoassay-based methods are able to diagnose symptomatic patients with similar detection limits as demonstrated in our assay since the sensitivity of the assays are dependent on binding constants of the antibodies employed and there is no amplification like in a molecular test. To show that our assay can detect COVID-19 positive patients similar to the currently available immunoassay tests, we evaluated nasal swab samples of individuals who were diagnosed with COVID-19 using the rapid antigen test. The bioluminescent assay using the fusion protein TA2-Gluc can detect viral spike antigen at concentration above the detection limit and correctly identified all (3) tested positive samples as positive in the assay (Fig. 7). This performance is similar to the currently available antigen tests.
10 dilution showed similar detection limit as the buffer. The levels of viral copies reported in saliva and nasopharyngeal swabs range from 104 to 107 copies per mL in infected individuals56,57 with symptoms. Although, RT-PCR-based analysis can yield much lower limit of detection due to the amplification process and allow early identification of COVID-19 infection, currently available immunoassay-based methods are able to diagnose symptomatic patients with similar detection limits as demonstrated in our assay since the sensitivity of the assays are dependent on binding constants of the antibodies employed and there is no amplification like in a molecular test. To show that our assay can detect COVID-19 positive patients similar to the currently available immunoassay tests, we evaluated nasal swab samples of individuals who were diagnosed with COVID-19 using the rapid antigen test. The bioluminescent assay using the fusion protein TA2-Gluc can detect viral spike antigen at concentration above the detection limit and correctly identified all (3) tested positive samples as positive in the assay (Fig. 7). This performance is similar to the currently available antigen tests.
4. Conclusions
Encapsulation of biological materials such as DNA, RNA, and proteins within metal organic frameworks improve their stability significantly. MOF encapsulated biologics are stable at room temperature therefore they do not require any refrigeration or cold-chain infrastructure. Since refrigeration is not required to store and ship these materials, these assays can be useful in low resource settings where access to power is very limited. In this study, we have encapsulated a fusion protein between tamavidin2 and Gaussia luciferase, which can be used as a universal reporter based on biotin–tamavidin-binding and was demonstrated that it can be stored at room temperature up to 6 months. Using MOF to enclose and encapsulate the bioluminescent protein allows us to store it for a longer period of time which provides a comparable or better preservation efficacy than freezing liquid samples at −20 °C. This is an energy-efficient and environmentally friendly approach that works to reduce the need for cold chain and temperature-controlled handling and testing of biospecimens.24 This encapsulated fusion protein then can be released, on demand, from its metal organic framework, and used as a reporter in bioluminescent assays. We also showed that this reporter can be used in the design of bioluminescent immunoassay by using SARS-CoV-2 spike antigen as a model target and showed its usefulness in detecting virus in real samples from the physiological fluids similar to the currently available antigen tests.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank NIGMS (R01GM114321 and R01GM127706), the National Science Foundation (CBET-1841419/EAGER-2041413), Dr. John T. McDonald Foundation, and University of Miami CTSI for funding support. S. D. thanks the Miller School of Medicine of the University of Miami for the Lucille P. Markey Chair in Biochemistry and Molecular Biology. The authors would like to thank Dr. Sean P. J. Whelan of Washington University School of Medicine for providing us the rVSV-SARS-CoV-2 viral clone.References
- O. Villard, et al., Evaluation of the usefulness of six commercial agglutination assays for serologic diagnosis of toxoplasmosis, Diagn. Microbiol. Infect. Dis., 2012, 73(3), 231–235 CrossRef PubMed.
- S. Edouard, et al., Evaluating the serological status of COVID-19 patients using an indirect immunofluorescent assay, France, Eur. J. Clin. Microbiol. Infect. Dis., 2021, 40(2), 361–371 CrossRef CAS PubMed.
- A. K. Malan, et al.,Detection of IgG and IgM to West Nile virus: development of an immunofluorescence assay, Am. J. Clin. Pathol., 2003, 119(4), 508–515 CrossRef CAS.
- H. R. Hill and J. M. Matsen, Enzyme-linked immunosorbent assay and radioimmunoassay in the serologic diagnosis of infectious diseases, J. Infect. Dis., 1983, 147(2), 258–263 CrossRef CAS PubMed.
- D. N. Sharmila, Serological Investigation in Viral Infection. A Review, Ann. Romanian Soc. Cell Biol., 2021, 826–840 Search PubMed.
- S. Aydin, A short history, principles, and types of ELISA, and our laboratory experience with peptide/protein analyses using ELISA, Peptides, 2015, 72, 4–15 CrossRef CAS PubMed.
- M. E. Goldberg and L. Djavadi-Ohaniance, Methods for measurement of antibody/antigen affinity based on ELISA and RIA, Curr. Opin. Immunol., 1993, 5(2), 278–281 CrossRef CAS PubMed.
- J. M. Faupel-Badger, et al., Comparison of liquid chromatography-tandem mass spectrometry, RIA, and ELISA methods for measurement of urinary estrogens, Cancer Epidemiol. Biomark. Prev., 2010, 19(1), 292–300 CrossRef CAS.
- D. B. Broyles, et al., Facile Synthesis and Characterization of a Novel Tamavidin-Luciferase Reporter Fusion Protein for Universal Signaling Applications, Adv. Biosyst., 2020, 4(4), 1900166 CrossRef CAS.
- R. Ladj, et al., Polymer encapsulation of inorganic nanoparticles for biomedical applications, Int. J. Pharm., 2013, 458(1), 230–241 CrossRef CAS PubMed.
- E. Villemin, et al., Polymer encapsulation of ruthenium complexes for biological and medicinal applications, Nat. Rev. Chem., 2019, 3(4), 261–282 CrossRef CAS.
- R. J. Kuppler, et al., Potential applications of metal-organic frameworks, Coord. Chem. Rev., 2009, 253(23–24), 3042–3066 CrossRef CAS.
- G. Lu, et al., Imparting functionality to a metal–organic framework material by controlled nanoparticle encapsulation, Nat. Chem., 2012, 4(4), 310–316 CrossRef CAS.
- C. Wang, et al., Metal-Organic Framework Encapsulation Preserves the Bioactivity of Protein Therapeutics, Adv. Healthcare Mater., 2018, 7(22), 1800950 CrossRef PubMed.
- S. Kitagawa, Metal–organic frameworks (MOFs), Chem. Soc. Rev., 2014, 43(16), 5415–5418 RSC.
- J. Zhuang, A. P. Young and C. K. Tsung, Integration of biomolecules with metal–organic frameworks, Small, 2017, 13(32), 1700880 CrossRef PubMed.
- L. E. Kreno, et al., Metal–organic framework materials as chemical sensors, Chem. Rev., 2012, 112(2), 1105–1125 CrossRef CAS PubMed.
- Q.-L. Zhu and Q. Xu, Metal–organic framework composites, Chem. Soc. Rev., 2014, 43(16), 5468–5512 RSC.
- W. Liang, et al., Metal–organic framework-based enzyme biocomposites, Chem. Rev., 2021, 121(3), 1077–1129 CrossRef CAS.
- Y. Feng, et al., Antibodies@ MOFs: an in vitro protective coating for preparation and storage of biopharmaceuticals, Adv. Mater., 2019, 31(2), 1805148 CrossRef.
- Y. Sun, et al., Metal–organic framework nanocarriers for drug delivery in biomedical applications, Nano-Micro Lett., 2020, 12(1), 1–29 CrossRef CAS.
- K. Liang, et al., Biomimetic mineralization of metal-organic frameworks as protective coatings for biomacromolecules, Nat. Commun., 2015, 6(1), 1–8 Search PubMed.
- C. Wang, et al., Metal–organic framework encapsulation for biospecimen preservation, Chem. Mater., 2018, 30(4), 1291–1300 CrossRef CAS.
- X. Lin, et al., Encapsulation of strongly fluorescent carbon quantum dots in metal–organic frameworks for enhancing chemical sensing, Anal. Chem., 2014, 86(2), 1223–1228 CrossRef CAS PubMed.
- D. Zou, D. Liu and J. Zhang, From Zeolitic Imidazolate Framework-8 to Metal-Organic Frameworks (MOF s): Representative Substance for the General Study of Pioneering MOF Applications, Energy Environ. Mater., 2018, 1(4), 209–220 CrossRef CAS.
- M. G. Campbell, et al., Chemiresistive sensor arrays from conductive 2D metal–organic frameworks, J. Am. Chem. Soc., 2015, 137(43), 13780–13783 CrossRef CAS PubMed.
- D. Wang, D. Jana and Y. Zhao, Metal–organic framework derived nanozymes in biomedicine, Acc. Chem. Res., 2020, 53(7), 1389–1400 CrossRef CAS PubMed.
- H. Li, et al., Design and synthesis of an exceptionally stable and highly porous metal-organic framework, Nature, 1999, 402(6759), 276–279 CrossRef CAS.
- M. Ataei-Kachouei, S. Shahrokhian and M. Ezzati, Bimetallic CoZn-MOFs easily derived from CoZn-LDHs, as a suitable platform in fabrication of a non-enzymatic electrochemical sensor for detecting glucose in human fluids, Sens. Actuators, B, 2021, 130254 CrossRef CAS.
- Z. Sun, et al., Sensor array for rapid pathogens identification fabricated with peptide-conjugated 2D metal-organic framework nanosheets, Chem. Eng. J., 2021, 405, 126707 CrossRef CAS.
- P. Sharanyakanth and M. Radhakrishnan, Synthesis of metal-organic frameworks (MOFs) and its application in food packaging: A critical review, Trends Food Sci. Technol., 2020, 104, 102–116 CrossRef CAS.
- Y.-R. Lee, et al., ZIF-8: A comparison of synthesis methods, Chem. Eng. J., 2015, 271, 276–280 CrossRef CAS.
- H. K. Jeong, Metal–organic framework membranes: Unprecedented opportunities for gas separations, AIChE J., 2021, 67(6), e17258 CrossRef CAS.
- Z. Lai, Development of ZIF-8 membranes: opportunities and challenges for commercial applications, Curr. Opin. Chem. Eng., 2018, 20, 78–85 CrossRef.
- Y.-R. Lee, J. Kim and W.-S. Ahn, Synthesis of metal-organic frameworks: A mini review, Korean J. Chem. Eng., 2013, 30(9), 1667–1680 CrossRef CAS.
- C. Wang, et al., Metal-Organic Framework as a Protective Coating for Biodiagnostic Chips, Adv. Mater., 2017, 29(7), 1604433 CrossRef PubMed.
- C. Doonan, et al., Metal–organic frameworks at the biointerface: synthetic strategies and applications, Acc. Chem. Res., 2017, 50(6), 1423–1432 CrossRef CAS PubMed.
- Q. Jiang, et al., Rapid, point-of-care, paper-based plasmonic biosensor for Zika virus diagnosis, Adv. Biosyst., 2017, 1(9), 1700096 CrossRef.
- Y. Takakura, et al., Tamavidin, a versatile affinity tag for protein purification and immobilization, J. Biotechnol., 2010, 145(4), 317–322 CrossRef CAS PubMed.
- Y. Takakura, et al., Lentiavidins: Novel avidin-like proteins with low isoelectric points from shiitake mushroom (Lentinula edodes), J. Biosci. Bioeng., 2016, 121(4), 420–423 CrossRef CAS PubMed.
- R. F. Delgadillo, et al., Detailed characterization of the solution kinetics and thermodynamics of biotin, biocytin and HABA binding to avidin and streptavidin, PLoS One, 2019, 14(2), e0204194 CrossRef CAS PubMed.
- Z. M. Kaskova, A. S. Tsarkova and I. V. Yampolsky, 1001 lights: luciferins, luciferases, their mechanisms of action and applications in chemical analysis, biology and medicine, Chem. Soc. Rev., 2016, 45(21), 6048–6077 RSC.
- B. Hu, et al., Characteristics of SARS-CoV-2 and COVID-19, Nat. Rev. Microbiol., 2021, 19(3), 141–154 CrossRef CAS.
- M. A. Shereen, et al., COVID-19 infection: Origin, transmission, and characteristics of human coronaviruses, J. Adv. Res., 2020, 24, 91 CrossRef CAS PubMed.
- L. Morawska, et al., How can airborne transmission of COVID-19 indoors be minimised?, Environ. Int., 2020, 142, 105832 CrossRef CAS PubMed.
- Q. Liu, K. Xu, X. Wang and W. Wang, From Sars to Covid-19: What Lessons Have We Learned?, J. Infect. Public Health, 2020, 13(11), 1611–1618 CrossRef PubMed.
- T. P. Velavan and C. G. Meyer, The COVID-19 epidemic, Trop. Med. Int. Health, 2020, 25(3), 278 CrossRef CAS.
- M. M. Lamers and B. L. Haagmans, SARS-CoV-2 pathogenesis, Nat. Rev. Microbiol., 2022, 20(5), 270–284 CrossRef CAS PubMed.
- S.-H. Tseng, et al., A novel pseudovirus-based mouse model of SARS-CoV-2 infection to test COVID-19 interventions, J. Biomed. Sci., 2021, 28(1), 1–8 CrossRef PubMed.
- J. Malicoat, S. Manivasagam, S. Zuñiga, I. Sola, D. McCabe, L. Rong, S. Perlman, L. Enjuanes and B. Manicassamy, Development of a Single-Cycle Infectious SARS-CoV-2 Virus Replicon Particle System for Use in Biosafety Level 2 Laboratories, J. Virol., 2022, 96(3), e01837-21 CrossRef PubMed.
- M. Chen and X.-E. Zhang, Construction and applications of SARS-CoV-2 pseudoviruses: a mini review, Int. J. Biol. Sci., 2021, 17(6), 1574 CrossRef CAS PubMed.
- J. B. Case, P. W. Rothlauf, R. E. Chen, Z. Liu, H. Zhao, A. S. Kim, L. M. Bloyet, Q. Zeng, S. Tahan, L. Droit and M. X. G. Ilagan, Neutralizing antibody and soluble ACE2 inhibition of a replication-competent VSV-SARS-CoV-2 and a clinical isolate of SARS-CoV-2, Cell Host Microbe, 2020, 28(3), 475–485 CrossRef CAS PubMed.
- H. Xia, et al., Metal-organic frameworks: a potential platform for enzyme immobilization and related applications, Front. Bioeng. Biotechnol., 2020, 8, 695 CrossRef.
- A. Fleiss and K. S. Sarkisyan, A brief review of bioluminescent systems (2019), Curr. Genet., 2019, 65(4), 877–882 CrossRef CAS.
- X. Wan, et al., Cascaded amplifying circuits enable ultrasensitive cellular sensors for toxic metals, Nat. Chem. Biol., 2019, 15(5), 540–548 CrossRef CAS PubMed.
- J. Zhu, et al., Viral dynamics of SARS-CoV-2 in saliva from infected patients, J. Infect., 2020, 81(3), e48–e50 CrossRef CAS PubMed.
- Y. Pan, et al., Viral load of SARS-CoV-2 in clinical samples, Lancet Infect. Dis., 2020, 20(4), 411–412 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sd00145d |
| This journal is © The Royal Society of Chemistry 2022 |