Open Access Article
Open Access ArticleElectrochemical aptasensor for Salmonella detection using Nafion-doped reduced graphene oxide†
Shalini
Muniandy
a,
Kwai Lin
Thong
a,
Jimmy Nelson
Appaturi
 b,
Chin Wei
Lai
b,
Chin Wei
Lai
 a and
Bey Fen
Leo
a and
Bey Fen
Leo
 *ac
*ac
aNanotechnology and Catalysis Research Centre, Institute of Graduate Studies, University of Malaya, 50603 Kuala Lumpur, Malaysia. E-mail: beyfenleo@um.edu.my
bSchool of Chemical Sciences, Universiti Sains Malaysia, 11800 Penang, Malaysia
cDepartment of Molecular Medicine, Faculty of Medicine, University of Malaya, 50603 Kuala Lumpur, Malaysia
First published on 26th August 2022
Abstract
A highly conductive nanocomposite composed of reduced graphene oxide (rGO)–Nafion (Nf) was successfully prepared via a chemical reduction method in this study. The nanocomposite was used to develop an electrochemical aptasensor for the detection of Salmonella enterica serovar typhimurium (STM) by immobilizing the single-stranded DNA (ssDNA) aptamer on the nanocomposite-coated glassy carbon electrode (GCE). The developed ssDNA/rGO–Nf sensing platform can reproducibly detect S. typhimurium with a detection limit of 101 cfu mL−1 and requires only 1–2 μl of bacteria contaminated sample. The electrochemical aptasensor was also effective in assessing the specificity of the aptamer against different types of bacteria, indicating the developed platform can be used to specifically screen Salmonella bacteria from food samples in 10 min. Besides, the changes in electronic properties due to transduction of electrons after the interaction of the biorecognition element and the bacterial target were also characterized using photoluminescence spectroscopy (PL) in the study. These findings suggest the potential of developing high performance graphene-related sensors for food safety applications.
1. Introduction
Salmonella enterica is the most frequently occurring pathogen in food products that affect human health globally and accounts for about 80 million cases of food poisoning associated with 155![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 deaths each year.1 Although early detection of Salmonella is crucial to mitigate food safety, the speed, sensitivity, complexity, and cost of detection are the biggest challenge.2
000 deaths each year.1 Although early detection of Salmonella is crucial to mitigate food safety, the speed, sensitivity, complexity, and cost of detection are the biggest challenge.2
The development of highly sensitive and specific detection tools, such as biosensors for S. enterica, is the top priority of researchers to curb foodborne illnesses worldwide. Electrochemical biosensors offer simple, direct and real-time detection of various types of foodborne pathogens present in complex food matrixes.3 Coupling electrochemical biosensors with an aptamer, as a biorecognition element, and surface modification with graphene-based nanomaterials significantly improve their analytical performance.4 The remarkable physical and electrical characteristics of graphene, including its unique structure, high specific surface area, excellent conductivity, electron transfer kinetics and extraordinary mechanical strength make it an ideal interfacial material for quick development of functional nanocomposites.5 Nonetheless, graphene has some shortcomings (e.g., high hydrophobicity and lack of chemical functional groups) that limit its use as a stable biosensing interface.6
In comparison, reduced graphene oxide (rGO) can partially overcome the aforementioned limitations because rGO is decorated with various functional groups, such as epoxy, carbonyl, carboxyl and hydroxyl groups which can be employed as an anchoring point for immobilization of molecules.7 Conducting polymers pose high electron affinity, electronic conductivity, and good mechanical strength, and surface modifications of polymers further enhance the electrochemical performance.8,9 Nafion (Nf) is the most widely used cation exchange conducting polymer for electrochemical applications due to its remarkable proton conductivity and stability.10 Thus, coupling graphene oxide with the Nafion polymer and reducing it into a functionalized nanocomposite provides good mechanical strength and enhanced conductivity comparable to pure graphene with improved biocompatibility. Moreover, this modification also increases the electrochemical efficiency and conductivity of graphene because the low capacitance of graphene is improved by compositing it with other conducting polymers.11,12
Even though rGO–polymer nanocomposite-based electrochemical biosensors have been studied extensively, there are limited studies involving the rGO–Nf nanocomposite for bacterial detection.13 Recently,14 chitosan–Nafion®-coated sensors have been prepared for the successful detection of wound bacteria with a concentration of 102 colony forming units (CFU) mL−1 and −88.1 ± 6.3 mV pH−1 over a pH range of 1–13. Nevertheless, the mechanistic understanding of the changes that occur on the surface of rGO–Nf and their interaction with bacterial targets, which is crucial for further development and optimization of aptasensors for various applications, has not been well studied. Herein, we aim to develop a novel rGO–Nf nanocomposite-based electrochemical aptasensor for whole-cell detection of Salmonella typhimurium. The changes in the electronic properties of the transducer element upon interaction with the aptamer–bacteria complex at the interface of the working electrode were further studied using an electrochemical approach and a photoluminescence analytical tool. The developed rGO–Nf nanocomposite-based aptasensor exhibits great potential as a promising sensing platform for label-free whole cell detection of foodborne pathogens for food safety due to its excellent electronic conductivity, stability, biocompatibility, and sensitivity.
2. Materials and methods
2.1 Apparatus
The morphology of the rGO–Nf sensing platform was studied using a Quanta FEG 650 (FEI, USA) scanning electron microscope (SEM) equipped with an energy dispersive X-ray (EDX) detector and a Carl Zeiss (LEO LIBRA 120) transmission electron microscope (TEM). The chemical and physical properties of rGO–Nf were studied using Fourier transform infrared spectroscopy (FTIR) and Raman spectroscopy. Both FTIR and Raman spectra of the rGO–Nf nanocomposite were obtained using a Bruker IFS 66V/S (USA) and a Renishaw inVia Raman microscope, respectively. For photoluminescence analysis (PL), the same Renishaw inVia Raman microscope was used by changing the lens to 325 nm. The bandgap energy of the materials was calculated using Planck's equation:where E = energy, λ = wavelength, h = Plank's constant (6.63 × 10−34 J s), and C = speed of light 3.0 × 108 m s−1. The electrochemical behavior and properties of the working electrode were analysed using a PGSTAT302N electrochemical workstation (Metrohm AG, Switzerland). The electrochemical studies were conducted using silver/silver chloride (Ag/AgCl), a platinum wire and a glassy carbon electrode (GCE, diameter = 3 mm) as the reference electrode, the counter electrode and the working electrode, respectively.
2.2 Reagents and chemicals
Graphite flakes, sulfuric acid (H2SO4), phosphoric acid (H3PO4), 80 wt% hydrazine hydrate (N2H4), hydrochloric acid (HCl), 25 wt% of ammonia solution (NH3), hydrogen peroxide (H2O2), potassium permanganate (KMnO4), iron(III) chloride hexahydrate (FeCl3·6H2O), and iron(II) chloride tetrahydrate (FeCl2·4H2O) were purchased from Sigma-Aldrich. All the chemicals were used as received. The electrochemical properties of the fabricated electrode were studied using Zobell's solution [3 mM potassium ferricyanide, K3Fe(CN)6 and 0.1 M potassium chloride, KCl] as the electrolyte. The DNA aptamer sequences (5′-TATGGCGGCGTCACCCGACGGGGACTTGACATTATGACAG3′) purchased from First Base Laboratory Sdn. Bhd. (Malaysia) were adapted from Joshi et al.15 The bacterial cultures of Salmonella enterica serovar typhimurium (STM), Escherichia coli, Shigella dysenteriae, Vibrio cholerae, Staphylococcus aureus and Klebsiella pneumoniae used in this study were obtained from the culture collection of Biomedical Science Laboratory, University of Malaya, Malaysia.2.3 Preparation of rGO–Nf
GO was prepared from graphite powder using the modified Hummer's method.16,17 To synthesize rGO–Nf, 0.5 mg mL−1 was prepared by dissolving Nafion powder in ethanol![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) water (1
water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). Then, 10 mL of Nafion solution was added into 10 mL of the GO solution and stirred at room temperature for 30 min, followed by the addition of 100 μL of ammonia solution to adjust the pH to neutral. The GO–Nf was reduced to form rGO–Nf by adding 10 μL of hydrazine solution. Then, the mixture was refluxed at 80 °C for 12 h, followed by filtration of the product with a nylon membrane (0.22 μm). The rGO–Nf nanocomposite obtained by filtration can be readily dispersed in water by ultra-sonication to obtain a series of concentrations ranging from 1 mg mL−1 to 8 mg mL−1.
1). Then, 10 mL of Nafion solution was added into 10 mL of the GO solution and stirred at room temperature for 30 min, followed by the addition of 100 μL of ammonia solution to adjust the pH to neutral. The GO–Nf was reduced to form rGO–Nf by adding 10 μL of hydrazine solution. Then, the mixture was refluxed at 80 °C for 12 h, followed by filtration of the product with a nylon membrane (0.22 μm). The rGO–Nf nanocomposite obtained by filtration can be readily dispersed in water by ultra-sonication to obtain a series of concentrations ranging from 1 mg mL−1 to 8 mg mL−1.
2.4 Preparation of the electrochemical aptasensor
Prior to each experiment, the GCE was carefully polished to a mirror-like finish using 0.05 μm alumina slurry, followed by rinsing with distilled water before the electrode was sonicated in an acetone/ethanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
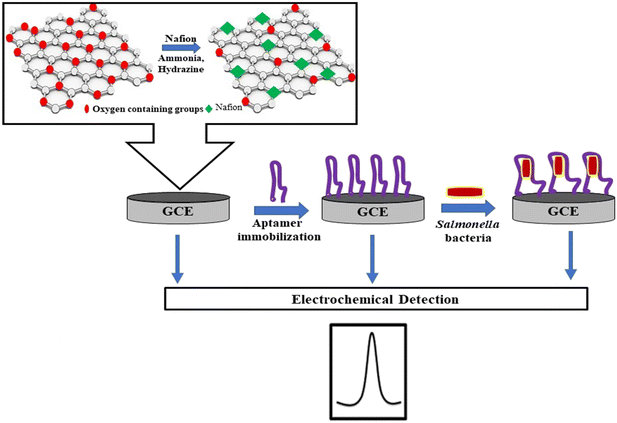
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) mixture for 3 min. The electrodes were dried at room temperature. After the cleaning process, the GCE was coated with rGO–Nf solution by drop casting 10 μL of the aqueous dispersion of the nanocomposite on the electrode surface and allowed to air-dry to form rGO–Nf/GCE. Subsequently, 5 μL of the aptamer (5 μmol L−1) was immobilized on the rGO–Nf/GCE electrode surface. The electrode was then air-dried to obtain ssDNA/rGO–Nf/GCE. This sensing platform was stored at 4 °C until use.
1, v/v) mixture for 3 min. The electrodes were dried at room temperature. After the cleaning process, the GCE was coated with rGO–Nf solution by drop casting 10 μL of the aqueous dispersion of the nanocomposite on the electrode surface and allowed to air-dry to form rGO–Nf/GCE. Subsequently, 5 μL of the aptamer (5 μmol L−1) was immobilized on the rGO–Nf/GCE electrode surface. The electrode was then air-dried to obtain ssDNA/rGO–Nf/GCE. This sensing platform was stored at 4 °C until use.
2.5 Electrochemical characterization of working electrodes and electrochemical detection of bacterial cells
The electrochemical redox behavior of the rGO–Nf/GCE electrode was studied using cyclic voltammetry (CV), electrochemical impedance spectroscopy (EIS) and differential pulse voltammetry (DPV) in Zobell's solution at a scan rate of 100 mV s−1. For bacterial detection, ssDNA/rGO–Nf/GCE was incubated in bacterial concentrations ranging from 101–108 cfu mL−1 for 5 min and this was followed by DPV electrochemical detection at a pulse amplitude of 0.025 V, a pulse width of 0.05 s and a pulse period of 0.05 s.3. Results and discussion
3.1 The surface chemistry and fabrication of the electrodes
The Nafion polymer along with rGO serves as an ideal transducer material for the construction of a biosensor. Its robust electronic and mechanical properties along with a large surface area and good biocompatibility are added advantages of the sensing platform.18Fig. 1 illustrates the fabrication process of the aptasensor. The presence of hydrophilic functional groups such as epoxy, carboxyl, and hydroxyl groups creates negative charge environments on the surface of GO.19 In the presence of water, the sulfonic group of the Nafion forms a hydrogen bond with GO.20 During the GO reduction process, some of the oxygenated groups were removed, whereas the bond between rGO and Nafion was retained by the hydrophobic interaction of the perfluoroalkyl backbone of the polymer on rGO.21 Furthermore, the Nafion layer is a conductive polymer providing steric repulsion between rGO sheets that overcome van der Waals force at contact. The nanocomposite was drop-cast on the GCE to obtain rGO–Nf/GCE. Subsequently, the DNA aptamer was immobilized on the surface of the electrode through a simple π–π stacking interaction between the nucleobases and rGO to obtain the sensing platform of ssDNA/rGO–Nf/GCE. This interaction causes a decrease in the DPV signal due to the inhibition of electron transfer by the nucleobases at the electrode–electrolyte interface.22 ssDNA/rGO–Nf/GCE can capture bacterial cells (STM) introduced into the system resulting in a dramatic decrease in its peak currents due to the inhibition of the electron transfer at the electrode–electrolyte interface.3.2 Structural and morphological characterization of the rGO–Nafion nanocomposite
The morphological characteristics of rGO and the rGO–Nafion nanocomposite were studied using FESEM and TEM. In Fig. 2A, wrinkled flakes and the sheet-like structure of rGO were observed. In the presence of Nafion (Fig. 2B), the polymer introduces more crumples and makes the rGO surface rougher which is consistent with a study reported previously,23 showing the successful formation of the rGO–Nf nanocomposite. The EDX results shown in Fig. 2B (inset) indicate the presence of key elements of Nafion, namely, S (2.2%), O (0.8%), and F (11.2%) occurring together with carbon (60.8%) of rGO. This further confirms the successful immobilization of Nafion on the rGO surface. Fig. 2C shows the TEM images of rGO and the rGO–Nf nanocomposite, respectively. rGO exhibits wrinkled transparent flakes of the ultra-thin film along with a few thin ripples within the sheets, whereas the rGO–Nf nanocomposite exhibits a rough surface with an increase in the thickness of the layer due to the deposition of the Nafion polymer. Both SEM and TEM showed consistent results.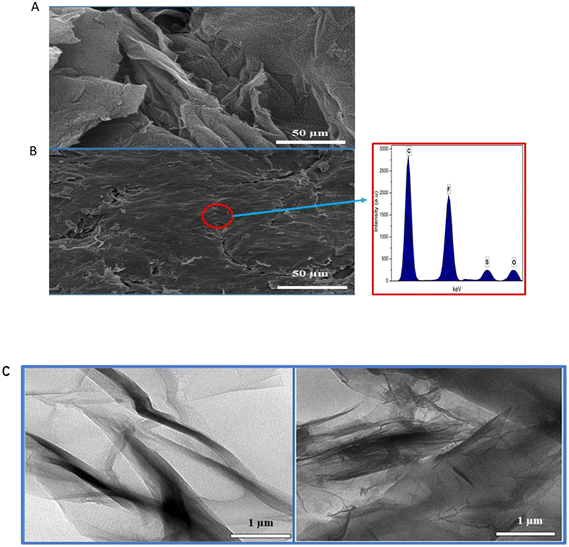 | ||
| Fig. 2 FESEM images of (A) rGO and (B) the rGO–Nf nanocomposite; the inset shows the EDX analysis of the rGO–Nf nanocomposite. (C) TEM images of rGO (left) and the rGO–Nf nanocomposite (right). | ||
3.3 Chemical characterization of the rGO–Nf nanocomposite
In this study, the FTIR technique was used to study the interfacial interactions between rGO and the Nafion polymer, as shown in Fig. 3A. The FTIR spectra of GO, rGO and the rGO–Nafion nanocomposite were elucidated. Theoretically, GO has several essential acute peaks that can be detected by FTIR such as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O stretching at 1730 cm−1, C–O–C stretching at 1214 cm−1 and C–O stretching at 1100 cm−1. In this study, similar results were observed and the presence of oxygen molecules (O) on GO was confirmed. Furthermore, a broad and intense band of the hydroxyl group (–OH) at 3300 cm−1 was observed from the FTIR pattern. Interestingly, broadening of the transmittance band was also observed, indicating the increased diffusion rate of oxygen molecules into the graphite flakes to form a carbonyl group, a carboxylic group, and an epoxy group in the GO sample.
O stretching at 1730 cm−1, C–O–C stretching at 1214 cm−1 and C–O stretching at 1100 cm−1. In this study, similar results were observed and the presence of oxygen molecules (O) on GO was confirmed. Furthermore, a broad and intense band of the hydroxyl group (–OH) at 3300 cm−1 was observed from the FTIR pattern. Interestingly, broadening of the transmittance band was also observed, indicating the increased diffusion rate of oxygen molecules into the graphite flakes to form a carbonyl group, a carboxylic group, and an epoxy group in the GO sample.
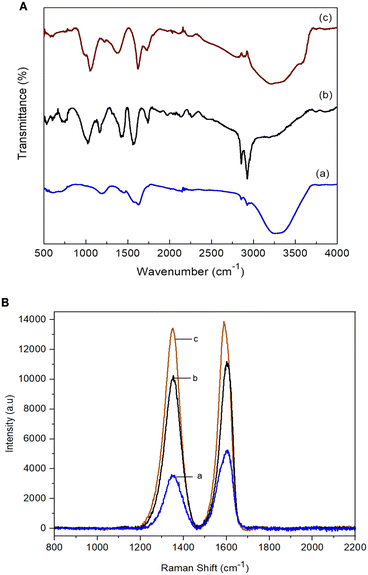 | ||
| Fig. 3 (A) FTIR spectra of (a) GO, (b) rGO and (c) rGO–Nf. (B) Raman spectra of (a) GO (ID/IG: 0.66), (b) rGO (ID/IG: 0.96) and (c) the rGO–Nf (ID/IG: 1.24) nanocomposite. | ||
However, after the chemical reduction, the intensity of all the essential characteristic peaks of the GO samples weakened while the peaks of some hydroxyl functional groups significantly decreased or disappeared. This phenomenon clearly illustrates that oxygen functional groups such as C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–O–C, and C–O, and hydroxyl groups were mostly removed to form rGO during the chemical reduction aided by hydrazine. In addition, the C
O, C–O–C, and C–O, and hydroxyl groups were mostly removed to form rGO during the chemical reduction aided by hydrazine. In addition, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C conjugation at 1525 cm−1 was detected on the rGO sample due to the sp2 hybridization. Meanwhile, the disappearance of the peak of the –OH group in the rGO sample at 3300 cm−1 was also observed and its intensity approached zero when the GO was fully reduced to rGO.
C conjugation at 1525 cm−1 was detected on the rGO sample due to the sp2 hybridization. Meanwhile, the disappearance of the peak of the –OH group in the rGO sample at 3300 cm−1 was also observed and its intensity approached zero when the GO was fully reduced to rGO.
After the surface functionalization, the spectra of the rGO–Nf nanocomposite showed all the characteristic bands of both rGO and Nafion. The nanocomposite showed C–O–C symmetric stretching bands at 965 and 980 cm−1. Moreover, the presence of S–O stretching in SO3H at 1056 cm−1 was also prominent in the nanocomposite. The presence of S–O stretching is attributed to the hydrogen bonding between the sulfonate moieties of the Nafion polymer and the dispersed rGO which enhances electron conductivity.24
Raman spectroscopy was conducted to evaluate the structural features and compositions of the prepared materials, as shown in Fig. 3B. The representative peaks of graphene (D and G bands) were observed in the nanocomposite. The ID/IG band ratio increased from 0.96 (rGO) to 1.24 for rGO–Nf. The increase in the ID/IG band ratio for the rGO–Nf nanocomposite may be due to the occurrence of chemical doping (p-type doping) on carbon materials.25 The CF2 and sulfonic acid groups of Nafion introduce holes on the carbon surface shifting the Fermi level of rGO to the valence band and inevitably increasing the conductivity of the rGO–Nf nanocomposite.21,26 Both FTIR and Raman spectra confirm the surface functionalization of the as-synthesized rGO–Nf.
3.4 Electrochemical analysis of the aptasensor
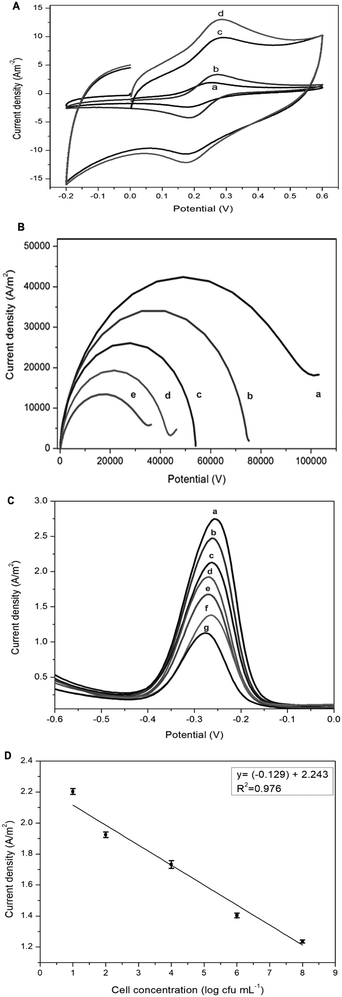
The electrochemical response of the fabricated electrodes was studied using CV, EIS, and DPV analytical techniques. Fig. 4A and B show the CV curves with quasi-reversible redox reaction peaks and EIS resistance obtained for bare GCE, rGO/GCE, rGO–Nf/GCE and ssDNA/rGO–Nf/GCE electrodes in Zobell's solution at a scan rate of 100 mV s−1, respectively. Bare GCE exhibited the lowest peak current and the highest resistance, but the peak current increased slightly after the coating with rGO. After the surface modification with rGO–Nf, a prominent increase in current could be observed with the presence of a pair of visible redox peaks at 0.25 V in the CV curve and the resistance was greatly reduced, as shown in the EIS as well. The peak current increased and the resistance decreased due to the presence of graphene sheets and Nafion enhanced the electrochemical response which was facilitated by the electron transfer between the negatively charged electrochemical probe [Fe(CN)6]3−/4− and the rGO electrode surface.27 The synergistic effects exerted by the functional groups of rGO and the electron-withdrawing groups of Nafion caused a significant improvement in both proton and electron conductivities.28 However, after the immobilization of the aptamer on the electrode's surface, a significant decrease in both conductivities and an increase in charge transfer resistance were observed. The presence of the rGO–Nf/aptamer complex (polyplex) creates a barrier for the diffusion of the electrochemical probe [Fe(CN)6]3−/4− at the electrode interface leading to a decrease in the electrochemical response of ssDNA/rGO–Nf/GCE, but the resistance increased, as shown in the EIS.29 Additionally, EIS was conducted in the presence of the bacterial cells. When the ssDNA/rGO–Nf/GCE electrode was exposed to the bacterial cells, the current resistance further increased. The negatively charged bacterial cell membrane repels the negatively charged redox probe [Fe(CN)6]3−/4−, which inhibits the diffusion on the redox molecules on the graphene's surface.The redox behavior of ssDNA/rGO–Nf/GCE at different concentrations of bacteria cells ranging from 101 to 108 cfu mL−1 was investigated by using DPV in 0.1 M PBS (pH 6.5), as shown in Fig. 4C. When the ssDNA/E-mail: rGO–Nf/GCE electrode was exposed to the bacterial cells, the peak current further decreased proportionally with increasing bacterial concentration. The negatively charged bacterial cell membrane repels the negatively charged redox probe [Fe(CN)6]3−/4−, which inhibits the diffusion on the redox molecules on the graphene's surface and this phenomenon further decreases the peak current and increases the charge transfer resistance.
The peak current was plotted with the logarithm of the target bacterial concentration as shown in Fig. 4D and the relationship can be described as follows:
I = −0.13(±0.03)log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c + 2.24(±0.05) c + 2.24(±0.05) |
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) c is the logarithm of the cell concentration. The binding of the bacterial target affects the interfacial electron-transfer kinetics and decreases the conductivity of the surface-bound aptamer–bacteria constructs.
c is the logarithm of the cell concentration. The binding of the bacterial target affects the interfacial electron-transfer kinetics and decreases the conductivity of the surface-bound aptamer–bacteria constructs.
The selectivity of the aptasensor was also studied as shown in the ESI† (S1) by exposing the fabricated electrodes to different types of non-Salmonella bacteria (E. coli, Shi. dysenteriae, V. cholerae, Staph. aureus and K. pneumoniae). As shown in S1,† there were no significant changes in the conductivity when exposed to non-Salmonella bacteria as compared to the Salmonella bacteria. This result indicates that the fabricated aptasensor exhibits high selectivity towards Salmonella bacteria. The results obtained using the aptasensor in the sensitivity and selectivity tests were validated with PCR assay, as shown in the ESI† (S2).
3.5 Electronic properties of the rGO–Nf nanocomposite in the presence of the aptamer–bacteria complex
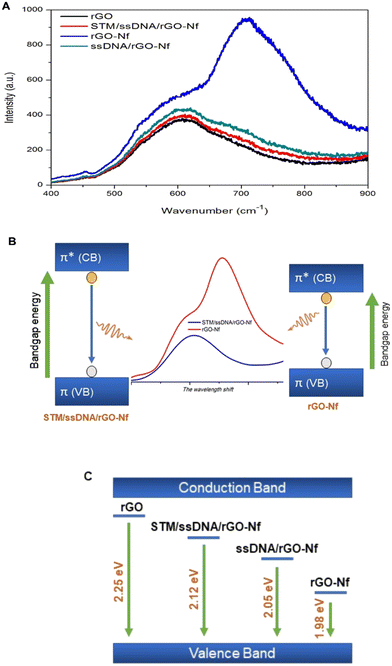
Photoluminescence spectroscopy was employed in this study to monitor the perturbation of the electric field on the material surface in the presence of the aptamer and bacterial cells. The electrical field on the material surface is determined by holes and electrons which causes spatial separation of charge carriers associated with the change in photoluminescence emission.30 Graphene oxide is a gapless semiconductor in which the bad gap can be increased through reduction to rGO to exhibit good absorption efficiency. Thus, rGO behaves like a semi-metal or semiconductor and its electrical conductivity may be tuned by controlling its oxygen content through a chemical reduction method.31 In addition, rGO combined with Nafion rGO exhibits photoluminescence properties due to the presence of a sulfonic group.32In Fig. 5A, the PL peak of rGO forms in the blue region of the spectrum due to the recovery of the sp2 carbon network during the reduction process which widens the bandgap energy (2.25 eV), and its intensity is the lowest among the samples due to surface defects created as a result of the removal of oxygen. In the presence of perfluorinated Nf, the PL intensity increases with a band gap of 2.08 eV. This is due to the increase in electron–hole pair recombination mediated by the electron-withdrawing property of sulfonic acid groups on the surface of graphene that causes a drastic increase in the conductivity of the graphene nanocomposite.33
The changes in conductivity resulting from the aptamer immobilization and bacterial interaction on the sensing platform can also be studied based on the changes in PL emission. Both the negatively charged aptamer and bacterial cells lead to the spatial separation of charge carriers and reduce the photoluminescence emission, with band gap energy values calculated to be 2.05 eV and 2.12 eV, respectively. These negatively charged aptamer and bacterial cells often create a higher defect density that can widen the band gap of graphene, whereas the defects may act as scattering centers which decrease the conductivity of graphene.34,35 The changes in the rGO–Nafion electronic properties due to electron movements proposed through PL studies can further be elucidated from Fig. 5B and the changes in the band gap or energy levels of the materials during each surface functionalization step are shown in Fig. 5C. Similar studies involving PL characterization for bacterial detection were reported in the past years.36,37
3.6 Reproducibility and shelf life of the aptasensor
The reproducibility (data not shown) of the fabricated electrodes was studied using successive CV measurements. Under optimal conditions, the RSD obtained for the oxidation peak was 0.71% (n = 5). The shelf life of the electrodes was experimentally determined by measuring the CV curve of an electrode for 28 days (4 °C). The peak current of the electrode was observed to decrease slightly, with an RSD value of 1.15%. The low RSD values obtained for rGO–Nf/GCE indicated the excellent reproducibility and stability of the electrode.3.7 Sensor comparative study
The constructed rGO–Nf/GCE aptasensor was compared to prior studies for bacteria detection, and the key characteristics are described in Table 1. The developed rGO–Nf/GCE aptasensor provides a unique and cost-effective approach for detecting foodborne pathogens. The usage of rGO–Nf/GCE removes the costly labelling step for an aptamer, resulting in stable and oriented immobilization of the aptamer, which improves the aptasensor's overall repeatability. The produced rGO–Nf/GCE aptasensor has a high sensitivity and an enhanced detection limit, as shown in the comparative table. Furthermore, the developed aptasensor demonstrated a low detection limit for whole-cell Salmonella bacteria in a shorter detection period. Although the use of electrolyte in the aptasensor function may limit its utility as a point-of-care device, the rGO–Nf/GCE nanocomposite appears to be a potential platform for fabricating large quantities of low-cost aptasensors for the S. typhimurium pathogen.| Detection method | Sensing platform | Targeted bacteria | Limit of detection | Assay time | Ref. |
|---|---|---|---|---|---|
| Amperometric aptasensor | rGO–TiO2 | S. typhimurium | 101 cfu ml−1 | 10 min | 38 |
| DPV | rGO–CHI with glutaraldehyde and thiol–DNA aptamer | S. typhimurium | 101 cfu ml−1 | 39 | |
| DPV | rGO/Si-10Co-aptamer/GCE | Salmonella spp. | 101 cfu ml−1 | 40 | |
| Potentiometric aptasensor | rGO–azophloxine | S. typhimurium | 101 cfu ml−1 | 10 min | 41 |
| DPV | rGO–CNT | S. typhimurium | 101 cfu ml−1 | 10 min | 42 |
| Electrochemical immunosensor | rGO–ant-TiO2 | V. cholerae | 0.12 nM L−1 of target antigen | Not stated | 43 |
| EIS | rGO–Au | S. aureus | 10 | Not stated | 44 |
| Multiplex-PCR | — | Salmonella spp. | 103 cfu ml−1 | 3 h | 45 |
| Electrochemical immunosensor | Graphene–copper(II) oxide-cysteine | E. coli O157:H7 | 101 cfu ml−1 but high detection limit for food samples | Not stated | 46 |
| Immunological assay | — | S. typhimurium | 103 cfu ml−1 | 2 h | 47 |
| Electrochemical genosensor | GO–iron oxide–chitosan nanocomposite | E. coli | 1 × 10−14 M of DNA target | 1 h | 48 |
| Aptasensor-DPV&EIS | rGO–Nf-label free DNA aptamer | S. typhimurium | 101 cfu ml−1 | 10 min | This work |
4. Conclusion
In summary, we demonstrated a facile method of fabricating rGO–Nf nanocomposite-based electrodes for the detection of Salmonella enterica. The aptasensor showed rapid, sensitive and selective detection of S. typhimurium with a low detection limit of 101 cfu mL−1 at room temperature. This approach is promising and can be applied as a point-of-care detection device for foodborne pathogens in the future. The study on the interactions of the aptamer–bacterial cells with the rGO–Nf interface via the photoluminescence method has further improved our understanding of the changes of the nanomaterial's energy levels associated with conductivity. The understanding gained from this study will provide caveats for the better design and optimization of graphene-based biosensors for future applications.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors acknowledge the financial support from the UMSC C.A.R.E Fund Research Grant (PV044-2021), Technology Development Fund 1 from the Ministry of Science, Technology and Innovation (MOSTI004-2022TED1) and the Universiti Malaya Research Grant – SATU (ST003-2020).References
- S. M. Jajere, Vet. World, 2019, 12, 504–521 Search PubMed.
- S. Cinti, G. Volpe, S. Piermarini, E. Delibato and G. Palleschi, Sensors, 2017, 17(8), 1910 CrossRef.
- A. Ahmed, J. V. Rushworth, N. A. Hirst and P. A. Millner, Clin. Microbiol. Rev., 2014, 27, 631–646 CrossRef CAS PubMed.
- F. Jia, N. Duan, S. Wu, R. Dai, Z. Wang and X. Li, Microchim. Acta, 2016, 183, 337–344 CrossRef CAS.
- W. Yi, Z. Li, C. Dong, H.-W. Li and J. Li, Microchem. J., 2019, 148, 774–783 CrossRef CAS.
- E. Povedano, F. H. Cincotto, C. Parrado, P. Díez, A. Sánchez, T. C. Canevari, S. A. S. Machado, J. M. Pingarrón and R. Villalonga, Biosens. Bioelectron., 2017, 89, 343–351 CrossRef CAS PubMed.
- D. Chauhan, Pooja, V. Nirbhaya, C. M. Srivastava, R. Chandra and S. Kumar, Microchem. J., 2020, 155, 104697 CrossRef CAS.
- J. M. S. Alshawi, M. Q. Mohammed, H. F. Alesary, H. K. Ismail and S. Barton, ACS Omega, 2022, 7, 20405–20419 CrossRef CAS.
- M. Q. Mohammed, H. K. Ismail, H. F. Alesary and S. Barton, Chem. Pap., 2022, 76, 715–729 CrossRef CAS.
- N. Yusoff, in Graphene-Based Electrochemical Sensors for Biomolecules, ed. A. Pandikumar and P. Rameshkumar, Elsevier, 2019, pp. 155–186, DOI:10.1016/B978-0-12-815394-9.00007-8.
- Y. Gao, Nanoscale Res. Lett., 2017, 12, 387 CrossRef PubMed.
- H. Rasouli, L. Naji and M. G. Hosseini, RSC Adv., 2017, 7, 3190–3203 RSC.
- Q. Gong, Y. Wang and H. Yang, Biosens. Bioelectron., 2017, 89, 565–569 CrossRef CAS.
- R. Sheybani and A. Shukla, Biosens. Bioelectron., 2017, 92, 425–433 CrossRef CAS PubMed.
- R. Joshi, H. Janagama, H. P. Dwivedi, T. S. Kumar, L.-A. Jaykus, J. Schefers and S. Sreevatsan, Mol. Cell. Probes, 2009, 23, 20–28 CrossRef CAS PubMed.
- W. S. Hummers Jr and R. E. Offeman, J. Am. Chem. Soc., 1958, 80, 1339 CrossRef.
- A. Singh, G. Sinsinbar, M. Choudhary, V. Kumar, R. Pasricha, H. Verma, S. P. Singh and K. Arora, Sens. Actuators, B, 2013, 185, 675–684 CrossRef CAS.
- F. He, J. Fan, D. Ma, L. Zhang, C. Leung and H. L. Chan, Carbon, 2010, 48, 3139–3144 CrossRef CAS.
- M. Vinothkannan, C. Karthikeyan, G. Gnana Kumar, A. R. Kim and D. J. Yoo, Spectrochim. Acta, Part A, 2015, 136, 256–264 CrossRef CAS PubMed.
- M. Vinothkannan, A. R. Kim, G. Gnana Kumar and D. J. Yoo, RSC Adv., 2018, 8, 7494–7508 RSC.
- Y. Liu, L. Gao, J. Sun, Y. Wang and J. Zhang, Nanotechnology, 2009, 20, 465605 CrossRef PubMed.
- Y. Guo, Y. Guo and C. Dong, Electrochim. Acta, 2013, 113, 69–76 CrossRef CAS.
- B. A. Aragaw, W.-N. Su, J. Rick and B.-J. Hwang, RSC Adv., 2013, 3, 23212–23221 RSC.
- J.-H. Jung, J.-H. Jeon, V. Sridhar and I.-K. Oh, Carbon, 2011, 49, 1279–1289 CrossRef CAS.
- B. A. Aragaw, W. N. Su, J. Rick and B. J. Hwang, RSC Adv., 2013, 3(45), 23212–23221 RSC.
- H. L. Poh, F. Šaněk, A. Ambrosi, G. Zhao, Z. Sofer and M. Pumera, Nanoscale, 2012, 4, 3515–3522 RSC.
- X. Xi and L. Ming, Anal. Methods, 2012, 4, 3013–3018 RSC.
- X. Li, F. Gittleson, M. Carmo, R. C. Sekol and A. D. Taylor, ACS Nano, 2012, 6, 1347–1356 CrossRef CAS PubMed.
- M. M. Mohsen, A. M. Wael, M. E. G. Nadia and A. E. Anwar, Int. J. Phys. Sci., 2011, 6(32), 7328–7334 Search PubMed.
- V. Duplan, E. Frost and J. J. Dubowski, Detection of bacteria using a photoluminescence-based quantum semiconductor device, in 2011 ICO International Conference on Information Photonics, IEEE, 2011, pp. 1–2 Search PubMed.
- E. C. Mattson, J. E. Johns, K. Pande, R. A. Bosch, S. Cui, M. Gajdardziska-Josifovska, M. Weinert, J. H. Chen, M. C. Hersam and C. J. Hirschmugl, J. Phys. Chem. Lett., 2014, 5, 212–219 CrossRef CAS PubMed.
- N. Bunkin, A. V. Shkirin, V. Kozlov, B. Ninham, E. V. Uspenskaya and S. Gudkov, J. Chem. Phys., 2018, 149, 164901 CrossRef CAS PubMed.
- S.-J. Kwon, T.-H. Han, T. Y. Ko, N. Li, Y. Kim, D. J. Kim, S.-H. Bae, Y. Yang, B. H. Hong, K. S. Kim, S. Ryu and T.-W. Lee, Nat. Commun., 2018, 9, 2037 CrossRef PubMed.
- W. Zhang, C.-T. Lin, K.-K. Liu, T. Tite, C.-Y. Su, C.-H. Chang, Y.-H. Lee, C.-W. Chu, K.-H. Wei, J.-L. Kuo and L.-J. Li, ACS Nano, 2011, 5, 7517–7524 CrossRef CAS PubMed.
- W. J. Yu, L. Liao, S. H. Chae, Y. H. Lee and X. Duan, Nano Lett., 2011, 11, 4759–4763 CrossRef CAS.
- R. Viter, V. Khranovskyy, N. Starodub, Y. Ogorodniichuk, S. Gevelyuk, Z. Gertnere, N. Poletaev, R. Yakimova, D. Erts and V. Smyntyna, IEEE Sens. J., 2014, 14, 2028–2034 CAS.
- S. Ahmadian-Fard-Fini, D. Ghanbari and M. Salavati-Niasari, Composites, Part B, 2019, 161, 564–577 CrossRef CAS.
- S. Muniandy, S. J. Teh, J. N. Appaturi, K. L. Thong, C. W. Lai, F. Ibrahim and B. F. Leo, Bioelectrochemistry, 2019, 127, 136–144 CrossRef CAS.
- I. J. Dinshaw, S. Muniandy, S. J. Teh, F. Ibrahim, B. F. Leo and K. L. Thong, J. Electroanal. Chem., 2017, 806, 88–96 CrossRef CAS.
- J. N. Appaturi, T. Pulingam, S. Muniandy, I. J. Dinshaw, L. B. Fen and M. R. Johan, Mater. Chem. Phys., 2019, 232, 493–505 CrossRef CAS.
- S. Muniandy, I. J. Dinshaw, S. J. Teh, C. W. Lai, F. Ibrahim, K. L. Thong and B. F. Leo, Anal. Bioanal. Chem., 2017, 409, 6893–6905 CrossRef CAS PubMed.
- J. N. Appaturi, T. Pulingam, K. L. Thong, S. Muniandy, N. Ahmad and B. F. Leo, Anal. Biochem., 2020, 589, 113489 CrossRef CAS.
- P. R. Solanki, S. Srivastava, M. A. Ali, R. K. Srivastava, A. Srivastava and B. D. Malhotra, RSC Adv., 2014, 4, 60386–60396 RSC.
- F. Jia, N. Duan, S. Wu, X. Ma, Y. Xia, Z. Wang and X. Wei, Microchim. Acta, 2014, 181, 967–974 CrossRef CAS.
- L. Wang, Y. Li and A. Mustaphai, J. Food Prot., 2007, 70, 1366–1372 CrossRef CAS.
- C. M. Pandey, I. Tiwari, V. N. Singh, K. N. Sood, G. Sumana and B. D. Malhotra, Sens. Actuators, B, 2017, 238, 1060–1069 CrossRef CAS.
- W. Wu, J. Li, D. Pan, J. Li, S. Song, M. Rong, Z. Li, J. Gao and J. Lu, ACS Appl. Mater. Interfaces, 2014, 6, 16974–16981 CrossRef CAS PubMed.
- I. Tiwari, M. Singh, C. M. Pandey and G. Sumana, Sens. Actuators, B, 2015, 206, 276–283 CrossRef CAS.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sd00098a |
| This journal is © The Royal Society of Chemistry 2022 |