Open Access Article
Open Access ArticleHost miRNAs as biomarkers of SARS-CoV-2 infection: a critical review
Kato
Pollet
 ab,
Nathalie
Garnier
ab,
Nathalie
Garnier
 ab,
Sabine
Szunerits
ab,
Sabine
Szunerits
 b,
Annemieke
Madder
b,
Annemieke
Madder
 c,
Didier
Hober
c,
Didier
Hober
 a and
Ilka
Engelmann†
a and
Ilka
Engelmann†
 *a
*a
aUniv. Lille, CHU Lille, Laboratoire de Virologie ULR3610, 59000 Lille, France. E-mail: ilka.engelmann@chu-montpellier.fr
bUniv. Lille, CNRS, Centrale Lille, Univ. Polytechnique Hauts-de-France, UMR 8520 – IEMN, 59000 Lille, France
cOrganic and Biomimetic Chemistry Research Group, Department of Organic and Macromolecular Chemistry, Ghent University, Krijgslaan 281-S4, 9000 Gent, Belgium
First published on 26th October 2022
Abstract
MicroRNAs (miRNAs), small non-coding RNAs that regulate gene expression, have received increasing attention as potential biomarkers of different diseases, including viral infections. Severe acute respiratory syndrome coronavirus 2 (SARS-CoV-2) infection and the disease it is causing, coronavirus disease (COVID-19), has affected health, society and life worldwide since its pandemic spread. Differential expression of miRNAs in COVID-19 patients compared to healthy controls and also between different severity grades of COVID-19 has been described in several recent studies. In this review, we discuss in detail studies that investigated miRNA expression in body fluids of COVID-19 patients. Several studies found a different miRNA expression profile in COVID-19 patients compared to controls but also in different severity grades of the disease. We compared the main findings of the studies in order to identify miRNAs that have been identified as differentially expressed by more than one study and could serve as diagnostic or prognostic biomarkers of COVID-19. Finally, we highlight the challenges and perspectives associated to the use of miRNAs as biomarkers of COVID-19.
Introduction
A novel coronavirus, later named severe acute respiratory syndrome coronavirus-2 (SARS-CoV-2), emerged in China at the end of 2019 and rapidly spread to cause a pandemic.1,2 SARS-CoV-2 belongs to the genus Betacoronavirus of the Coronaviridae family.2 Coronaviruses are enveloped viruses with a positive-sense single-stranded RNA genome of 26–32 kb. Other members of this family are responsible for infections in humans, including the severe acute respiratory syndrome coronavirus (SARS-CoV), the Middle East respiratory syndrome coronavirus (MERS-CoV), associated with severe disease, and viruses associated with clinically less severe infections, namely HCoV-OC43, HCoV-229E, HCoV-HKU1, HCoV-NL63.3SARS-CoV-2 causes COVID-19, a disease presenting symptoms including fever or chills, cough, shortness of breath, fatigue, nausea, muscle ache, headache, nausea or vomiting, diarrhoea, sore throat, congestion or running nose, loss of smell (anosmia) and loss of taste (ageusia). Severity ranges from asymptomatic to critical illness with multi-organ failure, respiratory failure or shock and possible fatal outcome.4
Severe COVID-19 is associated with severe pulmonary disease and can present acute respiratory distress syndromes (ARDS), cardiac impairment, liver dysfunction, acute kidney injury and coagulopathy.5 A hyperinflammatory state termed cytokine storm contributes further to severe COVID-19.6,7 An imbalanced host immune response characterized by reduced type I interferon production and high expression of other cytokines has been suggested to drive severe COVID-19.8,9 TNF-α, IL-6, IL-8 and IL-10 are significantly elevated in patients with severe COVID-19.6,7,9
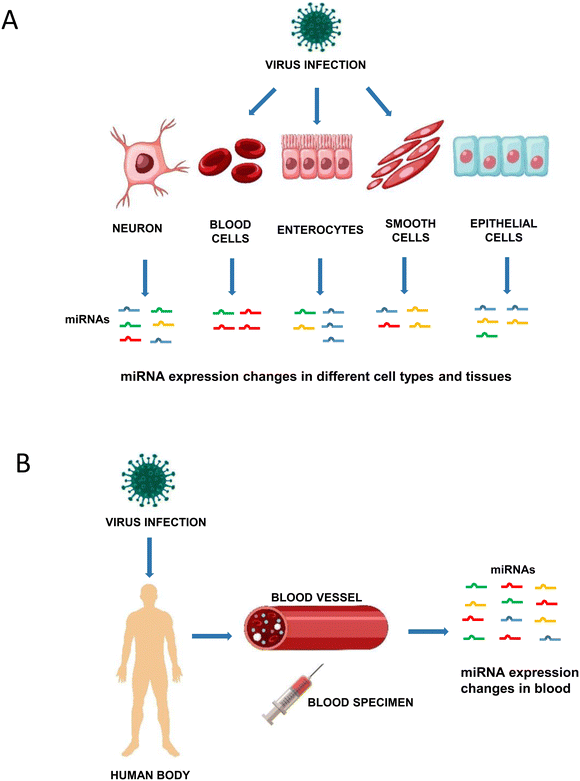
miRNAs are small, noncoding RNAs that regulate gene expression. They bind to sequences that are often localized in the 3'untranslated region (3'UTR) of a messenger RNA (mRNA) and inhibit translation or induce degradation of this mRNA.10–12 Biogenesis of miRNAs includes transcription by RNA polymerase II to pri-miRNAs that are further processed by Drosha giving rise to pre-miRNAs. Export to the cytoplasm is followed by Dicer processing, which generates miRNA duplexes that are loaded into an Argonaute protein to form a miRNA induced silencing complex (miRISC).12,13 miRNAs are involved in virtually all physiological and pathological processes, including viral infections and the antiviral immune response.13 As discussed in some recent work, viral infections change the expression profile of cellular miRNAs (Fig. 1A).14–16 We recently reviewed data showing that acute and persistent enterovirus infection induced distinct miRNA expression changes in infected cells.15,17 Furthermore, different enterovirus types induced different miRNA expression changes and these were dependent on the cell type and the time point after infection.15 Concerning respiratory virus infections, Martinez-Espinoza and colleagues showed that infections with influenza A virus, human respiratory syncytial virus (HRSV), human metapneumovirus (HMPV), adenovirus and SARS-CoV-2 influenced the miRNA expression profile of different cell types in vitro but also in the blood of patients.16 Wu and colleagues reviewed data of HRSV and HMPV infections, demonstrating that these viruses induced distinct miRNA expression changes in different cell types in vitro and in clinical samples.14 These findings all point to miRNAs being important biomarkers for viral infections and being involved in the pathophysiology of viral infections.
miRNAs as biomarkers
miRNAs can be detected in several body fluids including plasma, serum, urine, saliva and semen.18 They are remarkably stable in these body fluids and storage for 24 hours at room temperature and up to ten freeze–thawing cycles had minimal impact on plasma and serum miRNA levels.19–23 The concentration of miRNAs in body fluids is known to change during various cancers and infectious diseases23–26 making miRNAs attractive candidates as biomarkers for viral infections and possibly allowing to distinguish an infected person from healthy controls (Fig. 1B).20,26–28 miRNA expression changes were documented in several viral infections, such as hepatitis B, hepatitis C, enterovirus infection, varicella, or influenza virus infection.26–28 Six miRNAs indeed allowed discriminating patients with enterovirus infections from healthy controls with areas under curve (AUC) above 0.80. A combination of these miRNAs resulted in a sensitivity of 97.1% and a specificity of 92.7% demonstrating a promising diagnostic performance.29 miRNAs may also serve as prognostic biomarkers, i.e. distinguish between different grades of disease severity. For example, hsa-miR-150 levels were significantly higher in patients with severe influenza A/H1N1 disease and hsa-miR-876-5p expression was 9.5-fold higher in patients with severe enterovirus 71 infection when compared to patients with milder disease.30,31In the recent literature, studies reporting miRNA expression in COVID-19 patients have been published (Tables 1 and 2). Some of these aimed to identify miRNAs as biomarkers, whereas others included miRNA expression analysis as part of Omics approaches. Several studies also investigated a small number of preselected miRNA candidates. The objective of this review was to summarize data on the expression of miRNAs in body fluids of COVID-19 patients. These miRNAs have the potential to represent candidate biomarkers. We systematically searched for studies that describe human miRNA expression in body fluids of COVID-19 patients and reviewed studies that specifically investigated miRNAs as biomarkers for the diagnosis of SARS-CoV-2 infection and those that described miRNAs associated with different clinical courses of COVID-19.
| Studies with miRNA screening | ||||||||
|---|---|---|---|---|---|---|---|---|
| Main findings | Patient numbers | Specimen type | Pre-analytical procedures | Extraction methods | miRNA quantification methods | Normalization methods | Statistical methods | Ref. |
| hsa-miR-17-5p, hsa-miR-142-5p down-regulated hsa-miR-15a-5p, hsa-miR-19a-3p, hsa-miR-19b-3p, hsa-miR-23a-3p, hsa-miR-92a-3p and hsa-miR-320a up-regulated in SARS-CoV-2-infected patients | 33 COVID-19 patients, 10 healthy controls | Plasma | Centrifugation within 2 hours | miRNeasy serum/plasma isolation kit (Qiagen) | miRCURY LNA miRNome qPCR panels/miRCURY LNA miRNA PCR assay; validation by RT-qPCR | miR-502-5p | Unpaired student's t-test/ROC; not mentioned whether correction for multiple testing was performed | 40 |
| 20 miRNAs up-regulated in COVID-19: hsa-miR-103a-3p, hsa-let-7f-5p, hsa-miR-423-5p, hsa-miR-320a-3p, hsa-miR-92a-3p, hsa-let-7a-5p, hsa-miR-148a-3p, hsa-miR-142-3p, hsa-miR-30a-5p, hsa-miR-320c, hsa-miR-320b, hsa-let-7e-5p, hsa-miR-197-3p, hsa-miR-576-5p, hsa-miR-1290, hsa-miR-195-5p, hsa-miR-483-5p, hsa-miR-193a-5p, hsa-miR-6721-5p, hsa-miR-206, hsa-miR-27a-5p, hsa-miR-2116-3p, hsa-miR-4742-3p, hsa-miR-3125, hsa-miR-31-5p; 30 miRNAs down-regulated in COVID-19: hsa-miR-1275, hsa-miR-3617-5p, hsa-miR-500b-3p, hsa-miR-3684, hsa-miR-627-5p, hsa-miR-651-5p, hsa-miR-18a-3p, hsa-miR-3115, hsa-miR-589-3p, hsa-miR-664b-3p, hsa-miR-548k, hsa-miR-769-3p, hsa-miR-1226-3p, hsa-miR-873-5p, hsa-miR-5189-3p, hsa-miR-3198, hsa-miR-4772-3p, hsa-miR-6772-3p, hsa-miR-145-3p, hsa-miR-3913-5p, hsa-miR-6503-3p, hsa-miR-210-3p, hsa-miR-766-3p, hsa-miR-3065-3p, hsa-miR-551b-3p, hsa-miR-28-5p, hsa-miR-491-5p, hsa-let-7i-3p, hsa-miR-4662a-5p, hsa-miR-150-5p; differentially expressed in COVID-19 patients with and without oxygen requirement: hsa-let-7e-5p, hsa-miR-651-5p, hsa-miR-766-3p, hsa-miR-4433b-5p | 10 COVID-19 patients, 10 healthy controls | Plasma | Lysis with Qiazol | miRNeasy Micro Kit (Qiagen) + glycogen | Next-generation sequencing: QIAseq miRNA Library Kit and QIAseq miRNA NGS 48 Index IL (Qiagen) | DESeq2 package in R (not detailed) | DESeq2 package in R (not detailed), FDR; machine learning analysis using the scikit-learn module in python; ROC/AUC | 41 |
| hsa-miR-16-2-3p, hsa-miR-6501-5p, hsa-miR-618 upregulated in COVID-19; hsa-miR-183-5p, hsa-miR-627-5p, hsa-miR-144-3p downregulated in COVID-19 | 10 COVID-19 patients, 4 healthy controls | Whole blood | PAXgene blood RNA tubes (BD) | PAXgene Blood miRNA Kit (Qiagen) | High-throughput sequencing: model PE75 using Illumina Hiseq X Ten | Reads per million | Package DESeq2 of R software, not mentioned whether FDR was used | 42, 43 |
| hsa-miR-19b-1-3p, hsa-miR-96, hsa-miR-19b-2-5p, hsa-miR-451a, hsa-miR-451b, hsa-miR-194-1-5p, hsa-miR-144, hsa-miR-486-2-3p, hsa-miR-15a, hsa-miR-29c-3p downregulated in COVID-19; hsa-miR-3609, hsa-miR-1244-1, hsa-miR-663a, hsa-miR-3916, hsa-miR-3687-2, hsa-miR-7846-3p, hsa-miR-5047, hsa-miR-3184-5p, hsa-miR-1248, hsa-miR-6891-3p upregulated in COVID-19 | 5 COVID-19 patients, 3 healthy controls | Plasma | Centrifugation | TRIzol Reagent | PALM-seq protocol; BGISEQ-500 sequencer; single-end 100 base reads | Transcripts per kilobase of exon model per million mapped reads | R package (DESeq2), p-value adjusted < 0.1 (no details on p-value adjustment given) | 44 |
| hsa-miR-150-5p, hsa-miR-375, hsa-miR-122-5p, hsa-miR-494-3p down-regulated in COVID-19 patients; hsa-miR-3197, hsa-miR-4690-5p, hsa-miR-1915-3p, hsa-miR-3652 upregulated in COVID-19 patients | 12 COVID-19 patients with moderate–severe disease, 8 healthy controls | Plasma | Lysis buffer, proteinase-K with constant shaking at 50 °C for 3 h | Automated extraction-free chemistry of HTG EdgeSeq | NGS (HTG EdgeSeq miRNA Whole Transcriptome Assay, HTG Molecular Diagnostics, TruSeq Small RNA Prep kit, Illumina), validation by RT-qPCR | Not indicated | Student's t-test; (no details on p-value adjustment given) | 45 |
| Downregulated in COVID-19: hsa-let-7i, hsa-let-7g, hsa-let-7f2, hsa-let-7f1, hsa-let-7d, hsa-miR-103a1, hsa-miR-126, hsa-miR-139, hsa-miR-15b, hsa-miR-16-1, hsa-miR-16-2, hsa-miR-181b1, hsa-miR-191, hsa-miR-21, hsa-miR-221, hsa-miR-222, hsa-miR-224, hsa-miR-23a, hsa-miR-23b, hsa-miR-25, hsa-miR-26a1, hsa-miR-30c1, hsa-miR-30c2, hsa-miR-30d, hsa-miR-320a, hsa-miR-92a1, hsa-miR-92a2, hsa-miR-93, hsa-miR-98, hsa-miR-151a, hsa-miR-340, hsa-miR-423, hsa-miR-425, hsa-miR-451a, hsa-miR-409, hsa-miR-454, hsa-miR-374b, hsa-miR-1244-1, hsa-miR-3609, hsa-let7a2, hsa-let7a1; upregulated in COVID-19: hsa-miR-663b, hsa-miR-3687-2, hsa-miR-3648-2 | 19 mild and 18 severe COVID-19 patients, 8 controls | Plasma | 1600g for 10 min at 4 °C and 16![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 10 min at 4 °C 000g for 10 min at 4 °C |
HiPure Liquid RNA Mini kit (Magen) | RNA library constructed using the PALM-seq protocol, DNBSEQ platform | Transcripts per kilobase of exon model per million mapped reads | DESeq, no correction for multiple testing mentioned | 46 |
| Both NGS and RT-qPCR: upregulated in COVID-19 pneumonia compared to controls: hsa-miR-193a-5p, hsa-miR-197-3p; upregulated in COVID-19 ARDS compared to COVID-19 pneumonia: hsa-miR-206 | 18 healthy controls, 15 patients with COVID-19 pneumonia, 15 patients with COVID-19 ARDS | Extracellular vesicles isolated from serum | Centrifuged at 3400 × g for 10 min at room temperature | Extracellular vesicles isolation kit (miRCURY Exosome Isolation Kit, Qiagen) followed by RNA extraction | NEBNext Multiplex Small RNA Library Prep Set for Illumina (New England Biolabs); miRCURY LNA RT kit, miRCURY LNA SYBR Green PCR kit, LNA miRNA PCR Assays (Qiagen) | RNA seq: DESeq2; RT-qPCR: geometric mean of reference miRNAs (hsa-miR-30d-5p, hsa-miR-30e-5p, hsa-let-7i-5p, hsa-miR-148b-3p, hsa-miR-146b-5p, hsa-miR-425-5p, hsa-miR-24-3p, hsa-miR-125a-5p) | DESeq2, Benjamini–Hochberg FDR correction | 47 |
| hsa-miR-146a-5p, hsa-miR-21-5p, hsa-miR-142-3p downregulated in COVID-19 compared to healthy controls; hsa-miR-3605-3p upregulated in COVID-19 compared to healthy controls; hsa-miR-15b-5p, hsa-miR-486-3p, hsa-miR-486-5p upregulated only in patients with severe COVID-19; hsa-miR-181a-2-3p, hsa-miR-31-5p, hsa-miR-99a-5p downregulated only in severe cases | 6 severe COVID-19 patients, 6 moderate COVID-19 patients, 4 healthy controls | Whole blood | Erythrocyte removal: erythrocyte lysis buffer | NucleoSpin miRNA kit (Macherey-Nagel) | NEBNext Multiplex Small RNA library Prep set for Illumina (NEB) | Number of mapped reads per kilobase per million reads | One-way ANOVA/Tukey's test (no details on p-value adjustment given) | 54 |
| Down in severe COVID-19: hsa-miR-335-5p, hsa-miR-24-3p; down in COVID-19: hsa-miR-181a-5p | 6 moderate, 4 severe COVID-19 patients, 4 controls | Whole blood | Red blood cell lysis | TRIzol (Life Technologies) | NEXTflex Small RNA-Seq Kit v3 (Bio Scientific Corporation) | miRDeep2 reads per million mapped reads | Limma package in R, FDR | 56 |
| Downregulation of hsa-miR-320a, hsa-miR-320b and hsa-miR-320c, hsa-miR-4747-3p, hsa-miR-4429, hsa-miR-6729-3p and hsa-miR-1908-5p in patients with severe respiratory failure; upregulation of hsa-miR-374a-3p, hsa-miR-15a-3p, hsa-miR-3688-5p, hsa-miR-4721 in COVID-19 patients with severe respiratory failure | 11 COVID-19 patients with moderate respiratory failure, 10 COVID-19 patients with severe respiratory failure, 8 healthy controls | Whole blood | PAXgene Blood RNA System for miRNA | PAXgene Blood miRNA Kit (Qiagen) | QIAseq miRNA Library Kit (Qiagen)/next-generation sequencing: MiSeq Reagent kit v3, the PhiX Sequencing Control v3 and the MiSeq Desktop Sequencer | Not indicated | One-way ANOVA withBonferroni corrected post-hoc analysis | 58 |
| Discovery: downregulated in non survivors: hsa-miR-8061, hsa-miR-181c-3p, hsa-miR-410-3p, hsa-miR-101-5p, hsa-miR-339-3p, hsa-miR-28-5p, hsa-miR-17-3p; upregulated in non survivors: hsa-miR-1285-5p, hsa-miR-221-3p, hsa-miR-203a-3p, hsa-miR-100-5p; validation: upregulated in non-survivors: hsa-miR-320b, hsa-miR-483-5p | Discovery: 12 COVID-19 patients (6 survivors, 6 non survivors); validation: 116 COVID-19 patients (75 survivors, 41 non survivors) | Not clear whether plasma or serum | Not detailed | Not detailed | Discovery: TruSeq Small RNA Library Prep Kit v2 (Illumina); validation: TaqMan miRNA Reverse Transcription Kit (Life Technologies), miRNA assays (Applied Biosystems) | Discovery: DESeq2; validation: cel-miR-39-3p, hsa-miR-92a and hsa-miR-103 | Discovery: t-test with Benjamini–Hochberg correction; validation: unpaired Student t-test or Mann–Whitney U test, Kaplan–Meier curves, log-rank test for equality of survivor functions, Cox proportional hazards analysis | 59 |
| hsa-miR-155 and hsa-miR-130a higher in mild cases compared to severe/critical disease and healthy controls | 16 severe/critical and 30 mild/moderate recovered COVID-19 patients, 24 healthy controls | Whole blood | — | TRIzol Reagent | Not clearly indicated for miRNAs (RT2 Profiler PCR array (Qiagen)?) | Not indicated | One-way ANOVA/two-tailed t-test | 60 |
| hsa-miR-27a-3p, hsa-miR-27b-3p, hsa-miR-148a-3p, hsa-miR-199a-5p, hsa-miR-491-5p upregulated in ICU patients; hsa-miR-16-5p, hsa-miR-92a-3p, hsa-miR-150-5p, hsa-miR-451a; hsa-miR-486-5p downregulated in ICU patients; hsa-miR-16-5p, hsa-miR-92a-3p, hsa-miR-98-5p, hsa-miR-132-3p, hsa-miR-192-5p, hsa-miR-323a-3p downregulated in patients who did not survive the ICU stay | 43 COVID-19 patients (hospitalized bot not ICU), 36 COVID-19 patients (ICU) | Plasma | Centrifugation | miRNeasy serum/plasma Advanced Kit (Qiagen) | RT-qPCR: miRCURY LNA Universal RT microRNA PCR system (Qiagen) for 41 selected miRNAs | cel-miR-39-3p | Limma, ROC | 61 |
| hsa-miR-4454, hsa-miR-7975 upregulated in severe COVID-19; hsa-miR-451a, hsa-miR-323a-3p, hsa-miR-188-5p, hsa-miR-432-5p, hsa-miR-433-3p, hsa-miR-371a-5p downregulated in severe COVID-19 | Screening: 20 mild, 21 moderate, 17 severe COVID-19 patients | Plasma | Centrifugation | Plasma/Serum RNA Purification Mini kit (Norgen) | Nanostring human v3 miRNA for 827 human miRNAs (Nanostring) | osa-miR-414 | Mann–Whitney U test, FDR correction using the Benjamini–Hochberg method | 62 |
| hsa-miR-1246, hsa-miR-4532, hsa-miR-145-5p, hsa-miR-3651 upregulated in severe versus asymptomatic COVID-19; hsa-miR-3180-3p, hsa-let-7i-5p downregulated in severe versus asymptomatic COVID-19; hsa-miR-3609, hsa-miR-199a-5p, hsa-miR-139-5p, hsa-miR-145-5p, hsa-miR-3651, hsa-miR-1273h-3p upregulated in severe versus mild COVID-19; hsa-miR-4632-5p, hsa-miR-6861-5p, hsa-miR-6802-5p, hsa-miR-5196-5p, hsa-miR-92b-5p, hsa-miR-6805-5p, hsa-miR-98-5p, hsa-miR-3185, hsa-miR-572, hsa-miR-371b-5p, hsa-miR-3180, hsa-miR-8073, hsa-miR-4750-5p, hsa-miR-6075, hsa-let-7i-5p, hsa-miR-1231, hsa-miR-885-3p downregulated in severe versus mild COVID-19 | 9 severe COVID-19 patients, 10 mild COVID-19 patients, 10 asymptomatic COVID-19 patients | Blood | PAXgene Blood RNA Tubes (PreAnalytiX) | Blood miRNA Kit (Qiagen) | Affymetrix GeneChip miRNA 4.0 array using FlashTag Biotin RNA Labeling Kit (Genisphere) | Multiarray average (RMA) method | Limma, FDR | 63 |
| Upregulated in severe COVID-19: hsa-miR-4516, hsa-miR-362-5p, hsa-miR-548k, hsa-miR-320a-3p, hsa-miR-320b, hsa-miR-320c, hsa-miR-320d, hsa-miR-185-5p, hsa-miR-629-5p, hsa-miR-1180-3p, hsa-miR-502-3p; downregulated in severe COVID-19: hsa-miR-454-3p, hsa-miR-625-3p, hsa-miR-30b-5p, hsa-miR-192-5p, hsa-miR-451a, hsa-miR-197-3p, hsa-miR-29b-3p, hsa-miR-126-3p, hsa-miR-146b-5p, hsa-miR-30c-5p, hsa-miR-144-5p, hsa-miR-29a-3p, hsa-miR-363-3p, hsa-miR-99a-5p, hsa-miR-342-3p, hsa-miR-193b-3p, hsa-miR-190a-5p, hsa-miR-365b-3p, hsa-miR-122b-5p, hsa-miR-122-3p | 3 mild COVID-19 patients, 5 severe COVID-19 patients, 2 negative controls | Plasma | Centrifugation 15 min at 2000g, heat inactivation | Internal purification protocol of GenXPro GmbH, based on silica columns | TrueQuant small RNA kit (GenXPro GmbH), Illumina sequencing | Not indicated | DESeq, no correction for multiple testing mentioned | 64 |
| Large number of differentially expressed miRNAs between the different groups; hsa-miR-22-3p and hsa-miR-3180-3p associated with lower probability of survival at 90 days and hsa-let-7f-1-3p, hsa-let-7g-5p, hsa-miR-1255a, hsa-miR-140-3p, hsa-miR-20a-5p, hsa-miR-363-5p, hsa-miR-4510, hsa-miR-6130 associated with better prognosis | 32 severe, 52 moderate, 12 asymptomatic/mild COVID-19 patients, 13 controls | Plasma | Not indicated | miRNeasy Serum Plasma Advanced kit (Qiagen) | NEBNext Multiplex Small RNA Library Prep for Illumina (New England Biolabs), Illumina sequencing | DeSeq2 | Generalized linear model with negative binomial distribution, FDR correction using the Benjamini–Hochberg method | 65 |
| Up in COVID-19: hsa-miR-6780b-3p, hsa-miR-6883-3p, hsa-miR-4769-5p, hsa-miR-6873-3p, hsa-miR-320b, hsa-miR-7111-3p, hsa-miR-4755-3p, hsa-miR-320c, hsa-miR-6511a-3p, hsa-miR-320d, hsa-miR-5187-3p, hsa-miR-4508, hsa-miR-4659a-5p; down in COVID-19: hsa-miR-4433b-5p, hsa-miR-16-2-3p, hsa-miR126-3p, hsa-miR-150-5p, hsa-miR-224-5p | 4 mild COVID-19 patients, 4 severe/critical COVID-19, 4 controls | Plasma | Centrifugation at 2500 rpm at 4 °C for 10 min | miRNeasy Serum/Plasma Kit (Qiagen) | QIAseq miRNA Library Kit (Qiagen), Illumina sequencing | DESeq2 | No correction for multiple testing mentioned | 66 |
| Down in severe versus mild COVID-19: hsa-miR-451a, hsa-miR-101-3p, hsa-miR-185-5p, hsa-miR-30d-5p, hsa-miR-25-3p, hsa-miR-342-3p, hsa-miR-30e-5p, hsa-miR-150-5p, hsa-miR-15b-5p, hsa-miR-29c-3p, hsa-miR-10b-5p, hsa-miR-16-2-3p, hsa-miR-186-5p, hsa-miR-16-5p, hsa-miR-425-5p, hsa-miR-187-3p, hsa-miR-125a-5p, hsa-miR-106b-3p, hsa-miR-22-3p, hsa-miR-144-5p, hsa-miR-151a-3p, hsa-miR-30a-5p, hsa-miR-92a-3p, hsa-miR-15a-5p, hsa-miR-195-5p, hsa-miR-4508, hsa-miR-194-5p, hsa-miR-140-3p, hsa-miR-142-5p, hsa-miR-99a-5p, hsa-miR-363-3p, hsa-let-7g-5p, hsa-miR-20a-5p, hsa-miR-144-3p, hsa-miR-10a-5p, hsa-miR-378a-3p, hsa-let-7f-5p, hsa-miR-660-5p, hsa-miR-3135b, hsa-miR-96-5p, hsa-miR-125b-5p, hsa-let-7i-5p | 4 mild COVID-19 patients, 4 severe/critical COVID-19 | Plasma | Centrifugation at 2500 rpm at 4 °C for 10 min | miRNeasy Serum/Plasma Kit (Qiagen) | QIAseq miRNA Library Kit (Qiagen), Illumina sequencing | DESeq2 | No correction for multiple testing mentioned | 66 |
| Differentially expressed according to severity of COVID-19: hsa-miR-133a-3p, hsa-miR-122-5p, hsa-miR-126-3p, hsa-miR-21-5p, hsa-miR-197-3p, hsa-miR-320a-3p, hsa-miR-223-3p, hsa-miR-210-3p, hsa-miR-192-5p | Screening: 18 COVID-19 patients with mild to moderate disease, 18 with severe disease, 11 controls; validation: 6 mild, 39 moderate, 16 severe COVID-19 patients | Citrate/CTAD plasma | Centrifugation at 2000g for 15 min, 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g for 5 min 000g for 5 min |
RNAseq: Maxwell RSC miRNA Tissue kit (Promega), RT-qPCR: miRNeasy Mini kit (Qiagen) | RNA-seq: RealSeq-Biofluids Plasma/Serum miRNA Library kit for Illumina sequencing (RealSeq Biosciences); RT-qPCR: miRCURY LNA RT kit (Exiqon), miRCURY LNA miRNA PCR Assays (Qiagen) | RNA-seq: not detailed; RT-qPCR: cel-miR-39-3p (Qiagen) | RNA-seq: the quasi-likelihood negative binomial generalized log-linear model functions of edge R v3.2828, DESeq2, Benjamini–Hochberg FDR correction; RT-qPCR: Student's t-tests/Mann–Whitney U tests, using ANOVA tests for continuous variables, Benjamini–Hochberg FDR correction | 67 |
| Discrimination between mild and severe COVID-19, discovery: hsa-miR-122-5p, hsa-let-7c-5p, hsa-miR-21-5p, hsa-miR-140-3p | 31 mild COVID-19 patients (9 who progressed to severe/critical disease) | Serum | Not detailed | miRNeasy Mini Kit (Qiagen) | QIAseq miRNA Library Kit (Qiagen) | Not indicated | One-way ANOVA, Fisher linear discriminant analysis, ROC analysis, Kaplan–Meier curves | 68 |
| hsa-miR-146a-3p, hsa-miR-126-3p, hsa-miR-151-3p, hsa-miR-126-5p downregulated in severe COVID-19; hsa-miR-15a, hsa-miR-424, hsa-miR-627-5p, hsa-miR-145, hsa-miR-205-5p, hsa-miR-200c upregulated in severe COVID-19 | 13 mildly ill, 17 severely ill patients | Small extracellular vesicles isolated from serum | Specific isolation procedure “EV-CATCHER” | miRNeasy Serum/Plasma kit (Qiagen) | Small-RNA sequencing (laboratory developed protocol) | Read counts were normalized to total counts | DESeq2 and edgeR; one-way analysis of variance (ANOVA) with Bonferroni post hoc test | 74 |
| hsa-miR-550-5p, hsa-miR-629-3p upregulated in severe COVID-19 | 13 mildly ill, 17 severely ill patients | Serum | Centrifugation | miRNeasy Serum/Plasma kit (Qiagen) | Small-RNA sequencing (laboratory developed protocol) | Read counts were normalized to total counts | DESeq2 and edgeR; one-way analysis of variance (ANOVA) with Bonferroni post hoc test | 74 |
| hsa-let-7a-5p, hsa-let-7d-5p, hsa-let-7f-5p, hsa-miR-98-5p, hsa-miR-340-5p, hsa-miR-378a-3p predictive of clinical outcome | 50 mild COVID-19, 16 severe COVID-19 patients, 17 healthy controls | Whole blood | None | miRNeasy Serum/Plasma Advanced Kit (Qiagen) | Small RNA sequencing (NEXTflex Small RNA-seq Kit v3, PerkinElmer) | Not indicated | Student's t-test, no correction for multiple testing mentioned | 76 |
| Studies that measured selected miRNAs | ||||||||
|---|---|---|---|---|---|---|---|---|
| Main findings | Patient numbers | Specimen type | Pre-analytical procedures | Extraction methods | miRNA quantification methods | Normalization methods | Statistical methods | Ref. |
| ICU: intensive care unit. FDR: false discovery rate. ROC: receiver operating characteristics. AUC: area under the curve. | ||||||||
| Higher expression of hsa-miR-29a-3p, hsa-miR-146a-3p, hsa-miR-155-5p, hsa-let-7b-3p in COVID-19 patients | 18 COVID-19 patients, 15 healthy controls | PBMC | Ficoll Hypaque density gradient centrifugation | miRNeasy Mini Kit (Qiagen) | RT-qPCR: miScript® II RT Kit (Qiagen), miScript SYBR Green PCR Kit (Qiagen) | SNORD47 RNA | t-Test, Mann–Whitney U test, ROC, FDR (Benjamini and Hochberg procedure) | 48 |
| hsa-miR-21-5p, hsa-miR-155-5p, hsa-miR-499-5p upregulated in COVID-19 patients compared to healthy controls and influenza-ARDS patients. hsa-miR-126-3p downregulated in COVID-19 patients compared to healthy controls and influenza-ARDS patients. hsa-miR-208a-3p upregulated in both severe COVID-19 and influenza-ARDS groups compared to healthy controls | Discovery cohort: mechanically-ventilated COVID-19 (n = 18) and healthy controls (n = 15); validation cohort: mechanically-ventilated COVID-19 patients (n = 20), influenza-ARDS patients (n = 13), healthy controls (n = 32) | Serum | — | miRNeasy Serum/Plasma Advanced Kit (Qiagen) | TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems), RT-qPCR with specific TaqMan miR assays (Applied Biosystems) | cel-miR-39-3p | Mann–Whitney U test, Dunn's multiple comparisons test, ROC | 49 |
| hsa-miR-10b-5p downregulated in COVID-19 patients | 33 patients with COVID-19, 29 controls | Plasma | Centrifugation 2500 rpm for 10 min | miRNeasy Serum/Plasma Kit (Qiagen) | miScript II RT Kit, miScript SYBR Green PCRKit (Qiagen) | U6 snRNA | Mann–Whitney U-test | 50 |
| hsa-miR-29b-3p and hsa-miR-1246 levels higher in COVID-19 patients; hsa-miR-186-5p and hsa-miR-15a-5p lower in COVID-19 patients | 29 COVID-19 patients; 29 healthy controls | Plasma | Centrifugation at 3000g for 10 min; plasmas were inactivated in a water bath at 56 °C for 30 min | RNA extraction and purification kit (Machery-Nagel) | miDETECTA Track miRNA qRT-PCR Starter Kit and primers (RiboBio) | cel-miR-39 | Mann–Whitney U test | 51 |
| hsa-miR-2392 higher in COVID-19 (serum and urine); hsa-miR-1-3p and hsa-miR-155-5p lower in the serum of COVID-19 patients | Serum: 20 COVID-19, 10 non COVID-19; urine: 25 COVID-19, 21 non COVID-19 patients | Serum, urine | Centrifugation | QIAGEN miRNeasy Serum/Plasma kit, Norgen urine microRNA Purification Kit | QIAGEN miRCURY LNA RT Kit, droplet digital PCR | Not detailed | Student's t-test (unadjusted), ROC | 52 |
| Downregulation of miR-200c-3p and miR-421-5p in COVID-19 | 30 COVID-19 patients, 18 controls | Blood (not detailed) | Not indicated | Total RNA isolation kit (Yekta Tajhiz Azama, Tehran, Iran) | Laboratory developed RT-qPCRs; SYBR Green Master Mix (Ampliqon) | U6 | Student t-test | 53 |
| Elevated in COVID-19: hsa-miR-146a-5p, hsa-miR-122-5p | 14 COVID-19 patients, 10 controls | Serum | Not indicated | Total RNA Purification Kit (Norgen) | TaqMan MicroRNA Reverse Transcription Kit (Applied Biosystems), Taqman assays (Applied Biosystems) | RNU6B | Mann–Whitney test | 55 |
| hsa-miR-155-5p upregulated in COVID-19 versus controls, severe versus moderate COVID-19 and non-survivors versus survivors | 98 moderate and 52 severe COVID-19 patients, 50 controls | Plasma | Centrifugation 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min 000 rpm for 10 min |
miRNeasy Mini Kit (Qiagen) | MiScript II reverse transcription Kit, miScript primer assay for miR-155, MiScript SYBR Green PCR Kit (Qiagen) | RNU6-2 | Post hoc test (LSD) for normal distributed variables, ROC | 69 |
| Up in COVID-19: hsa-miR-146b; down in COVID-19: hsa-miR-155 | 22 mild, 15 severe COVID-19 patients, 15 controls | Plasma | Centrifugation at 4 °C for 10 min at 1200g, and for 10 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g 000g |
TRI-Reagent-LS (Sigma) | TaqMan MicroRNA-RT-Kit and Taqman assays (Thermo Fisher Scientific) | cel-miR39 | Univariate and multivariate logistic regression analysis, ROC | 70 |
| Down in patients who died: hsa-miR-155 | 6 COVID-19 patients who died and 31 who survived | Plasma | Centrifugation at 4 °C for 10 min at 1200g, and for 10 min at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000g 000g |
TRI-Reagent-LS (Sigma) | TaqMan MicroRNA-RT-Kit and Taqman assays (Thermo Fisher Scientific) | cel-miR39 | Univariate and multivariate logistic regression analysis, ROC | 70 |
| hsa-miR-31-3p, hsa-miR-29a-3p, and hsa-miR-126-3p decreased in more severe disease grades; hsa-miR-17-3p increased in more severe disease grades | Grade 1 (n = 21), grade 2 (n = 20), grade 3 (n = 20), grade 4 (n = 21), grade 5 (n = 21) COVID-19 patients, 20 healthy controls | Serum | Not indicated | mirPremier microRNA isolation kit (Sigma-Aldrich) | Mir-X miRNA First-Strand Synthesis kit (Takara Bio Inc.), Mir-X miRNA qPCR SYBR (Invitrogen) | RNU 48 | One-way ANOVA | 71 |
| hsa-mir-21, hsa-mir-124, hsa-mir-146a downregulated, hsa-mir-326, hsa-mir-155, hsa-mir-27b upregulated in COVID-19 patients with increase of disease grade | Grade 1 (n = 21), grade 2 (n = 20), grade 3 (n = 20), grade 4 (n = 21), grade 5 (n = 21) COVID-19 patients, 20 healthy controls | Serum | Centrifugation at 3000 rpm for 10 min | mirPremier microRNA isolation kit (Sigma-Aldrich) | Mir-X miRNA First-Strand Synthesis kit (Takara Bio Inc.), Mir-X miRNA qPCR SYBR (Invitrogen) | RNU 48 | One-way ANOVA | 72 |
| hsa-miR-4257 downregulated in COVID-19 versus control and in severe versus mild COVID-19 | 59 mild COVID-19, 41 severe COVID-19, 100 healthy controls | Serum | Centrifugation at 4000 rpm for 20 min | miRNeasy extraction kit (Qiagen) | TaqMan MicroRNA Reverse Transcription Kit (Thermo Fisher Scientific), miR-4257 TaqMan probe, universal TaqMan master mix | U6 snRNA | One-way ANOVAs, Mann–Whitney test, ROC, multivariable logistic regression analysis | 73 |
| Main findings | Patient numbers | Specimen type | Pre-analytical procedures | Extraction methods | miRNA quantification methods | Normalization methods | Statistical methods | Ref. |
|---|---|---|---|---|---|---|---|---|
| Associated with severe COVID-19: hsa-miR-125a-5p, hsa-miR-1290, hsa-miR-15b-5p, hsa-miR-491-5p, hsa-miR-532-3p, hsa-miR-200b-3p, hsa-miR-629-5p, hsa-miR-103a-2-5p, hsa-miR-340-5p, hsa-miR-455-5p | 21 non severe COVID-19 patients, 20 severe COVID-19 patients, 20 controls | Nasopharyngeal swabs | None | MagMAX mirVana Total RNA Isolation Kit (Thermo Fisher Scientific) | TaqMan Advanced miRNA cDNA Synthesis Kit (Thermo Fisher Scientific), TaqMan Fast Advanced Master Mix, TaqMan™ Advanced miRNA Human A and B Cards (Thermo Fisher Scientific) | Global mean normalization | Mann–Whitney U test, sparse partial least squares-discriminant analysis, ROC, FDR (Benjamini and Hochberg procedure) | 39 |
| hsa-miR-200c-3p overexpressed in severe COVID-19 | 39 patients without COVID-19, 37 symptomatic not hospitalized COVID-19 patients, 21 hospitalized COVID-19 patients, 14 patients with severe COVID-19 | Saliva/sublingual smear (severe group) | Collecting tube with saline solution for sublingual smears | EasyExtract DNA–RNA Kit (Interprise) | TaqMan miRNA reverse transcription kit (Applied Biosystems), hsa-miR-200c-3p (478351_mir) qPCR with HOT FIRE Pol Probe Universal qPCR Mix (Solis BioDyne) | miR-191 | Students' t-test/Mann–Whitney test, logistic regression | 77 |
| Deregulation of five miRNA ratios: hsa-miR-122-5p/hsa-miR-199a-5p, hsa-miR-125a-5p/hsa-miR-133a-3p, hsa-miR-155-5p/hsa-miR-486-5p, hsa-miR-214-3p/hsa-miR-222-3p, hsa-miR-221-3p/hsa-miR-27a-3p between COVID-19 and non-COVID-19 ICU patients | 18 COVID-19 ICU patients; 14 non-COVID-19 ICU patients | Bronchial aspirates | None | miRNeasy Mini Kit (Qiagen) | miRCURY LNA RT Kit (Qiagen); miRCURY LNA SYBR Green PCR Kit (Qiagen) in 384-well miRCURY LNA miRNA Custom PCR Panels (Qiagen) | Ratios | Linear models for arrays, not mentioned whether FDR was used | 78 |
| Deregulation of five miRNA ratios: hsa-miR-1-3p/hsa-miR-124-3p, hsa-miR-125b-5p/hsa-miR-34a-5p, hsa-miR-126-3p/hsa-miR-16-5p, hsa-miR-199a-5p/hsa-miR-9-5p, hsa-miR-221-3p/hsa-miR-491-5p between COVID-19 ICU survivors and nonsurvivors | 39 COVID-19 ICU survivors and 18 nonsurvivors | Bronchial aspirates | None | miRNeasy Mini Kit (Qiagen) | miRCURY LNA RT Kit (Qiagen); miRCURY LNA SYBR Green PCR Kit (Qiagen) in 384-well miRCURY LNA miRNA Custom PCR Panels (Qiagen) | Ratios | Linear models for arrays, not mentioned whether FDR was used | 78 |
| Upregulated in COVID-19: hsa-miR-142-3p, hsa-miR-93-5p, hsa-miR-486-5p, hsa-miR-451a, hsa-miR-19a-3p; downregulated in COVID-19: hsa-miR-3065-3p, hsa-miR-3065-5p, hsa-miR-628-3p | 12 COVID-19 positive, 8 uninfected patients | Anterior nares swabs | None | miRNeasy micro kit (Qiagen) | QIAseq miRNA Library Kit with QIAseq miRNA NGS 48 Index IL (Qiagen), sequencing on the NovaSeq 6000 (Illumina) | Not detailed | DESeq2; adjusted FDR of a p-value of < 0.05, machine learning | 79 |
| Upregulated in COVID-19: hsa-miR-100, hsa-miR-34b-5p, hsa-miR-200a, hsa-miR-34c-5p, hsa-miR-342-5p, hsa-let-7i, hsa-miR-29a | 10 SARS-CoV-2-positive and 10 SARS-CoV-2-negative patients | Nasopharyngeal swabs | None | In-house method using Sera-Mag beads (GE Healthcare) | Small RNA library construction: NEBNext® Small RNA Library Prep Set for Illumina® (NEB), purification: TailorCut Gel Extraction Tool Set (SeqMatic); MiniSeq High Output Reagent Kit, 75 cycles (Illumina); validation by RT-qPCR (TaqMan MicroRNA Assays, Applied Biosystems) | Normalized to 1 million reads (RPM); hsa-mir-148a miRNA for RT-qPCR | Principal component analysis, Kruskal–Wallis test, not mentioned whether FDR was used | 80 |
| Up-regulated in COVID-19: hsa-miR-4443, hsa-miR-12116, hsa-miR-765, hsa-miR-1224-3p, hsa-miR-6880-3p, hsa-miR-6886-3p, hsa-miR-6758-5p, hsa-miR-4716-5p, hsa-miR-1281, hsa-miR-6741-3p, hsa-miR-769-5p, hsa-miR-877-3p, hsa-miR-1469, hsa-miR-10401-5p, hsa-miR-7111-3p, hsa-miR-4646-3p, hsa-miR-204-3p, hsa-miR-7107-5p, hsa-miR-6510-5p, hsa-miR-6823-3p, hsa-miR-3196, hsa-miR-665, hsa-miR-7847-3p, hsa-miR-1268b, hsa-miR-139-3p; downregulated: hsa-miR-34b-3p | 4 SARS-CoV-2-positive and 4 SARS-CoV-2-negative patients | Nasopharyngeal swabs | None | mirVana PARIS kit (Invitrogen) | Small RNA libraries were prepared using NEB Next Multiplex Small RNA Library Prep Set (Illumina) | DEseq2 median of ratios method | Unpaired two-tailed Mann–Whitney U test, not mentioned whether FDR was used | 81 |
Methods
For the elaboration of this review the PRISMA 2020 guidelines were followed.32Eligibility criteria
Studies that reported host miRNA expression in body fluids of COVID-19 patients and compared miRNA expression to healthy controls or between different severity groups of COVID-19 were included. Studies focusing on comparison of convalescent versus acute phase in COVID-19, after vaccination, compared to other diseases or between treatment responders and non-responders were not included.33–36 Studies with investigation of miRNA expression in tissues37 were excluded because this specimen type would not be useful in routine for biomarker measurements. Studies that analyzed miRNA expression in COVID-19 restricted to a particular subpopulation, e.g. pregnant women,38 were not included. Studies were grouped into studies that compared miRNA expression in COVID-19 patients versus controls and those that compared miRNA expression between different severity grades of COVID-19. Studies investigating miRNA expression in other than blood-derived specimen types were reported separately.Information sources and search strategy
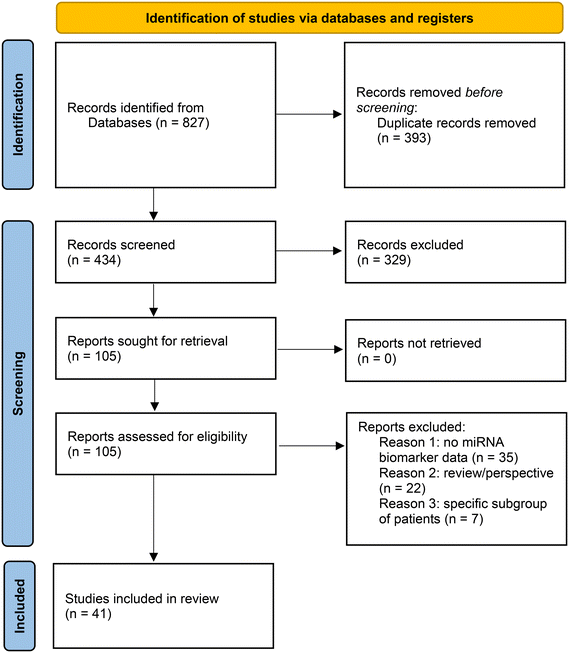
IE searched Pubmed (https://pubmed.ncbi.nlm.nih.gov/) on January 7th, 2022 with the search terms: “miRNA and COVID-19” and “miRNA and SARS-CoV-2”. These searches were performed in order to be exhaustive in the identification of potentially eligible studies. The two searches retrieved 380 and 312 articles respectively. Duplicate studies were removed, resulting in 379 studies for screening. Furthermore, four additional articles were identified by searching the reference lists of publications eligible for full-text review. Screening of studies was performed independently by two researchers (IE and KP) based on the content of the title and abstract. 75 full text articles were retrieved and 26 included in the review. Inconsistencies were discussed until consensus was obtained. A second search with the same search terms and same procedures was performed on April 6th, 2022, with the date limit January first, 2022 to April 6th, 2022. This search retrieved 75 and 54 articles for screening. Duplicates were removed resulting in 54 studies for screening of which 29 were retrieved and 14 were included. Furthermore, data of our study was included.39Fig. 2 presents the flow diagram combined for both searches. Data collection process: data was collected by one researcher (IE) and checked by two other researchers (KP and NG). Data collected was on differentially expressed miRNAs between COVID-19 patients and controls or different severity grades of COVID-19 patients. Furthermore, pre-analytical data, data on experimental procedures, normalization and statistical analysis was collected. Main differentially expressed miRNAs and study details were reported in Tables 1 and 2. A comparative analysis of differentially expressed miRNAs was performed and the miRNAs identified in the different studies reported in Table 1 were compared. Whenever possible, the information of the full name of the mature miRNA was retrieved from the articles, supplementary information, or from technical information (namely, the Thermo Fisher Scientific Taqman miRNA assay ID). The direction of regulation (up- or downregulation) was also retrieved whenever possible. For studies that reported screening followed by validation, we took into account only validated miRNAs. For studies that only performed screening, the screening results were taken into account. Some studies lacked the information on whether the 3p or 5p form of the miRNA was differentially expressed, in which case we considered both forms. Some studies did not report whether miRNAs were up- or downregulated but only whether they were differentially expressed. These cases were still included in the analysis.Figures
Figures were made by using images designed by Freepik (https://fr.freepik.com).Results and discussion
A total of 41 studies met the inclusion criteria and were included in the review. Tables 1 and 2 show the main findings with pre-analytical and experimental details of these studies concerning miRNA expression measurements in blood-derived specimens (Table 1) and respiratory specimens (Table 2) given.miRNAs in blood as biomarkers of SARS-CoV-2 infection
Several studies have investigated the correlation of miRNA dysregulation in patients suffering from severe to mild COVID-19 symptoms. Mainly blood-derived specimens from patients with different disease severity were used. The aim was to examine the differences in circulating miRNA expression in mild and severe disease as well as in healthy controls in order to identify miRNAs associated with COVID-19 or COVID-19 severity. These studies have found several significantly dysregulated miRNAs (Table 1).Diagnostic biomarkers
Several studies performed screening of miRNA expression and analyzed differential expression of miRNAs in COVID-19 patients compared to controls. In the study conducted by Fayyad-Kazan et al., miRCURY LNA miRNome qPCR panels were used to identify differentially expressed miRNAs and a validation of miRNA candidates was performed by RT-qPCR (Table 1). hsa-miR-17-5p, hsa-miR-142-5p were both down-regulated whereas hsa-miR-15a-5p, hsa-miR-19a-3p, hsa-miR-19b-3p, hsa-miR-23a-3p, hsa-miR-92a-3p and hsa-miR-320a were up-regulated in 33 SARS-CoV-2-infected patients compared to 10 healthy controls.40 The study contains a detailed description of methods used for quantification and normalization with the exception that it is not mentioned whether correction for multiple testing was performed (Table 1).Sequencing was used in several studies to profile miRNA expression. Farr et al. reported 50 differentially expressed miRNAs between COVID-19 patients and healthy controls. They proposed three miRNAs (hsa-miR-423-5p, hsa-miR-23a-3p, hsa-miR-195-5p) that classified early SARS-CoV-2 infection accurately with an AUC = 1.0.41 A limitation of the study is the small patient number, especially for the comparison of COVID-19 with and without oxygen that was only based on 3 and 4 patients, respectively (Table 1). An additional comparison between COVID-19 patients and healthy controls was published by Li et al. in two articles.42,43 As far as miRNA expression is concerned, both articles seem to show the same data. The most differentially expressed miRNAs were hsa-miR-16-2-3p, hsa-miR-6501-5p, hsa-miR-618, hsa-miR-183-5p, hsa-miR-627-5p and hsa-miR-144-3p. However, fold-changes were moderate ranging from 1.3 to 2.3.42 This study is further limited by the small patient number and lack of description of whether a correction for multiple testing was used (Table 1).
Yang et al.44 found differentially expressed miRNAs in COVID-19 patients compared to healthy controls: hsa-miR-19b-1-3p, hsa-miR-96, hsa-miR-19b-2-5p, hsa-miR-451a, hsa-miR-451b, hsa-miR-194-1-5p, hsa-miR-144, hsa-miR-486-2-3p, hsa-miR-15a, hsa-miR-29c-3p were downregulated in COVID-19 whereas hsa-miR-3609, hsa-miR-1244-1, hsa-miR-663a, hsa-miR-3916, hsa-miR-3687-2, hsa-miR-7846-3p, hsa-miR-5047, hsa-miR-3184-5p, hsa-miR-1248, hsa-miR-6891-3p were upregulated in COVID-19 patients. The small patient number remains a limitation of this study (Table 1).
Akula et al. found 8 differentially expressed miRNAs in plasma of COVID-19 patients compared to healthy controls by using sequencing and validation by RT-qPCR (Table 1).45 Interestingly, hsa-miR-150-5p was subsequently shown to inhibit SARS-CoV-2 replication. Therefore, the observed downregulation of hsa-miR-150-5p in COVID-19 patients may promote SARS-CoV-2 replication.45
A study with relatively large patient numbers identified several differentially expressed miRNAs between COVID-19 patients and controls (Table 1). Of note, the vast majority was downregulated in COVID-19: hsa-let-7i, hsa-let-7g, hsa-let-7f-2, hsa-let-7f-1, hsa-let-7d, hsa-miR-103a1, hsa-miR-126, hsa-miR-139, hsa-miR-15b, hsa-miR-16-1, hsa-miR-16-2, hsa-miR-181b1, hsa-miR-191, hsa-miR-21, hsa-miR-221, hsa-miR-222, hsa-miR-224, hsa-miR-23a, hsa-miR-23b, hsa-miR-25, hsa-miR-26a1, hsa-miR-30c1, hsa-miR-30c2, hsa-miR-30d, hsa-miR-320a, hsa-miR-92a1, hsa-miR-92a2, hsa-miR-93, hsa-miR-98, hsa-miR-151a, hsa-miR-340, hsa-miR-423, hsa-miR-425, hsa-miR-451a, hsa-miR-409, hsa-miR-454, hsa-miR-374b, hsa-miR-1244-1, hsa-miR-3609, hsa-let-7a2, hsa-let-7a1. Only three miRNAs were upregulated: hsa-miR-663b, hsa-miR-3687-2 and hsa-miR-3648-2.46
Meidert et al. used NGS of serum-derived extracellular vesicles and identified several miRNAs dysregulated in patients with COVID-19 pneumonia compared to healthy controls. When RT-qPCR was used to validate the findings, hsa-miR-193a-5p and hsa-miR-197-3p were upregulated in COVID-19 pneumonia patients (Table 1).47
Four studies investigated the expression of selected miRNAs by RT-qPCR: Donyavi et al. found that four specific miRNAs, hsa-miR-29a-3p, hsa-miR-146-3p, hsa-miR-155-5p and hsa-let-7b-3p were upregulated in PBMCs of COVID-19 patients compared to healthy controls.48 ROC curve analysis revealed that hsa-miR-29a-3p, hsa-miR-146a-3p and hsa-miR-155-5p allowed discrimination between controls and COVID-19 patients. On the other hand, to distinguish post-acute from acute phase COVID-19, hsa-miR-29a-3p and hsa-miR-146-3p may be used.48
Garg et al. included influenza infected patients in addition to COVID-19 and healthy controls. They found hsa-miR-21-5p, hsa-miR-155-5p, hsa-miR-499-5p upregulated in COVID-19 patients compared to healthy controls and influenza-ARDS patients. Hsa-miR-208a-3p was upregulated in both severe COVID-19 and influenza-ARDS groups compared to healthy controls. This shows that some miRNAs may be commonly dysregulated by different viral infections whereas others are dysregulated specifically.49
In a large study, hsa-miR-10b-5p was found downregulated in the plasma of COVID-19 patients.50 Wu et al. analyzed expression of 12 IFN signaling-related miRNAs in plasma of 29 COVID-19 patients and 29 controls and found higher levels of hsa-miR-29b-3p and hsa-miR-1246 levels in COVID-19 patients and lower hsa-miR-186-5p and hsa-miR-15a-5p lower in COVID-19 patients.51 One should note that plasma samples were heated in this study. These two studies report differential expression but did not perform ROC analysis to evaluate diagnostic performance of these miRNAs as biomarkers.
McDonald et al. measured hsa-miR-2392 expression by droplet digital PCR and found it upregulated in COVID-19 patients, in both serum and urine, and hsa-miR-2392 expression was higher in serum of COVID-19 patients necessitating ICU care.52 Hsa-miR-1-3p and hsa-miR-155-5p were downregulated in the serum of COVID-19 patients.52
Abdolahi and colleagues used laboratory-developed RT-qPCR assays to quantify miRNA expression and found a downregulation of miR-200c-3p and miR-421-5p in COVID-19 patients.53
miRNAs in blood associated with COVID-19 disease severity
Because miRNA expression not only differs between COVID-19 patients and healthy controls but also between severe and mild or moderate COVID-19, miRNAs may be useful as prognostic biomarkers for disease severity (Table 1).Using sequencing, Tang et al. found that hsa-miR-146a-5p, hsa-miR-21-5p and hsa-miR-142-3p were downregulated whereas hsa-miR-3605-3p was upregulated in COVID-19 compared to healthy controls in whole blood after red blood cell lysis.54 In contrast, hsa-miR-146a-5p was found upregulated in the serum of COVID-19 patients in another study.55 Some miRNAs were upregulated only in patients with severe COVID-19, including hsa-miR-15b-5p, hsa-miR-486-3p and hsa-miR-486-5p, and some miRNAs were downregulated only in severe cases, including hsa-miR-181a-2-3p, hsa-miR-31-5p, and hsa-miR-99a-5p.54 Another study limited by the small patient numbers and performing red blood cell lysis and sequencing, found hsa-miR-335-5p and hsa-miR-24-3p downregulated in severe COVID-19 (ref. 56) (Table 1). This leads to release of red blood cell miRNAs into the blood57 and the authors thus measured a combination of miRNAs in blood cells, red blood cells and extracellular miRNAs.
Two studies used NGS and validation by RT-qPCR to identify miRNAs differentially expressed in different severity grades of COVID-19: a striking downregulation of the miR-320 family members including hsa-miR-320a, hsa-miR-320b and hsa-miR-320c was found in whole blood of patients with severe respiratory failure. Other strongly downregulated miRNAs were hsa-miR-4747-3p, hsa-miR-4429, hsa-miR-6729-3p and hsa-miR-1908-5p, with hsa-miR-374a-3p, hsa-miR-15a-3p, hsa-miR-3688-5p, hsa-miR-4721 being strongly upregulated in COVID-19 patients with severe respiratory failure.58 The second study found that high serum levels of hsa-miR-320b and hsa-miR-483-5p were associated with a higher risk of mortality.59 The discordance of the results concerning hsa-miR-320b may be due to the use of whole blood versus serum for miRNA expression measurements. This shows that the choice of the specimen type is very important and that expression of miRNAs differs between different blood fractions.
More so, Li et al. made a comparison between severe/critical and mild/moderate COVID-19 patients and investigated COVID-19 patients after recovery. This resulted in the observation that hsa-miR-155 and hsa-miR-130a were remarkably higher in the mild cases compared to severe/critical disease and healthy controls.60 This study lacks on the other hand a detailed description of methods used for quantification and normalization of miRNA expression.
A study performed on 79 COVID-19 patients found that five miRNAs were up- and five miRNAs were downregulated in ICU COVID-19 patients compared to non ICU patients by using RT-qPCR61 (Table 1). However, the fold changes were modest. Hsa-miR-148a-3p, hsa-miR-486-5p and hsa-miR-451a were associated with ICU stay in multivariable analysis. Hsa-miR-16-5p, hsa-miR-92a-3p, hsa-miR-98-5p, hsa-miR-132-3p, hsa-miR-192-5p, hsa-miR-323a-3p were downregulated in patients who did not survive the ICU stay. The expression of two miRNAs, hsa-miR-192-5p and hsa-miR-323a-3p, predicted mortality during the ICU stay with an AUC of 0.80.61
Wilson et al. analyzed miRNA expression in plasma of 58 patients with mild, moderate or severe COVID-19 with Nanostring technology. A dysregulation of several miRNAs in severe COVID-19 was found, of which most were downregulated in severe COVID-19 (Table 1).62
Parray et al. compared differential expression of miRNAs between three different COVID-19 severity groups. Multiple miRNAs were found upregulated and downregulated in severe versus asymptomatic COVID-19 as well as between severe and mild COVID-19 (Table 1).63
Grehl and colleagues found 11 upregulated and 20 downregulated miRNAs in severe COVID-19 cases by next generation sequencing (Table 1).64 This study was performed on a small number of patients and no correction for multiple testing was detailed in the article.
Another study compared the expression of miRNAs in severe, moderate, mild/asymptomatic COVID-19 patients and controls. They performed several comparisons between the different groups and found a large number of differentially expressed miRNAs. They further built a score based on the expression of ten miRNAs (hsa-miR-22-3p, hsa-miR-3180-3p, hsa-let-7f-1-3p, hsa-let-7g-5p, hsa-miR-1255a, hsa-miR-140-3p, hsa-miR-20a-5p, hsa-miR-363-5p, hsa-miR-4510 and hsa-miR-6130) that allowed to classify patients into low or high risk for mortality.65
de Nicoletti et al. found several differentially expressed miRNAs between COVID-19 patients and uninfected controls as well as depending on COVID-19 severity (Table 1).66 Only four patients per group were analyzed and it was not mentioned whether correction for multiple testing was applied.
A thoroughly conducted study that included a screening and a validation cohort found several differentially expressed miRNAs in COVID-19 patients with different disease severities67 (Table 1). After adjustment for age and sex, hsa-miR-122-5p, and hsa-miR-133a-3p were the only miRNAs besides platelet- and endothelium-derived hsa-miR-126-3p that showed a significantly different expression in mild, moderate and severe COVID-19.67 RT-qPCR measurements of hsa-miR-122-5p and hsa-miR-133a-3p classified patients into severe (n = 16) and non-severe (n = 45) COVID-19 with AUCs of 0.75 and 0.79, respectively.67 Interestingly, hsa-miR-122-5p was also found in extracellular vesicles of COVID-19 patients and allowed to differentiate mild COVID-19 patients from those who progressed to severe or critical disease with an AUC = 0.81.68
Two studies specifically investigated the expression of hsa-miR-155. One found that hsa-miR-155 was overexpressed in COVID-19 versus controls, severe versus moderate COVID-19 and non-survivors versus survivors. AUC to distinguish COVID-19 patients from controls was 0.986 whereas for the distinction of severe and moderate patients AUC = 0.75.69 The other study found hsa-miR-155 underexpressed in patients with COVID-19 compared to controls and underexpressed in patients who died compared to survivors with an AUC = 0.83 to differentiate survivors and non survivors.70 These studies seem to have observed an opposite regulation of the same miRNA.
Keikha and coworkers published two articles on the expression of selected miRNAs in COVID-19. The first article reports that hsa-miR-31-3p, hsa-miR-29a-3p and hsa-miR-126-3p were decreased in more severe COVID-19 disease grades, whereas hsa-miR-17-3p was increased in more severe disease grades.71 Expression in healthy controls is not shown in the article. The second article, probably on the same cohort, showed that hsa-miR-21, hsa-miR-124, hsa-miR-146a were downregulated, whereas hsa-miR-326, hsa-miR-155, hsa-miR-27b were upregulated in COVID-19 patients with increased of disease severity.72
Hsa-miR-4257 was downregulated in COVID-19 patients versus controls and in severe versus mild COVID-19 as reported by Agwa and colleagues.73 ROC analysis revealed that hsa-miR-4257 expression could be used as biomarker to distinguish COVID-19 patients from controls with an AUC of 0.911.73
Differential expression of two miRNAs was found by using whole serum of patients with severe and non-severe COVID-19, whereas ten distinct differentially expressed miRNAs were identified when using extracellular vesicles isolated from sera (Table 1).74 This shows that the choice of the specimen type is very important and that expression of miRNAs differs between different blood fractions. Direct comparison of miRNA expression in different specimen types is therefore not recommended.75
By using RNA-seq, another study found 20 differentially expressed miRNAs in serum-derived extracellular vesicles from patients with COVID-19 pneumonia compared to patients with COVID-19 ARDS. However, only one of these (hsa-miR-206) showed the same differential expression when RT-qPCR was used (Table 1).47
In a relatively large study, six miRNAs, namely hsa-let-7a-5p, hsa-let-7d-5p, hsa-let-7f-5p, hsa-miR-98-5p, hsa-miR-340-5p, hsa-miR-378a-3p were found predictive of clinical outcome of COVID-19 among other biomarkers.76
miRNAs in respiratory specimens
Lately, a few studies investigated miRNA expression in respiratory specimens of COVID-19 patients. This is of interest because it may be useful to identify biomarkers in the same specimen type that is used for the diagnosis of SARS-CoV-2 infection by RT-PCR, i.e. nasopharyngeal specimens. This would avoid obtaining a blood specimen for biomarker measurement.One study measured expression of hsa-miR-200c-3p in saliva of symptomatic non-hospitalized, hospitalized COVID-19 patients and patients without COVID-19 as well as in sublingual smears of ICU COVID-19 patients (severe group).77 A higher expression of hsa-miR-200c-3p was found in the severe group (Table 2). However, given the difference in specimen types used in the different groups, this difference may be due to the different specimen type rather than the COVID-19 severity.77 Of note, hsa-miR-200c-3p was found downregulated in the blood of COVID-19 patients in an independent study.53
Molinero et al. studied miRNA expression in bronchial aspirate specimens of COVID-19 and non COVID-19 ICU patients. Rather than comparing the expression of single miRNAs, they compared expression ratios and found several differences of the expression of certain miRNA ratios when COVID-19 and non-COVID-19 as well as ICU survivors and nonsurvivors were compared (Table 2). They constructed a model for ICU mortality prediction and found that the best combination of miRNA ratios (hsa-miR-125b-5p/hsa-miR-34a-5p, hsa-miR-199a-5p/hsa-miR-9-5p, and hsa-miR-221-3p/hsa-miR-491-5p) had an AUC = 0.85 for predicting ICU mortality. The hsa-miR-199a-5p/hsa-miR-9-5p ratio showed an AUC = 0.80 for predicting ICU mortality.78
Farr et al. investigated miRNA expression in anterior nares swabs and found six differentially expressed miRNAs between COVID-19 and uninfected patients (Table 2). Supervised machine learning identified a three-miRNA signature consisting of hsa-miR-30c-2-3p, hsa-miR-628-3p and hsa-miR-93-5p independently classified COVID-19 cases with 100% accuracy.79
Eichmeier and colleagues identified 7 miRNAs in nasopharyngeal swabs by sequencing that were differentially expressed in COVID-19 positive and negative patients (Table 2). However, RT-qPCR comparison of the miRNA expression yielded different results.80
Wu et al. studied miRNA expression in nasopharyngeal swabs specimens of 4 COVID-19 patients and 4 controls and identified several differentially expressed miRNAs, where most of them were upregulated in COVID-19 compared to controls (Table 2).81
We recently investigated miRNA expression in COVID-19 patients and found ten miRNAs associated with severe COVID-19 (Table 2). Interestingly, expression of most of these miRNAs was lower in severe than in non-severe COVID-19 patients. ROC analysis revealed that three of these miRNAs are promising candidate biomarkers for severe COVID-19, namely hsa-miR-125a-5p, hsa-miR-491-5p and hsa-miR-200b-3p. These miRNAs discriminated severe from non-severe cases with AUCs ranging from 0.76 to 0.79.39
Comparison and overlap of miRNAs in different studies
When comparing the miRNAs identified in the different studies, the overlap is surprisingly small. As far as differential expression between COVID-19 patients and controls is concerned, 20 miRNAs were identified in at least two studies with the same deregulation when this was described. For studies, comparing the differential expression between different severity grades of COVID-19, the overlap was bigger: 35 miRNAs were identified in at least two studies with the same direction of deregulation when this was described. Applying more strict criteria, i.e. at least three studies with the same direction of deregulation when this was described, resulted in three miRNAs for the studies investigating differential expression between COVID-19 patients and controls, namely hsa-miR-126, hsa-miR-150-5p, hsa-miR-155. Interestingly, hsa-miR-155 was found downregulated in plasma and serum of COVID-19 patients in two studies,52,70 whereas it was found upregulated in plasma, serum, whole blood and PBMCs in four studies.48,49,60,69 Applying these stricter criteria for the studies investigating differential expression between different severity grades of COVID-19 resulted in six miRNAs, namely hsa-miR-126-3p, hsa-miR-140-3p, hsa-miR-192-5p, hsa-miR-451a, hsa-miR-98-5p, hsa-miR-99a-5p. These miRNAs can be considered as most robust and thus most promising but to be used as biomarkers.The reasons for the small overlap of miRNAs identified as differentially expressed in the different studies are multiple. First, the choice of the patient population differs. Next, the definition of mild versus severe COVID-19 also is not uniform in the different studies. Another main factor is certainly the choice of the initial specimen: the use of whole blood, plasma, serum or extracellular vesicles. miRNAs found associated with severe COVID-19 were distinct when using serum or extracellular vesicles from serum.74 Pre-analytical aspects and techniques used for miRNA extraction and expression profiling were also different (Tables 1 and 2). Normalization strategies and statistical methods were also diverse. Many studies did not report correction for multiple analyses and most did not validate their results in a validation cohort or by using a different technique. All these factors influence on the results.
Future perspectives and challenges
Before miRNA expression can be used in routine patient management, some challenges need to be overcome. These challenges are detailed in recent reviews75,82 and range from standardization of the specimen type, pre-analytical issues like specimen processing and storage, to data analysis and interpretation. The choice of the specimen type, including the blood fraction (plasma or serum) and anticoagulant (EDTA, heparin or citrate) influence both miRNA levels.57 Impacts of potential confounders, including physiological factors (i.e. age and sex), body mass index, underlying diseases and pharmacological treatments, on miRNA expression need to be taken into account.75,83,84 Pre-analytical factors, such as centrifugation and storage duration and temperature should be standardized. As far as analytical procedures are concerned, different RNA isolation protocols may impact the efficacy of small RNA isolation.75,82 Furthermore the technique chosen for miRNA quantification can impact on the results.82 The importance of controls used for monitoring of RNA extraction quality cannot be underestimated. Finally, the methods of normalization and statistical analyses also influence the results and should be critically reviewed.Whereas the published studies reported relative quantification of miRNAs, the question of quantification for routine diagnostic use arises. Potentially, absolute quantification of miRNAs would be more useful in the routine diagnostic setting with cut-offs of miRNA expression levels associated with diagnostic or prognostic performance of the given miRNA to be defined.
The next steps may be to investigate influence of physiological factors on the expression of candidate biomarker miRNAs in order to determine whether age or sex influence their expression. The impact of infections with other viruses on their expression level also needs to be investigated in order to ascertain that the observed changes are specific to SARS-CoV-2 infection.
While a vast amount of data on miRNA expression in body fluids of COVID-19 patients is available, before the application of miRNAs as diagnostic or prognostic biomarkers, a number of technical and analytical issues have to be resolved first. Further studies with larger patient cohorts allowing for multivariable analysis taking into account confounding factors are necessary.
Author contributions
Conceptualization: Ilka Engelmann; data curation: Kato Pollet, Nathalie Garnier, Ilka Engelmann; formal analysis: Kato Pollet, Nathalie Garnier, Ilka Engelmann; funding acquisition: Annemieke Madder, Sabine Szunerits, Didier Hober, Ilka Engelmann; investigation: Kato Pollet, Nathalie Garnier, Ilka Engelmann; project administration: Ilka Engelmann; supervision: Annemieke Madder, Sabine Szunerits, Didier Hober, Ilka Engelmann; visualization: Kato Pollet, Nathalie Garnier, Ilka Engelmann; writing – original draft: Kato Pollet, Nathalie Garnier, Ilka Engelmann; writing – review & editing: Annemieke Madder, Sabine Szunerits, Didier Hober.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
This research was supported by I-SITE ULNE, the Centre Hospitalier Universitaire de Lille and Université de Lille. NG wants to thank the Ecole Doctorale Biologie-Santé for a PhD fellowship.References
- N. Zhu, D. Zhang, W. Wang, X. Li, B. Yang and J. Song, et al., A Novel Coronavirus from Patients with Pneumonia in China, 2019, N. Engl. J. Med., 2020, 382(8), 727–733 CrossRef CAS.
- Coronaviridae Study Group of the International Committee on Taxonomy of Viruses, The species Severe acute respiratory syndrome-related coronavirus: classifying 2019-nCoV and naming it SARS-CoV-2, Nat. Microbiol., 2020, 5(4), 536–544 CrossRef.
- H. Yang and Z. Rao, Structural biology of SARS-CoV-2 and implications for therapeutic development, Nat. Rev. Microbiol., 2021, 19(11), 685–700 CrossRef CAS PubMed.
- M. S. Sofi, A. Hamid and S. U. Bhat, SARS-CoV-2: A critical review of its history, pathogenesis, transmission, diagnosis and treatment, Biosaf. Health, 2020, 2(4), 217–225 CrossRef PubMed.
- Y. Yazdanpanah and French COVID cohort investigators and study group, Impact on disease mortality of clinical, biological, and virological characteristics at hospital admission and overtime in COVID-19 patients, J. Med. Virol., 2021, 93(4), 2149–2159, DOI:10.1002/jmv.26601.
- A. Copaescu, O. Smibert, A. Gibson, E. J. Phillips and J. A. Trubiano, The role of IL-6 and other mediators in the cytokine storm associated with SARS-CoV-2 infection, J. Allergy Clin. Immunol., 2020, 146(3), 518–534 CrossRef CAS , e1.
- S. S. Cabler, A. R. French and A. Orvedahl, A Cytokine Circus with a Viral Ringleader: SARS-CoV-2-Associated Cytokine Storm Syndromes, Trends Mol. Med., 2020, 26(12), 1078–1085 CrossRef CAS PubMed.
- D. Blanco-Melo, B. E. Nilsson-Payant, W. C. Liu, S. Uhl, D. Hoagland and R. Møller, et al., Imbalanced Host Response to SARS-CoV-2 Drives Development of COVID-19, Cell, 2020, 181(5), 1036–1045 CrossRef CAS PubMed , e9.
- J. Hadjadj, N. Yatim, L. Barnabei, A. Corneau, J. Boussier and N. Smith, et al., Impaired type I interferon activity and inflammatory responses in severe COVID-19 patients, Science, 2020, 369(6504), 718–724 CrossRef CAS PubMed.
- B. Berkhout and J. Haasnoot, The interplay between virus infection and the cellular RNA interference machinery, FEBS Lett., 2006, 580, 2896–2902 CrossRef CAS PubMed.
- A. Esquela-Kerscher and F. J. Slack, Oncomirs - microRNAs with a role in cancer, Nat. Rev. Cancer, 2006, 6, 259–269 CrossRef CAS.
- P. M. Frédérick and M. J. Simard, Regulation and different functions of the animal microRNA-induced silencing complex. WIREs, RNA, 2021, e1701 Search PubMed.
- D. P. Bartel, Metazoan MicroRNAs, Cell, 2018, 173(1), 20–51 CrossRef CAS PubMed.
- W. Wu, E. J. Choi, I. Lee, Y. S. Lee and X. Bao, Non-Coding RNAs and Their Role in Respiratory Syncytial Virus (RSV) and Human Metapneumovirus (hMPV) Infections, Viruses, 2020, 12(3), 345 CrossRef CAS.
- I. Engelmann, E. K. Alidjinou, A. Bertin, F. Sane and D. Hober, miRNAs in enterovirus infection, Crit. Rev. Microbiol., 2018, 44(6), 701–714 CrossRef CAS PubMed.
- I. Martinez-Espinoza, M. D. R. Banos-Lara and A. Guerrero-Plata, The Importance of miRNA Identification During Respiratory Viral Infections, J. Cell. Immunol., 2021, 3(4), 207–214 Search PubMed.
- I. Engelmann, E. K. Alidjinou, A. Bertin, J. Bossu, C. Villenet and M. Figeac, et al., Persistent coxsackievirus B4 infection induces microRNA dysregulation in human pancreatic cells, Cell. Mol. Life Sci., 2017, 74, 3851–3861 CrossRef CAS.
- J. A. Weber, D. H. Baxter, S. Zhang, D. Y. Huang, K. H. Huang and M. J. Lee, et al., The microRNA spectrum in 12 body fluids, Clin. Chem., 2010, 56, 1733–1741 CrossRef CAS.
- P. S. Mitchell, R. K. Parkin, E. M. Kroh, B. R. Fritz, S. K. Wyman and E. L. Pogosova-Agadjanyan, et al., Circulating microRNAs as stable blood-based markers for cancer detection, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 10513–10518 CrossRef CAS PubMed.
- X. Chen, Y. Ba, L. Ma, X. Cai, Y. Yin and K. Wang, et al., Characterization of microRNAs in serum: a novel class of biomarkers for diagnosis of cancer and other diseases, Cell Res., 2008, 18, 997–1006 CrossRef CAS PubMed.
- A. S. Ho, X. Huang, H. Cao, C. Christman-Skieller, K. Bennewith and Q. T. Le, et al., Circulating miR-210 as a Novel Hypoxia Marker in Pancreatic Cancer, Transl. Oncol., 2010, 3, 109–113 CrossRef.
- S. Gilad, E. Meiri, Y. Yogev, S. Benjamin, D. Lebanony and N. Yerushalmi, et al., Serum microRNAs are promising novel biomarkers, PLoS One, 2008, 3, e3148 CrossRef PubMed.
- G. Reid, M. B. Kirschner and N. van Zandwijk, Circulating microRNAs: Association with disease and potential use as biomarkers, Crit. Rev. Oncol. Hematol., 2011, 80, 193–208 CrossRef.
- A. Etheridge, I. Lee, L. Hood, D. Galas and K. Wang, Extracellular microRNA: a new source of biomarkers, Mutat. Res., 2011, 717, 85–90 CrossRef CAS.
- S. K. Ajit, Circulating microRNAs as Biomarkers, Therapeutic Targets, and Signaling Molecules, Sensors, 2012, 12, 3359–3369 CrossRef CAS PubMed.
- C. N. Correia, N. C. Nalpas, K. E. McLoughlin, J. A. Browne, S. V. Gordon and D. E. MacHugh, et al., Circulating microRNAs as Potential Biomarkers of Infectious Disease, Front. Immunol., 2017, 8, 118 CrossRef.
- J. F. Wang, M. L. Yu, G. Yu, J. J. Bian, X. M. Deng and X. J. Wan, et al., Serum miR-146a and miR-223 as potential new biomarkers for sepsis, Biochem. Biophys. Res. Commun., 2010, 394, 184–188 CrossRef CAS.
- N. Schöler, C. Langer, H. Döhner, C. Buske and F. Kuchenbauer, Serum microRNAs as a novel class of biomarkers: a comprehensive review of the literature, Exp. Hematol., 2010, 38(12), 1126–1130 CrossRef.
- L. Cui, Y. Qi, H. Li, Y. Ge, K. Zhao and X. Qi, et al., Serum microRNA expression profile distinguishes enterovirus 71 and coxsackievirus 16 infections in patients with hand-foot-and-mouth disease, PLoS One, 2011, 6, e27071 CrossRef CAS.
- J. Morán, G. Ramírez-Martínez, L. Jiménez-Alvarez, A. Cruz, S. Pérez-Patrigeon and A. Hidalgo, et al., Circulating levels of miR-150 are associated with poorer outcomes of A/H1N1 infection, Exp. Mol. Pathol., 2015, 99(2), 253–261 CrossRef PubMed.
- R. Y. Wang, K. F. Weng, Y. C. Huang and C. J. Chen, Elevated expression of circulating miR876-5p is a specific response to severe EV71 infections, Sci. Rep., 2016, 6, 24149 CrossRef CAS PubMed.
- M. J. Page, D. Moher, P. M. Bossuyt, I. Boutron, T. C. Hoffmann and C. D. Mulrow, et al., PRISMA 2020 explanation and elaboration: updated guidance and exemplars for reporting systematic reviews, BMJ [Br. Med. J.], 2021, 372, n160 Search PubMed.
- H. Y. Zheng, M. Xu, C. X. Yang, R. R. Tian, M. Zhang and J. J. Li, et al., Longitudinal transcriptome analyses show robust T cell immunity during recovery from COVID-19, Signal Transduction Targeted Ther., 2020, 5(1), 1–12 CrossRef PubMed.
- J. Sabbatinelli, A. Giuliani, G. Matacchione, S. Latini, N. Laprovitera and G. Pomponio, et al., Decreased serum levels of the inflammaging marker miR-146a are associated with clinical non-response to tocilizumab in COVID-19 patients, Mech. Ageing Dev., 2021, 193, 111413 CrossRef CAS PubMed.
- P. Martínez-Fleta, P. Vera-Tomé, M. Jiménez-Fernández, S. Requena, E. Roy-Vallejo and A. Sanz-García, et al., A Differential Signature of Circulating miRNAs and Cytokines Between COVID-19 and Community-Acquired Pneumonia Uncovers Novel Physiopathological Mechanisms of COVID-19, Front. Immunol., 2022, 12, 815651 CrossRef PubMed.
- Y. Miyashita, T. Yoshida, Y. Takagi, H. Tsukamoto, K. Takashima and T. Kouwaki, et al., Circulating extracellular vesicle microRNAs associated with adverse reactions, proinflammatory cytokine, and antibody production after COVID-19 vaccination, npj Vaccines, 2022, 7(1), 16 CrossRef CAS.
- A. Centa, A. S. Fonseca, S. G. da S Ferreira, M. L. V. Azevedo, C. B. Vaz de Paula and S. Nagashima, et al., Deregulated miRNA expression is associated with endothelial dysfunction in post-mortem lung biopsies of COVID-19 patients, Am. J. Physiol., 2020, 320(3), L405–L412 Search PubMed.
- I. Saulle, M. Garziano, C. Fenizia, G. Cappelletti, F. Parisi and M. Clerici, et al., MiRNA Profiling in Plasma and Placenta of SARS-CoV-2-Infected Pregnant Women, Cells, 2021, 10(7), 1788 CrossRef CAS PubMed.
- N. Garnier, K. Pollet, M. Fourcot, M. Caplan, G. Marot and J. Goutay, et al., Altered microRNA expression in severe COVID-19: Potential prognostic and pathophysiological role, Clin. Transl. Med., 2022, 12(6), e899 CAS.
- M. Fayyad-Kazan, R. Makki, N. Skafi, M. El Homsi, A. Hamade and R. El Majzoub, et al., Circulating miRNAs: Potential diagnostic role for coronavirus disease 2019 (COVID-19), Infect., Genet. Evol., 2021, 94, 105020 CrossRef CAS PubMed.
- R. J. Farr, C. L. Rootes, L. C. Rowntree, T. H. O. Nguyen, L. Hensen and L. Kedzierski, et al., Altered microRNA expression in COVID-19 patients enables identification of SARS-CoV-2 infection, PLoS Pathog., 2021, 17(7), e1009759 CrossRef CAS PubMed.
- C. Li, X. Hu, L. Li and J. Li, Differential microRNA expression in the peripheral blood from human patients with COVID-19, J. Clin. Lab. Anal., 2020, 34(10), e23590 CAS.
- C.-X. Li, J. Chen, S.-K. Lv, J.-H. Li, L.-L. Li and X. Hu, Whole-Transcriptome RNA Sequencing Reveals Significant Differentially Expressed mRNAs, miRNAs, and lncRNAs and Related Regulating Biological Pathways in the Peripheral Blood of COVID-19 Patients, Mediators Inflammation, 2021, 2021, e6635925 Search PubMed.
- P. Yang, Y. Zhao, J. Li, C. Liu, L. Zhu and J. Zhang, et al., Downregulated miR-451a as a feature of the plasma cfRNA landscape reveals regulatory networks of IL-6/IL-6R-associated cytokine storms in COVID-19 patients, Cell. Mol. Immunol., 2021, 18(4), 1064–1066 CrossRef CAS PubMed.
- S. M. Akula, P. Bolin and P. P. Cook, Cellular miR-150-5p may have a crucial role to play in the biology of SARS-CoV-2 infection by regulating nsp10 gene, RNA Biol., 2021, 19(1), 1–11 Search PubMed.
- Y. Wang, J. Li, L. Zhang, H. X. Sun, Z. Zhang and J. Xu, et al., Plasma cell-free RNA characteristics in COVID-19 patients, Genome Res., 2022, 32(2), 228–241 CrossRef.
- A. S. Meidert, S. Hermann, F. Brandes, B. Kirchner, D. Buschmann and J. N. Billaud, et al., Extracellular Vesicle Associated miRNAs Regulate Signaling Pathways Involved in COVID-19 Pneumonia and the Progression to Severe Acute Respiratory Corona Virus-2 Syndrome, Front. Immunol., 2021, 12, 784028 CrossRef CAS.
- T. Donyavi, F. Bokharaei-Salim, H. B. Baghi, K. Khanaliha, M. Alaei Janat-Makan and B. Karimi, et al., Acute and post-acute phase of COVID-19: Analyzing expression patterns of miRNA-29a-3p, 146a-3p, 155-5p, and let-7b-3p in PBMC, Int. Immunopharmacol., 2021, 97, 107641 CrossRef CAS.
- A. Garg, B. Seeliger, A. A. Derda, K. Xiao, A. Gietz and K. Scherf, et al., Circulating cardiovascular microRNAs in critically ill COVID-19 patients, Eur. J. Heart Failure, 2021, 23(3), 468–475 CrossRef CAS PubMed.
- Z. Bagheri-Hosseinabadi, H. Ostad Ebrahimi, F. Bahrehmand, G. Taghipour and M. Abbasifard, The relationship between serum levels of interleukin-2 and IL-8 with circulating microRNA-10b in patients with COVID-19, Iran. J. Immunol., 2021, 18(1), 65–73 Search PubMed.
- J. Wu, X. Liu, J. Shao, Y. Zhang, R. Lu and H. Xue, et al., Expression of plasma IFN signaling-related miRNAs during acute SARS-CoV-2 infection and its association with RBD-IgG antibody response, Virol. J., 2021, 18(1), 244 CrossRef CAS.
- J. T. McDonald, F. J. Enguita, D. Taylor, R. J. Griffin, W. Priebe and M. R. Emmett, et al., Role of miR-2392 in driving SARS-CoV-2 infection, Cell Rep., 2021, 37(3), 109839 CrossRef CAS , Available from: https://www.sciencedirect.com/science/article/pii/S2211124721013036.
- S. Abdolahi, M. Hosseini, R. Rezaei, S. R. Mohebbi, M. Rostami-Nejad and E. N. Mojarad, et al., Evaluation of miR-200c-3p and miR-421-5p levels during immune responses in the admitted and recovered COVID-19 subjects, Infect., Genet. Evol., 2022, 98, 105207 CrossRef CAS PubMed.
- H. Tang, Y. Gao, Z. Li, Y. Miao, Z. Huang and X. Liu, et al., The noncoding and coding transcriptional landscape of the peripheral immune response in patients with COVID-19, Clin. Transl. Med., 2020, 10(6), e200 CAS.
- C. Pinacchio, M. Scordio, L. Santinelli, F. Frasca, L. Sorrentino and C. Bitossi, et al., Analysis of serum microRNAs and rs2910164 GC single-nucleotide polymorphism of miRNA-146a in COVID-19 patients, J. Immunoassay Immunochem., 2022, 43(4), 347–364 CrossRef CAS.
- X. Liu, Y. Z. Wen, Z. L. Huang, X. Shen, J. H. Wang and Y. H. Luo, et al., SARS-CoV-2 causes a significant stress response mediated by small RNAs in the blood of COVID-19 patients, Mol. Ther.--Nucleic Acids, 2022, 27, 751–762 CrossRef CAS.
- L. Moldovan, K. E. Batte, J. Trgovcich, J. Wisler, C. B. Marsh and M. Piper, Methodological challenges in utilizing miRNAs as circulating biomarkers, J. Cell. Mol. Med., 2014, 18(3), 371–390 CrossRef CAS.
- R. P. Duecker, E. H. Adam, S. Wirtz, L. Gronau, Y. Khodamoradi and F. J. Eberhardt, et al., The MiR-320 Family Is Strongly Downregulated in Patients with COVID-19 Induced Severe Respiratory Failure, Int. J. Mol. Sci., 2021, 22(19), 10351 CrossRef CAS PubMed.
- A. Giuliani, G. Matacchione, D. Ramini, M. Di Rosa, A. R. Bonfigli and J. Sabbatinelli, et al., Circulating miR-320b and miR-483-5p levels are associated with COVID-19 in-hospital mortality, Mech. Ageing Dev., 2022, 202, 111636 CrossRef CAS PubMed.
- S. Li, X. Duan, Y. Li, M. Li, Y. Gao and T. Li, et al., Differentially expressed immune response genes in COVID-19 patients based on disease severity, Aging, 2021, 13(7), 9265–9276 CrossRef CAS PubMed.
- D. de Gonzalo-Calvo, I. D. Benítez, L. Pinilla, A. Carratalá, A. Moncusí-moix and C. Gort-Paniello, et al., Circulating microRNA profiles predict the severity of COVID-19 in hospitalized patients, Transl. Res., 2021, 236, 147–159 CrossRef PubMed.
- J. C. Wilson, D. Kealy, S. R. James, T. Plowman, K. Newling and C. Jagger, et al., Integrated miRNA/cytokine/chemokine profiling reveals severity-associated step changes and principal correlates of fatality in COVID-19, iScience, 2022, 25(1), 103672 CrossRef CAS PubMed.
- A. Parray, F. A. Mir, A. Doudin, A. Iskandarani, I. M. M. Danjuma and R. A. T. Kuni, et al., SnoRNAs and miRNAs Networks Underlying COVID-19 Disease Severity, Vaccines, 2021, 9(10), 1056 CrossRef CAS.
- C. Grehl, C. Schultheiß, K. Hoffmann, M. Binder, T. Altmann and I. Grosse, et al., Detection of SARS-CoV-2 Derived Small RNAs and Changes in Circulating Small RNAs Associated with COVID-19, Viruses, 2021, 13(8), 1593 CrossRef CAS.
- A. Fernández-Pato, A. Virseda-Berdices, S. Resino, P. Ryan, O. Martínez-González and F. Pérez-García, et al., Plasma miRNA profile at COVID-19 onset predicts severity status and mortality, Emerging Microbes Infect., 2022, 11(1), 676–688 CrossRef.
- A. S. de Nicoletti, M. B. Visacri, C. R. da SC da Ronda, P. E. do NS Vasconcelos, J. C. F. Quintanilha and R. N. de Souza, et al., Differentially expressed plasmatic microRNAs in Brazilian patients with Coronavirus disease 2019 (COVID-19): preliminary results, Mol. Biol. Rep., 2022, 49(7), 6931–6943 CrossRef.
- C. Gutmann, K. Khamina, K. Theofilatos, A. B. Diendorfer, S. A. Burnap and A. Nabeebaccus, et al., Association of cardiometabolic microRNAs with COVID-19 severity and mortality, Cardiovasc. Res., 2022, 118(2), 461–474 CrossRef CAS.
- Y. Fujita, T. Hoshina, J. Matsuzaki, Y. Yoshioka, T. Kadota and Y. Hosaka, et al., Early prediction of COVID-19 severity using extracellular vesicle COPB2, J. Extracell. Vesicles, 2021, 10(8), e12092 CAS.
- R. A. H. Haroun, W. H. Osman, R. E. Amin, A. K. Hassan, W. S. Abo-Shanab and A. M. Eessa, Circulating plasma miR-155 is a potential biomarker for the detection of SARS-CoV-2 infection, Pathology, 2022, 54(1), 104–110 CrossRef CAS PubMed.
- R. Kassif-Lerner, K. Zloto, N. Rubin, K. Asraf, R. Doolman and G. Paret, et al., miR-155: A Potential Biomarker for Predicting Mortality in COVID-19 Patients, J. Pers. Med., 2022, 12(2), 324 CrossRef PubMed.
- R. Keikha, S. M. Hashemi-Shahri and A. Jebali, The relative expression of miR-31, miR-29, miR-126, and miR-17 and their mRNA targets in the serum of COVID-19 patients with different grades during hospitalization, Eur. J. Med. Res., 2021, 26(1), 75 CrossRef CAS PubMed.
- R. Keikha, S. M. Hashemi-Shahri and A. Jebali, The miRNA neuroinflammatory biomarkers in COVID-19 patients with different severity of illness, Neurología, 2021 DOI:10.1016/j.nrl.2021.06.005 , Available from: https://www.sciencedirect.com/science/article/pii/S0213485321001201.
- S. H. A. Agwa, H. Elghazaly, M. S. E. Meteini, S. M. Shawky, M. Ali and A. M. Abd Elsamee, et al., In Silico Identification and Clinical Validation of a Novel Long Non-Coding RNA/mRNA/miRNA Molecular Network for Potential Biomarkers for Discriminating SARS CoV-2 Infection Severity, Cells, 2021, 10(11), 3098 CrossRef CAS PubMed.
- M. I. Mitchell, I. Z. Ben-Dov, C. Liu, K. Ye, K. Chow and Y. Kramer, et al., Extracellular Vesicle Capture by AnTibody of CHoice and Enzymatic Release (EV-CATCHER): A customizable purification assay designed for small-RNA biomarker identification and evaluation of circulating small-EVs, J. Extracell. Vesicles, 2021, 10(8), e12110 CAS.
- L. Pinilla, I. D. Benitez, J. González, G. Torres, F. Barbé and D. de Gonzalo-Calvo, Peripheral blood microRNAs and the COVID-19 patient: methodological considerations, technical challenges and practice points, RNA Biol., 2021, 1–8 Search PubMed.
- Y. Chen, Y. Zheng, Y. Yu, Y. Wang, Q. Huang and F. Qian, et al., Blood molecular markers associated with COVID-19 immunopathology and multi-organ damage, EMBO J., 2020, 39(24), e105896 CrossRef CAS PubMed.
- R. Pimenta, N. I. Viana, G. A. dos Santos, P. Candido, V. R. Guimarães and P. Romão, et al., MiR-200c-3p expression may be associated with worsening of the clinical course of patients with COVID-19, Mol. Biol. Res. Commun., 2021, 10(3), 141–147 CAS.
- M. Molinero, I. D. Benítez, J. González, C. Gort-Paniello, A. Moncusí-Moix and F. Rodríguez-Jara, et al., Bronchial Aspirate-Based Profiling Identifies MicroRNA Signatures Associated With COVID-19 and Fatal Disease in Critically Ill Patients, Kusuri no Chishiki, 2022, 8, 756517 Search PubMed.
- R. J. Farr, C. L. Rootes, J. Stenos, C. H. Foo, C. Cowled and C. R. Stewart, Detection of SARS-CoV-2 infection by microRNA profiling of the upper respiratory tract, PLoS One, 2022, 17(4), e0265670 CrossRef CAS PubMed.
- A. Eichmeier, T. Kiss, M. Kocanova, E. Hakalova, M. Spetik and J. Cechova, et al., Conserved MicroRNAs in Human Nasopharynx Tissue Samples from Swabs Are Differentially Expressed in Response to SARS-CoV-2, Genes, 2022, 13(2), 348 CrossRef CAS PubMed.
- W. Wu, E. J. Choi, B. Wang, K. Zhang, A. Adam and G. Huang, et al., Changes of Small Non-coding RNAs by Severe Acute Respiratory Syndrome Coronavirus 2 Infection, Front. Mol. Biosci., 2022, 9, 821137 CrossRef CAS PubMed.
- D. de Gonzalo-Calvo, J. Pérez-Boza, J. Curado and Y. Devaux, Challenges of microRNA-based biomarkers in clinical application for cardiovascular diseases, Clin. Transl. Med., 2022, 12(2), e585 CAS.
- S. Ameling, T. Kacprowski, R. K. Chilukoti, C. Malsch, V. Liebscher and K. Suhre, et al., Associations of circulating plasma microRNAs with age, body mass index and sex in a population-based study, BMC Med. Genomics, 2015, 8(1), 61 CrossRef PubMed.
- S.-S. Zhou, J.-P. Jin, J.-Q. Wang, Z.-G. Zhang, J. H. Freedman and Y. Zheng, et al., miRNAS in cardiovascular diseases: potential biomarkers, therapeutic targets and challenges, Acta Pharmacol. Sin., 2018, 39(7), 1073–1084 CrossRef CAS.
Footnote |
| † Present address: Pathogenesis and Control of Chronic and Emerging Infections, INSERM, EFS, Univ Antilles, Univ Montpellier, Laboratoire de Virologie, CHU Montpellier, France. |
| This journal is © The Royal Society of Chemistry 2023 |