 Open Access Article
Open Access ArticleInsect olfactory system inspired biosensors for odorant detection
Yanli
Lu
and
Qingjun
Liu
 *
*
Biosensor National Special Laboratory, Key Laboratory for Biomedical Engineering of Education Ministry, Department of Biomedical Engineering, Zhejiang University, Hangzhou, 310027, P. R. China. E-mail: qjliu@zju.edu.cn
First published on 8th October 2022
Abstract
With their accurate olfactory system, insects can detect and discriminate thousands of odorants at very low concentrations in a complicated chemical environment. Inspired by this remarkable olfactory ability, physicochemical transducers integrated with different olfactory derived materials or biomimetic elements were studied and developed, which are collectively called olfactory biosensors. Widely used biological materials include insect antennae, odorant-binding proteins, chemosensory proteins, olfactory receptors, and even sensitive peptides. Based on the physiological properties of these biological materials, they can be incorporated with different analytical techniques, including electrochemical impedance spectroscopy, localized surface plasmon resonance, field-effect transistors, quartz crystal microbalance, surface acoustic waves, and fluorescence imaging, for odorant detection. This paper reviews the development of olfactory biosensors along with typical biochemical detecting cases. The roles of the olfactory system are highlighted to show the design, construction, and detection of olfactory biosensors. Meanwhile, the performance and advantages of insect olfactory system inspired biosensors are introduced with their applications in food evaluation, environmental monitoring, and healthcare diagnosis. With advances in olfactory sensing mechanisms, sensing technologies, and miniaturized electronics, olfactory biosensors, especially those biosensors based on olfactory sensing systems, will eventually become effective detection and analytical tools in the future.
1. Introduction
Olfaction is one of the most important chemical sensing abilities for insects, which plays an essential role throughout their life cycles. It has evolved great sensitivity and discriminatory power that could precisely detect and simultaneously analyze the tremendous amount of chemical signals from the environment and other sources.1–3 This ability helps insects to locate food sources, identify mates, select oviposition sites, and avoid predators.4–6 The primary olfactory appendages of insects are the antennae and maxillary palps. They are covered in numerous hollow sensory hairs called sensilla, which can be divided into basiconic, coeloconic and trichoid.7 The cuticular wall of an olfactory sensillum is covered with many pores, which provide an access route for odor molecules from the environment into the olfactory system. To reach the olfactory receptors (ORs) and OR co-receptor (Orco) ion channels on the ciliated sensory endings of olfactory sensory neuron dendrites, these hydrophobic odorant molecules should first be solubilized and transported. This is performed by small soluble proteins, odorant binding proteins (OBPs).8–10 OBPs are extracellular proteins secreted by support cells that surround olfactory sensory neurons, and are present at high concentrations in the lymph.Based on the strong odor-evoked behaviors and change in physiological states in response to volatile chemicals, olfaction by different insects has been widely investigated.11,12 Insect olfactory systems emerged as prominent models in neuroscience. Moreover, inspired by the sophisticated abilities of olfaction systems to detect chemical signatures, researchers tried to train insects such as honeybees to detect different odors, especially those of poisonous and harmful substances.13–15 Insect-sniffers seemed like potentially useful tools for the quantitative and qualitative analysis of odors in medical, forensic, and food safety applications.16 However, the high cost of training, time consumption, and the short lifetime of insects, have greatly limited further application and development.
Over the last decade, to mimic animals' sense of smell, novel artificial olfaction systems in vitro have been studied and developed, named smell or olfactory biosensors.17,18 Biosensors are analytical devices that combine biological or biological-derived sensitive components with different physicochemical transducers, which could be used for detecting target analytes.19,20 For olfactory biosensors, the biological components could be tissue, organs, olfactory cells, proteins, or even peptides, all of which are responsible for interacting with, binding with, or recognizing different odors.21–24 With the help of Micro-Electro-Mechanical Systems (MEMSs) and nanotechnology, olfactory biosensors have been vigorously developed, which also show great potential in numerous fields, such as environmental monitoring, food quality control, and clinical diagnostics.
Specifically, olfactory tissue- or organ-based biosensors have been developed based on the physiological activity of olfactory receptor neurons on the olfactory tissue or organs. For instance, antenna-based biosensors were mainly developed with electroantennography, which could measure the average potential output of the insect antenna to its brain for a given odorant.25 Apart from natural odorants, studies also indicated that an insect-antenna-based biosensor could also detect odors emitted from different cancer cells.26 For cell-based biosensors, olfactory cells can be treated as encapsulated arrays of receptors and ion channels, which are maintained in a physiologically stable manner, and are responsive to analytes via native cellular machinery.27 One of the most widespread sensing signals emitted by cells is electrophysiological signals, which offer a method of physiological monitoring in situ.23,28 Compared to tissue, organs, and cells, proteins are much easier to obtain, preserve, operate, and incorporate with different sensors, including field-effect transistors (FETs), electrochemical impedance spectra (EIS) sensors, and surface plasmon resonance (SPR) sensors.29–32 Widely used proteins include ORs, OBPs, and peptide sequences derived from olfactory proteins. Through directly interacting with chemicals at the molecular level, these proteins show promising potential for odorant detection, and even for the recognition of chiral molecules.9,33 It is suggested that olfactory proteins are competitive candidates in developing ultra-sensitive biosensors to satisfy the needs of practical applications.
This review summarizes the latest development of olfactory biosensors, especially those inspired by insect olfactory systems. Based on different olfactory bio-materials, biosensors are introduced according to insect antennae, olfactory receptors, odorant-binding proteins, and even olfaction-inspired biomaterials. Firstly, the principles and fabrication strategies of different olfactory biosensors are described. Then, the performances and advantages of these biosensors are discussed with their applications in various fields for odorant detection. Finally, the current limits and challenges facing olfactory biosensors are highlighted in a brief discussion, while the prospects for future directions of development and potential opportunities are described.
2. Insect-antenna-based olfactory biosensors
Apart from using insect-sniffers to detect various odors, insect olfactory organs, especially the antennae, have also been used to construct olfactory biosensors. There are hundreds of peripheral olfactory receptor neurons (ORNs, also called olfactory sensory neurons) in the chemosensory organs of insects, which can be activated by different odors.5,34 When they are incorporated with sensing techniques, the integrated olfactory neurons and olfactory receptors are finely retained. Based on the properties of insect antennae, the typical detectable signals are action potentials or calcium imaging signals. Typical insect-antennae-based biosensors are summarized in Table 1. With the help of electroantennography, field-effect transistors, and fluorescence, different volatile organic compounds could be sensitively detected.| Measurement techniques | Insect species | Target substances | Detection range | Detection limit | Ref. |
|---|---|---|---|---|---|
| Electroantennogram (EAG) | Calliphora vicina (blowfly) | 1,4-Diaminobutane | 4.4 ng–44 μg | 1 ng | 35 |
| Leptinotarsa decemlineata | (Z)-3-Hexen-1-ol, (E)-2-hexenal, and linalool | 10 pg–100 ng | 1 pg | 36 | |
| Calliphora vicina | 1,4-Diaminobutane, 1-hexanol, butanoic acid | 1 ppb–100 ppm (1,4-diaminobutane) | — | 37 | |
| 20–200 ppm (1-hexanol) | |||||
| 8–500 ppm (butanoic acid) | |||||
| Drosophila melanogaster | cis-11-Hexadecenal, cis-3-hexenol, hexanoic acid, benzyl acetate, 2-methyl-5-nitroaniline, cyclohexanone, α-pinene, cisnerolidol, trans-nerolidol, β-caryophyllene, β-ocimene, (R)-(+)-limonene, methyl jasmonate, 2-diisopropylaminoethanol, indole, 2,2-thiodiethanol, 1-heptanol, 1-octanol, 1-nonanol, 1-decanol | 10–100 μg μl−1 | — | 38 | |
| Heliothis virescens | |||||
| Helicoverpa zea | |||||
| Ostrinia nubilalis | |||||
| Microplitis croceipes | |||||
| Electroantennogram (EAG) | Helicoverpa zea | Z-11-Hexadecenal | 1–100 μg | 1 μg | 39 |
| Male Anticarsia gemmatalis, male Trichoplusia ni, male Heliothis virescens, male Helicoverpa zea | Z-11-Hexadecenal, Z-11-tetradecenyl acetate, E,E-8,10-dodecadien-1-ol, E-11-tetradecen-1-ol | 100 μg | — | 40 | |
| Agrotis ipsilon | Z-7-Dodecenyl acetate | 1 μg, 10 μg | — | 40 | |
| Bombyx mori (silkmoth) | Bombykol | 1–1000 ng | — | 41 | |
| Field-effect transistor (FET) | Leptinotarsa decemlineata | cis-3-Hexen-1-ol | 1 ppb–100 ppm | 1 ppt | 42, 43 |
| Leptinotarsa decemlineata | cis-3-Hexen-1-ol, guaiacol, 1-octen | 1 ppb–100 ppm (cis-3-hexen-1-ol) | 1 ppb (cis-3-hexen-1-ol) | 44 | |
| 5–500 ppb (guaiacol) | 100 ppb (guaiacol, 1-octen) | ||||
| 15 ppb–1.5 ppm (1-octen) | |||||
| Phaenops cyanea | cis-3-Hexen-1-ol, guaiacol | 1–100 ppt (cis-3-hexen-1-ol) | 1 ppt (cis-3-hexen-1-ol) | 44 | |
| 50 ppt–500 ppb (guaiacol) | 50 ppt (guaiacol) | ||||
| Fluorescence | Drosophila melanogaster | Volatile organic compounds of cancer cells | — | — | 26 |
2.1 Insect-antenna-based biosensors with electroantennography
Olfactory biosensors based on insect antennae began with the detection of electroantennograms (EAGs) by Schneider.45,46 As the olfactory system of the insect is completely preserved, an EAG offers an easy way to measure the average potential output of the insect antenna to its brain. Thus, it plays important roles not only in physiological studies to explore the mechanism of odorant transduction in olfactory systems, but also in odorant detection. Based on this, a number of commercial insect antennal potential measurement systems have been developed, which provide great convenience for researchers to carry out research into insect antennal related physiological functions. Electroantennograms are usually recorded from insect antennae with micropipette electrodes. The signals can be assessed and analyzed with other software.47 With the help of advanced data acquisition analysis systems, electroantennograms have been successfully employed to establish olfactory biosensors for a wide variety of odorant identifications.35–37 For instance, using the antenna of the Colorado potato beetle, an electroantennographic detector was successfully used to detect volatile organic compounds emitted by its host plants, which could also be used to explore the plant–insect interactions.36 However, the electroantennograms summed antennal activities with only a single feature. In order to read out signals from multiple olfactory receptor neurons for odor discrimination, artificial sensor arrays composed of four antennae from four different species of insects (a multi-channel electroantennogram) were investigated.38,39 With the discriminating electroantennogram biosensors, subtle differences in the electroantennogram responses of the antennae were sensitively interpreted, which could be used for distinguishing different volatile compounds in real time.To meet the demands of practical applications, insect electroantennogram sensors have been integrated with autonomous robots or drones for efficient olfactory search.40,41Fig. 1A gives an example of an autonomous biohybrid drone with an insect-antenna-based sensor device. The antennae of adult male silkmoths (Bombyx mori) were used as olfactory organs. A self-developed miniaturized EAG device as a portable biosensor can be installed on a small drone. The mountable EAG device is composed of the sensing part that includes amplifiers and filters and a processing part that includes a microcontroller (Fig. 1A(ii)). The antennae can be mounted on it with two Ag/AgCl electrodes. The sensing property of the developed EAG device was tested with different glass cartridges containing 1, 10, 100, and 1000 ng of bombykol dissolved in 50 μL of hexane on filter paper. The detected signal intensities increase as the bombykol concentration increases (Fig. 1A(iii)). Equipped with a sensor enclosure, this olfactory sensor integrated with a bio-hybrid drone can identify differences in odor concentration and direction in real time in a pseudo-open environment. Due to the real-time response characteristics of an EAG to odor molecules, the combination of EAG sensor and autonomous robot proposes an efficient technology platform for developing highly sensitive bionic autonomous odor detection equipment.
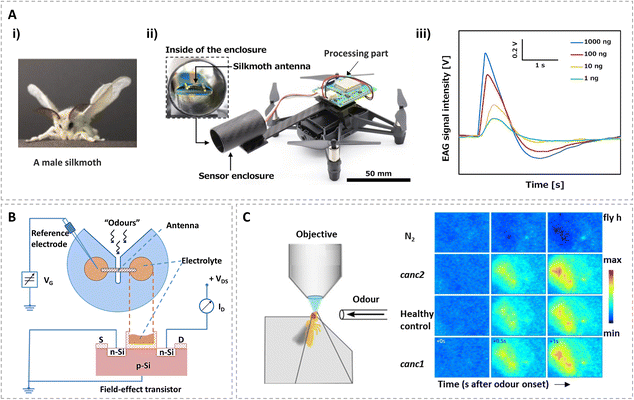 | ||
| Fig. 1 Biosensors based on insect antennae. (A) The electroantennogram (EAG) recorded from an isolated antenna of an adult male silkmoth (Bombyx mori); (i) picture of a male silkmoth; (ii) picture of the bio-hybrid drone and an enlarged view of the sensing part with a silkmoth antenna; (iii) sensing results of the EAG device stimulated by bombykol. Reprinted with permission from ref. 41; Copyright 2021, Elsevier. (B) Experimental set-up of an isolated-antenna field-effect transistor (FET). Reprinted with permission from ref. 42; Copyright 1999, Elsevier. (C) Spatial odor response patterns (fluorescence changes) on the antenna differ for different cancer cell lines (canc1 refers to SKBR3 cells, cancl2 refers to BT474 cells). Reprinted with permission from ref. 26; Copyright 2014, the authors. | ||
With the olfactory system of the insect completely preserved, electroantennogram biosensors with well-designed hardware/software create practical values for olfactory biosensors. It also provides the possibility for the development of an intelligent olfactory perception system. However, electrophysiological experiments cannot simulate the exact intensity of insect response to odors under natural conditions.48 In addition, the limited lifetime is one of the most fragile aspects of an insect-antenna-based biosensor that uses living tissues as the sensing elements.42 Different research showed considerable variations in the lifetimes of isolated antennae, usually less than 2 h.38,49 Although the lifetime of an insect-antenna-based biosensor can be considerably extended to 8 h,50,51 it still needs to further extend its longevity to make full use of the activity of insect antennae, which is important for meeting the needs for the practical application of olfactory sensors.
2.2 Insect-antenna-based biosensors with field-effect transistor
Beyond electroantennogram testing, the electrophysiological signals of insect antennae can also be detected by a field-effect transistor, which is a kind of transistor that uses an electric field to control the electrical behaviors of the device.52 A fundamental structure of the field-effect transistor is composed of substrate, drain electrode, source electrode, and gate electrode. With their innate signal amplification function, field-effect transistors are suitable for detecting weak interactions when the surface carrier changes. This offers a label-free and rapid method for electrophysiological signal detection.Taking advantage of the high sensitivity, a single isolated antenna of a Colorado potato beetle (Leptinotarsa decemlineata Say) was connected with a typical field-effect transistor for odorant detection.42,53,54 The isolated antenna was mounted onto an antenna holder and connected to the FET gate via an electrolyte (Fig. 1B). With the stimulation of its host plant odor (cis-3-hexen-1-ol), the electrophysiological signals generated in insect antennae could be sensitively detected. Meanwhile, impedance spectroscopy was also performed to further characterize the insect antenna as the receptor part of the biosensor.54
Through detecting the impedance of the antenna that vary with applied bias voltages and the amount of applied odors, it is suggested that ion channels in the cell membranes of olfactory neurons might be influenced by the applied voltages or odor concentrations. Although there are still some intrinsic shortcomings of insect-antenna-based biosensors, such as the short lifetime of isolated insect antennae, field-effect transistors provide a sensitive and easy way to develop olfactory biosensors.
2.3 Insect-antenna-based biosensors with fluorescence detection
Fluorescence detection is a widely used optical method to evaluate binding affinities between proteins and their ligands. More importantly, the intensities of fluorescence can be used for cell imaging,55,56 which provides a useful way to study the space distribution information of cells and tissues, such as insect antennae.Inspired by the olfactory ability of insects that could discriminate thousands of substances, a multidimensional analysis of antenna responses to volatile organic compounds emitted from cancer cells and non-cancer cells was performed.26 Using in vivo calcium imaging, an array of olfactory receptor neurons could be recorded on a fruit fly's antenna. As a proxy for neuronal activity, calcium imaging that relies on intracellular calcium concentrations was correlated with action potential rates. Unlike signals from electroantennograms and field-effect transistors, spatial odorant response patterns could be observed through detecting calcium imaging. Through detecting the fluorescence changes in response to odor stimulation, it could be used to discriminate healthy mammary epithelial cells from human breast cancer cell lines (Fig. 1C). This study provided a platform for utilizing the fruit fly's olfactory system to detect cancer cells, which showed promising potential in the early screening of tumors in clinical diagnostics. Fluorescence detection can provide multidimensional information, but it needs a versatile fluorescent probe capable of binding the olfactory proteins and allowing them to compete with the odorants.57
Using insect antennae as the biological recognition elements of olfactory biosensors provided a simple and easy method for odorant detection. However, all of the antennae were extracted from live insects. The short usable life severely limited applications of insect-antenna-based olfactory biosensors. Further research needs to address these limitations. Meanwhile, due to the adaptation of the antenna, the sensor signals might be significantly reduced during detection.43 This may easily lead to errors in odorant detection, analysis, and identification. Protein interactions are essential for cells to realize their functions and could reflect information about cellular metabolism.58 Thus, proteins are promising sensing components in developing biosensors to increase stability, sensitivity and selectivity.
3. Odorant binding protein-based biosensors
For the sense of smell, hydrophobic odorant molecules have first to be solubilized and transported from the lymph of chemosensilla to the membranes of olfactory receptor neurons.10 This is performed by different extracellular soluble proteins, including OBPs and CSPs.2,59 The dissociation constants, Kd, of these extracellular soluble proteins are in the upper nanomolar or lower micromolar range for hydrophobic molecules. The OBP family includes two kinds of proteins: pheromone binding proteins (PBPs) and general odorant binding proteins (GOBPs).60 Although the exact function of OBPs is still unclear, studies summarize them as follows: those acting as transporters of hydrophobic odorant molecules that protect the odorants from degradation before activation of the receptors, the essential cofactors in activating ORs, and scavengers or deactivators that remove odorants from the sensillum lymph.61 In general, the tertiary structure of insect OBPs contains a central hydrophobic cavity, which provides an optimal environment for binding odorants through hydrophobic, polar, electrostatic, and π-stacking interactions. The structures of OBPs can encode different binding properties for the wide spectrum of odorant molecules, such as aromatic molecules and aliphatic compounds.Compared with membrane proteins (e.g. ORs), OBPs are small globular proteins of about 10–30 kDa, and are easier to isolate and purify.60,62,63 More importantly, studies have shown that OBPs were robust enough to stand up to wide ranges of pH and temperature (even to 80 °C) for substantial mistreatment, without denaturing and losing their binding properties.64 All these suggested that insect extracellular soluble proteins were ideal materials to develop olfactory biosensors. Due to the fact that OBPs were easy to obtain and preserve, they could be easily incorporated with different analytical techniques, including electrochemical impedance spectroscopy, localized surface plasmon resonance, field-effect transistors, and surface acoustic waves. Typical insect odorant binding protein-based biosensors are summarized in Table 2.
| Measurement techniques | Biosensing elements | Target substances | Detection range | Ref. |
|---|---|---|---|---|
| Electrochemical impedance spectroscopy (EIS) | OBPs from honeybee (Apis cerana cerana) | Linalool, geraniol, β-ionone, 4-allylveratrole, phenylacetaldehyde, dibutyl phthalate, isoamyl acetate, methy-p-hydroxyl benzoate | 10−6–10−3 M | 64 |
| OBPs from honeybee (Apis cerana cerana) | Isoamyl acetate | 10−9–10−4 M | 66 | |
| OBPs from oriental fruit fly (Bactrocera dorsalis) | Isoamyl acetate, β-ionone, benzaldehyde | 10−7–10−4 M | 67 | |
| OBPs from oriental fruit fly (Bactrocera dorsalis) | Benzaldehyde | 10−7–10−3 M | 68 | |
| CSPs from honeybee (Apis cerana cerana) | Isoamyl acetate, geraniol, phenylacetaldehyde | 10−7–10−4 M | 30 | |
| OBPs from fruit fly (Drosophila melanogaster) | Denatonium, quinine, berberine | 10−9–10−6 mg mL−1 | 69 | |
| OBPs from honeybee (Apis cerana cerana) | Methyl p-hydroxy-benzoate, vanillyl alcohol | 10−7–10−4 M | 70 | |
| Localized surface plasmon resonance (LSPR) | OBPs from honeybee (Apis cerana cerana) | β-Ionone, 2,4,6-trinitrotoluene (TNT) | 0.01–1 mM | 32 |
| 2,4-Dinitrotoluene (DNT) | ||||
| 3-Mononitrotoluene (1NT) | ||||
| Field-effect transistor (FET) | OBP 14 from honeybee (Apis mellifera) | Homovanillic acid, methyl vanillate, eugenol, citral, methyl eugenol, geraniol | 0.1–200 μM | 71 |
| OBP14 from honeybee (Apis mellifera) | Homovanillic acid | 0.1–100 μM | 72 | |
| OBP14 from honeybee (Apis mellifera) | Eugenol, homovanillic acid | 0.1–200 μM | 57 | |
| OBPs from fruit fly (Drosophila), LUSH | Eugenol, homovanillic acid | 0.001–1% | 73 |
3.1 OBP-based biosensors with electrochemical techniques
Electrochemical impedance spectroscopy is one of the most powerful and sensitive electrochemical techniques to characterize the properties of biosensors during electrode fabrication and target molecule detection. It is usually conducted by applying a small sinusoidal potential to an electrochemical cell, and then measuring the current signals. Thus, electrochemical impedance spectroscopy can sensitively characterize and reflect biochemical processes occurring at the electrode–solution sensing interface.65 It is also the most commonly used technique in OBP-based biosensors.With a uniform electric field and high electrode coverage, interdigitated electrodes were widely used in impedance detection. Fig. 2A shows a typical sensing electrode. Using the interdigitated electrodes, different olfactory biosensors with OBPs from honeybee (Apis cerana cerana) and oriental fruit fly (Bactrocera dorsalis) were established.64,66–68 The impedance spectra of isoamyl acetate are displayed in Fig. 2B as an example. The results could be analyzed with the Randles circuit (inset of Fig. 3B). There are four elements: solution resistance (Rs), charge transfer resistance (Rct), Warburg impedance (Zw), and constant phase element (CPE). Normally, Rct was chosen as the sensing parameter to interpret protein–ligand interactions.
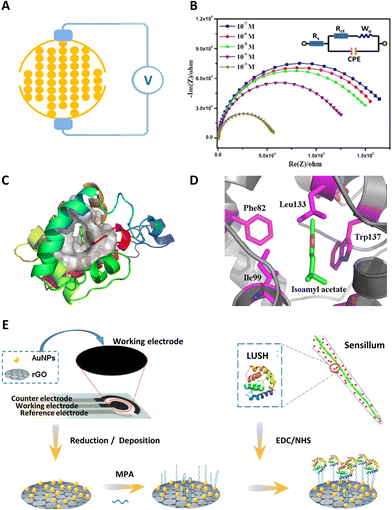 | ||
| Fig. 2 OBP-based electrochemical biosensors. (A) Schematic diagram of an interdigitated electrode device and impedance measurement. (B) Impedance spectra of isoamyl acetate at different concentrations. (C) Structure of OBPs of oriental fruit fly (Bactrocera dorsalis) and the binding mode of an OBP-isoamyl acetate complex. (D) Isoamyl acetate in the binding pocket of OBPs. Reprinted with permission from ref. 67; Copyright 2015, Elsevier. (E) Schematic diagram of an OBP-based biosensor with gold nanoparticles (AuNPs) and reduced graphene oxide (rGO) deposited on screen-printed working electrodes, 3-mercaptopropionic acid (MPA) was the linker, LUSH was the OBP. Reprinted with permission from ref. 69; Copyright 2020, American Chemical Society. | ||
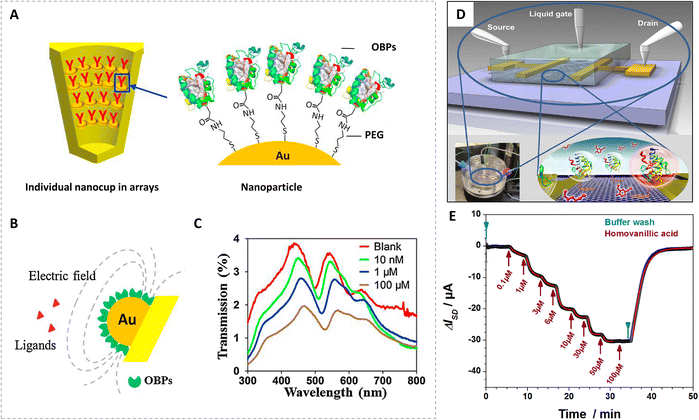 | ||
| Fig. 3 Odorant binding protein-based localized surface plasmon resonance biosensors and field-effect transistor biosensors. (A) Schematic diagram of nanocup arrays functionalized by self-assembled OBPs. (B) Biosensing mechanism of OBP-functionalized nanocup arrays. (C) Transmission spectrum of OBP-modified nanocup arrays for 2,4,6-trinitrotoluene (TNT) at different concentrations. Reprinted with permission from ref. 32; Copyright 2015, Elsevier. (D) Schematic illustration of a graphene field-effect transistor biosensor with a gold source and drain electrodes and an Ag/AgCl gate electrode. (E) Real-time biosensor measurement of the binding of methyl vanillate to OBP14 from the honey bee Apis mellifera. Reprinted with permission from ref. 72; Copyright 2016, the authors. | ||
With increasing concentration of isoamyl acetate, charge transfer resistance gradually decreased. This indicated a recovery in the efficiency of the mass transfer phenomenon related to the dielectric and conductive properties of the sensing membrane on the electrodes.67 Through analyzing the impedance changes, different odorants could be sensitively detected by the OBP-based biosensors. Additionally, the interactions between OBPs and different odorants were also investigated by molecular docking in these studies. As shown in Fig. 2C and D, isoamyl acetate lay exactly in the hydrophobic middle of several hydrophobic groups of OBPs. Amino acid residues of Phe82, Trp137, Ile99, and Leu133 might play important roles in the binding process. With the help of molecular docking, the impedance sensing results were validated. Moreover, the functional amino acid residues or peptides could be interpreted from the natural sequences of OBPs, which might advance the practical applications of OBP-based biosensors by designing highly specific recognition elements.
Moreover, studies showed that OBPs were also expressed in other tissues, such as gustatory sensilla.74,75 Thus, OBPs were also used to detect substances with a bitter taste.69 Through one-step reduction, gold nanoparticles (AuNPs) and reduced graphene oxide (rGO) could be deposited on the carbon working electrode (Fig. 2E). Through 3-mercaptopropionic acid (MPA), the sensing proteins could be immobilized on disposable screen-printed electrodes. Denatonium, quinine, and berberine were sensitively detected with this biosensor, which provided a simple approach for chemical molecular sensing at low concentrations. There were many other electrochemical biosensors that took OBPs of mammals, such as rat OBP-1F, porcine OBP (pOBP), and human OBPs, as sensing membranes. For instance, nanopores or screen-printed electrodes that functionalized with human OBPs have been used to detect aldehydes and fatty acids, which showed potential in discriminating fatty acids with different lengths of carbon chains.76,77 They exhibited good ability not only in odorant and biomolecule detection, but also in disease diagnosis. Inspired by these biosensors that used mammalian OBPs, we know that insect OBPs still have promising potential for exploration due to the high sensitivity of the insect olfactory system.
Although electrochemical impedance spectroscopy had advantages of good stability, precise quantification, and a low detection limit, its repeatability is poor and it is time-consuming, which hindered its practical applications. At present, there are only a few studies on using other electrochemical techniques, such as cyclic voltammetry, to develop insect olfactory biosensors.76 However, electrochemical techniques, such as cyclic voltammetry and square wave voltammetry, can be used in quantitative detection for important analytes with the strengths of high reliability and simplicity. Thus, much work remains to be done to develop these kinds of olfactory biosensors.
3.2 OBP-based biosensors with optical technology
As a powerful label-free method, surface plasmon resonance could be used to study molecular binding processes through measuring changes in refractive index. When visible light stimulates nanoscale structures, the effects of surface plasmon resonance turned into nonpropagating or propagating within the limited volume on the device surface, which is named localized surface plasmon resonance.78,79 Moreover, localized surface plasmon resonance detection allows multiple-component analysis in an array without labels on targets. It can provide an excellent platform to quantify small molecules which elicit only small changes in bio-interactions. During the last decade, nano-plasmonic sensors based on various nanostructures (e.g. nanoparticles, nanorods, and nanoholes) have attracted a lot of attention in the field of chemical and biological sensing.80–83 Thus, interactions between OBPs and the ligands were also studied with sensors.Fig. 3A shows a typical OBP-based localized surface plasmon resonance biosensor.32 Through Au–S semi-covalent linkage, a self-assembled film of HS-PEG-COOH was used to fix OBPs of honeybees on the surface of nanocup arrays (nanoCA). The binding of small molecule ligands to OBPs on nanocups might lead to changes in protein properties (e.g., conformation and electron distribution), which modulate localized surface plasmon resonance in the electrical field near the nanocup array surface (Fig. 3B). Apart from floral odorants, nitro-compounds such as 2,4,6-trinitrotoluene (TNT), 2,4-dinitrotoluene (DNT), and 3-mononitrotoluene (1NT) were also detected. Transmission spectra of the bio-nanoCA with OBPs in presence of TNT at different concentrations are shown in Fig. 3C. There were significant wavelength shifts at concentrations of 1 μM and 100 μM, which indicated that the biosensor had application prospects in explosive detection.
As a powerful label-free method to study molecular binding processes on a sensor surface, localized surface plasmon resonance is especially helpful for monitoring binding processes and binding affinities between OBPs and ligands in real time. Moreover, studies showed that localized surface plasmon resonance could sensitively detect slight changes in peptides caused by interactions with small target molecules.84 This provided a promising approach to quantify small molecule–protein interactions in developing other olfactory biosensors.
3.3 OBP-based biosensors with field-effect transistors
As previously mentioned, a field-effect transistor is one type of widely used sensor, which is compatible with building up testing systems with expanded circuits and electronic technologies. In addition to testing electrophysiological signals, field-effect transistors can also be used to detect slight changes on the surface of electrodes. Thus, the charges induced by interactions between olfactory proteins and target analytes can influence the grid voltage and subsequently cause changes in the source–drain current measured in the test. With their high sensitivity, field-effect transistors are one of the best candidates for developing OBP-based biosensors.A dual-gate sensing system with an ion-sensitive field-effect transistor (ISFET) with OBPs of Drosophila (LUSH) was designed for ethanol detection.73 In addition, a research group from Austria focused on the study of a reduced graphene oxide field-effect transistor (rGO-FET) in biochemical sensing.57,71,72 As shown in Fig. 3D, the rGO-FET was fabricated on a silicon wafer base covered with an oxide layer and reduced graphene oxide. Taking 1-pyrenebutanoic acid succinimidyl ester as the linker, the odorant binding protein (OBP14) from the honeybee, Apis mellifera, was modified on the reduced graphene oxide covered base substrate. Obvious variations in source–drain current response were observed when detecting a proved ligand, homovanillic acid, of different concentrations (Fig. 3E). Altogether the research group measured fourteen kinds of floral chemicals binding to OBP14, and sorted these targets into groups of compounds with similar structure. Among the 14 targets tested, the best ligands for wild-type OBP14 were eugenol, homovanillic acid, and similar compounds with a phenol-methoxy backbone.71 Besides wild-type OBPs, two different mutants of OBP14 were also applied for odorant detection.57 One with a His-Tag at N-terminus showed weaker binding capacity while the other with an additional disulfide bond could improve the sensor's sensitivity for specific analytes. This finding inspired further exploration into mutant design for desired specificity.
Combinations between the external environment and ORs or OBPs could be used as promising sensing materials for a new generation of olfactory sensors with their increasingly clear sensing properties. As mentioned previously, in addition to research into insects' OBPs, mammalian OBPs have also been extensively studied for developing olfactory biosensors.76,77,85 Porcine OBPs (pOBPs) with a water-gated organic field-effect transistor could sensitively detect S-(+)-carvone in the picomolar concentration range.33 Along with the different sensors mentioned above, OBPs could also be integrated with surface acoustic wave sensors for odorant detection.86,87 Each of the sensing techniques mentioned above could be an ideal tool for odorant detection. However, the specificity of OBP-based biosensors still needs to improve due to the broad binding spectra of OBPs.
4. Olfactory receptor-based biosensors
Over the past decade, the molecular basis of odorant reception in insects has been widely investigated.34,88–90 In general, olfactory receptor proteins (ORs) together with OR co-receptor (Orco) ion channels play essential roles both in detecting a wide variety of chemical signals amid a cacophony of chemical noise, and in converting the recognition of an odorant molecule into electrical signals in peripheral olfactory neurons. As important molecular interfaces between the chemical world and the brain, ORs and Orco have attracted the attention of many experts and scholars to explore and use specific physiological functions and binding capabilities for odorant detection.91–93 With advances in biosynthesis and separation techniques, receptors and ion channels were expressed on homologous cells, heterologous cells, and even nanometer vesicles or nanodiscs constructed by the phospholipid bilayer.94–97 These cells with specific receptor proteins are potential biomaterial candidates for sensing. Nowadays, the expression and purification of olfactory receptors in vitro have been made possible by the establishment of a cell-free expression system.62,98 This provided broad prospects for developing olfactory receptor-based biosensors with high sensitivity and high specificity.Over the past two decades, different insect olfactory receptors, such as GPROR2 and OR56a, have been successfully expressed on heterologous cells to maintain their biological activity. Based on the remarkable binding function of olfactory receptors to different odorants, researchers have developed various technologies for transducing OR–ligand interactions into readable formats, and thereby produced insect olfactory receptor protein-based biosensors (Table 3).
| Olfactory receptors | Species | Expression system | Measurement techniques | Target substances | Detection range | Ref. |
|---|---|---|---|---|---|---|
| BmOR1 | Bombyx mori (silkmoth) | Insect Sf 21 (Spodoptera frugiperda) | Fluorescence | Bombykol | 30 nM–30 μM | 100 |
| BmOR3 | Bombykal | |||||
| OR56a | Drosophila melanogaster | Insect Sf 21 (Spodoptera frugiperda) | Fluorescence | Geosmin | 0.2–20 mM | 101 |
| GPROR2 | Anopheles gambiae (malaria vector mosquito) | Human embryonic kidney cells (HEK293T) | Electrophysiology | Benzaldehyde | 0.0001–1 mM (solution) | 99 |
| 2-Methylphenol | 0.01–100 mM (vapor) | |||||
| BmOR1 | Bombyx mori (silkmoth) | Xenopus leaevis oocytes | Two-electrode voltage clamping method | Bombykol | 10 nM–10 μM | 105 |
| BmOR3 | Plutella xylostella (diamondback moth) | Bombykal | ||||
| PxOR1; | Drosophila melanogaster (fruit fly) | (Z)-11-Hexadecenal | ||||
| DOr85b | 2-Heptanone | |||||
| ORs (OR8) and OR co-receptor (OR7) proteins | Aedes aegypti (yellow fever mosquito) | Bilayer lipid membranes | Amplifier | Octenol | 0.01–0.2 ppm | 106 |
| ORs (OR8) and OR co-receptor (OR7) proteins | Aedes aegypti (yellow fever mosquito) | Liposomes | Patch clamp amplifier | Octenol | 0.5–5000 ppb | 102 |
| OR10a | Drosophila melanogaster | Lipid nanodiscs | Field-effect transistor (FET) | Methyl salicylate | 1 fM–100 pM | 97 |
| OR22a | Methyl hexanoate | |||||
| OR35a | Trans-2-hexen-1-al | |||||
| OR71a | 4-Ethylguaiacol | |||||
| OR10a | Drosophila melanogaster | Lipid nanodiscs or liposomes | FET | Methyl salicylate | 1 fM–100 pM | 96 |
| OR22a | Methyl hexanoate | |||||
| OR35a | Drosophila melanogaster | Liposome | Electrochemical impedance spectroscopy (EIS) | E2-Hexenal | 1 fM–1 μM | 103 |
| OR10a | Drosophila melanogaster | Liposome | EIS | Methyl salicylate | 100 fM–100 nM | 107 |
| OR22a | Methyl hexanoate | 1 fM–100 nM | ||||
| OR71a | 4-Ethylguaiacol | 0.01 fM–1 nM | ||||
| OR22a | Drosophila melanogaster | Lipid nanodiscs | EIS | Ethyl hexanoate | 1 fM–100 pM | 104 |
To detect chemical vapors, researchers proposed a novel electrophysiology technique with a reconstituted insect olfactory receptor complex.99 As shown in Fig. 4A, both the malaria vector mosquito (Anopheles gambiae) protein GPROR2, a receptor for 2-methylphenol (2-MP), and the fruit fly (Drosophila melanogaster) protein Or47a, a receptor for pentyl acetate, were expressed in human embryonic kidney cells (HEK293T). The insect OR-expressing HEK293T spheroids were produced through culturing these transfected cells in a hydrogel microchamber array. After verifying the olfactory responses of the receptors, the extracellular field potential of spheroids upon olfactory stimulation was recorded. Through integrating cell assembly and heterologous gene expression techniques, this OR-based olfactory biosensor was applicable to functional analysis and odorant detection.
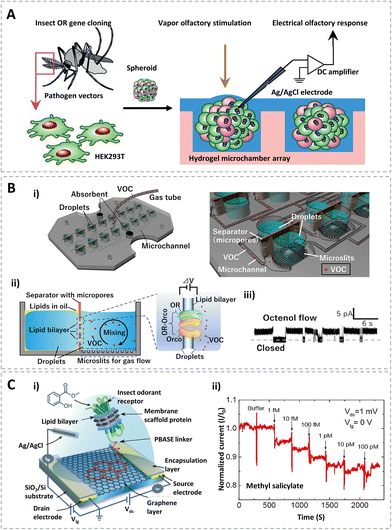 | ||
| Fig. 4 Olfactory receptor-based biosensors. (A) HEK293T cells that were expressed in insect odor-gated ion channels dispersed to form OR expressing spheroids, and the extracellular field potentials which were recorded with odor stimulation. Reprinted with permission from ref. 99; Copyright 2014, Wiley. (B) Schematic diagram of the biosensor with microchannels for volatile organic compound (VOC) detection (i), structure and sensing mechanism of the lipid bilayer array with OR-Orco (ii), and the representative current traces of the OR-Orco in response to octenol (iii). Reprinted with permission from ref. 102; Copyright 2021, the authors. (C) Device schematic and circuit connections of an OR nanodisc-immobilized graphene field-effect transistor (i), normalised real-time sensing response of an OR10a nanodisc field-effect transistor with the addition of positive ligands (methyl salicylate) with increasing concentrations (ii). Reprinted with permission from ref. 96; Copyright 2020, American Chemical Society. | ||
Based on biological living cells expressing insect olfactory receptors, biosensors based on fluorescence were also studied. The insect odorant receptor and olfactory receptor co-receptor of Drosophila melanogaster were expressed in Spodoptera frugiperda (sf) 21 cells.100,101 They worked as ligand-gated ion channels of sf21 cells. When odors were bound to the ligand-gated ion channels of sf21 cells, calcium ions including the fluorescent calcium ion indicator protein (GCaMP3) would flow into the cells. Through fluorescent measurement, the intensity changes of the fluorescent light could be used as biosensor responses.
In addition, to reducing unnecessary biochemical reactions of OR-expressing cells and maintaining the structure of seven transmembrane helices, insect ORs and Orco have been reconstituted into different types of phospholipid bilayer, such as liposomes and lipid nanodiscs to develop cell-free biosensors.93,102 As shown in Fig. 4B, an olfactory biosensor array was constructed with a specifically designed gas flow system including microchannels, hydrophobic microslits, and ORs with OR co-receptors (OR-Orco) in a lipid bilayer. The microchannels and microslits enable gas to be quickly introduced into the droplet and interact with receptors. Besides, ORs (OR8) and OR co-receptor (OR7) proteins of the yellow fever mosquito (Aedes aegypti) in the lipid bilayer can form a ligand-gated ion channel. When the target gas interacts with the receptors, open–closed current signals are generated. Octenol at the parts per billion (ppb) level, which is a biomarker in human breath, was sensitively detected with this system.
Recent studies showed that insect olfactory receptors without co-receptors can also be used for odorant detection.96,103,104 A typical biosensor is shown in Fig. 4C. Self-assembled nanodiscs composed of an insect olfactory receptor, a phospholipid bilayer and a membrane scaffold protein was immobilized on a graphene field effect transistor with the help of a 1-pyrenebutyric acid N-hydroxysuccinimide ester (PBASE) linker (Fig. 4C(i)). The transistor was fabricated on a 300 nm SiO2/Si substrate with an encapsulated source and drain electrodes for sensing measurements.
During the test, an Ag/AgCl standard electrode was used as the gate electrode for liquid-gate measurements. OR10a from the fruit fly Drosophila melanogaster in nanodiscs showed a concentration-dependent response to its respective positive ligand, methyl salicylate (Fig. 4C(ii)). The limit of detection was as low as 1 fM. Meanwhile, OR22a in nanodiscs was also investigated with this transistor, which also showed a selective electrical response to methyl hexanoate. Additionally, this study also indicated that the sensitivity can be enhanced by the introduction of an OR co-receptor. With the high specificity and sensitivity of olfactory receptors, receptor-based biosensors could detect their respective odorants down to the femtomolar level, which makes it possible for use in environmental monitoring, food quality analysis, and medical diagnostics.
Besides the olfactory receptor-based biosensors mentioned above, biosensors with ORs from vertebrates have also attracted the attention of researchers. Through engineering a yeast strain with rat olfactory receptors and green fluorescent proteins, fluorescent biosensors were studied.108 When olfactory receptors “smell” an explosive residue mimic, DNT, the biosensor turned fluorescent green. Meanwhile, using carbon nanotube transistors, nanovesicles with cells expressing canine ORs, were used to develop olfactory biosensors for the detection of volatile odorants related to the freshness of foods, such as hexanal.109 When hexanal was bound to the vesicles, an influx of Ca2+ flowed into the vesicles and increased the potential of nanovesicles in the vicinity of the transistors. Through mimicking the way that receptors bind to odorants in the canine nose, the sensor could detect hexanal down to 1 fM, even when it was mixed with its analogs of pentanal, heptanal, and octanal. This presented a promising prospect in assessing food quality. All of these biosensors could be references for developing future insect olfactory receptor protein-based biosensors. More importantly, insect receptors might be more robust and could form ion channels themselves, which means they need fewer biological components to function.110 Thus, the sensitivities of insect olfactory receptors make them prime candidates for biological detectors for volatile odorants in bio-sensing.
5. Peptide-based biosensors for odorant detection
Based on standard synthetic protocols, peptides are much easier to prepare than proteins. As specific amino acid sequences extracted from proteins, peptides can retain certain functions of the proteins. Meanwhile, the great chemical and conformational stability of peptides mean they can be preserved for a long time and applied in harsh conditions.111 Therefore, the application of a synthetic peptide sequence as a sensing material could be an advanced technique for developing a highly sensitive and selective biosensor to detect low concentrations of target molecules.112–114 In recent years, some specific peptides derived from insect olfactory proteins have been explored for incorporation with various sensors, such as electrochemical sensors, and optical sensors. Table 4 summarizes some typical peptide-based biosensors for the detection of different odorants.| Peptide sequence | Source | Measurement techniques | Target substances | Detection range | Detection limit | Ref. |
|---|---|---|---|---|---|---|
| SLMAGTVNKKGEFC | An OBP from Drosophila, LUSH | Quartz crystal microbalance (QCM) | 3-Methyl-1-butanol, 1-hexanol | 0–100 ppm | 1–3 ppm | 113 |
| TKCVSLMAGTVNKKGEFFFF | An OBP from Drosophila | Field-effect transistor (FET) | 3-Methyl-1-butanol | 10 nM–1 fM | 1 fM | 21 |
| WFVI | The antennal-specific protein-1 (ASP1), an OBP from honeybee, Apis mellifera | FET | 2,4,6-Trinitrotoluene (TNT) | — | 12 ppb | 29, 115 |
| NQLSNLSFSDLCFFF | — | FET | Trimethylamine | 10 fM–1 μM | 10 fM | 120 |
| WHWQRPLMPVS | — | Impedance | TNT | 1 μM–1 mM | 7 × 10−7 M | 121 |
| NQLSNLSFSDLC | — | Resistance | Trimethylamine | 0.01 ppt–10 ppb | 0.01 ppt | 122 |
| WHWQRPLMPVSI | — | Optical spectroscopy | TNT | 10−9–10−4 mg ml−1 | 4.4 × 10−12 mM | 118 |
| WHWQRPLMPVSIC | — | Optical spectroscopy | TNT | 2 × 10−7–10−4 M | — | 119 |
Utilizing the peptide sequence (SLMAGTVNKKGEFC) extracted from Drosophila odorant binding protein (LUSH), a piezoelectric sensor was proposed for detecting volatile organic compounds indicative of Salmonella contamination in packaged beef.113 The sensor was sensitive to alcohols (3-methyl-1-butanol and 1-hexanol) with estimated lower detection limits of 5 ppm. After that, another bioelectronic nose, which can be used for detecting Salmonella contamination in ham, using Drosophila odorant binding protein-derived peptide (TKCVSLMAGTVNKKGEFFFF) and a carbon nanotube field-effect transistor (CNT-FET), was developed (Fig. 5A).21 Three additional phenylalanine residues at the C-terminus of the peptide were synthesized for directly immobilizing the peptides onto the sensors via π–π interactions. The peptide-based biosensor could not only sensitively detect 3-methyl-1-butanol at a concentration of 1 fM, but also selectively distinguished the target odor molecules from other compounds with similar structures (Fig. 5B). All of these studies indicated that olfactory protein-derived peptides which specifically bind with different food contaminants could be applied for assessing various foods.
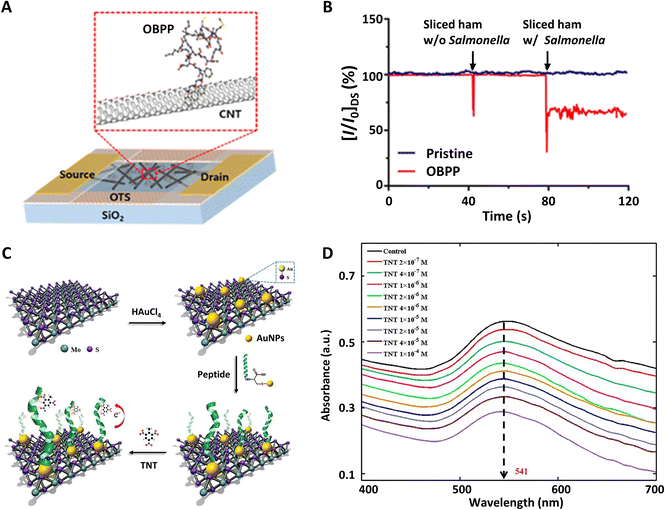 | ||
| Fig. 5 Peptide-based biosensors. (A) Schematic diagram of olfactory biosensors using a carbon nanotube field-effect transistor functionalized with a Drosophila odorant binding protein-derived peptide. (B) Real-time detection signals of Salmonella contamination in sliced ham. Reprinted with permission from ref. 21; Copyright 2016, American Chemical Society. (C) The construction process of a biosensor based on nanocomposites of molybdenum disulfide (MoS2) and gold nanoparticles (AuNPs) for 2,4,6-trinitrotoluene (TNT) detection. (D) Absorption spectra of the biosensor with TNT at different concentrations. Reprinted with permission from ref. 119; Copyright 2018, Elsevier. | ||
As a widely used insect research model, trained honeybees were found to have abilities to detect trace amounts of explosives. Inspired by their super sensitive sense of smell, antennal-specific protein-1 (ASP1C), an OBP from Apis mellifera, was investigated to detect TNT. This showed that four amino acid residues (WFVI) at the C-terminus of ASP1C played crucial roles in binding to TNT.29,115 Combined with the single-wall carbon nanotube binding peptide P1 (HSSYWYAFNNKT), the ASP1C-derived peptide could be modified on a single-wall carbon nanotube field-effect transistor and exhibited selective responses to 12 ppb TNT.
Compared with other biological components, such as cells and proteins, peptides could be chemically engineered to bind specific targets with long-term stability and sensitivity.116 Meanwhile, the peptide sequences could be specially designed to bond with different nanomaterials, which allowed them to achieve growing interest as bio-sensitive elements.117 For instance, combined with gold nanoparticles (AuNPs) and two-dimensional nano-materials such as graphene oxide (GO) and molybdenum disulfide (MoS2), peptide chains (WHWQRPLMPVS) were successfully used to detect explosives by optical spectroscopy.118,119
As shown in Fig. 5C, AuNPs@MoS2 nanocomposites could be synthesized by one-step conjugation.119 AuNPs could be used to immobilize polypeptide (WHWQRPLMPVSIC) through gold–sulfur (Au–S) bonds and generate detectable spectral signals. MoS2 could enhance the plasmonic resonance of the nanocomposites. When different concentrations of 2,4,6-trinitrotoluene (TNT) were detected, the absorption peak value at the wavelength of 541 nm changed significantly (Fig. 5D). This indicated that there is specific binding between peptides and TNT molecules. It provides a fast responsive and label-free assay for explosive detection.
Taken together, with the help of molecular dynamics simulations and phage display, more and more specific and sensitive amino acid residues could be determined from olfactory proteins, which are ideal sensitive elements for developing biosensors.123 With tunable physical and chemical properties, polypeptides have been playing an increasingly important role in the field of biosensors. Peptide-based biosensors could provide a model platform for applications ranging from noninvasive breath monitoring to food spoilage or biological and chemical threat detectors.
6. Conclusions and future perspectives
Integrated with different detection techniques and transducers, various olfactory biosensors that utilized the ultrasensitive abilities of olfactory systems were developed. This review has presented a brief summary of how these insect olfactory systems inspired smell biosensors. Until now, based on the sensing materials, they can be roughly classified into: (i) olfactory biosensors based on insect antennae; (ii) olfactory biosensors based on OBPs; (iii) olfactory biosensors based on olfactory receptors; (iv) olfactory biosensors based on peptides. With the further exploration of insect olfactory sensing mechanisms and the development of physical transducers, the inspired biosensors can achieve more bio-sensitive materials to improve their sensitivities and specificities. They can provide excellent platforms for odorant detection in various areas, such as environmental monitoring, food inspection and evaluation, and disease screening and detection.With their extremely sensitive systems of olfaction, insects can sensitively and selectively detect thousands of odorants at very low concentrations, even in a very complicated chemical environment. Emerging information about insect olfactory detection abilities and learning abilities revealed that insect olfactory systems are potentially useful tools for the quantitative and qualitative analysis of odorants.16,124,125 Thus, some insect-sniffers were trained to detect chemicals associated with medicine, security, forensics, agriculture, and food safety applications. Compared to sniffer dogs, an insect sniffer system might be low-cost and could be conditioned with impressive speed for specific chemical detection tasks.16,126 Moreover, with the functionalities of various components in insect olfaction systems discovered through studies in anatomy, neurophysiology, and molecular biology, different olfaction-related biomaterials, including ORNs, ORs, OBPs and peptides, have become popular in developing olfactory biosensors.2,17,58 Although biosensors based on the olfactory elements of mammals or vertebrates are not described in detail in this review, research into this kind of biosensor has also received extensive attention. Mammalian olfactory epithelia, olfactory cells, olfactory receptors, OBPs and other sensitive elements have been successfully combined with different transducers for odorant detection. Although great progress has been made, almost all of the olfactory biosensors are still at the stage of lab research because the specificity, sensitivity, and service life of these biosensors still cannot meet the demands of practical applications. There are sustainable challenges in exploring the ability of insect olfaction to design ultrasensitive biosensors.
6.1 Specificity of olfactory biosensors
For olfactory biomaterials of insects, much further effort should be made to find novel sensing proteins with high specificity. On the one hand, more efficient methods for insect OR expression, stabilization, and purification should be investigated to obtain specific receptors in large quantities: for instance, finding an optimal class of detergent in membrane protein studies that could solubilize proteins and maintain their stability and function.127,128 It had been demonstrated that peptides could form various nanostructures, such as nanovesicles, nanotubes, and nanodiscs, to solubilize and stabilize diverse multi-transmembrane proteins.129,130 Thus, with self-assembled peptide surfactants coupled with cell-free technology, stabilized insect ORs might be available,127 which is essential for developing olfactory receptor protein-based biosensors with high selectivity. On the other hand, mutant proteins with novel functions should be explored by computational methods and experiments. Studies showed that binding specificities of proteins could be changed drastically by a computational design algorithm.131 Moreover, different mutants of OBPs had already been applied to improve a sensor's sensitivity for specific analytes.57,132 With more functional amino acid residues or peptides interpreted from the natural sequences of olfactory proteins, we believe that mutant proteins or peptide chains can be synthesized for use as promising recognition elements in future biosensors for practical applications.6.2 Sensitivity of olfactory biosensors
Similarly, in order to improve the sensitivity of olfactory biosensors, new solutions for detecting techniques should be studied. One of the most widely used solutions is using different nanomaterials, such as nanopores, two-dimensional materials, and even hybrid nanomaterials.133–135 For example, functionalized with embedded biosensing elements, nanopores with a field-effect transistor could even realize single-molecule detection.136 Additionally, in recent years, two-dimensional transition metal dichalcogenide (TMD) nanosheets and their composites have aroused great research interest in using them to construct sensing devices, such as electrochemical and optical devices. Owing to the intriguing physical, chemical, electronic, and optical properties of TMD nanomaterials, the performance of biosensors functionalized with these kinds of materials could be greatly enhanced.137–139 Therefore, apart from exploring novel olfactory sensing elements, different nanomaterials are also worth studying to improve the recognition and transduction processes of olfactory biosensors.6.3 Practical applications of olfactory biosensors
Imitating the biological olfactory system, both electronic noses and olfactory biosensors can be used to detect odorants. Depending on complicated data processing and classification algorithms, electronic gas sensors usually with metal oxide can discriminate different volatile analytes. Meanwhile, as physical and chemical sensors, they have a long service life and good stability. Thus, electronic gas sensors show promising potential in practical application. However, intrinsic shortcomings, such as low sensitivity, high power consumption, susceptibility to poisoning, and complex signal processing, still hinder their rapid development. The olfactory biosensor in use today replaces neither complex analytical equipment nor electronic gas sensors, but supplements both.140,141 Although it has several limitations, advantages regarding fast detection speed, high sensitivity and selectivity make its entry into our daily life for practical application promising. One of the most difficult challenges for olfactory biosensors based on bio-elements, whether extracted from mammals or insects, is achieving their practical application. Now, numerous olfactory biosensors have been investigated for detection of different odorants. However, the concept of olfactory biosensors is still demonstrated with standard tests under optimal laboratory conditions. In order to further promote the practical process, three issues should be considered. Firstly, the stability and lifetime of biological materials incorporated into electronic devices need to be improved. Cells, olfactory receptors, odorant-binding proteins, and peptides can undergo long-term storage at low temperature (−80 °C).142 However, the lifetime is uncertain after these biomaterials are immobilized on a sensor. The sensing properties of biomaterials may lose functional activity due to degradation, which also greatly reduces the lifetime of the developed biosensors.38,49 Recent advances showed that biomaterials stored in a sealed vessel could help to extend their service life to several weeks with the help of nanoscale structures.31,143 Secondly, establishing multichannel or multiplexed biosensors is a good alternative. Through comprehensive analysis of multiple data similar to an olfactory perception process, it could discriminate targeted odors in a complex mixture with a reduced detection error rate.31 Therefore, further research should focus on the exploration of sensor arrays with high-throughput detection and analysis techniques. Thirdly, portable equipment for detection and analysis should be developed to meet the demands of on-site testing. Among the different alternatives, smart mobile terminals such as tablets and smartphones are potential solutions due to their powerful computing capability, advanced hardware system, and open-source operating system. Nowadays, the portability and ubiquitous availability of smartphones make them widely integrated with sensors for biochemical detection.144,145 Meanwhile, they can control, perform, analyze, and display the whole sensing process. Therefore, the combination of mobile terminals and olfactory biosensors can provide a technical means for the real-time and portable monitoring of human exhaled air and other gas molecules. It can provide a rapid tool for human health monitoring and disease diagnosis. Although there are still many difficulties to overcome, such as the relationship between disease and marker molecules not being established, an olfactory biosensor is a promising and powerful technical means to solve this problem.Research into biosensors based on biological components is a feasible technology to build a bionic olfactory sensing platform. With the continuous development of biosynthetic technology, detection technology, data analysis and artificial intelligence, olfactory biosensors may show potential to detect odor molecules beyond the ability of natural perception. It is convincing that olfactory biosensors based on insect or mammalian olfactory sensing systems will eventually become effective detection and analytical tools for disease diagnosis.
Author contributions
Y. L. wrote the draft of the manuscript. Q. L. critically reviewed the manuscript and gave his inputs to improve the manuscript draft. All the authors thoroughly read the final manuscript draft and gave permission for its submission.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the National Natural Science Foundation of China (Grant No. 81971703, 81801793).References
- C.-Y. Su, K. Menuz and J. R. Carlson, Cell, 2009, 139, 45–59 CrossRef CAS PubMed.
- E. Suh, J. D. Bohbot and L. J. Zwiebel, Curr. Opin. Insect Sci., 2014, 6, 86–92 CrossRef PubMed.
- M. Renou and S. Anton, Curr. Opin. Insect Sci., 2020, 42, 1–7 CrossRef.
- A. Dahanukar, E. A. Hallem and J. R. Carlson, Curr. Opin. Neurobiol., 2005, 15, 423–430 CrossRef CAS PubMed.
- U. B. Kaupp, Nat. Rev. Neurosci., 2010, 11, 188–200 CrossRef CAS PubMed.
- J. P. Martin, A. Beyerlein, A. M. Dacks, C. E. Reisenman, J. A. Riffell, H. Lei and J. G. Hildebrand, Prog. Neurobiol., 2011, 95, 427–447 CrossRef PubMed.
- R. Benton, Trends Neurosci., 2007, 30, 512–519 CrossRef CAS.
- B. Antony, J. Johny and S. A. Aldosari, Front. Physiol., 2018, 9, 252 CrossRef PubMed.
- P. Pelosi, I. Iovinella, A. Felicioli and F. R. Dani, Front. Physiol., 2014, 5, 320 Search PubMed.
- P. Pelosi, I. Iovinella, J. Zhu, G. Wang and F. R. Dani, Biol. Rev., 2018, 93, 184–200 CrossRef PubMed.
- M. Laska, C. G. Galizia, M. Giurfa and R. Menzel, Chem. Senses, 1999, 24, 429–438 CrossRef CAS PubMed.
- A. F. Carey and J. R. Carlson, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 12987–12995 CrossRef CAS PubMed.
- M. Giurfa, S. Zhang, A. Jenett, R. Menzel and M. V. Srinivasan, Nature, 2001, 410, 930–933 CrossRef CAS PubMed.
- P. J. Rodacy, S. Bender, J. Bromenshenk, C. Henderson and G. Bender, in Detection and Remediation Technologies for Mines and Minelike Targets VII, SPIE, 2002, vol. 4742, pp. 474–481 Search PubMed.
- M. Schott, B. Klein and A. Vilcinskas, PLoS One, 2015, 10, e0128528 CrossRef PubMed.
- G. C. Rains, J. K. Tomberlin and D. Kulasiri, Trends Biotechnol., 2008, 26, 288–294 CrossRef CAS PubMed.
- T. Wasilewski, J. Gębicki and W. Kamysz, Sens. Actuators, B, 2018, 257, 511–537 CrossRef CAS.
- T. Wasilewski, J. Gębicki and W. Kamysz, Biosens. Bioelectron., 2017, 87, 480–494 CrossRef CAS PubMed.
- B. A. Cornell, V. Braach-Maksvytis, L. King, P. Osman, B. Raguse, L. Wieczorek and R. Pace, Nature, 1997, 387, 580–583 CrossRef CAS PubMed.
- R. Edmondson, J. J. Broglie, A. F. Adcock and L. Yang, Assay Drug Dev. Technol., 2014, 12, 207–218 CrossRef CAS PubMed.
- M. Son, D. Kim, J. Kang, J. H. Lim, S. H. Lee, H. J. Ko, S. Hong and T. H. Park, Anal. Chem., 2016, 88, 11283–11287 CrossRef CAS PubMed.
- Q. Liu, W. Ye, N. Hu, H. Cai, H. Yu and P. Wang, Biosens. Bioelectron., 2010, 26, 1672–1678 CrossRef CAS PubMed.
- Q. Liu, W. Ye, L. Xiao, L. Du, N. Hu and P. Wang, Biosens. Bioelectron., 2010, 25, 2212–2217 CrossRef CAS PubMed.
- J. H. Ahn, J. H. Lim, J. Park, E. H. Oh, M. Son, S. Hong and T. H. Park, Sens. Actuators, B, 2015, 210, 9–16 CrossRef CAS.
- A. M. Fraser, W. L. Mechaber and J. G. Hildebrand, J. Chem. Ecol., 2003, 29, 1813–1833 CrossRef CAS PubMed.
- M. Strauch, A. Lüdke, D. Münch, T. Laudes, C. G. Galizia, E. Martinelli, L. Lavra, R. Paolesse, A. Ulivieri and A. Catini, Sci. Rep., 2014, 4, 1–9 Search PubMed.
- L. Du, L. Zou, Q. Wang, L. Zhao, L. Huang, P. Wang and C. Wu, Sens. Actuators, B, 2015, 217, 186–192 CrossRef CAS.
- J. Y. Lee, H. J. Ko, S. H. Lee and T. H. Park, Enzyme Microb. Technol., 2006, 39, 375–380 CrossRef CAS.
- Z. Kuang, S. N. Kim, W. J. Crookes-Goodson, B. L. Farmer and R. R. Naik, ACS Nano, 2010, 4, 452–458 CrossRef CAS PubMed.
- Q. Liu, H. Wang, H. Li, J. Zhang, S. Zhuang, F. Zhang, K. J. Hsia and P. Wang, Biosens. Bioelectron., 2013, 40, 174–179 CrossRef CAS PubMed.
- O. S. Kwon, H. S. Song, S. J. Park, S. H. Lee, J. H. An, J. W. Park, H. Yang, H. Yoon, J. Bae and T. H. Park, Nano Lett., 2015, 15, 6559–6567 CrossRef CAS PubMed.
- D. Zhang, Y. Lu, Q. Zhang, Y. Yao, S. Li, H. Li, S. Zhuang, J. Jiang, G. L. Liu and Q. Liu, Sens. Actuators, B, 2015, 221, 341–349 CrossRef CAS.
- M. Y. Mulla, E. Tuccori, M. Magliulo, G. Lattanzi, G. Palazzo, K. Persaud and L. Torsi, Nat. Commun., 2015, 6, 1–9 Search PubMed.
- M. Pellegrino and T. Nakagawa, J. Exp. Biol., 2009, 212, 1973–1979 CrossRef CAS PubMed.
- M. Huotari and M. Mela, Sens. Actuators, B, 1996, 34, 240–244 CrossRef CAS.
- B. Weißbecker, S. Schütz, A. Klein and H. E. Hummel, Talanta, 1997, 44, 2217–2224 CrossRef.
- M. J. Huotari, Sens. Actuators, B, 2000, 71, 212–222 CrossRef CAS.
- K. C. Park, S. A. Ochieng, J. Zhu and T. C. Baker, Chem. Senses, 2002, 27, 343–352 CrossRef PubMed.
- K. C. Park and T. C. Baker, J. Insect Physiol., 2002, 48, 1139–1145 CrossRef CAS PubMed.
- A. Myrick, K. Park, J. Hetling and T. Baker, Bioinspiration Biomimetics, 2008, 3, 046006 CrossRef CAS PubMed.
- D. Terutsuki, T. Uchida, C. Fukui, Y. Sukekawa, Y. Okamoto and R. Kanzaki, Sens. Actuators, B, 2021, 339, 129770 CrossRef CAS.
- P. Schroth, M. J. Schöning, S. Schütz, Ü. Malkoc, A. Steffen, M. Marso, H. Hummel, P. Kordos and H. Lüth, Electrochim. Acta, 1999, 44, 3821–3826 CrossRef CAS.
- M. J. Schöning, P. Schroth and S. Schütz, Electroanalysis, 2000, 12, 645–652 CrossRef.
- P. Schroth, M. J. Schöning, H. Lüth, B. Weißbecker, H. Hummel and S. Schütz, Sens. Actuators, B, 2001, 78, 1–5 CrossRef CAS.
- D. Schneider, Experientia, 1957, 13, 89–91 CrossRef.
- D. Schneider, J. Insect Physiol., 1962, 8, 15–30 CrossRef.
- T. Morawo, M. Burrows and H. Fadamiro, F1000Research, 2016, 5, 2725 Search PubMed.
- D. Lu, F. Lu, L. Geng and G. Pang, Sens. Mater., 2018, 30, 67–87 CrossRef CAS PubMed.
- J. Visser and P. Piron, Entomol. Exp. Appl., 1995, 77, 37–46 CrossRef CAS.
- K. Park and J. Hardie, J. Insect Physiol., 1998, 44, 919–928 CrossRef CAS.
- K. Park, D. Elias, B. Donato and J. Hardie, J. Insect Physiol., 2000, 46, 597–604 CrossRef CAS.
- J.-J. Xu, X.-L. Luo and H.-Y. Chen, Front. Biosci., 2005, 10, 420–430 CrossRef CAS PubMed.
- M. J. Schöning, S. Schütz, P. Schroth, B. Weißbecker, A. Steffen, P. Kordoš, H. Hummel and H. Lüth, Sens. Actuators, B, 1998, 47, 235–238 CrossRef.
- P. Schroth, H. Lüth, H. Hummel, S. Schütz and M. J. Schöning, Electrochim. Acta, 2001, 47, 293–297 CrossRef CAS.
- P. Ravichandiran, S. A. Subramaniyan, A. P. Bella, P. M. Johnson, A. R. Kim, K. S. Shim and D. J. Yoo, Anal. Chem., 2019, 91, 10095–10101 CrossRef CAS PubMed.
- J. Jana, H. J. Lee, J. S. Chung, M. H. Kim and S. H. Hur, Anal. Chim. Acta, 2019, 1079, 212–219 CrossRef CAS PubMed.
- C. Kotlowski, M. Larisika, P. M. Guerin, C. Kleber, T. Kröber, R. Mastrogiacomo, C. Nowak, P. Pelosi, S. Schütz and A. Schwaighofer, Sens. Actuators, B, 2018, 256, 564–572 CrossRef CAS.
- R. Glatz and K. Bailey-Hill, Prog. Neurobiol., 2011, 93, 270–296 CrossRef PubMed.
- J. Fan, F. Francis, Y. Liu, J. Chen and D. Cheng, GMR, Genet. Mol. Res., 2011, 10, 3056–3069 CrossRef CAS PubMed.
- E. Lescop, L. Briand, J.-C. Pernollet and E. Guittet, Biochemistry, 2009, 48, 2431–2441 CrossRef CAS PubMed.
- S. W. Kruse, R. Zhao, D. P. Smith and D. N. Jones, Nat. Struct. Mol. Biol., 2003, 10, 694–700 CrossRef CAS PubMed.
- L. Kaiser, J. Graveland-Bikker, D. Steuerwald, M. Vanberghem, K. Herlihy and S. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2008, 105, 15726–15731 CrossRef CAS PubMed.
- P. Pelosi, R. Mastrogiacomo, I. Iovinella, E. Tuccori and K. C. Persaud, Appl. Microbiol. Biotechnol., 2014, 98, 61–70 CrossRef CAS PubMed.
- Y. Lu, H. Li, S. Zhuang, D. Zhang, Q. Zhang, J. Zhou, S. Dong, Q. Liu and P. Wang, Sens. Actuators, B, 2014, 193, 420–427 CrossRef CAS.
- E. Katz and I. Willner, Electroanalysis, 2003, 15, 913–947 CrossRef CAS.
- Y. Yao, Y. Lu, Q. Zhang, D. Zhang, S. Zhuang, H. Li, J. Shan and Q. Liu, Int. J. Electrochem. Sci., 2015, 10, 5548–5560 CAS.
- Y. Lu, Y. Yao, Q. Zhang, D. Zhang, S. Zhuang, H. Li and Q. Liu, Biosens. Bioelectron., 2015, 67, 662–669 CrossRef CAS PubMed.
- Y. Lu, Y. Yao, S. Li, Q. Zhang and Q. Liu, Sens. Rev., 2017, 37(4), 396–403 CrossRef.
- Z. Chen, Q. Zhang, J. Shan, Y. Lu and Q. Liu, ACS Omega, 2020, 5, 27536–27545 CrossRef CAS PubMed.
- J.-F. Zhang, F. Wu, M.-Z. Tang, D.-X. Chen, J. Tan, Q.-J. Liu and H.-L. Li, IEEE Sens. J., 2021, 21, 8855–8860 CAS.
- M. Larisika, C. Kotlowski, C. Steininger, R. Mastrogiacomo, P. Pelosi, S. Schütz, S. F. Peteu, C. Kleber, C. Reiner-Rozman and C. Nowak, Angew. Chem., Int. Ed., 2015, 54, 13245–13248 CrossRef CAS PubMed.
- C. Reiner-Rozman, C. Kotlowski and W. Knoll, Biosensors, 2016, 6, 17 CrossRef PubMed.
- C.-M. Lim, J. Y. Kwon and W.-J. Cho, ACS Appl. Mater. Interfaces, 2017, 9, 14051–14057 CrossRef CAS PubMed.
- E. R. Liman, Y. V. Zhang and C. Montell, Neuron, 2014, 81, 984–1000 CrossRef CAS PubMed.
- Y. T. Jeong, J. Shim, S. R. Oh, H. I. Yoon, C. H. Kim, S. J. Moon and C. Montell, Neuron, 2013, 79, 725–737 CrossRef CAS PubMed.
- Y. Lu, Y. Huang, S. Li, Q. Zhang, J. Wu, Z. Xiong, L. Xiong, Q. Wan and Q. Liu, Sens. Actuators, B, 2017, 252, 973–982 CrossRef CAS.
- Y. Lu, D. Zhang, Q. Zhang, Y. Huang, S. Luo, Y. Yao, S. Li and Q. Liu, Biosens. Bioelectron., 2016, 79, 251–257 CrossRef CAS PubMed.
- K. A. Willets and R. P. Van Duyne, Annu. Rev. Phys. Chem., 2007, 58, 267–297 CrossRef CAS PubMed.
- J. Wang, H. Z. Zhang, R. S. Li and C. Z. Huang, TrAC, Trends Anal. Chem., 2016, 80, 429–443 CrossRef CAS.
- P. Wang, M. E. Nasir, A. V. Krasavin, W. Dickson, Y. Jiang and A. V. Zayats, Acc. Chem. Res., 2019, 52, 3018–3028 CrossRef CAS PubMed.
- H.-M. Kim, M. Uh, D. H. Jeong, H.-Y. Lee, J.-H. Park and S.-K. Lee, Sens. Actuators, B, 2019, 280, 183–191 CrossRef CAS.
- Z. Chen, F. Zhang, Y. Li, J. Shan, Y. Lu and Q. Liu, Biosens. Bioelectron., 2022, 201, 113956 CrossRef CAS PubMed.
- D. Zhang, Y. Lu, J. Jiang, Q. Zhang, Y. Yao, P. Wang, B. Chen, Q. Cheng, G. L. Liu and Q. Liu, Biosens. Bioelectron., 2015, 67, 237–242 CrossRef CAS PubMed.
- D. Zhang, Q. Zhang, Y. Lu, Y. Yao, S. Li, J. Jiang, G. L. Liu and Q. Liu, Nano-Micro Lett., 2016, 8, 36–43 CrossRef PubMed.
- J. Vidic, J. Grosclaude, R. Monnerie, M.-A. Persuy, K. Badonnel, C. Baly, M. Caillol, L. Briand, R. Salesse and E. Pajot-Augy, Lab Chip, 2008, 8, 678–688 RSC.
- X. Zhao, G. M. Ashley, L. Garcia-Gancedo, H. Jin, J. Luo, A. J. Flewitt and J. R. Lu, Sens. Actuators, B, 2012, 163, 242–246 CrossRef CAS.
- F. Di Pietrantonio, M. Benetti, V. Dinca, D. Cannatà, E. Verona, S. D'auria and M. Dinescu, Appl. Surf. Sci., 2014, 302, 250–255 CrossRef CAS.
- E. A. Hallem, A. Dahanukar and J. R. Carlson, Annu. Rev. Entomol., 2006, 51, 113–135 CrossRef CAS PubMed.
- R. M. Joseph and J. R. Carlson, Trends Genet., 2015, 31, 683–695 CrossRef CAS PubMed.
- G. M. Pask and A. Ray, in Chemosensory Transduction, Elsevier, 2016, pp. 101–122 Search PubMed.
- K. Sato, M. Pellegrino, T. Nakagawa, T. Nakagawa, L. B. Vosshall and K. Touhara, Nature, 2008, 452, 1002–1006 CrossRef CAS PubMed.
- J. Fleischer, P. Pregitzer, H. Breer and J. Krieger, Cell. Mol. Life Sci., 2018, 75, 485–508 CrossRef CAS PubMed.
- R. Khadka, C. Carraher, C. Hamiaux, J. Travas-Sejdic and A. Kralicek, Biosens. Bioelectron., 2020, 153, 112040 CrossRef CAS PubMed.
- H. J. Ko and T. H. Park, Biosens. Bioelectron., 2005, 20, 1327–1332 CrossRef CAS PubMed.
- N. Montagné, T. Chertemps, I. Brigaud, A. François, M. C. François, A. De Fouchier, P. Lucas, M. C. Larsson and E. Jacquin-Joly, Eur. J. Neurosci., 2012, 36, 2588–2596 CrossRef PubMed.
- T. Murugathas, C. Hamiaux, D. Colbert, A. V. Kralicek, N. O. Plank and C. Carraher, ACS Appl. Electron. Mater., 2020, 2, 3610–3617 CrossRef CAS.
- T. Murugathas, H. Y. Zheng, D. Colbert, A. V. Kralicek, C. Carraher and N. O. Plank, ACS Appl. Mater. Interfaces, 2019, 11, 9530–9538 CrossRef CAS PubMed.
- L. T. Tegler, K. Corin, J. Hillger, B. Wassie, Y. Yu and S. Zhang, Sci. Rep., 2015, 5, 1–7 Search PubMed.
- K. Sato and S. Takeuchi, Angew. Chem., Int. Ed., 2014, 53, 11798–11802 CrossRef CAS PubMed.
- H. Mitsuno, T. Sakurai, S. Namiki, H. Mitsuhashi and R. Kanzaki, Biosens. Bioelectron., 2015, 65, 287–294 CrossRef CAS PubMed.
- T. Nakamoto, M. Kakizaki, Y. Suzuki, H. Mitsuno and R. Kanzaki, in Sensors, 2014 IEEE., 2014, pp. 1491–1494 Search PubMed.
- T. Yamada, H. Sugiura, H. Mimura, K. Kamiya, T. Osaki and S. Takeuchi, Sci. Adv., 2021, 7, eabd2013 CrossRef CAS PubMed.
- R. Khadka, N. Aydemir, C. Carraher, C. Hamiaux, P. Baek, J. Cheema, A. Kralicek and J. Travas-Sejdic, Electroanalysis, 2019, 31, 726–738 CrossRef CAS.
- J. A. Cheema, N. Aydemir, C. Carraher, R. Khadka, D. Colbert, H. T. Lin, A. Nelson, A. Kralicek and J. Travas-Sejdic, Sens. Actuators, B, 2021, 329, 129243 CrossRef CAS.
- N. Misawa, H. Mitsuno, R. Kanzaki and S. Takeuchi, Proc. Natl. Acad. Sci. U. S. A., 2010, 107, 15340–15344 CrossRef CAS PubMed.
- N. Misawa, S. Fujii, K. Kamiya, T. Osaki, T. Takaku, Y. Takahashi and S. Takeuchi, ACS Sens., 2019, 4, 711–716 CrossRef CAS PubMed.
- R. Khadka, N. Aydemir, C. Carraher, C. Hamiaux, D. Colbert, J. Cheema, J. Malmström, A. Kralicek and J. Travas-Sejdic, Biosens. Bioelectron., 2019, 126, 207–213 CrossRef CAS PubMed.
- V. Radhika, T. Proikas-Cezanne, M. Jayaraman, D. Onesime, J. H. Ha and D. N. Dhanasekaran, Nat. Chem. Biol., 2007, 3, 325–330 CrossRef CAS PubMed.
- J. Park, J. H. Lim, H. J. Jin, S. Namgung, S. H. Lee, T. H. Park and S. Hong, Analyst, 2012, 137, 3249–3254 RSC.
- B. Marshall, C. G. Warr and M. De Bruyne, Chem. Senses, 2010, 35, 613–625 CrossRef CAS PubMed.
- X. Liu, M. Marrakchi, D. Xu, H. Dong and S. Andreescu, Biosens. Bioelectron., 2016, 80, 9–16 CrossRef CAS PubMed.
- K. S. Hwang, M. H. Lee, J. Lee, W.-S. Yeo, J. H. Lee, K.-M. Kim, J. Y. Kang and T. S. Kim, Biosens. Bioelectron., 2011, 30, 249–254 CrossRef CAS PubMed.
- S. Sankaran, S. Panigrahi and S. Mallik, Biosens. Bioelectron., 2011, 26, 3103–3109 CrossRef CAS PubMed.
- Q. Liu, J. Wang and B. J. Boyd, Talanta, 2015, 136, 114–127 CrossRef CAS PubMed.
- Y. Cui, S. N. Kim, R. R. Naik and M. C. McAlpine, Acc. Chem. Res., 2012, 45, 696–704 CrossRef CAS PubMed.
- A. Lakshmanan, S. Zhang and C. A. Hauser, Trends Biotechnol., 2012, 30, 155–165 CrossRef CAS PubMed.
- V. J. Ruigrok, M. Levisson, M. H. Eppink, H. Smidt and J. Van Der Oost, Biochem. J., 2011, 436, 1–13 CrossRef CAS PubMed.
- Q. Zhang, D. Zhang, Y. Lu, Y. Yao, S. Li and Q. Liu, Biosens. Bioelectron., 2015, 68, 494–499 CrossRef CAS PubMed.
- J. Wu, Y. Lu, Z. Wu, S. Li, Q. Zhang, Z. Chen, J. Jiang, S. Lin, L. Zhu and C. Li, Sens. Actuators, B, 2018, 261, 279–287 CrossRef CAS.
- J. H. Lim, J. Park, J. H. Ahn, H. J. Jin, S. Hong and T. H. Park, Biosens. Bioelectron., 2013, 39, 244–249 CrossRef CAS PubMed.
- D. Zhang, J. Jiang, J. Chen, Q. Zhang, Y. Lu, Y. Yao, S. Li, G. L. Liu and Q. Liu, Biosens. Bioelectron., 2015, 70, 81–88 CrossRef CAS PubMed.
- Z. Wang, W. Ma, J. Wei, K. Lan, S. Yan, R. Chen and G. Qin, Biosens. Bioelectron., 2022, 195, 113673 CrossRef CAS PubMed.
- J. W. Jaworski, D. Raorane, J. H. Huh, A. Majumdar and S.-W. Lee, Langmuir, 2008, 24, 4938–4943 CrossRef CAS PubMed.
- G. C. Rains, D. Kulasiri, Z. Zhou, S. Samarasinghe, J. K. TomBERLIN and D. M. Olson, Biotechnol. Genet. Eng. Rev., 2009, 26, 179–204 CrossRef PubMed.
- G. M. Fernández-Grandon, R. D. Girling and G. M. Poppy, J. Plant Interact., 2011, 6, 109–112 CrossRef.
- G. C. Rains, S. L. Utley and W. J. Lewis, Biotechnol. Prog., 2006, 22, 2–8 CrossRef CAS PubMed.
- K. Corin, P. Baaske, D. B. Ravel, J. Song, E. Brown, X. Wang, C. J. Wienken, M. Jerabek-Willemsen, S. Duhr and Y. Luo, PLoS One, 2011, 6, e25067 CrossRef CAS PubMed.
- R. Sachse, S. K. Dondapati, S. F. Fenz, T. Schmidt and S. Kubick, FEBS Lett., 2014, 588, 2774–2781 CrossRef CAS PubMed.
- S. Vauthey, S. Santoso, H. Gong, N. Watson and S. Zhang, Proc. Natl. Acad. Sci. U. S. A., 2002, 99, 5355–5360 CrossRef CAS PubMed.
- S. R. Midtgaard, M. C. Pedersen, J. J. K. Kirkensgaard, K. K. Sørensen, K. Mortensen, K. J. Jensen and L. Arleth, Soft Matter, 2014, 10, 738–752 RSC.
- L. L. Looger, M. A. Dwyer, J. J. Smith and H. W. Hellinga, Nature, 2003, 423, 185–190 CrossRef CAS PubMed.
- K. Cali, E. Scorsone and K. Persaud, in 2017 ISOCS/IEEE International Symposium on Olfaction and Electronic Nose (ISOEN), 2017, pp. 1–3 Search PubMed.
- X. Li, J. Zhu and B. Wei, Chem. Soc. Rev., 2016, 45, 3145–3187 RSC.
- X. Li, Y. Lu and Q. Liu, Talanta, 2021, 235, 122726 CrossRef CAS PubMed.
- J. D. Spitzberg, A. Zrehen, X. F. van Kooten and A. Meller, Adv. Mater., 2019, 31, 1900422 CrossRef PubMed.
- R. Ren, Y. Zhang, B. P. Nadappuram, B. Akpinar, D. Klenerman, A. P. Ivanov, J. B. Edel and Y. Korchev, Nat. Commun., 2017, 8, 1–9 CrossRef CAS PubMed.
- J. Huang, L. Ye, X. Gao, H. Li, J. Xu and Z. Li, J. Mater. Chem. B, 2015, 3, 2395–2401 RSC.
- S. Mao, J. Chang, H. Pu, G. Lu, Q. He, H. Zhang and J. Chen, Chem. Soc. Rev., 2017, 46, 6872–6904 RSC.
- Z. Wang, T.-Y. Lv, Z.-B. Shi, S.-S. Yang and Z.-Y. Gu, Dalton Trans., 2021, 50, 13608–13619 RSC.
- F. Röck, N. Barsan and U. Weimar, Chem. Rev., 2008, 108, 705–725 CrossRef PubMed.
- T. Wasilewski, J. Gębicki and W. Kamysz, TrAC, Trends Anal. Chem., 2017, 93, 23–36 CrossRef CAS.
- J. W. Cave, J. Kenneth and A. N. Mitropoulos, Biosens. Bioelectron., 2019, 123, 211–222 CrossRef CAS PubMed.
- S. J. Park, O. S. Kwon, S. H. Lee, H. S. Song, T. H. Park and J. Jang, Nano Lett., 2012, 12, 5082–5090 CrossRef CAS PubMed.
- Y. Lu, Z. Shi and Q. Liu, Curr. Opin. Food Sci., 2019, 28, 74–81 CrossRef.
- D. Zhang and Q. Liu, Biosens. Bioelectron., 2016, 75, 273–284 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |
