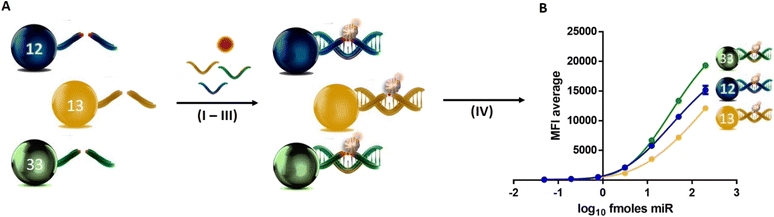Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Open a new window in the world of circulating microRNAs by merging ChemiRNA Tech with a Luminex platform†
Antonio
Marín-Romero
 a,
Mavys
Tabraue-Chávez
a,
Mavys
Tabraue-Chávez
 a,
James W.
Dear
a,
James W.
Dear
 b,
Juan José
Díaz-Mochón
b,
Juan José
Díaz-Mochón
 *cde and
Salvatore
Pernagallo
*cde and
Salvatore
Pernagallo
 *a
*a
aDESTINA Genomica S.L. Parque Tecnológico Ciencias de la Salud (PTS), Avenida de la Innovación 1, Edificio BIC, Armilla, Granada, 18100, Spain. E-mail: salvatore@destinagenomics.com
bPharmacology, Therapeutics and Toxicology, Centre for Cardiovascular Science, University of Edinburgh, The Queen's Medical Research Institute, 47, Little France Crescent, Edinburgh, EH16 4TJ, UK
cGENYO, Centre for Genomics and Oncological Research: Pfizer/University of Granada, Andalusian Regional Government, PTS Granada, Avenida de la Ilustración, 114 - 18016, Granada, Spain. E-mail: juandiaz@ugr.es
dUniversidad de Granada, Facultad de Farmacia, Departamento de Quimica Farmacéutica y Orgánica, Campus Cartuja s/n, 18071, Granada, Spain
eBiosanitary Research Institute of Granada (ibs.GRANADA), University Hospitals of Granada-University of Granada, Granada, Spain
First published on 5th October 2022
Abstract
microRNAs are small noncoding RNA molecules showing huge promise as biomarkers and diagnostic tools for illnesses, as significant changes in their expression occur in response to pathological states. However, multiplex detection of microRNAs represents a major challenge towards the use of microRNA signatures. Hence, our group has tackled this need by developing an accurate and PCR-free approach to detect multiple microRNAs in serum samples. ChemiRNA Tech has been refined to be merged with a Luminex xMAP system. The combination of these two technologies has created a unique bead-based multiplex assay capable of simultaneously measuring miR-122-5p, miR-451a-5p and miR-193a-5p in only 10 μL of serum. This novel multiplex assay for the direct quantitative measurement of circulating microRNA signatures offers benefits over gold standard singleplex assay techniques in terms of cost, time savings and ease of use.
Introduction
microRNAs (miRs) are small non-coding RNAs of 19–24 nucleotides in length that regulate gene expression by base pairing with the 3′-untranslated region of target gene's messenger RNAs (mRNAs), leading to degradation and/or translational repression of those genes. miRs are implicated in many biological events, and their deregulation is associated with serious illnesses.1–4miRs circulate in biological fluids (e.g., serum, plasma, urine and saliva) in a stable fashion and the unique expression patterns can be used as fingerprints for various diseases.5–8 They fulfil most of the characteristics to be considered as ideal biomarkers. They are remarkably stable in body fluids while in circulation, resisting ribonucleases and severe physicochemical conditions such as extreme pH levels. Furthermore, they are (a) specific to the disease or pathology of interest; (b) reliable to indicate the disease's emergence before clinical symptoms appear (early detection); (c) sensitive to changes in the pathology stage (disease progression or therapeutic response); (d) easy to obtain from biological fluids; (e) easily translatable from model systems to humans. All those characteristics are greatly enhancing miR's translation in clinical applications attracting more and more interest in the medical scientific community with tens of thousands of peer-reviewed articles.8–15
Traditional methods for miR analysis include amplification based strategies, such as the RT-qPCR,16 rolling circle amplification,17 exponential amplification reaction,18 and duplex-specific nucleic signal amplification,19 and also amplification free techniques, such as hybridization chain reaction20 and catalysed hairpin amplification.21 Unfortunately, most of the above-mentioned methods are time-consuming, have demanding assay workflows and, mainly, lack multiplexing capability. Thus, despite the extensive academic research, the current analytical methods remain less than satisfactory, and their complexity limits their use of miRs as diagnostic tools.22–25 Hence, it is necessary to provide novel, simple, sensitive and reliable methods for the direct detection of such molecules from body fluids.
Due to the need, over the last years, our group has been extensively working on the development of ChemiRNA Tech.26 This is a novel chemical approach for Nucleic Acid Testing (NAT) particularly well suited to directly quantify circulating miRs. It combines SMART-Base with a biotin tag and modified peptide nucleic acid (PNA) capture probes with an abasic position (DGL-Probes) to interrogate miRs. This technology uses the power of the Watson–Crick base pairing rules (C-G/A-T) that enable the dynamic covalent chemical reaction between the aldehyde group of the SMART Bases and the secondary amine on the abasic position of the DGL-Probe.25,27–39
In our previous studies, ChemiRNA Tech has been combined with some of the most advanced ultra-sensitive immunoassay platforms (such as Merck SMCxPro, Quanterix SIMOA and Luminex xMAP technology) to develop the so-called LiverAce™, the first platform-agnostic and PCR-free molecular assay for the direct detection and quantification of miR-122-5p, a biomarker for hepatotoxicity and Drug-Induced Liver Injury (DILI) diagnostics.25,38–40
However, and as also stated above, multiplex detection of miRs remains a major challenge in the use of miR signatures. Therefore, in this proof of concept (PoC) study, our group has faced this need by implementing the “multi-ChemiRNA Tech” by the merging of ChemiRNA Tech with Luminex xMAP technology. Luminex technology uses different color-coded microspheres that allow the simultaneous detection of multiple analytes using a fluorescent reporter. This technology has been widely used for multiplex detection of proteins and genomic DNAs,41–45 although reports on the application of this technology to miR analysis have been sparse.
The multi-ChemiRNA Tech is a unique tool capable of detecting and quantifying simultaneously the circulating miRs. In this PoC study, to develop the multi-ChemiRNA Tech, three miRs with important roles in diverse biological processes were interrogated, namely miR-122-5p, miR-451a and miR-193a-5p from few microliter volumes of serum samples. miR-122-5p is a liver specific miR, involved in various processes of liver development, differentiation, metabolism and stress responses.46,47 Compared with conventional hepatotoxic markers, circulating miR-122-5p can effectively and consistently distinguish intrahepatic damage from extrahepatic damage with higher sensitivity and specificity. miR-122-5p is expected to be a valuable pre-clinical and clinical biomarker of DILI.48–51 Our group has broadly demonstrated the feasibility of miR-122-5p as a biomarker for DILI.25,38–40 miR-451a-5p is an erythroid cell-specific miRNA and is associated with human erythroid and maturation.52–56 Several studies have provided evidence about the potential use of miR-451a-5p as a biomarker for cancer diagnosis, prognosis, and treatment.57–60 In 2019, our group has reported the direct singleplex detection of miR-451a-5p in haemolysed plasma using ChemiRNA Tech in combination with a conventional microplate reader.32 miR-193a-5p is dysregulated in the tumour cells. Several studies have evaluated the expression level of miR-193a-5p such that it is up-regulated in prostate cancer while it has a decreased expression in colorectal cancer cells which shows its dual role in carcinogenesis.61,62 In addition, our group has recently reported that the circulating miR-193a-5p level increases in the serum of patients with liver injury.63
Experimental
Apparatus
The Luminex MAGPIX was used for multiplex assaying and fluorescence signal detection (Mean of Fluorescence Intensity – MFI). The platform was associated with dedicate software xPONENT. An automatic 96-well plate washer was used for washing (Biotek 405 TS). A 96-well plate shaker was used to perform incubations (VWR® Microplate Shaker).Materials and reagents
Carboxylated MagPlex® (Luminex beads) were purchased from Luminex Corporation (1.25 × 107 mL−1). DGL-Probe 122, DGL-Probe 451 and DGL-Probe 193 and aldehyde-modified biotinylated cytosine nucleobase (SMART-C Biotin) were provided by DESTINA Genomica S.L. (Table S1 and Fig. S1†). A wash buffer and Stabiltech buffer were prepared as described elsewhere.38 An assay buffer and bead diluent were purchased from Merck. Chemicals for beads' functionalization were purchased from Sigma-Aldrich. Synthetic mimic miR-122-5p, miR-451a-5p and miR-193a-5p DNA oligomers were purchased from Integrated DNA Technologies (Table S1†). Concentrations of DNA oligomer solutions were determined using a ThermoFisher NanoDrop1000 spectrophotometer. Streptavidin-R-phycoerythrin (SA-PE) was purchased from Moss Biotech (SA-PE-001, SA-PE-001E, SA-PE-001 16P, SA-PE-001 4P, SA-PE-003) and ThermoFisher Inc (#S21388) at the concentration of 1 mg mL−1.Clinical samples
The DILI sample was provided by Professor James W. Dear from the University of Edinburgh (UK). A sample was collected from a DILI patient (over 16 years old). Full informed consent was obtained from the participant and ethical approval was given by the South East Scotland Research Ethics Committee and the East of Scotland Research Ethics Committee via the South East Scotland Human Bioresource. Blood sample was taken and centrifuged immediately at 11![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000× g for 15 min at 4 °C. The serum was separated into aliquots and stored at −80 °C. The primary endpoint for the study was acute liver injury, pre-defined as a peak hospital stay serum ALT activity greater than 100 U L−1 as shown elsewhere.64
000× g for 15 min at 4 °C. The serum was separated into aliquots and stored at −80 °C. The primary endpoint for the study was acute liver injury, pre-defined as a peak hospital stay serum ALT activity greater than 100 U L−1 as shown elsewhere.64
Blood samples were collected from a healthy volunteer (HV). Full informed consent was obtained from the HV ahead of the study. As shown in Fig. S2-A,† the samples were chemically haemolysed to liberate intra-erythropoietic miR-451a-5p. Briefly, 2 mL of whole blood was incubated for 10 min with 2 mL of RBC lysis buffer (dilution 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2) to break up red blood cells. The solution was then centrifuged at 1500 g for 10 min at room temperature to obtain the haemolysed serum (HS).
2) to break up red blood cells. The solution was then centrifuged at 1500 g for 10 min at room temperature to obtain the haemolysed serum (HS).
Pool samples (PS 1–PS 3) were prepared by mixing DILI and haemolysed samples (Table S2†). Control samples were prepared using whole blood from the HV without haemolysis. In this case, 2 mL of whole blood was centrifuged at 1500 g for 10 min at room temperature. The supernatant serum was collected and diluted 2 times with RBC lysis buffer to obtain the non-haemolysed serum (Fig. S2-B†).
DGL-Probes coupling to beads
Magplex® (Luminex) beads from colour regions 12, 13 and 33 (ref. 44) were functionalized respectively with DGL 122, DGL 451 and DGL 193 abasic PNAs as described in the ESI,† section S2, to generate DGL 122 beads, DGL 451 beads and DGL 193 beads.Coupling confirmation
Luminex beads' coupling efficiency was determined by hybridization with complementary synthetic biotinylated DNA oligomers. This study was carried out using DGL 122 beads. Briefly, in a 96-well plate format, 45 μL of assay buffer containing 1250 DGL 122 beads was pipetted into the wells. A volume of 5 μL containing 15 fmols of biotinylated DNA oligomer (positive) and 5 μL of water (blank) were added into the wells (positives and negatives were run in duplicate). The 96-well plate was incubated for 1 h at 40 °C using a microplate shaker shaking at 700 rpm. DGL 122 beads were washed 3 times with the wash buffer and incubated with 50 μL of 2 μg mL−1 SA-PE for 30 min at 30 °C while being shaken at 700 rpm. DGL 122 beads were washed 3 times with the wash buffer and finally resuspended in a volume of 120 μL of wash buffer to be analysed with the Luminex MAGPIX system by using an injection volume of 100 μL. MFI values were recorded.Singleplex DGL bead sensitivity study
DGL beads for each miR were validated with ChemiRNA Tech with complementary oligomers to analyse coupling efficiency and sensitivity.The following protocol was used to generate three parallel calibration curves associated respectively with DGL 122 beads, DGL 451 beads and DGL 193 beads. In a 96-well plate format, 55 μL of Stabiltech buffer containing a pool of complementary synthetic DNA oligomer mimicking miR-122-5p, miR-451a-5p and miR-193a-5p (0.2 to 200 fmols) and 55 μL of only Stabiltech buffer as the blank were pipetted into the wells. A volume of 10 μL of DGL beads at 125 beads per μL (1250 beads in total) was added to the wells, followed by the addition of 10 μL of serum matrix (control samples). The 96-well plate was incubated for 2 h at 30 °C using a microplate shaker shaking at 700 rpm. After the incubation, DGL beads were washed 3 times with the wash buffer. DGL beads were resuspended in 50 μL of assay buffer containing 5 μM SMART-C Biotin and 1 mM sodium cyanoborohydride and incubated for further 1 h at 40 °C under 700 rpm shaking. Beads were washed 3 times with the wash buffer and incubated with 50 μL of 2 μg mL−1 SA-PE for 30 min at 30 °C while being shaken at 700 rpm. Beads were washed 3 times with the wash buffer and finally resuspended in a volume of 120 μL of wash buffer to be analysed on the Luminex MAGPIX system to determine the MFI values (injection volume of 100 μL). Experiments were performed in duplicate.
Multiplex DGL bead sensitivity study
The three sets of DGL beads were mixed to perform the multiplex testing. A volume of 100 μL of DGL 122 beads (3750 beads per μL) was mixed respectively with 100 μL of DGL 451 beads (3750 beads per μL) and 100 μL of DGL 193 beads (3750 beads per μL). The 300 μL bead mixture was added to 700 μL of bead diluent. The following protocol was used to generate the multiplex calibration curve. In a 96-well plate format, 55 μL of Stabiltech buffer containing a pool of complementary synthetic miR mimic miR-122-5p, miR-451a-5p and miR-193a-5p (0.2 to 200 fmols) and 55 μL of only Stabiltech buffer as the blank were pipetted into the wells. A volume of 10 μL of DGL bead mixture (125 beads per μL for each set of DGL bead) was added to the wells (total 1250 × 3 = 3750), followed by the addition of 10 μL of serum matrix (control samples). The protocol is described in “Singleplex DGL bead sensitivity study”.Multiplex assay in clinical samples
The three sets of DGL beads were mixed as described in the “Multiplex DGL beads sensitivity study”. The following protocol was used to interrogate clinical samples. In a 96-well plate format, 55 μL of Stabiltech buffer was pipetted into the wells. A volume of 10 μL of DGL bead mixture (125 beads per μL for each set of DGL bead) was added to the wells (total 1250 × 3 = 3750), followed by the addition of 10 μL of the clinical sample (DILI, HS or PS). The protocol followed is described in “Singleplex DGL bead sensitivity study”.Results and discussion
Reagents for multi-ChemiRNA Tech
Three DGL-Probes complementary to the three miRs (DGL-Probe 122, DGL-Probe 451 and DGL-Probe 193) were prepared with propanoic acid at the gamma position modifications in the backbone of sequences (ESI,† Table S1). As described elsewhere by our group,30 the presence of these modifications enhanced DGL-Probes' performance being more readily available to hybridise complementary miRs, hence, allowing more efficient dynamic incorporation of the aldehyde-modified biotinylated nucleobase (SMART-Base). DGL-Probe 122, DGL-Probe 451 and DGL-Probe 193 were conjugated to Luminex beads with colour regions 12, 13 and 33 respectively to generate DGL 122 beads, DGL 451 beads and DGL 193 beads using the conjugation chemistry reported elsewhere.30 The coupling efficiency was determined by running a hybridization with a complementary synthetic biotinylated DNA oligomer that mimics the target miR-122-5p sequence (data not shown). SMART-Base with a biotin tag was used for the study (Fig. S1†).Selection of SA-PE
To improve the detection sensitivity, different streptavidin-R-phycoerythrin (SA-PE) conjugations were tested on DGL 122 beads in a singleplex format. The study was carried out with six different commercially available SA-PE molecules: a) SA-PE from ThermoFisher and b) from MossBio, namely SA-PE 001, SA-PE 001E, SA-PE 001 16P, SA-PE 001 4P and SA-PE 003. Performances were determined by interrogating spike-in oligomers mimicking miR-122-5p (as the positive) and only water (as the blank). The protocol used for the study is described in the experimental section ‘Singleplex DGL bead sensitivity study’. As shown in Fig. S3,† all SA-PEs from MossBio improved the S/B ratio when compared with the SA-PE from ThermoFisher. Due to the overall good performances of all the MossBio SA-PEs, SA-PE 001 16P was selected for the present study because it is slightly better in terms of performance (Fig. S3†).Singleplex assay
DGL beads were used in a singleplex format to perform analytical sensitivity studies by creating three parallel calibration curves using known quantities of DNA oligomers mimicking miR targets (miR-122-5p, miR-451a-5p and miR-193a-5p). A pool of miR targets in the serum was prepared by spiking seven known quantities of oligomers, respectively, with the calibration points: 200.00, 50.00, 12.50, 3.13, 0.78, 0.20 and 0.05 fmols (each in duplicate). No spike-in oligomers were added to the blank (water instead). As shown in Fig. 1A–C, the singleplex workflow is a multistep comprised of four main steps (I–IV): I) DGL beads hybridise to a complementary DNA oligomer; II) SMART-C Biotin is specifically incorporated into the duplex by a chemical dynamic reaction. This specific incorporation is ensured by the three events required: (a) perfect hybridisation between the DGL-Probe and complementary oligomer; (b) generation of a reversible iminium species between the secondary amine of the “abasic position” and the aldehyde group of a cytosine SMART-Base with a biotin tag (SMART-C Biotin), which is driven by the templating “G” nucleotide on the complementary oligomer; (c) reduction of the iminium species by sodium cyanoborohydride to lock-up the SMART-C Biotin covalently into the duplex; III) DGL beads are labelled with SA-PE; IV) read-out with the Luminex MAGPIX system.Experiments were performed in duplicate, resulting in a coefficient of variance for each assay below 5% (Tables S3–S5†). Curves were prepared by plotting the mean of fluorescence intensity (MFI) values versus the logarithm base 10 quantity of oligomers (Fig. 1D–F). MFI values are reported in the ESI,† Tables S3–S5 and curve equations are reported in the ESI,† section S3.
Determination of the signal-to-blank ratio was performed by comparing measured signals from spike-in calibration points with those of blanks and establishing the minimum concentration at which the spike-in calibration point can be reliably detected. A signal-to-blank ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 was considered for estimating the limit of detection (LOD), as described elsewhere.65 As a result, DGL 122 beads and DGL 451 beads show LODs between 0.05 and 0.20 fmols while DGL 193 beads show below 0.05 fmols.
1 was considered for estimating the limit of detection (LOD), as described elsewhere.65 As a result, DGL 122 beads and DGL 451 beads show LODs between 0.05 and 0.20 fmols while DGL 193 beads show below 0.05 fmols.
Multiplex assay
The three singleplex assays described in Fig. 1A–C were combined to deliver a multiplex assay capable of profiling miR-122-5p, miR-451a-5p and miR-193a-5p simultaneously.The three sets of DGL beads were mixed to perform the multiplex testing. This study allowed the multiplex and singleplex assays to be compared for the detection of ultra-low prevalence miRs in spiked serums.
The analytical sensitivity study was carried out by creating calibration curves to determine the sensitivity of DGL beads in a multiplex format. The same calibration points used for the singleplex assay were interrogated: 200.00, 50.00, 12.50, 3.13, 0.78, 0.20 and 0.05 fmols of each DNA oligomer, mimicking respectively miR-122-5p, miR-451a-5p and miR-193a-5p. The workflow described in Fig. 2A for the multiplex is essentially the same as described for the singleplex assay with the only difference that instead of a single DGL bead (1250 beads), an equal mix of DGL beads corresponding to the three target DNA oligomers (1250 × 3 = 3750 beads) was used. As for the singleplex assay, experiments were performed in duplicate, resulting in a coefficient of variance for each condition also below 5% (Table S6†). The curve was prepared by plotting the MFI values versus logarithm base 10 quantity of DNA oligomers (Fig. 2B). MFI values are reported in the ESI,† Table S6. DGL 122 beads and DGL 451 beads show an LOD between 0.05 and 0.20 fmols and DGL 193 beads show below 0.05 fmols.
Inter-assay variability
Table S7 in the ESI† shows the comparative analysis between the singleplex and multiplex assays. By comparing the MFI average signals obtained for each calibration curve, it was possible to determine the inter-assay variability between the two assays. Inter-CV values were below 10% for miR-122-5p and miR-451a-5p measurements and 15% for miR-193a-5p measurement. This shows the robustness of the multiplex assay in comparison with the singleplex assays. In addition, no signal loss nor cross reactivity was observed when the multiplex analysis was performed.Assay validation using serum samples
The multiplex assay was challenged by profiling different levels of miR-122-5p, miR-451a-5p and miR-193a-5p in serum samples. For the study, five types of serums were analysed:a) DILI – a serum from a DILI patient rich in miR-122-5p;
b) HS – a serum from haemolysed blood rich in miR-451a-5p;
c) PS 1 – a pool sample generated by the mixture of a + b with a ratio of 75%/25%;
d) PS 2 – a pool sample generated by the mixture of a + b with a ratio of 50%/50%;
e) PS 3 – a pool sample generated by the mixture of a + b with a ratio of 25%/75%.
A serum sample obtained from a healthy volunteer (HV) without significant levels of circulating miR-122-5p, miR-451a-5p or miR-193a-5p at the time of the study was used as a control. DGL beads were mixed to interrogate the five types of serums described from a to e (see above).
The multiplex assay has included the Stabiltech buffer. As described by our group elsewhere,25 this buffer has been developed to lysate the serum samples and lead to the miR release from protein complexes and/or extracellular vesicles. MFI values obtained from samples (Table S8†) were used to make an absolute quantification of miRNA-122-5p, miR-451a-5p and miR-193a-5p by extrapolating the fmols from the calibration curves in Fig. 2B. As shown in Table S9,† the assay obtained the profiling levels (quantity in fmols) of: a) the only miRNA-122-5p in the DILI patient (quantity n.d. of miR-451a); b) the only miR-451a-5p in the HS sample (quantity n.d. of miR-122-5p); c–e) both miRNA-122-5p and miR-451a-5p in PS 1–3 samples (see Table S9†). None of the above shows levels of miR-193a-5p (quantity n.d.), because either it is not present or its amounts fall below the assay's LOD. As shown in Fig. 3, PS 1–PS 3 show levels of miR-122-5p versus mir-451a-5p coherent with volume ratios (% described in c to e) of DILI versus HS serums. In Fig. 3, % of miR is calculated assuming: a) 100% of miR-122-5p = the average quantity measured for DILI (6.27 fmols, see Table S9†); b) 100% of miR-451a-5p = the average quantity measured for HS (78.40 fmols, see Table S9†). % of PS 1, PS 2 and PS 3 was calculated dividing the average quantities of miR-122-5p and miR-451a-5p respectively for PS 1 (4.58 and 17.12 fmols, see Table S9†), PS 2 (3.09 and 33.50 fmols, see Table S9†) and PS 3 (1.71 and 52.31 fmols, see Table S9†) by the average quantities measured respectively for DILI (6.27 fmols, see Table S9†) and HS (78.40 fmols, see Table S9†).
 | ||
| Fig. 3 Serum sample analysis (n = 2). miR-122-5p and miR-451a-5p detection in DILI, PS1, PS2, PS3 and HS samples. y-Axis: % of miR is calculated as described in the main text. | ||
Conclusions
Many studies have shown that circulating miRNAs could play potential roles as diagnostic and prognostic biomarkers. Combinations of miR biomarkers can identify diseases, predict the course of pathology and help in assigning drug treatment.However, and as stated above, despite the extensive academic research the translation of miR biomarkers to clinical practice remains a challenge. Current analysis platforms require workflows which include extraction, elongation, reverse transcription and quantitative PCR which make the quantification of miRs with the standard analytical platforms very challenging and not scalable. On top of that, multiplexing is another complexity that current systems cannot easily overcome. Hence, there is a need to develop new technologies to accurately detect and measure multi-miR signatures in clinical settings.
To tackle the issue, in this PoC study, our group has developed multi-ChemiRNA Tech, a novel assay capable of interrogating multi-miRs simultaneously without the need of extracting or amplifying target miRs. The novel assay was created by merging the ChemiRNA Tech with the Luminex xMAP system.
The manuscript describes an overview of the PoC for testing the integration of the two technologies and its validation using serum samples. All the key PoC objectives were achieved, including effective DGL-Probe coupling with Luminex beads and ChemiRNA Tech reactions in singleplex and multiplex formats, demonstrating successful translation of ChemiRNA Tech reagents onto the Luminex xMAP platform.
The novel assay was evaluated and validated for detecting miR-122-5p, miR-451a-5p and miR-193a-5p. The presence of miR-122-5p as a hepatic toxicity biomarker enables the assay to be used as a drug development tool with wide applications in the field of drug development.66–69 In fact, miR-122-5p has received an endorsement from the Food and Drug Administration (FDA) as a biomarker of liver specific injury based on its performance in patients with acute DILI, including patients with DILI due to acetaminophen overdose, compared to non-DILI controls.70
In the first instance, both singleplex and multiplex formats were implemented by spiking DNA oligomers (mimicking the three miRs) in serum samples successfully. MFI values between both singleplex and multiplex were compared, and no signal differences were observed, thus demonstrating the high compatibility of the multi-ChemiRNA Tech reagents (see Table 1). It has demonstrated that the multiplexed measurement does not differ from the singleplex assay in terms of sensitivity and accuracy.
| miR ID | Singleplex assay | Multiplex assay | ||
|---|---|---|---|---|
| Sensitivity | Precision | Sensitivity | Precision | |
| miR-122-5p | 0.05–0.20 | <5% | 0.05–0.20 | <5% |
| miR-451a-5p | 0.05–0.20 | <5% | 0.05–0.20 | <5% |
| miR-193a-5p | Below 0.05 | <5% | Below 0.05 | <5% |
Once the development with DNA oligomers was completed, the novel multi-ChemiRNA Tech was validated using biological samples. A serum from (a) a patient with liver injury, (b) haemolysed blood and (c) pool of serums of a + b at three different ratios (PS 1–PS 3) were interrogated to determine the levels of miRs. miR-122-5p and miR-451a-5p were detected and quantified successfully. Meanwhile, miR-193a-5p was not detected and this is, perhaps, because its level falls below the LOD of the assay.
The novel assay has included the Stabiltech buffer, a lysing buffer that allowed the liberation and capture of miRs in serum samples.25 As shown in Fig. 3, miR-122-5p and miR-451a-5p were detected and resulted in quantities corresponding to the ratio of the percentage of DILI versus HS samples. No miR detection was reported for the control HV.
To the best of our knowledge, this is the first use of Luminex xMAP technology for a direct (PCR-free) multi miR profiling from serum samples (without pre-extraction of RNAs). This novel assay used a small sample volume (10 μL), which is particularly useful for testing paediatric samples, precious samples such as cerebrospinal fluid or even samples from small animals such as rats in preclinical testing.
The multi-ChemiRNA Tech is very promising and potentially suitable for developing a highly innovative next-generation system to multiplex the analysis of miRs. It shows the way forward to simplified, more cost effective and robust multiplex tests in the future, to interrogate multi-miR biomarkers with predictive value in pathologies such as liver injury, cancer, but as well as to evaluate drug-induced injuries affecting other organs such as kidney and heart.
Author contributions
S. P., J. J. D. M. and A. M. R. contributed to the design of the experiments of this work. A. M. R. and M. T. C. performed the experiments. S. P. and A. M. R. wrote the manuscript. J. W. D. and J. J. D. M. critically revised the manuscript.Conflicts of interest
J. J. D. M. is a shareholder and director of DESTINA Genomica S.L. S. P. is a shareholder of DESTINA Genomica S.L.Acknowledgements
The authors thank the staff of DESTINA Genomica S.L. for their important technical support. This study was funded by the Estars 2 programme under the grant number 114589.Notes and references
- D. P. Bartel, MicroRNAs: genomics, biogenesis, mechanism, and function, Cell, 2004, 116(2), 281–297 CrossRef CAS.
- D. P. Bartel, MicroRNAs: target recognition and regulatory functions, Cell, 2009, 136(2), 215–233 CrossRef CAS PubMed.
- A. Drusco and C. M. Croce, MicroRNAs and Cancer: A Long Story for Short RNAs, Adv. Cancer Res., 2017, 135, 1–24 CrossRef CAS.
- C. Price and J. Chen, MicroRNAs in Cancer Biology and Therapy: Current Status and Perspectives, Genes Dis., 2014, 1(1), 53–63 CrossRef PubMed.
- K. Kang, X. Peng, J. Luo and D. Gou, Identification of circulating miRNA biomarkers based on global quantitative real-time PCR profiling, J. Anim. Sci. Biotechnol., 2012, 3(1), 4 CrossRef CAS.
- A. Izzotti, S. Carozzo, A. Pulliero, D. Zhabayeva, J. L. Ravetti and R. Bersimbaev, Extracellular MicroRNA in liquid biopsy: applicability in cancer diagnosis and prevention, Am. J. Cancer Res., 2016, 6(7), 1461–1493 CAS.
- N. Kosaka, H. Iguchi and T. Ochiya, Circulating microRNA in body fluid: a new potential biomarker for cancer diagnosis and prognosis, Cancer Sci., 2010, 101(10), 2087–2092 CrossRef CAS PubMed.
- K. W. Witwer, Circulating microRNA biomarker studies: pitfalls and potential solutions, Clin. Chem., 2015, 61(1), 56–63 CrossRef CAS PubMed.
- J. Krauskopf, T. M. de Kok, S. J. Schomaker, M. Gosink, D. A. Burt and P. Chandler, et al., Serum microRNA signatures as “liquid biopsies” for interrogating hepatotoxic mechanisms and liver pathogenesis in human, PLoS One, 2017, 12(5), e0177928 CrossRef PubMed.
- G. H. Liu, Z. G. Zhou, R. Chen, M. J. Wang, B. Zhou and Y. Li, et al., Serum miR-21 and miR-92a as biomarkers in the diagnosis and prognosis of colorectal cancer, Tumor Biol., 2013, 34(4), 2175–2181 CrossRef CAS PubMed.
- J. C. McCrae, N. Sharkey, D. J. Webb, A. D. Vliegenthart and J. W. Dear, Ethanol consumption produces a small increase in circulating miR-122 in healthy individuals, Clin. Toxicol., 2016, 54(1), 53–55 CrossRef CAS PubMed.
- P. J. Starkey Lewis, J. Dear, V. Platt, K. J. Simpson, D. G. Craig and D. J. Antoine, et al., Circulating microRNAs as potential markers of human drug-induced liver injury, Hepatology, 2011, 54(5), 1767–1776 CrossRef CAS PubMed.
- H. Ogata-Kawata, M. Izumiya, D. Kurioka, Y. Honma, Y. Yamada and K. Furuta, et al., Circulating exosomal microRNAs as biomarkers of colon cancer, PLoS One, 2014, 9(4), e92921 CrossRef PubMed.
- X. Zhang, S. Shi, B. Zhang, Q. Ni, X. Yu and J. Xu, Circulating biomarkers for early diagnosis of pancreatic cancer: facts and hopes, Am. J. Cancer Res., 2018, 8(3), 332–353 CAS.
- L. Nandagopal and G. Sonpavde, Circulating Biomarkers in Bladder Cancer, Bladder Cancer, 2016, 2(4), 369–379 Search PubMed.
- C. Chen, R. Tan, L. Wong, R. Fekete and J. Halsey, Quantitation of microRNAs by real-time RT-qPCR, Methods Mol. Biol., 2011, 687, 113–134 CrossRef CAS PubMed.
- N. I. Goo and D. E. Kim, Rolling circle amplification as isothermal gene amplification in molecular diagnostics, BioChip J., 2016, 10(4), 262–271 CrossRef CAS PubMed.
- R. D. Li, B. C. Yin and B. C. Ye, Ultrasensitive, colorimetric detection of microRNAs based on isothermal exponential amplification reaction-assisted gold nanoparticle amplification, Biosens. Bioelectron., 2016, 86, 1011–1016 CrossRef CAS.
- E. Tan, J. Wong, D. Nguyen, Y. Zhang, B. Erwin and L. K. Van Ness, et al., Isothermal DNA amplification coupled with DNA nanosphere-based colorimetric detection, Anal. Chem., 2005, 77(24), 7984–7992 CrossRef CAS PubMed.
- Z. Ge, M. Lin, P. Wang, H. Pei, J. Yan and J. Shi, et al., Hybridization chain reaction amplification of microRNA detection with a tetrahedral DNA nanostructure-based electrochemical biosensor, Anal. Chem., 2014, 86(4), 2124–2130 CrossRef CAS PubMed.
- Y. H. Yuan, Y. D. Wu, B. Z. Chi, S. H. Wen, R. P. Liang and J. D. Qiu, Simultaneously electrochemical detection of microRNAs based on multifunctional magnetic nanoparticles probe coupling with hybridization chain reaction, Biosens. Bioelectron., 2017, 97, 325–331 CrossRef CAS PubMed.
- L. Moldovan, K. E. Batte, J. Trgovcich, J. Wisler, C. B. Marsh and M. Piper, Methodological challenges in utilizing miRNAs as circulating biomarkers, J. Cell. Mol. Med., 2014, 18(3), 371–390 CrossRef CAS PubMed.
- B. Wang and Y. Xi, Challenges for MicroRNA Microarray Data Analysis, Microarrays, 2013, 2(2), 34–50 CrossRef CAS PubMed.
- H. Lin, L. E. Ewing, I. Koturbash, B. J. Gurley and I. R. Miousse, MicroRNAs as biomarkers for liver injury: Current knowledge, challenges and future prospects, Food Chem. Toxicol., 2017, 110, 229–239 CrossRef CAS.
- B. López-Longarela, E. E. Morrison, J. D. Tranter, L. Chahman-Vos, J. F. Léonard and J. C. Gautier, et al., Direct Detection of miR-122 in Hepatotoxicity Using Dynamic Chemical Labeling Overcomes Stability and isomiR Challenges, Anal. Chem., 2020, 92(4), 3388–3395 CrossRef PubMed.
- Destina GL. DESTINA - ChemiRNA Tech, YouTube, 2019, https://www.youtube.com/watch?v=7jX0uZn3YpM.
- F. R. Bowler, J. J. Diaz-Mochon, M. D. Swift and M. Bradley, DNA analysis by dynamic chemistry, Angew. Chem., Int. Ed., 2010, 49(10), 1809–1812 CrossRef CAS PubMed.
- F. R. Bowler, P. A. Reid, A. C. Boyd, J. J. Díaz-Mochón and M. Bradley, Dynamic chemistry for enzyme-free allele discrimination in genotyping by MALDI-TOF mass spectrometry, Anal. Methods, 2011, 3(7), 1656–1663 RSC.
- S. Pernagallo, G. Ventimiglia, C. Cavalluzzo, E. Alessi, H. Ilyine and M. Bradley, et al., Novel biochip platform for nucleic acid analysis, Sensors, 2012, 12(6), 8100–8111 CrossRef CAS PubMed.
- S. Venkateswaran, M. A. Luque-González, M. Tabraue-Chávez, M. A. Fara, B. López-Longarela and V. Cano-Cortes, et al., Novel bead-based platform for direct detection of unlabelled nucleic acids through Single Nucleobase Labelling, Talanta, 2016, 161, 489–496 CrossRef CAS PubMed.
- M. Angélica Luque-González, M. Tabraue-Chávez, B. López-Longarela, R. María Sánchez-Martín, M. Ortiz-González and M. Soriano-Rodríguez, et al., Identification of Trypanosomatids by detecting Single Nucleotide Fingerprints using DNA analysis by dynamic chemistry with MALDI-ToF, Talanta, 2018, 176, 299–307 CrossRef PubMed.
- A. Marín-Romero, A. Robles-Remacho, M. Tabraue-Chávez, B. López-Longarela, R. M. Sánchez-Martín and J. J. Guardia-Monteagudo, et al., A PCR-free technology to detect and quantify microRNAs directly from human plasma, Analyst, 2018, 143(23), 5676–5682 RSC.
- S. Detassis, M. Grasso, M. Tabraue-Chávez, A. Marín-Romero, B. López-Longarela and H. Ilyine, et al., New Platform for the Direct Profiling of microRNAs in Biofluids, Anal. Chem., 2019, 91(9), 5874–5880 CrossRef CAS PubMed.
- A. Delgado-Gonzalez, A. Robles-Remacho, A. Marin-Romero, S. Detassis, B. Lopez-Longarela and F. J. Lopez-Delgado, et al., PCR-free and chemistry-based technology for miR-21 rapid detection directly from tumour cells, Talanta, 2019, 200, 51–56 CrossRef CAS PubMed.
- M. Tabraue-Chávez, M. A. Luque-González, A. Marín-Romero, R. M. Sánchez-Martín, P. Escobedo-Araque and S. Pernagallo, et al., A colorimetric strategy based on dynamic chemistry for direct detection of Trypanosomatid species, Sci. Rep., 2019, 9(1), 3696 CrossRef PubMed.
- E. Garcia-Fernandez, M. C. Gonzalez-Garcia, S. Pernagallo, M. J. Ruedas-Rama, M. A. Fara and F. J. López-Delgado, et al., miR-122 direct detection in human serum by time-gated fluorescence imaging, Chem. Commun., 2019, 55(99), 14958–14961 RSC.
- A. Robles-Remacho, M. A. Luque-González, R. A. González-Casín, M. V. Cano-Cortés, F. J. Lopez-Delgado and J. J. Guardia-Monteagudo, et al., Development of a nanotechnology-based approach for capturing and detecting nucleic acids by using flow cytometry, Talanta, 2021, 226, 122092 CrossRef CAS PubMed.
- A. Marín-Romero, M. Tabraue-Chávez, B. López-Longarela, M. A. Fara, R. M. Sánchez Martín and J. W. Dear, et al., Simultaneous Detection of Drug-Induced Liver Injury Protein and microRNA Biomarkers Using Dynamic Chemical Labelling on a Luminex MAGPIX System, Analytica, 2021, 2, 130–139 CrossRef.
- A. Marín-Romero, M. Tabraue-Chávez, J. W. Dear, R. M. Sánchez-Martín, H. Ilyne and J. J. Guardia-Monteagudo, et al., Amplification-free profiling of microRNA-122 biomarker in DILI patient serums, using the luminex MAGPIX system, Talanta, 2020, 219, 121265 CrossRef PubMed.
- D. M. Rissin, B. López-Longarela, S. Pernagallo, H. Ilyine, A. D. B. Vliegenthart and J. W. Dear, et al., Polymerase-free measurement of microRNA-122 with single base specificity using single molecule arrays: Detection of drug-induced liver injury, PLoS One, 2017, 12(7), e0179669 CrossRef PubMed.
- F. Chowdhury, A. Williams and P. Johnson, Validation and comparison of two multiplex technologies, Luminex and Mesoscale Discovery, for human cytokine profiling, J. Immunol. Methods, 2009, 340(1), 55–64 CrossRef CAS PubMed.
- S. Esposito, A. Scala, S. Bianchini, M. L. Presicce, A. Mori and C. S. Sciarrabba, et al., Partial comparison of the NxTAG Respiratory Pathogen Panel Assay with the Luminex xTAG Respiratory Panel Fast Assay V2 and singleplex real-time polymerase chain reaction for detection of respiratory pathogens, Diagn. Microbiol. Infect. Dis., 2016, 86(1), 53–57 CrossRef CAS PubMed.
- J. Gray and L. J. Coupland, The increasing application of multiplex nucleic acid detection tests to the diagnosis of syndromic infections, Epidemiol. Infect., 2014, 142(1), 1–11 CrossRef CAS PubMed.
- R. Perraut, V. Richard, M. L. Varela, J. F. Trape, M. Guillotte and A. Tall, et al., Comparative analysis of IgG responses to Plasmodium falciparum MSP1p19 and PF13-DBL1α1 using ELISA and a magnetic bead-based duplex assay (MAGPIX®-Luminex) in a Senegalese meso-endemic community, Malar. J., 2014, 13, 410 CrossRef PubMed.
- N. G. Satterly, M. A. Voorhees, A. D. Ames and R. J. Schoepp, Comparison of MagPix Assays and Enzyme-Linked Immunosorbent Assay for Detection of Hemorrhagic Fever Viruses, J. Clin. Microbiol., 2017, 55(1), 68–78 CrossRef CAS PubMed.
- Y. Zhang, Y. Jia, R. Zheng, Y. Guo, Y. Wang and H. Guo, et al., Plasma microRNA-122 as a biomarker for viral-, alcohol-, and chemical-related hepatic diseases, Clin. Chem., 2010, 56(12), 1830–1838 CrossRef CAS PubMed.
- D. J. Antoine, J. W. Dear, P. S. Lewis, V. Platt, J. Coyle and M. Masson, et al., Mechanistic biomarkers provide early and sensitive detection of acetaminophen-induced acute liver injury at first presentation to hospital, Hepatology, 2013, 58(2), 777–787 CrossRef CAS PubMed.
- D. N. Bateman, Changing the Management of Paracetamol Poisoning, Clin. Ther., 2015, 37(9), 2135–2141 CrossRef PubMed.
- M. I. Danjuma, J. Sajid, H. Fatima and A. N. Elzouki, Novel biomarkers for potential risk stratification of drug induced liver injury (DILI): A narrative perspective on current trends, Medicine, 2019, 98(50), e18322 CrossRef CAS PubMed.
- S. Fu, D. Wu, W. Jiang, J. Li, J. Long and C. Jia, et al., Molecular Biomarkers in Drug-Induced Liver Injury: Challenges and Future Perspectives, Front. Pharmacol., 2019, 10, 1667 CrossRef CAS PubMed.
- D. J. Antoine and J. W. Dear, Transformative biomarkers for drug-induced liver injury: are we there yet?, Biomarkers Med., 2017, 11(2), 103–106 CrossRef CAS PubMed.
- S. Masaki, R. Ohtsuka, Y. Abe, K. Muta and T. Umemura, Expression patterns of microRNAs 155 and 451 during normal human erythropoiesis, Biochem. Biophys. Res. Commun., 2007, 364(3), 509–514 CrossRef CAS PubMed.
- L. Pase, J. E. Layton, W. P. Kloosterman, D. Carradice, P. M. Waterhouse and G. J. Lieschke, miR-451 regulates zebrafish erythroid maturation in vivo via its target gata2, Blood, 2009, 113(8), 1794–1804 CrossRef CAS PubMed.
- D. M. Patrick, C. C. Zhang, Y. Tao, H. Yao, X. Qi and R. J. Schwartz, et al., Defective erythroid differentiation in miR-451 mutant mice mediated by 14-3-3zeta, Genes Dev., 2010, 24(15), 1614–1619 CrossRef CAS PubMed.
- K. D. Rasmussen, S. Simmini, C. Abreu-Goodger, N. Bartonicek, M. Di Giacomo and D. Bilbao-Cortes, et al., The miR-144/451 locus is required for erythroid homeostasis, J. Exp. Med., 2010, 207(7), 1351–1358 CrossRef CAS PubMed.
- S. Svasti, S. Masaki, T. Penglong, Y. Abe, P. Winichagoon and S. Fucharoen, et al., Expression of microRNA-451 in normal and thalassemic erythropoiesis, Ann. Hematol., 2010, 89(10), 953–958 CrossRef CAS PubMed.
- H. B. Bian, X. Pan, J. S. Yang, Z. X. Wang and W. De, Upregulation of microRNA-451 increases cisplatin sensitivity of non-small cell lung cancer cell line (A549), J. Exp. Clin. Cancer Res., 2011, 30, 20 CrossRef CAS PubMed.
- M. Khordadmehr, F. Jigari-Asl, H. Ezzati, R. Shahbazi, S. Sadreddini and S. Safaei, et al., A comprehensive review on miR-451: A promising cancer biomarker with therapeutic potential, J. Cell. Physiol., 2019, 234(12), 21716–21731 CrossRef CAS PubMed.
- X. Pan, R. Wang and Z. X. Wang, The potential role of miR-451 in cancer diagnosis, prognosis, and therapy, Mol. Cancer Ther., 2013, 12(7), 1153–1162 CrossRef CAS PubMed.
- R. Wang, Z. X. Wang, J. S. Yang, X. Pan, W. De and L. B. Chen, MicroRNA-451 functions as a tumor suppressor in human non-small cell lung cancer by targeting ras-related protein 14 (RAB14), Oncogene, 2011, 30(23), 2644–2658 CrossRef CAS PubMed.
- M. Khordadmehr, R. Shahbazi, S. Sadreddini and B. Baradaran, miR-193: A new weapon against cancer, J. Cell. Physiol., 2019, 234(10), 16861–16872 CrossRef CAS PubMed.
- J. H. Xu, J. X. Zhao, M. Y. Jiang, L. P. Yang, M. L. Sun and H. W. Wang, MiR-193 promotes cell proliferation and invasion by ING5/PI3K/AKT pathway of triple-negative breast cancer, Riv. Eur. Sci. Med. Farmacol., 2020, 24(6), 3122–3129 Search PubMed.
- A. D. Vliegenthart, J. M. Shaffer, J. I. Clarke, L. E. Peeters, A. Caporali and D. N. Bateman, et al., Comprehensive microRNA profiling in acetaminophen toxicity identifies novel circulating biomarkers for human liver and kidney injury, Sci. Rep., 2015, 5, 15501 CrossRef CAS PubMed.
- J. W. Dear, J. I. Clarke, B. Francis, L. Allen, J. Wraight and J. Shen, et al., Risk stratification after paracetamol overdose using mechanistic biomarkers: results from two prospective cohort studies, Lancet Gastroenterol. Hepatol., 2018, 3(2), 104–113 CrossRef PubMed.
- Administration UFaD, Q2 (R1) Validation of Analytical Procedures: Text and Methodology Guidance for Industry, 2021 Search PubMed.
- W. J. Bailey and W. E. Glaab, Accessible miRNAs as Novel Toxicity Biomarkers, Int. J. Toxicol., 2018, 37(2), 116–120 CrossRef CAS PubMed.
- A. L. Schofield, J. P. Brown, J. Brown, A. Wilczynska, C. Bell and W. E. Glaab, et al., Systems analysis of miRNA biomarkers to inform drug safety, Arch. Toxicol., 2021, 95(11), 3475–3495 CrossRef CAS PubMed.
- E. Schraml, M. Hackl and J. Grillari, MicroRNAs and toxicology: A love marriage, Toxicol. Rep., 2017, 4, 634–636 CrossRef CAS PubMed.
- R. Blanco-Domínguez, R. Sánchez-Díaz, H. de la Fuente, L. J. Jiménez-Borreguero, A. Matesanz-Marín and M. Relaño, et al., A Novel Circulating MicroRNA for the Detection of Acute Myocarditis, N. Engl. J. Med., 2021, 384(21), 2014–2027 CrossRef PubMed.
- FDA, Letter of Support for Drug-Induced Liver Injury (DILI) Biomarker(s) to the “Safer and Faster Evidence-based Translation (SAFE-T) Consortium”, 2016, [Available from: https://www.fda.gov/media/99532/download].
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sd00111j |
| This journal is © The Royal Society of Chemistry 2022 |