Open Access Article
Open Access ArticleHighly bioresistant, hydrophilic and rigidly linked trityl-nitroxide biradicals for cellular high-field dynamic nuclear polarization†
Ru
Yao‡
 a,
David
Beriashvili‡
b,
Wenxiao
Zhang
a,
Shuai
Li
a,
Adil
Safeer
b,
Andrei
Gurinov
b,
Antal
Rockenbauer
c,
Yin
Yang
d,
Yuguang
Song
a,
David
Beriashvili‡
b,
Wenxiao
Zhang
a,
Shuai
Li
a,
Adil
Safeer
b,
Andrei
Gurinov
b,
Antal
Rockenbauer
c,
Yin
Yang
d,
Yuguang
Song
 a,
Marc
Baldus
a,
Marc
Baldus
 *b and
Yangping
Liu
*b and
Yangping
Liu
 *a
*a
aThe Province and Ministry Co-sponsored Collaborative Innovation Center for Medical Epigenetics, Tianjin Key Laboratory on Technologies Enabling Development of Clinical Therapeutics and Diagnostics, School of Pharmacy, Tianjin Medical University, Tianjin 300070. P. R. China. E-mail: liuyangping@tmu.edu.cn
bNMR Spectroscopy Group, Bijvoet Center for Biomolecular Research, Utrecht University, Padualaan 8, 3584 CH Utrecht, The Netherlands. E-mail: m.baldus@uu.nl
cInstitute of Materials and Environmental Chemistry, Hungarian Academy of Sciences And, Department of Physics, Budapest University of Technology and Economics, Budafoki Ut 8, 1111 Budapest, Hungary
dState Key Laboratory of Elemento-organic Chemistry, Collaborative Innovation Center of Chemical Science and Engineering, Nankai University, Tianjin 300071, China
First published on 17th November 2022
Abstract
Cellular dynamic nuclear polarization (DNP) has been an effective means of overcoming the intrinsic sensitivity limitations of solid-state nuclear magnetic resonance (ssNMR) spectroscopy, thus enabling atomic-level biomolecular characterization in native environments. Achieving DNP signal enhancement relies on doping biological preparations with biradical polarizing agents (PAs). Unfortunately, PA performance within cells is often limited by their sensitivity to the reductive nature of the cellular lumen. Herein, we report the synthesis and characterization of a highly bioresistant and hydrophilic PA (StaPol-1) comprising the trityl radical OX063 ligated to a gem-diethyl pyrroline nitroxide via a rigid piperazine linker. EPR experiments in the presence of reducing agents such as ascorbate and in HeLa cell lysates demonstrate the reduction resistance of StaPol-1. High DNP enhancements seen in small molecules, proteins and cell lysates at 18.8 T confirm that StaPol-1 is an excellent PA for DNP ssNMR investigations of biomolecular systems at high magnetic fields in reductive environments.
Introduction
Characterization of biomolecular structure and function within a native environment is highly desirable but technically challenging.1–3 While solution-state nuclear magnetic resonance (NMR) has been used to study small, fast-tumbling, and soluble proteins at atomic levels inside cells,1 solid-state NMR (ssNMR) provides an opportunity for in situ interrogation of large protein complexes and protein assemblies without the limitation of molecular tumbling times.3 However, cellular ssNMR approaches suffer from inherently low sensitivity making investigation of non-abundant biomolecules technically challenging.4–6 Coupling dynamic nuclear polarization (DNP7,8) with ssNMR overcomes this sensitivity limitation by transferring large electron spin polarization to NMR active nuclei.9–11 As a result, DNP-ssNMR has been utilized to study crude cell lysates,12,13 globular,10,14 and membrane15–17 proteins in their native settings as well as quantify drug concentrations directly within mammalian cells.18DNP-supported ssNMR experiments rely on tailored polarizing agents (PAs) that enhance the sensitivity of ssNMR by one to two orders of magnitude. However, development of PAs suitable for cellular DNP is still confronted with several challenges: (I) the unfavorable magnetic field dependence of many PAs which greatly reduces their DNP enhancements at high magnetic fields;19–21 (II) significantly lower DNP enhancements for biological macromolecules as compared to those for small molecules;10,12,22 (III) fast bioreduction of the currently available PAs in cellular environments.11,23–26 In the past years, intense efforts have been exerted to address the first two challenges and accordingly PAs with improved hydrophilicity and superior DNP performance at high magnetic fields (usually, ≥14.1 T) have been obtained. For example, water-soluble dinitroxide-based biradicals (e.g., AMUPol,27 AsymPolPOK28,29 and M-TinyPols30) and trityl-nitroxide (TN) hybrid biradicals (e.g., TEMTriPols31 and NATriPols22) have achieved relative DNP enhancements over 100 for [13C, 15N]-proline or 13C-urea, and ∼30 for soluble proteins in the DNP juice at 18.8 T. In particular, we have recently demonstrated that the hydrophilic TN biradical SNAPol-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 32 has outstanding DNP performance with enhancement factors of 133 for [13C, 15N]-proline, 100 for soluble proteins and 61 for membrane proteins at 18.8 T with favourable TB times. While these dinitroxide- and TN-based PAs are well suited for cellular DNP applications in which samples are rapidly prepared and cooled to cryogenic temperatures,10,32,33 their stability is compromised at ambient temperatures due to the cellular reducing environments.11,23,24,26 Thus, the above mentioned third challenge has not been well addressed yet. Especially, PAs with high biostability and excellent DNP performance for proteins at high magnetic fields have not been reported so far.
32 has outstanding DNP performance with enhancement factors of 133 for [13C, 15N]-proline, 100 for soluble proteins and 61 for membrane proteins at 18.8 T with favourable TB times. While these dinitroxide- and TN-based PAs are well suited for cellular DNP applications in which samples are rapidly prepared and cooled to cryogenic temperatures,10,32,33 their stability is compromised at ambient temperatures due to the cellular reducing environments.11,23,24,26 Thus, the above mentioned third challenge has not been well addressed yet. Especially, PAs with high biostability and excellent DNP performance for proteins at high magnetic fields have not been reported so far.
Nitroxide radicals used for synthesis of PAs are prone to bioreduction in cellular environments.11,23,24,26,34 Reduction of one of two spins in the biradical-based PAs inactivates cross-effect (CE) DNP which so far is the most effective DNP mechanism at high magnetic fields. The DNP enhancements of AMUPol which was electroporated to HEK293 cells decreased by more than 50% after incubation with the cells for 45 min at room temperature.26 Thus, attempts have been made in recent years to increase the intracellular stability of TOTAPol23 or AMUPol26 by addition of N-ethylmaleimide (NEM) to scavenge endogenous biothiols or regenerate TOTAPol by oxidation of its hydroxylamine forms with potassium ferricyanide before their potential use for in-cell DNP experiments. Although the addition of exogenous substances preserved nitroxides in cells, the stabilization of biradicals by these strategies did not translate into improved DNP performance.23,26 Alternatively, the use of the highly stable nitroxide radicals for construction of PAs presents itself as an attractive route to overcoming bioreduction. However, efforts on this front have so far been limited, possibly because neither relaxation time nor hydrophilicity are optimal for these stable nitroxides as compared to the spirocyclic nitroxides used in the previously reported PAs.32,34,35
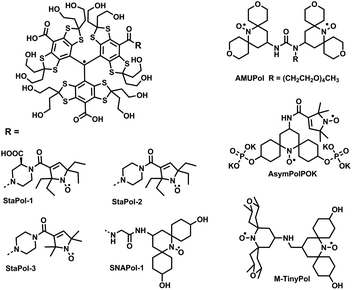
In the current study, we have synthesized a hydrophilic and highly stable TN biradical (StaPol-1, Fig. 1) and its analogue StaPol-2 in which the trityl radical OX063 is covalently conjugated with the highly stable gem-diethyl pyrroline nitroxide36 through rigid piperazine linkers. Both StaPol-1 and StaPol-2 are extraordinarily stable towards reducing agents such as ascorbic acid and in the lysate of HeLa cells. They also exhibit high DNP enhancements for [13C, 15N]-proline (117 for StaPol-1 and 84 for StaPol-2) and [13C, 15N]-ubiquitin (117 for StaPol-1) at 18.8 T. Furthermore, the high biostability and excellent DNP performance of StaPol-1 allowed us to record 1H–13C cross-polarization DNP ssNMR data on [13C,15N]-Ubiquitin in HeLa cells and cell lysates with DNP enhancements of 50 and 183 on aliphatic carbons, respectively.
Results and discussion
Rational design of stable hydrid biradical StaPol-1
In contrast to nitroxides which are mostly prone to bioreduction, trityl radicals are almost resistant to biological reducing agents such as ascorbic acid and exhibit high stability in cellular environments.37–39 Thus, the nitroxide part is key for development of stable TN-based PAs for cellular DNP. In this study, a gem-diethyl pyrroline nitroxide was used which exhibits extraordinarily high stability towards bioreduction.36 The use of the hydrophilic trityl radical OX063 instead of CT-03 prevents self-aggregation of PAs and is beneficial for DNP as demonstrated by our recent work.22Besides the radical moieties, the linker is also critical. The piperazine linker used in this study exhibits the following features: (1) similar to the amino acid linker used for SNAPols,32 TEMTriPols31,40 and NATriPols,22 the non-conjugated piperazine maintains moderate spin–spin interactions which are mandated for CE DNP. (2) The rigid piperazine linker potentially provides a narrow conformer distribution which is beneficial for DNP as shown by AsymPolPok28,29 and M-TinyPol.30 The presence of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bond in the pyrroline nitroxide further increases the linker rigidity.28,29 Moreover, the additional carboxylate group in StaPol-1 can further increase its hydrophilicity, thus improving its DNP performance, especially on proteins.22 Overall, the present molecular design not only enhances cellular stability of TN-based PAs but also takes into account other physicochemical properties such as hydrophilicity and spin–spin interactions which are critical for their DNP performance at high magentic fields.
C bond in the pyrroline nitroxide further increases the linker rigidity.28,29 Moreover, the additional carboxylate group in StaPol-1 can further increase its hydrophilicity, thus improving its DNP performance, especially on proteins.22 Overall, the present molecular design not only enhances cellular stability of TN-based PAs but also takes into account other physicochemical properties such as hydrophilicity and spin–spin interactions which are critical for their DNP performance at high magentic fields.
Synthesis
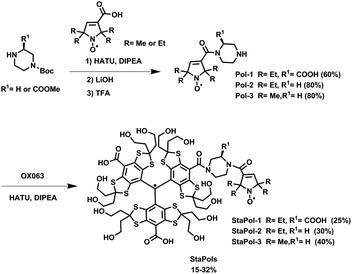
StaPol-1 and StaPol-2 which contain the same gem-diethyl pyrroline nitroxide were synthesized using our previously reported method with moderate modifications (see ESI†).32 For comparison, StaPol-3 with a gem-dimethyl pyrroline nitroxide (Fig. 1) was also synthesized. Two successive amide condensation reactions from the nitroxides, 4-Boc piperazine and OX063 conveniently afforded the TN biradicals as green solids in total yields of 15–32% over two steps (see Scheme 1). Their purity was determined by HPLC to be >97% and their molecular structures were confirmed by EPR and high-resolution mass spectrometry (see ESI†) (Table 1).| Polarizing agent | RT (min) | 13C εon/off | T B (s) | T 1 (s) |
|---|---|---|---|---|
| StaPol-1 | 10.9 | 117 ± 0.9 | 7.13 | 7.83 |
| StaPol-2 | 15.5 | 84 ± 0.75 | 3.68 | 4.10 |
| StaPol-3 | 11.2 | 105 ± 1.27 | 3.50 | 4.03 |
| SNAPol-1* | 7.6 | 133 | 4.20 | 4.20 |
DNP enhancement for [13C–15N] proline
With these stable PAs in hand, we first tested their DNP performance by 1H–13C cross polarization experiments using a sample containing 0.25 M [13C–15N] proline in “DNP juice” (d8-glycerol/D2O/H2O, 60/30/10, v/v/v) doped with 10 mM biradical in a 3.2 mm sapphire rotor at 18.8 T, 8 kHz MAS rate and 95 K. As shown in Table S7,† these PAs exhibit excellent DNP enhancements with εon/off values of 84 for StaPol-2, 105 for StaPol-3 and 117 for StaPol-1 which are significantly higher than TEMTriPol-1 (50) under similar experimental conditions.31 Especially, StaPol-1 has a comparable DNP performance as compared to SNAPol-1, the best preforming molecule at high field magnetic fields at present.32Hydrophilicity
For further analysis of DNP performance of StaPol-1, we measured its hydrophilicity which has been confirmed by us as a key factor controlling DNP properties of TN biradicals in the DNP juice.22 Since these PAs are almost insoluble in n-octanol, their octanol–water partition coefficients (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) P) were not measurable. As such, the HPLC method on a reverse-phase C18 column was used to evaluate their hydrophilicity.22,32,41 Due to the structural similarity of PAs used in this study, this HPLC approach can be utilized to reliably evaluate their relative hydrophilicity. StaPol-1 with a negatively-charged carboxylate group on the piperazine linker is more hydrophilic than StaPol-2 as evidenced by their retention times (RT) of 10.9 and 15.5 min, respectively, under the same HPLC conditions. Moreover, the four ethyl groups in StaPol-2 also make it less hydrophilic than StaPol-3 (RT = 11.2 min) with four methyl groups. Thus, hydrophilicity could mainly account for the excellent DNP enhancement of StaPol-1 for proline (εon/off = 117) as compared to StaPol-2 (84) since both have almost identical magnetic interactions as shown below. As observed in our recent study,22 the relatively low hydrophilicity induced the self-aggregation of TN biradical-based PAs and their inhomogeneous dispersion in the matrix which was detrimental to their DNP performance.42,43 Moreover, the self-aggregation of PAs may also accelerate their electron relaxation rates due to intermolecular spin–spin interactions and decrease their solvent accessibility,34 thus attenuating their DNP enhancements. As for StaPol-3 which showed very similar hydrophilicity to StaPol-1, the slightly smaller value (εon/off = 105) of StaPol-3 than that of StaPol-1 is possibly due to the relatively faster relaxation of gem-dimethyl pyrroline nitroxide than its gem-diethyl analogue.44 Previous studies consistently showed that slowing the relaxation of nitroxide radicals down improves the DNP performance of both dinitroxide- and TN-based PAs.32,34,42,45,46
P) were not measurable. As such, the HPLC method on a reverse-phase C18 column was used to evaluate their hydrophilicity.22,32,41 Due to the structural similarity of PAs used in this study, this HPLC approach can be utilized to reliably evaluate their relative hydrophilicity. StaPol-1 with a negatively-charged carboxylate group on the piperazine linker is more hydrophilic than StaPol-2 as evidenced by their retention times (RT) of 10.9 and 15.5 min, respectively, under the same HPLC conditions. Moreover, the four ethyl groups in StaPol-2 also make it less hydrophilic than StaPol-3 (RT = 11.2 min) with four methyl groups. Thus, hydrophilicity could mainly account for the excellent DNP enhancement of StaPol-1 for proline (εon/off = 117) as compared to StaPol-2 (84) since both have almost identical magnetic interactions as shown below. As observed in our recent study,22 the relatively low hydrophilicity induced the self-aggregation of TN biradical-based PAs and their inhomogeneous dispersion in the matrix which was detrimental to their DNP performance.42,43 Moreover, the self-aggregation of PAs may also accelerate their electron relaxation rates due to intermolecular spin–spin interactions and decrease their solvent accessibility,34 thus attenuating their DNP enhancements. As for StaPol-3 which showed very similar hydrophilicity to StaPol-1, the slightly smaller value (εon/off = 105) of StaPol-3 than that of StaPol-1 is possibly due to the relatively faster relaxation of gem-dimethyl pyrroline nitroxide than its gem-diethyl analogue.44 Previous studies consistently showed that slowing the relaxation of nitroxide radicals down improves the DNP performance of both dinitroxide- and TN-based PAs.32,34,42,45,46
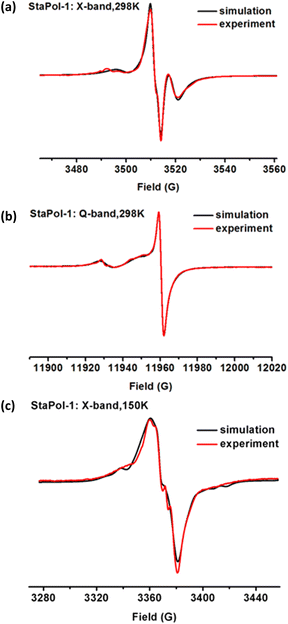
Magnetic interactions of StaPol-1 and StaPol-2
The electron–electron dipolar/exchange interactions play important roles in the high-field DNP performance of biradical-based PAs and are strongly dependent on the linkers used.28,30,31,47 To reveal the effect of the piperazine linker, we examined the magnetic interactions of StaPol-1 by EPR.Since the dipolar interaction (D) is averaged out at ambient temperature due to fast molecular tumbling, the exchange interaction (J) can be measured. Fig. 2a shows the X-band EPR spectrum of StaPol-1 in phosphate buffer (20 mM, pH 7.4) at room temperature. Spectral simulation indicates that StaPol-1 has only one stable conformer with a J value of 14.6 G at room temperature due to the rigid piperazine linker. To examine whether the relatively broad EPR lines hide the presence of two conformers in StaPol-1, we collected its Q-band EPR spectrum (Fig. 2b) since EPR lines at this band are better separated.48 One conformer was consistently observed at Q-band EPR with the similar J value (10.5 G) to that at X-band. Similar results were also observed for StaPol-2 which also has one conformer with J values of 11.2 G at X-band and 7.9 G at Q-band (Fig. S4, S5 and Tables S3, S4†). In contrast, two stable conformers were observed for SNAPol-1 with a glycine linker at Q-band (Table S4†). Similar results were also reported for the other TN biradicals with the relatively flexible amino acid linkers.21,47,48
Next, EPR spectra of both StaPol-1 and StaPol-2 were also recorded in the DNP juice at 150 K (Fig. 2c and S6†). Computer simulation shows that both of them have similar exchange and dipolar interactions (StaPol-1, J = 23 G and D = 9 G; StaPol-2, J = 20 G and D = 8 G). These moderate magnetic interactions are almost optimal for high-field DNP at 18.8 T.21,22,32 Thus, when using piperazine as linker, the optimal magnetic interactions can be also established for TN-based PAs by selecting 3-COOH pyrroline nitroxide (Fig. 1). In contrast, in the cases of TEMTriPols, NATriPols and SNAPols, 4-NH2 piperidine nitroxide was matched with amino acid linkers.
Stability towards ascorbic acid and in cell lysates
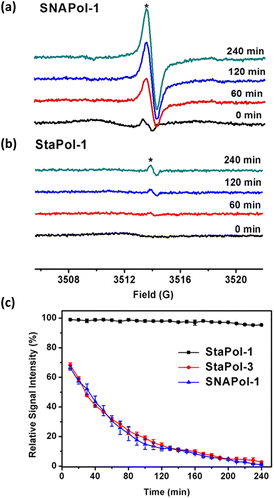
Despite growing interest in cellular DNP, the stability of PAs in cellular environments has not been well addressed. Nitroxide radicals used for synthesis of the currently available PAs are prone to bioreduction,34,38 while trityl radicals are relatively stable.37–39 Therefore, monitoring the signal of the trityl radical formed from reduction of the TN biradical was initially used to measure redox status and oxygenation.49,50Here we examined the stability of TN biradicals (StaPol-1, StaPol-2, StaPol-3 and SNAPol-1) to ascorbic acid and HeLa cell lysates, in which various nitroxides were conjugated with OX063. The reduction of both StaPol-3 and SNAPol-1 by ascorbate in PBS (20 mM, pH 7.4) led to gradual enhancement of EPR singlet signal due to the corresponding trityl radical, while the broad signal of the biradicals decreased over time (Fig. S18†). However, StaPol-1 and StaPol-2 with the stable nitroxide were almost inert to ascorbate with appearance of the very weak singlet signal. The second-order rate constants of StaPol-3 and SNAPol-1 with ascorbate were determined to be 1.3 ± 0.1 and 12.4 ± 0.5 M−1 s−1, respectively (see Fig. S21 and S24†). The almost 10 times higher reactivity of SNAPol-1 than StaPol-3 is consistent with the reactivity difference from the corresponding nitroxide radicals.38,51
Next, we explored the stability of the four TN biradicals in HeLa cell lysates by EPR. As shown in Fig. 3a and S25,† incubation of either StaPol-3 or SNAPol-1 with the cell lysates similarly led to the intense singlet EPR signals, whereas only a very small fraction of the singlet signal (∼3%) was observed for both StaPol-1 and StaPol-2 after 240 min incubation in the cell lysates (Fig. 3b and S25†). Fig. 3c shows their reduction kinetics in the cell lysates. Based on these data, the half-lives (t1/2) of StaPol-3 and SNAPol-1 were estimated to be t1/2 = 41.8 ± 1.4 and 66.0 ± 1.4 min, respectively (Fig. S26†). The smaller difference in the stability of StaPol-3 and SNAPol-1 in the cell lysates than that in the ascorbate solution may be due to the bulkiness of the hydrophilic spirocyclic nitroxide in SNAPol-1 which prevents its enzymatic reduction in the cell lysates.52
Taken together, the newly synthesized StaPol-1 and its analogue StaPol-2 both of which have the same nitroxide and trityl parts are highly stable in reducing environments, while the other TN biradicals tested in this study are sensitive to bioreduction.
Dynamic nuclear polarization studies on [13C–15N] ubiquitin in vitro, inside HeLa cells, and cell lysates
Considering that StaPol-1 is highly hydrophilic, exhibits excellent DNP enhancement for [13C,15N]-proline, and high stability in reducing environments, we investigated its DNP performance in different biological settings. Initially, 13C enhancements were determined for in vitro [13C,15N]-ubiquitin (Fig. 4); at 30 mM StaPol-1 and its parent compound SNAPol-1 exhibited enhancements of 105 and 110 (judged on the carbonyl signal, Fig. 4a) with TB values of 5.47 s and 4.35 s (Table 2).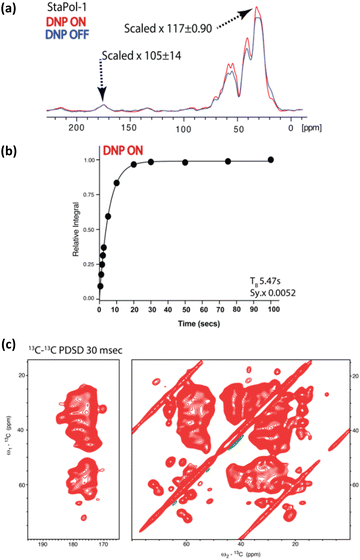 | ||
| Fig. 4 DNP performance of StaPol-1 on [13C–15N] ubiquitin at 800 MHz, 8 kHz, and 95 K. (a) 1H–13C cross-polarization on [13C–15N] ubiquitin in the DNP juice with/without DNP; enhancements on aliphatic and carbonyl signals. See Materials and methods in the ESI† for details of the error analysis. (b) TB was determined through saturation recovery 1H–13C cross-polarization experiments. Due to high NMR signal sensitivity, error bars are not visible and instead detailed in Table S14.† Sy.x denotes the s.d of the residual least squares. (c) Aliphatic and carbonyl cutouts from 13C–13C proton driven spin diffusion (PDSD) experiment with a 30 ms mixing conducted on [13C–15N] ubiquitin. | ||
| Polarizing agent | Sample | 13C εon/off | T B (s) |
|---|---|---|---|
| a Carbonyl (170–180 ppm). b Aliphatic (0–70 ppm). c Aliphatic (50–60 ppm). d Aliphatic (41 ppm). See Materials and methods in ESI for details of the error analysis. | |||
| StaPol-1 | In vitro | 105 ± 14a/117 ± 0.9b | 5.47a |
| SNAPol-1* | In vitro | 110a | 4.35a |
| StaPol-1 | In cell | 50 ± 1.65b | 2.4/24.5a |
| SNAPol-1** | In cell | 55b | 1.4/25.5a |
| StaPol-1 | Lysate | 183 ± 4.99c,d | 1.2/5.6d |
| SNAPol-1 | Lysate | 68 ± 5.20c/150 ± 1.96d | 0.34/4.3d |
With these encouraging in vitro results, StaPol-1 and SNAPol-1 were delivered by passive diffusion to HeLa cells containing endogenous concentrations (80 μM) of [13C,15N]-ubiquitin transferred via electroporation as described in the ref. 10. 1H–13C spectra revealed similar enhancements of roughly 50 times in cell for both PAs with polarization build up curves best fit by a biexponential (Fig. 5a and b). Taken together with the rapid way in which cell samples were prepared (less than 5 minutes between PA addition and plunge freezing), this result suggests that PA reduction does not play a determining role in modulating PA performance in cells.11
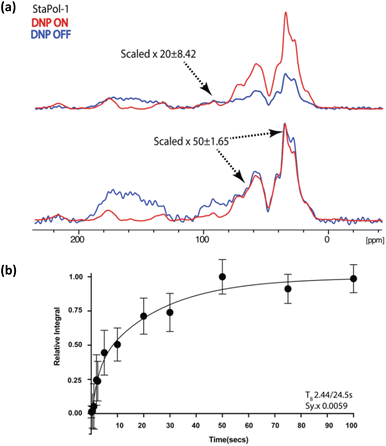 | ||
| Fig. 5 (a) 1D [1H–13C] cross polarization of HeLa cells containing electroporated [13C, 15N] ubiquitin doped with 30 mM StaPol-1. See ESI† for determination of enhancement error. (b) TB determined through saturation recovery 1H–13C cross-polarization experiments on the carbonyl signal. Error bars denote the standard deviation of the signal to noise. Sy.x denotes the s.d of the residual least squares. In all cases, the DNP OFF spectrum was phased to resemble the aliphatic region of DNP ON spectra for enhancement analysis. We attribute DNP OFF signal between 120 and 200 ppm to natural abundance signals (possibly from nuclei acids, see ref. 9) and to baseline imperfections. For scaling of these imperfections see (a). | ||
Next, the performance of StaPol-1 and SNAPol-1 was assayed in HeLa cell lysates (Fig. 6). HeLa cells were electroporated with [13C,15N]-ubiquitin as detailed above, lysed by passing through a 23 G needle, and finally incubated with 30 mM of either PA for 1.5 h at ambient temperature (Fig. 6a). In both cases, the resulting 1H–13C CP experiments had a strong non-ubiquitin signal centered at 41 ppm (Fig. 6b). The near disappearance of this signal in a double quantum single quantum (DQSQ)53 filtered experiment (Fig. S29†) would be consistent with natural abundance lipid tail signals seen in the CP experiment. We attribute the occurrence of these signals to the details of our lysis procedure. Briefly, shearing cells creates membrane vesicles (see, e.g. ref. 16) that could permit for PA intercalation at the highly polar membrane interface and possibly even encapsulation54 of the PA. Both effects will likely result in increased lipid signal enhancements.
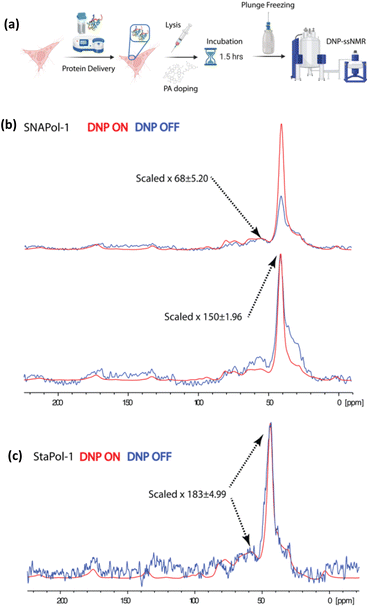 | ||
| Fig. 6 (a) Scheme showing HeLa cell lysate preparation. (b and c) 1D 1H–13C cross polarization of HeLa cell lysates doped with 30 mM SNAPol-1 (b) or 30 mM StaPol-1 (c). Enhancement errors are calculated as detailed in the section of Materials and methods of the ESI.† | ||
For SNAPol-1, the observed reduction in aliphatic DNP enhancements corroborates its increased susceptibility to bioreduction as compared to StaPol-1 as mentioned above. This notion would be in line with literature showing that the majority of ubiquitin is not membrane associated in cells.55 On the other hand, the almost constant enhancement of the lipid chain signals could point to a membrane-associated population of the PA in which reduction is less efficient due to steric hindrance effects.
Conclusions
In summary, we have introduced a new molecular design of PAs which exhibit extraordinarily high resistance to bioreduction, high hydrophilicity and excellent DNP performance at 18.8 T. StaPol-1, the best one among them, has achieved a uniform DNP enhancement of 183 at 18.8 T for [13C,15N]-ubiquitin in HeLa cell lysates even after an extended incubation time (1.5 h) at room temperature. The similar and moderate enhancements (∼50 times) of StaPol-1 and SNAPol-1 for [13C,15N]-ubiquitin in HeLa cells indicate that the efficiency of cellular uptake of the two PAs needs to be further optimized, possibly through direct electroporation25 or covalent linkage to cell permeable peptides.11To the best of our knowledge, StaPol-1 represents the first PA that combines excellent performance at high magnetic fields with long-term stability in cellular settings, making it highly attractive for a broad range of cellular DNP-based ssNMR applications. Our present work advances our understanding of TN biradical-based polarizing agents and provides new routes for optimization of high-field polarizing agents for biomolecular applications,56,57 particularly in bioreductive environments.
Data availability
Data supporting the findings of this study are available within the paper and the ESI,† as well as from the corresponding authors upon reasonable request.Author contributions
R. Y., D. B., Y. G. S., M. B. and Y. P. L. designed experiments, analyzed data, and wrote the manuscript. R. Y., W. X. Z. and S. L. conducted synthesis of StaPols and characterization of physicochemical properties. R. Y. and A. R. performed EPR spectral simulation. Y. Y. collected Q-band EPR spectra. A. S. assisted in Ub production. D. B. and A. G. conducted DNP experiments. All authors reviewed the manuscript and agreed to its publication.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded in part by the National Natural Science Foundation of China (No. 22174099, 21871210 and 21572161 to Y. P. L.; No. 31500684 to Y. G. S.), the Dutch Research Council (NWO) (No. 700.26.121, 700.10.443 and 718.015.001, and ECHO.018.067 to M. B.) and by uNMR-NL, the National Roadmap Large-Scale NMR Facility of the Netherlands (grant 184.032.207), Science & Technology Projects of Tianjin (No. 20JCZDJC00050 to Y. P. L.) and Hefei National Laboratory for Physical Sciences at the Microscale (KF2020001).Notes and references
- E. Luchinat and L. Banci, J. Biol. Chem., 2016, 291, 3776–3784 CrossRef CAS PubMed.
- F. X. Theillet, A. Binolfi, B. Bekei, A. Martorana, H. M. Rose, M. Stuiver, S. Verzini, D. Lorenz, M. van Rossum, D. Goldfarb and P. Selenko, Nature, 2016, 530, 45–50 CrossRef CAS PubMed.
- S. Narasimhan, C. Pinto, A. L. Paioni, J. van der Zwan, G. E. Folkers and M. Baldus, Nat. Protoc., 2021, 16, 893–918 CrossRef CAS PubMed.
- L. Banci, L. Barbieri, I. Bertini, E. Luchinat, E. Secci, Y. Zhao and A. R. Aricescu, Nat. Chem. Biol., 2013, 9, 297–299 CrossRef CAS PubMed.
- M. Renault, R. Tommassen-van Boxtel, M. P. Bos, J. A. Post, J. Tommassen and M. Baldus, Proc. Natl. Acad. Sci. U. S. A, 2012, 109, 4863–4868 CrossRef CAS PubMed.
- K. K. Frederick, V. K. Michaelis, B. Corzilius, T. C. Ong, A. C. Jacavone, R. G. Griffin and S. Lindquist, Cell, 2015, 163, 620–628 CrossRef CAS PubMed.
- Q. Z. Ni, E. Daviso, T. V. Can, E. Markhasin, S. K. Jawla, T. M. Swager, R. J. Temkin, J. Herzfeld and R. G. Griffin, Acc. Chem. Res., 2013, 46, 1933–1941 CrossRef CAS PubMed.
- D. A. Hall, D. C. Maus, G. J. Gerfen, S. J. Inati, L. R. Becerra, F. W. Dahlquist and R. G. Griffin, Science, 1997, 276, 930–932 CrossRef CAS PubMed.
- M. Renault, S. Pawsey, M. P. Bos, E. J. Koers, D. Nand, R. Tommassen-van Boxtel, M. Rosay, J. Tommassen, W. E. Maas and M. Baldus, Angew. Chem., Int. Ed., 2012, 51, 2998–3001 CrossRef CAS PubMed.
- S. Narasimhan, S. Scherpe, A. L. Paioni, J. van der Zwan, G. E. Folkers, H. Ovaa and M. Baldus, Angew. Chem., Int. Ed., 2019, 58, 12969–12973 CrossRef CAS PubMed.
- B. J. Albert, C. Gao, E. L. Sesti, E. P. Saliba, N. Alaniva, F. J. Scott, S. T. Sigurdsson and A. B. Barnes, Biochemistry, 2018, 57, 4741–4746 CrossRef CAS PubMed.
- T. Viennet, A. Viegas, A. Kuepper, S. Arens, V. Gelev, O. Petrov, T. N. Grossmann, H. Heise and M. Etzkorn, Angew. Chem., Int. Ed., 2016, 55, 10746–10750 CrossRef CAS PubMed.
- P. T. Judge, E. L. Sesti, L. E. Price, B. J. Albert, N. Alaniva, E. P. Saliba, T. Halbritter, S. T. Sigurdsson, G. B. Kyei and A. B. Barnes, J. Phys. Chem. B, 2020, 124, 2323–2330 CrossRef CAS PubMed.
- S. Chordia, S. Narasimhan, A. Lucini Paioni, M. Baldus and G. Roelfes, Angew. Chem., Int. Ed., 2021, 60, 5913–5920 CrossRef CAS PubMed.
- M. Kaplan, A. Cukkemane, G. C. van Zundert, S. Narasimhan, M. Daniels, D. Mance, G. Waksman, A. M. Bonvin, R. Fronzes, G. E. Folkers and M. Baldus, Nat. Methods, 2015, 12, 649–652 CrossRef CAS PubMed.
- M. Kaplan, S. Narasimhan, C. de Heus, D. Mance, S. van Doorn, K. Houben, D. Popov-Celeketic, R. Damman, E. A. Katrukha, P. Jain, W. J. C. Geerts, A. J. R. Heck, G. E. Folkers, L. C. Kapitein, S. Lemeer, P. M. P. van Bergen En Henegouwen and M. Baldus, Cell, 2016, 167, 1241–1251 CrossRef CAS PubMed.
- M. R. Elkins, I. V. Sergeyev and M. Hong, J. Am. Chem. Soc., 2018, 140, 15437–15449 CrossRef CAS PubMed.
- A. Bertarello, P. Berruyer, M. Artelsmair, C. S. Elmore, S. Heydarkhan-Hagvall, M. Schade, E. Chiarparin, S. Schantz and L. Emsley, J. Am. Chem. Soc., 2022, 144, 6734–6741 CrossRef CAS PubMed.
- Y. Hovav, I. Kaminker, D. Shimon, A. Feintuch, D. Goldfarb and S. Vega, Phys. Chem. Chem. Phys., 2015, 17, 226 RSC.
- D. Mance, P. Gast, M. Huber, M. Baldus and K. L. Ivanov, J. Chem. Phys., 2015, 142, 234201 CrossRef PubMed.
- F. Mentink-Vigier, G. Mathies, Y. P. Liu, A. L. Barra, M. A. Caporini, D. Lee, S. Hediger, R. G. Griffin and G. De Paepe, Chem. Sci., 2017, 8, 8150–8163 RSC.
- W. X. Zhai, A. L. Paioni, X. Y. Cai, S. Narasimhan, J. Medeiros-Silva, W. X. Zhang, A. Rockenbauer, M. Weingarth, Y. G. Song, M. Baldus and Y. P. Liu, J. Phys. Chem. B, 2020, 124, 9047–9060 CrossRef CAS PubMed.
- K. M. McCoy, R. Rogawski, O. Stovicek and A. E. McDermott, J. Magn. Reson., 2019, 303, 115–120 CrossRef CAS PubMed.
- S. A. Overall, L. E. Price, B. J. Albert, C. Gao, N. Alaniva, P. T. Judge, E. L. Sesti, P. A. Wender, G. B. Kyei and A. B. Barnes, Int. J. Mol. Sci., 2020, 21, 4649 CrossRef CAS PubMed.
- R. Ghosh, Y. L. Xiao, J. Kragelj and K. K. Frederick, J. Am. Chem. Soc., 2021, 143, 18454–18466 CrossRef CAS PubMed.
- R. Ghosh, R. Dumarieh, Y. Xiao and K. K. Frederick, J. Magn. Reson., 2022, 336, 107150 CrossRef CAS PubMed.
- C. Sauvee, M. Rosay, G. Casano, F. Aussenac, R. T. Weber, O. Ouari and P. Tordo, Angew. Chem., Int. Ed., 2013, 52, 10858–10861 CrossRef CAS PubMed.
- F. Mentink-Vigier, I. Marin-Montesinos, A. P. Jagtap, T. Halbritter, J. van Tol, S. Hediger, D. Lee, S. T. Sigurdsson and G. De Paepe, J. Am. Chem. Soc., 2018, 140, 11013–11019 CrossRef CAS PubMed.
- R. Harrabi, T. Halbritter, F. Aussenac, O. Dakhlaoui, J. van Tol, K. K. Damodaran, D. Lee, S. Paul, S. Hediger, F. Mentink-Vigier, S. T. Sigurdsson and G. De Paepe, Angew. Chem., Int. Ed., 2022, 61, e202114103 CrossRef CAS PubMed.
- A. Lund, G. Casano, G. Menzildjian, M. Kaushik, G. Stevanato, M. Yulikov, R. Jabbour, D. Wisser, M. Renom-Carrasco, C. Thieuleux, F. Bernada, H. Karoui, D. Siri, M. Rosay, I. V. Sergeyev, D. Gajan, M. Lelli, L. Emsley, O. Ouari and A. Lesage, Chem. Sci., 2020, 11, 2810–2818 RSC.
- G. Mathies, M. A. Caporin, V. K. Michaelis, Y. P. Liu, K. N. Hu, D. Mance, J. L. Zweier, M. Rosay, M. Baldus and R. G. Griffin, Angew. Chem., Int. Ed., 2015, 54, 11770–11774 CrossRef CAS PubMed.
- X. Y. Cai, A. L. Paioni, A. Adler, R. Yao, W. X. Zhang, D. Beriashvili, A. Safeer, A. Gurinov, A. Rockenbauer, Y. G. Song, M. Baldus and Y. P. Liu, Chem.–Eur. J., 2021, 27, 12758–12762 CrossRef CAS PubMed.
- P. T. Judge, E. L. Sesti, N. Alaniva, E. P. Saliba, L. E. Price, C. Gao, T. Halbritter, S. T. Sigurdsson, G. B. Kyei and A. B. Barnes, J. Magn. Reson., 2020, 313, 106702 CrossRef CAS PubMed.
- G. Stevanato, G. Casano, D. J. Kubicki, Y. Rao, L. E. Hofer, G. Menzildjian, H. Karoui, D. Siri, M. Cordova, M. Yulikov, G. Jeschke, M. Lelli, A. Lesage, O. Ouari and L. Emsley, J. Am. Chem. Soc., 2020, 142, 16587–16599 CrossRef CAS PubMed.
- A. P. Jagtap, M. A. Geiger, D. Stoppler, M. Orwick-Rydmark, H. Oschkinat and S. T. Sigurdsson, Chem. Commun., 2016, 52, 7020–7023 RSC.
- Y. Wang, J. T. Paletta, K. Berg, E. Reinhart, S. Rajca and A. Rajca, Org. Lett., 2014, 16, 5298–5300 CrossRef CAS PubMed.
- Y. P. Liu, F. A. Villamena and J. L. Zweier, Chem. Commun., 2008, 46, 4336–4338 RSC.
- A. P. Jagtap, I. Krstic, N. C. Kunjir, R. Hansel, T. F. Prisner and S. T. Sigurdsson, Free Radical Res., 2015, 49, 78–85 CrossRef CAS PubMed.
- N. Fleck, C. Heubach, T. Hett, S. Spicher, S. Grimme and O. Schiemann, Chem. - Eur. J., 2021, 27, 5292–5297 CrossRef CAS PubMed.
- Y. P. Liu, F. A. Villamena, A. Rockenbauer, Y. G. Song and J. L. Zweier, J. Am. Chem. Soc., 2013, 135, 2350–2356 CrossRef CAS PubMed.
- C. Rizzi, S. Marque, F. Belin, J. C. Bouteiller, R. Lauricella, B. Tuccio, V. Cerri and P. Tordo, J. Chem. Soc., 1997, 12, 2513–2518 Search PubMed.
- A. Zagdoun, G. Casano, O. Ouari, M. Schwarzwalder, A. J. Rossini, F. Aussenac, M. Yulikov, G. Jeschke, C. Coperet, A. Lesage, P. Tordo and L. Emsley, J. Am. Chem. Soc., 2013, 135, 12790–12797 CrossRef CAS PubMed.
- A. B. Barnes, G. D. Paepe, P. C. van der Wel, K. N. Hu, C. G. Joo, V. S. Bajaj, M. L. Mak-Jurkauskas, J. R. Sirigiri, J. Herzfeld, R. J. Temkin and R. G. Griffin, Appl. Magn. Reson., 2008, 34, 237–263 CrossRef CAS PubMed.
- A. A. Kuzhelev, R. K. Strizhakov, O. A. Krumkacheva, Y. F. Polienko, D. A. Morozov, G. Y. Shevelev, D. V. Pyshnyi, I. A. Kirilyuk, M. V. Fedin and E. G. Bagryanskaya, J. Magn. Reson., 2016, 266, 1–7 CrossRef CAS PubMed.
- A. Zagdoun, G. Casano, O. Ouari, G. Lapadula, A. J. Rossini, M. Lelli, M. Baffert, D. Gajan, L. Veyre, W. E. Maas, M. Rosay, R. T. Weber, C. Thieuleux, C. Coperet, A. Lesage, P. Tordo and L. Emsley, J. Am. Chem. Soc., 2012, 134, 2284–2291 CrossRef CAS PubMed.
- D. Wisser, G. Karthikeyan, A. Lund, G. Casano, H. Karoui, M. Yulikov, G. Menzildjian, A. C. Pinon, A. Purea, F. Engelke, S. R. Chaudhari, D. Kubicki, A. J. Rossini, I. B. Moroz, D. Gajan, C. Coperet, G. Jeschke, M. Lelli, L. Emsley, A. Lesage and O. Ouari, J. Am. Chem. Soc., 2018, 140, 13340–13349 CrossRef CAS PubMed.
- W. X. Zhai, Y. L. Feng, H. Q. Liu, A. Rockenbauer, D. Mance, S. Y. Li, Y. G. Song, M. Baldus and Y. P. Liu, Chem. Sci., 2018, 9, 4381–4391 RSC.
- W. Moore, R. Yao, Y. P. Liu, S. S. Eaton and G. R. Eaton, J. Magn. Reson., 2021, 332, 107078 CrossRef CAS PubMed.
- Y. P. Liu, F. A. Villamena, A. Rockenbauer and J. L. Zweier, Chem. Commun., 2010, 46, 628–630 RSC.
- Y. P. Liu, F. A. Villamena, Y. G. Song, J. Sun, A. Rockenbauer and J. L. Zweier, J. Org. Chem., 2010, 75, 7796–7802 CrossRef CAS PubMed.
- S. Huang, H. Zhang, J. T. Paletta, S. Rajca and A. Rajca, Free Radical Res., 2018, 52, 327–334 CrossRef CAS PubMed.
- S. Okazaki, M. A. Mannan, K. Sawai, T. Masumizu, Y. Miura and K. Takeshita, Free Radical Res., 2007, 41, 1069–1077 CrossRef CAS PubMed.
- M. Hohwy, C. M. Rienstra, C. P. Jaroniec and R. G. Griffin, J. Chem. Phys., 1999, 110, 7983–7992 CrossRef CAS.
- C. Fernandez-de-Alba, H. Takahashi, A. Richard, Y. Chenavier, L. Dubois, V. Maurel, D. Lee, S. Hediger and G. De Paepe, Chem.–Eur. J., 2015, 21, 4512–4517 CrossRef CAS PubMed.
- S. E. Kaiser, B. E. Riley, T. A. Shaler, R. S. Trevino, C. H. Becker, H. Schulman and R. R. Kopito, Nat. Methods, 2011, 8, 691–696 CrossRef CAS PubMed.
- T. Biedenbander, V. Aladin, S. Saeidpour and B. Corzilius, Chem. Rev., 2022, 122, 9738–9794 CrossRef PubMed.
- W. Y. Chow, G. De Paepe and S. Hediger, Chem. Rev., 2022, 122, 9795–9847 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthesis, characterization and experimental details. See DOI: https://doi.org/10.1039/d2sc04668g |
| ‡ Equal first author contribution. |
| This journal is © The Royal Society of Chemistry 2022 |