 Open Access Article
Open Access ArticleThe sequestration mechanism as a generalizable approach to improve the sensitivity of biosensors and bioassays†
Alejandro
Chamorro-Garcia
 ab,
Claudio
Parolo
ab,
Claudio
Parolo
 c,
Gabriel
Ortega
de,
Andrea
Idili
c,
Gabriel
Ortega
de,
Andrea
Idili
 b,
Joshua
Green
a,
Francesco
Ricci
b,
Joshua
Green
a,
Francesco
Ricci
 b and
Kevin W.
Plaxco
b and
Kevin W.
Plaxco
 *a
*a
aDepartment of Chemistry and Biochemistry University of California Santa Barbara (UCSB), Santa Barbara, CA 93106, USA. E-mail: kpw@ucsb.edu
bDipartimento di Scienze e Tecnologie Chimiche, University of Rome, Tor Vergata, Via della Ricerca Scientifica, 00133 Rome, Italy
cISGlobal–Barcelona Institute for Global Health, Carrer del Rosselló 132, 08036 Barcelona, Spain
dIkerbasque, Basque Foundation for Science, 48013 Bilbao, Spain
ePrecision Medicine and Metabolism Laboratory, CIC BioGUNE, Basque Research and Technology Alliance, Parque Tecnológico de Bizkaia, 48160 Derio, Spain
First published on 23rd September 2022
Abstract
Biosensors and bioassays, both of which employ proteins and nucleic acids to detect specific molecular targets, have seen significant applications in both biomedical research and clinical practice. This success is largely due to the extraordinary versatility, affinity, and specificity of biomolecular recognition. Nevertheless, these receptors suffer from an inherent limitation: single, saturable binding sites exhibit a hyperbolic relationship (the “Langmuir isotherm”) between target concentration and receptor occupancy, which in turn limits the sensitivity of these technologies to small variations in target concentration. To overcome this and generate more responsive biosensors and bioassays, here we have used the sequestration mechanism to improve the steepness of the input/output curves of several bioanalytical methods. As our test bed for this we employed sensors and assays against neutrophil gelatinase-associated lipocalin (NGAL), a kidney biomarker for which enhanced sensitivity will improve the monitoring of kidney injury. Specifically, by introducing sequestration we have improved the responsiveness of an electrochemical aptamer based (EAB) biosensor, and two bioassays, a paper-based “dipstick” assay and an enzyme-linked immunosorbent assay (ELISA). Doing so we have narrowed the dynamic range of these sensors and assays several-fold, thus enhancing their ability to measure small changes in target concentration. Given that introducing sequestration requires only the addition of the appropriate concentration of a high-affinity “depletant,” the mechanism appears simple and easily adaptable to tuning the binding properties of the receptors employed in a wide range of biosensors and bioassays.
1. Introduction
Due to the extraordinary versatility, affinity, and specificity of biomolecular recognition, biosensors and bioassays (sensors and assays employing biological recognition elements) have seen significant academic and commercial exploration over the last 50 years.1,2 Widely used examples include the home glucose meter,3 enzyme linked immunosorbent assays (ELISA),4,5 and lateral flow immunoassays.6 Another, more recent example is electrochemical aptamer-based (EAB) sensors,7–9 a platform technology capable of performing continuous, high-frequency molecular measurements not only in unmodified clinical samples, but even in situ in the living body.10–13The many promising attributes of biological recognition notwithstanding, biosensors nevertheless suffer from a sometimes-significant limitation. Specifically, the physics of saturable binding creates a hyperbolic relationship (the “Langmuir isotherm”) between receptor occupancy and ligand concentration that renders the useful dynamic range of most biosensors rather broad, or relatively insensitive to small changes in target concentration (an effect amusingly termed the “tyranny of the Langmuir isotherm”14,15). For example, to shift from 10% to 90% occupancy (a span that is often used to arbitrarily define a “useful dynamic range”) of a single-site receptor the ligand concentration must change by 81-fold.16 This broad dynamic range greatly reduces the precision with which technologies that employ such receptors can monitor small relative changes in target concentration.
Because of the broad dynamic range associated with the Langmuir isotherm, a 10% change in ligand concentration produces at most a 2.5% change in receptor occupancy. This can be a major limitation. For example, the entire healthy physiological ranges of many ions and metabolites are less than 2-fold, including potassium at 1.5-fold (physiological range 3.5 to 5 mM),17 sodium at 1.3-fold (physiological range 120 to 160 mM),18 and glucose at 2-fold (5.5 to 11 mM).19 The therapeutic windows of many pharmaceuticals are likewise narrow,20–23 including vancomycin (clinical range 6 to 21 μM)24 and doxorubicin (35 to 65 μM).25 Given the narrowness of many physiological or clinical concentration ranges, relatively small changes in the level of a ion, metabolite, drug or biomolecule can have profound clinical implications, thus driving the need for high-precision measurements. A sensor based on a receptor with a set dynamic range spanning over a much broader 81-fold change in target concentration, however, offers poor sensitivity and precision. In response, the ability to engineer sensors with narrower dynamic ranges matching the clinically relevant range of their targets will optimize the precision of their measurements.
Nature also faces the tyranny of the Langmuir isotherm. For example, blood oxygen, must be delivered efficiently over only 5-fold difference of partial pressures between the lungs and peripheral tissues.26 How does nature maintain such tight control over molecular concentrations using receptors that, nominally, would exhibit only trivially small changes in occupancy over such narrow ranges? To achieve this, evolution has invented a number of mechanisms by which biomolecular receptors can achieve much higher sensitivity (i.e., a larger change in output signal for a given change in relative target concentration) than that afforded by a single, saturable binding site.27 These include, for example, cooperativity,28,29 sequestration30,31 and amplification cascades/allosteric activators,27,32 mechanisms that are employed in the regulation of numerous biological pathways,33–35 hormone responses,36 across-membrane transporters,37,38 and transcriptional activation.39,40
Given the ubiquity with which nature uses the above-described mechanisms to “fine-tune” the binding properties of naturally occurring receptors, we have argued that these mechanisms might also be of value in improving the measurement precision of artificial biosensors.41–43 Specifically, we have made the case that steepening concentration/occupancy curves can enhance the precision of biomolecule-based measurements. Such steepening would likewise benefit scenarios, such as HIV status, and pregnancy, in which a qualitative, “yes/no” answer is sufficient.44 In these cases, steeper binding curves minimize false positive results by converting the response into a more binary, “on-off” output on either side of a desired threshold concentration. Thus motivated, we explore here the application of one such mechanism, sequestration (Fig. 1), to the problem of improving the sensitivity of biosensors and bioassays.
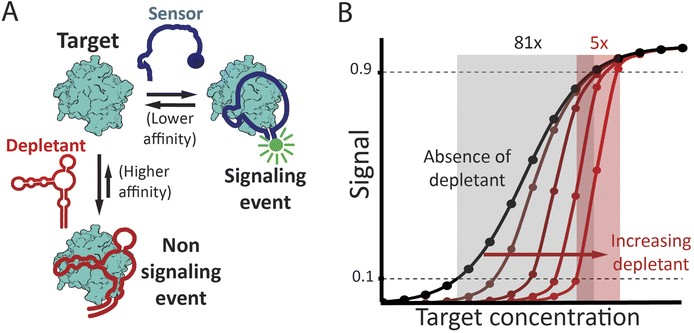 | ||
| Fig. 1 (A) The sequestration mechanism employs a depletant: biomolecule that binds its target with high affinity but without producing any significant signal. This depletant acts as a “sink,” ensuring that the concentration of free target remains low until the target concentration exceeds the sink's capacity, which is principally defined by the concentration of the depletant. As the target concentration rises above this threshold, the relative concentration of free target increases dramatically. This, in turn activates a second, lower affinity “receptor” that generates a signal change. (Right) Sequestration leads to a binding curve that is steeper, improving the response to small changes in target concentration, but at the cost of being shifted to higher target concentrations, leading to poorer limits of detection. The theoretical curves presented in panel B were determined using the model of Buchler and Cross31 (presented as eqn S6† in the ESI†), with an nH of 1, Kddep 10-fold below Kdreceptor, and depletant concentrations of 5x, 20x, 50x and 100x Kddep. | ||
2. Experimental
2.1. Reagents
Sodium hydroxide, sulfuric acid, sodium chloride, potassium chloride, sodium borate, potassium dihydrogen phosphate, tris(hydroxymethyl)aminomethane (tris) (Pittsburg, PA), ethylendiaminetetraacetic acid (EDTA), 6-mercapto-1-hexanol, tris(2-carboxyethyl) phosphine (TCEP), sodium hydrogen phosphate, biotinylated peroxidase (Biotin-HRP), 1-step Ultra 3,3′,5,5′-Tetramethylbenzidine (TMB) ELISA substrate, Tween 20, sucrose, and Bovine serum albumin (BSA) were purchased from sigma Aldrich (St. Louis, MO). Artificial urine solution (SURINE) was purchased from DTI (Lenexa, KS). Phosphate buffered (PBS; 137 mM NaCl, 2.7 mM KCl, 10 mM Na2HPO4, 1.8 mM KH2PO4 pH 7.4) was prepared at 10x stock and diluted to 1x before use. The assembling buffer was composed of 10 mM Na2HPO4 1 mM MgCl2 1 M NaCl pH 7.0.CH multipotentiostat Model 1000C, polycrystalline gold disk electrodes (2 mm diameter), reference electrode Ag/AgCl in saturated 3 M KCl and a platinum wire counter electrode were purchased CH instruments Inc., Austin, TX. MicroCloth (2.875 inches), 1 μm monocrystalline diamond suspension (oil-based), and 0.05 μm aluminum slurry were purchased from Buehler, Lake Bluff, IL.
Nitrocellulose for the dipstick assays, Unisart CN 180 backed, was purchased from Sartorius (Göttingen, Lower Saxony, Germany). Absorbent pads, grade MF1, purchased from Millipore-sigma (Temecula, CA). Backing cards to assemble the pads were purchased from Kenosha (Amstelveen, the Netherlands). Streptavidin coated plates (Pierce high binding high capacity, HBC, SuperBlock™ blocking buffer), HRP labelled streptavidin were purchased from thermofisher (Rockford, IL). Multi-well plates, clear flat bottom, half area, non-treated polystyrene, were purchased from Corning (Corning, NY).
The DNA sequences of the aptamers used to fabricate the sensors were: NGAL Full aptamer (80-base): 5′-GAATTCCGCCCTCGTCCCATCTCGGCTTGGTATGGCGGAGCT-GGATAGTATAGTCGGAACACCAACCGAGAACGGAATTC–3’ (purchased from Sigma Aldrich Saint Louis, MO).
NGAL truncated aptamer (26-base): 5′ thiol S–ATGGCGGAGCTGGATAGTATAGTCGG – Methylene Blue–3’ (purchased from LGC Biosearch technologies; Petaluma, CA).
NGAL truncated aptamer biotinylated (26-base): 5′ Biotin–ATGGCGGAGCTGGATAGTATAGTCGG-3’ (purchased from Integrated DNA technologies; IDT, Coralville, IA).
All aptamers were purchased as dual-HPLC purification grade (and used as received), dissolved to 100 μM in a 1 mM EDTA, 10 mM Tris–HCl, pH 8 buffer upon receipt and stored at −20 °C.
The target (Neutrophil gelatinase-associated lipocalin, NGAL) was produced recombinantly in E. coli following a previously reported procedure,45 and briefly described in the ESI.† Monoclonal antibody against NGAL (unlabeled, NGAL-4C10A7-, mouse mAb) was purchased from GenScript (Piscataway, NJ). Chicken HRP labeled Polyclonal antibody against mouse IgG was purchased from Life Technologies Corporation (Carlsbad, CA). Goat anti mouse polyclonal IgG, unlabeled was purchased from Abcam (Waltham, MA).
2.2. Electrochemical sensor fabrication
We fabricated the aptamer based electrochemical sensors (EAB sensors) on gold electrodes, following previously described procedures.7,45,46 In brief, we cleaned the polycrystalline gold disk electrodes by polishing them with 1 μm monocrystalline diamond suspension on a MicroCloth, sonicated them in ethanol, polished again using this time a 0.05 μm aluminium slurry, and finally sonicated in ethanol. Subsequently, we rinsed the electrodes with double distilled water and cleaned them electrochemically. This first entailed oxidation and reduction cycling in 0.5 M in NaOH at −0.4 to −1.35 V (all potentials versus Ag/AgCl) for 1000 scans at 2 V s−1, 0.01 V interval. Next, we rinsed the electrodes and transferred them to a 0.5 M H2SO4 solution in which we applied oxidation (2 V for 5 s) and reduction steps (−0.35 V for 10 s), followed by oxidation and reduction scans over −0.35 to 1.5 V for 20 scans at 4 V s−1 scan rate and 0.01 V sample interval. Finally, we applied 4 more scans at scan rate 0.1 V s−1 and 0.01 V sample interval.We fabricated the EAB sensors as follows. Cleaned electrodes were rinsed with double distilled water and kept immersed in distilled water until fabrication of the sensor. To reduce the thiol in the DNA we mixed the aptamers at a ratio of 5 μL of TCEP10 mM in double distilled water per 1 μL of DNA at 100 μM and incubated for 60 min at room temperature in the dark. Next, we diluted the reduced DNA in PBS 1x buffer to a concentration of 100 nM before immersing the clean electrodes in this solution for 1 h at room temperature in the dark. After the incubation we rinsed the electrodes with double distilled water and immersed them in a 2.8 mM 6-mercaptohexanol in PBS buffer overnight at room temperature in the dark.
2.3. Electrochemical measurements
All electrochemical measurements were performed with a CH multipotentiostat Model 1000C. Polycrystalline gold disk electrodes (2 mm diameter; CH instruments Inc., Austin, Tx) were used as working electrodes. An external reference electrode Ag/AgCl in saturated 3 M KCl and a platinum wire counter electrode were used in all electrochemical experiments.We removed the sensors from the 6-mercapohexanol solution, rinsed them with double distilled water and placed them in a standard electrochemical cell with external reference and counter electrodes, and containing a 2.5 mL of artificial urine. At this point, we added the corresponding amount of depletant to achieve the desired working concentration. We used the same measurement parameters for square wave voltammetry as previously reported for the specific aptamer used:45 50 mV amplitude, potential step of 4 mV and frequency of 100 Hz. Prior to performing the NGAL titrations we applied 50 scan cycles until the voltammetry peak stabilized. When titrating NGAL, we mixed and allow stabilization for 10 min after every addition. We calculated relative signal change as follows:
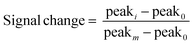 | (1) |
2.4. Dipstick strip fabrication and measurement
We fabricated our dipstick assay using the typical lateral flow strip fabrication process47 with a few variations. Specifically, we opted for not using neither sample pad nor conjugation pad and instead we lyophilized gold-nanoparticle-labelled antibodies (Ab-NPs) in an Eppendorf tube. When adding the sample and depletant to the tube the Ab-NPs are reconstituted and mixed with the sample, then we inserted the strip in upright position (for details see the “dipstick assay” section). Given this we call the resulting assay a “dipstick” rather than a “lateral flow assay.”![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 by volume) and thoroughly rinsed with double distilled water. First, we heated a 0.25 mM aqueous solution of HAuCl4·3H2O in double distilled water to boiling and then added vigorous stirring. Next, we added 1.25 mL of a 34 mM trisodium citrate while maintaining heating and stirring until the colourless solution turn into violet and later red solution (∼10 min). We characterized the resulting nanoparticles by UV-vis to confirm the presence of the absorbance peak at 520 nm (Fig. SI2 in the ESI†) characteristic of 20 nm diameter gold nanoparticles.49 The resulting AuNP solution was stored at 4 °C in the dark.
1 by volume) and thoroughly rinsed with double distilled water. First, we heated a 0.25 mM aqueous solution of HAuCl4·3H2O in double distilled water to boiling and then added vigorous stirring. Next, we added 1.25 mL of a 34 mM trisodium citrate while maintaining heating and stirring until the colourless solution turn into violet and later red solution (∼10 min). We characterized the resulting nanoparticles by UV-vis to confirm the presence of the absorbance peak at 520 nm (Fig. SI2 in the ESI†) characteristic of 20 nm diameter gold nanoparticles.49 The resulting AuNP solution was stored at 4 °C in the dark.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g for 30 min at 4 °C, discarded the supernatant, and resuspended the pellet in 2 mM pH 7.4 borate buffer. We repeated the centrifugation step a second time and resuspended the resulting pellet in 2 mM pH 7.4 borate buffer at ¼ the original volume. We then, split the resulting Ab-NPs in Eppendorf tubes and lyophilized them. Each tube contained sufficient “mix” for one dipstick assay: 60 μL of Ab-AuNPs at optical density of 1.2 absorbance units in 10 mM pH 7.4 PBS buffer containing 10% (w/v) sucrose 0.05% (v/v) Tween 20 and 10 mg mL−1 BSA. We spun these down and froze them at −80 °C for 1 h prior to lyophilizing overnight (Fig. SI3†).
000 g for 30 min at 4 °C, discarded the supernatant, and resuspended the pellet in 2 mM pH 7.4 borate buffer. We repeated the centrifugation step a second time and resuspended the resulting pellet in 2 mM pH 7.4 borate buffer at ¼ the original volume. We then, split the resulting Ab-NPs in Eppendorf tubes and lyophilized them. Each tube contained sufficient “mix” for one dipstick assay: 60 μL of Ab-AuNPs at optical density of 1.2 absorbance units in 10 mM pH 7.4 PBS buffer containing 10% (w/v) sucrose 0.05% (v/v) Tween 20 and 10 mg mL−1 BSA. We spun these down and froze them at −80 °C for 1 h prior to lyophilizing overnight (Fig. SI3†).
For the assay we mixed a volume of 60 μL of NGAL in synthetic urine with the appropriate amount of depletant and then added to a tube containing the lyophilized Ab-AuNPs with 5 min gentle stirring. We then inserted the dipstick strips into this tube. After the strip absorbed the sample, we added an additional 30 μL of buffer to wash the excess of reagents in the nitrocellulose pad. When necessary, we replaced the absorbent pads to ensure complete flow. Once the 30 μL of washing buffer had been absorbed into the strip we imaged the strips by scanning them in a Chemidoc, MP Imaging system (Bio-Rad technologies), determined the intensity of the relevant spots using the software Image J, and normalized against the background signal and correcting by the pixel size of the area measured. The integrated density of the pixels corresponding to the detection dot were subtracted the integrated density of a section of plain nitrocellulose with the same area size as the detection dot. The resulting value is divided by the size of the area used:
 | (2) |
2.5. ELISA plate preparation and assays
We deposited 100 μL of 0.5 μM biotinylated truncated aptamer in pH 7.4 PBS in the wells of streptavidin coated plates and incubated at room temperature for 1 h. We washed the plates 3 times using 200 μL of 0.05% (v/v) Tween 20 in pH 7.4 PBS per well. Since the streptavidin coated plates are already pre-blocked, and no other components in the assay contain biotin, further blocking was unnecessary. We titrated NGAL in artificial urine containing the corresponding concentration of depletant, using a mouse monoclonal antibody against NGAL (GenScript, Piscataway, New Jersey) at 5 μg mL−1, a total volume of 100 μL per well. After 1 hour incubation at room temperature, the plates were washed 3 times using 200 μL per well of 0.05% (v/v) Tween 20 pH 7.4 PBS. Next, we added chicken anti-mouse IgG HRP-conjugated at 200 ng mL−1 in 0.5% (w/v) BSA 0.05% (v/v) pH 7.4 PBS. Following, we performed 3 washings cycles by adding 200 μL per well of 0.05% (v/v) Tween 20 pH 7.4 before performing a last washing step using pH 7.4 PBS. To generate the color in response to target we added 100 μL per well of 3,3′,5,5′-tetramethylbenzidine (TMB) reagent. After 3 min we stopped the reaction by adding 50 μL of a 0.25 M H2SO4 per well and after 15 min we measured absorbance at 450 nm.3. Results
Sequestration relies on the use of two biorecognition elements: a higher affinity, non-signaling “depletant,” which binds the target but does not produce any output signal, and a lower affinity, signal-generating “receptor,” which alters its output signal upon target binding (Fig. 1, left). At target concentrations above the dissociation constant of the depletant, but below the dissociation constant of the receptor, near stoichiometric binding to the depletant ensures the low concentration of the free target. After this “sink” is saturated, however, further increases in total target concentration produce a concomitantly large increase in the relative concentration of free target. This, in turn, causes occupancy of the receptor to rise rapidly with any increase in total target concentration, generating an output signal that is a steeper function of that concentration (Fig. 1, right). The resulting enhancement in responsiveness improves our ability to measure small relative changes in target concentration (i.e., improves relative precision). It does so, however, at the cost of reducing sensitivity at lower target concentrations.Here we have applied sequestration to improve the sensitivity of one biosensor and two bioassays all targeting the protein neutrophil gelatinase-associated lipocalin52,53 (NGAL): an EAB sensor, a paper-based dipstick assay, and an ELISA. Plasma and urine NGAL levels above various threshold values are indicative of acute kidney damage,54–56 with the relevant reported cut-offs in urine of 2 nM for patients with IgA nephropathy,57 5 nM for patients with chronic heart failure,56 6 nM infants after cardiac surgery,55 and 85 nM for acute kidney damage (AKI) in adults.58 As a consequence, a range of assays and sensors have been reported for the measurement of NGAL, including ELISAs,59 nanoparticle aggregation tests,60 lateral flow assays61 and electrochemical biosensors.45,62,63 Given that the clinical indicators here are specific threshold concentration values, the availability of higher sensitivity, more all-or-none detection systems would be of value in these applications, thus motivating our use of this biomarker as our test bed for studies of the adaptation of sequestration strategies in biosensors and bioassays in complex samples.
We have employed two variants of a previously reported, NGAL-binding, DNA aptamer45,64 as the depletant and receptor in our studies. Specifically, our depletant is the full-length, 80-base “parent” aptamer, which has a reported dissociation constant (KD) of 0.92 nM.64 For the lower-affinity, signaling receptor we employed a 26-base long truncation of this parent sequence, which is destabilized enough that it is unfolded in the absence of target and presents a reported KD of 58 nM.45,65 Target binding must thus overcome the truncated aptamer's unfavourable folding free energy, reducing its affinity (relative to that of the parent sequence) by a factor of 38 (Fig. SI1†). Conveniently, the binding-induced folding of the truncated aptamer also provides the conformational change necessary to support EAB signaling.66
As our first demonstration of the utility of sequestration we employed our receptor/depletant pair in an NGAL-detecting EAB sensor (Fig. 2A). The conformation-linked signaling mechanism of EAB sensors renders them both convenient (reagentless and single step) and, because it mimics the signal transduction employed by chemoperception systems in nature, selective enough to deploy directly in complex clinical samples.45 For example, EAB sensors have been shown to work well when challenged in complex media such as blood serum,67,68 undiluted blood,69,70 and even circulating blood in a live animal.10,11,71
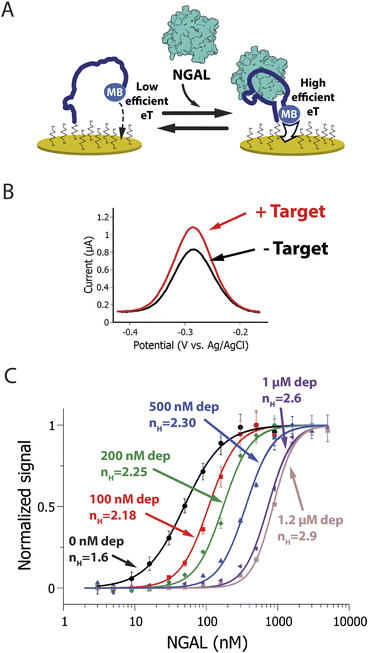 | ||
| Fig. 2 (A) Our neutrophil gelatinase associated lipocalin (NGAL)-detecting EAB sensor consists of a redox-reporter-modified, NGAL-binding aptamer attached to an interrogating electrode. Target binding induces a conformational change in this aptamer, which alters the electron transfer kinetics of its attached redox reporter, methylene blue (MB), thus producing (B) an easily detected change in square wave voltametric signal. (C) Here we have used sequestration to improve the sensitivity (change in signal for a given change in relative target concentration) of this EAB sensor in synthetic urine samples. To do so we added a high affinity NGAL-binding aptamer as a depletant to the sample. By thus introducing sequestration, we narrow the sensor's useful dynamic range, steepening its binding curve and improving sensitivity. For example, the sensor's pseudo-Hill (nH) coefficient, a measure of binding curve steepness, shifts from 1.6 at 0 nM depletant to 2.9 at 1.2 μM depletant, narrowing the concentration change required to transition from 10% occupied to 90% occupied from 15.6-fold to just 4.5-fold. Note: error bars in this figure reflect the standard deviation of 4 replicate measurements using independent sensors. The solid curves are fits to the Hill equation (eqn (3)); the Chi-square values are, for the curves from low to high depletant, 0.004, 0.006, 0.008, 0.007, 0.009 and 0.003. | ||
We fabricated an NGAL-detecting EAB sensor by linking a redox-reporter-modified version of the signalling receptor via its 5′ terminus to the surface of a gold electrode using thiol-on-gold self-assembled monolayer chemistry.7,46,72 Binding induced folding of this receptor alters the rate of electron transfer from the reporter (here methylene blue), producing an easily measurable change when the sensor is interrogated using square wave voltammetry.8,73 To introduce sequestration to this sensor we added depletant to the NGAL-containing sample in a complex matrix (synthetic urine) prior to measurement. Doing so we find that, as expected, the sensor's useful dynamic range systematically narrows as the concentration of added depletant is increased (Fig. 2B).
To quantify the extent to which sequestration improves sensitivity we employ two equivalent approaches. The first is to denote the width of the useful dynamic range; any steepening of the binding curve will concomitantly narrow the range of concentration required to transition from 10% to 90% of the signal change seen at saturating target. In parallel, we also fit the output of our sensors and assays to a cooperative binding curve, the equation first used by A.V. Hill in 1910 to describe the cooperative binding74 of oxygen to haemoglobin:
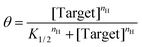 | (3) |
In this, the fraction of receptors occupied, θ, is given as a function of the free target concentration, the concentration at which half of the receptors are occupied (K1/2, which in single binding site receptors corresponds to KD), and the Hill coefficient (nH). The latter parameter reflects the steepness of the receptor transition from unbound to bound, with Hill coefficients greater than 1 associated with the steep transitions found in systems with enhanced sensitivity. Here we use the resulting pseudo-Hill coefficient (“pseudo” because, while our systems mimic cooperativity, they are not actually cooperative) as a convenient, empirical way to compare sensitivities.30 Applying these metrics to the NGAL-detecting EAB sensor, we find that, even in the absence of depletant, the sensor achieves an nH of 1.60 ± 0.08 (the reported confidence intervals reflect the 95% confidence intervals calculated from the fits) and a dynamic range of just 16-fold (Fig. 2B, black line). Previously we have shown that the higher-than-unity pseudo-Hill coefficient of the NGAL-detecting EAB sensor is caused by inter-chain interactions at the high density with which the aptamer receptor is packed on the electrode surface; e.g., this effect is lost at lower packing densities.45 Upon the addition of depletant, however, the sensor's dynamic range narrows still more, reaching just 4.6-fold (with an nH of 2.90 ± 0.10) when the depletant concentration reaches 1.2 μM (Fig. 2B brown curve). As expected, this increasing steepness comes at the cost of higher binding midpoints, which shifts the minimum amount of target required to achieve a statistically-significant signal (we define this limit of detection as the concentration for which the signal change reaches 3 times the standard deviation of a blank). In the absence of depletant the EAB sensor's limit of detection is 12 nM, which rises to 198 nM at the highest depletant concentration we have employed.
To explore the versatility of the sequestration mechanism we next introduced it into an NGAL-detecting dipstick assay. This commonly employed class of point-of-care assays detects an analyte that has been driven through a membrane by capillarity.5 During this migration, the target binds to receptors attached to the membrane at a specific “detection region”. Detection typically relies on the formation of a sandwich in which a second reporter-modified receptor binds to a second epitope on the target, causing it to accumulate at the detection region and create an output signal.75 Here we attached the lower affinity, truncated aptamer to the detection region of the membrane (Fig. 3A). As a control we also spotted a goat anti-mouse IgG at a second site farther along the direction of flow on the membrane. Finally, as our depletant we again employed the full-length anti-NGAL aptamer, and as our reporter-modified receptor we employed a commercial mouse monoclonal anti-NGAL antibody modified with 20 nm diameter gold nanoparticles.
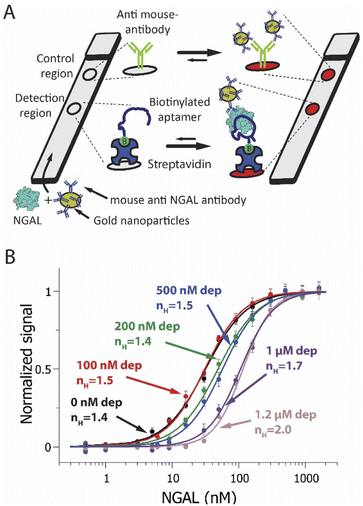 | ||
| Fig. 3 (A) A NGAL-detecting dipstick (lateral flow-like) assay. In this, a gold-nanoparticle-modified anti-NGAL antibody binds to any NGAL in the sample before the resulting complex is driven through a paper strip by capillarity. An NGAL aptamer attached to the membrane at the detection region binds a second epitope on NGAL, creating an aptamer-NGAL-antibody “sandwich”. This causes gold nanoparticles to accumulate at the detection region, generating a characteristic red/pink color. Any antibody not bound to NGAL flows past the detection region, ultimately binding an anti-IgG antibody placed at the control region. (B) Using the sequestration mechanism, we have improved the sensitivity of an NGAL-detecting dipstick assay to small changes in target concentration. To do so we fabricated dipstick assays that included varying amounts of depletant (the full-length anti-NGAL aptamer) in the sample/antibody mixture. This results in a steeper binding curve and a narrower dynamic range. Shown are NGAL dipstick assays achieving pseudo-Hill coefficients ranging from 1.4 at 0 nM depletant to 2.0 at 1.2 μM depletant, in synthetic urine samples. Note: error bars in this figure reflect the standard deviation of 4 replicate measurements using independent devices. The solid curves are fits to the Hill equation (eqn (3)); the Chi-square values are, for the curves from low to high depletant, 0.011, 0.010, 0.008, 0.006, 0.005 and 0.002. | ||
Sequestration increases the sensitivity of the dipstick assay to small changes in target concentration. Specifically, in the absence of depletant (black line) we observed a dynamic range of 23-fold and an nH of 1.4 ± 0.1 (Fig. 3B, black curve). Upon the addition of depletant to the sample prior to analysis, however, the binding curve steepens, ultimately narrowing the dynamic range to just 9-fold and producing a pseudo-Hill coefficient of 2.0 ± 0.1 at a depletant concentration of 1.2 μM (Fig. 3B, pink curve). Over this same change in depletant concentration, the assay's limit of detection increased from 2.9 nM to 58 nM.
As our final example we deployed the sequestration mechanism in an NGAL-detecting ELISA (Fig. 4). The working principle underlying this widely-used technique76,77 relies on the attachment of the target to the surface of a plastic well. In the type of ELISA we explored, a sandwich ELISA, this attachment is mediated by a “capture reagent” (here our truncated aptamer) immobilized on the well surface (here using a biotinylated aptamer and streptavidin coated wells). We incubated the samples containing the target and the corresponding concentration of depletant (again, the full-length aptamer) in the wells, where the target binds the capture reagent. Detection takes place via a second receptor, here an anti-NGAL mouse monoclonal antibody, that binds the target at a second epitope. To generate a signal, we used a horseradish peroxidase-modified anti-mouse IgG that binds to the mouse anti-NGAL antibody and, upon reaction with tetramethylbenzidine, produces a colorimetric output.
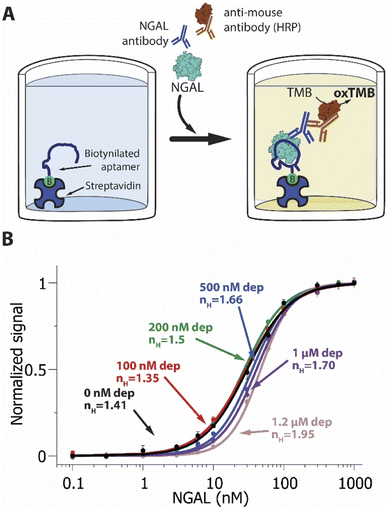 | ||
| Fig. 4 (A) A NGAL-detecting enzyme linked immunosorbent assay (ELISA). The target, NGAL, binds a low affinity receptor attached to the bottom of a well. The subsequent addition of an anti-NGAL antibody and horse-radish-peroxidase-modified secondary antibody produces an enzymatic signal, allowing the detection and quantification of NGAL. (B) ELISA assays that included varying amounts of depletant in the sample result in steeper binding curves and narrower dynamic ranges. Shown are NGAL-detecting ELISA assays achieving pseudo-Hill coefficients ranging from 1.41 ± 0.06 at 0 nM depletant to 1.95 ± 0.05 at 1.2 μM depletant. Note: error bars in this figure reflect the standard deviation of 4 replicate measurements using independent wells. The solid curves are fits to the Hill equation (eqn (3)); The Chi-square values are, for the curves from low to high depletant, 0.005, 0.001, 0.001, 0.004, 0.002 and 0.001. | ||
As we observed for our EAB sensor and lateral flow examples, sequestration increases the sensitivity of the NGAL-detecting ELISA. Specifically, whereas in the absence of depletant, the useful dynamic range of the assay is of 22.5-fold, corresponding to an nH of 1.41 ± 0.06 (Fig. 4B, black line), the addition of depletant steepens this binding curve, narrowing the dynamic range to 9-fold and producing a pseudo-Hill coefficient of 1.95 ± 0.05 at a depletant concentration of 1.2 μM (Fig. 4B, pink line). Conversely, over this same range of depletant concentrations the limit of detection increased from 1.5 nM to 9.2 nM.
4. Discussion
Here we have used sequestration, a mechanism previously used to tune the binding properties of DNA receptors30,50,78 and enzymatic79 and protein80 networks, to improve the sensitivity in three different detection techniques: an EAB sensor,45 a dipstick assay, and an ELISA. Doing so we find that, for the EAB sensor, the effective dynamic range of the sensor narrowed from 15.6-fold to just 4.5-fold. In the dipstick assay, the binding curve contracted from 23 to 9-fold. And for the ELISA, the useful dynamic range contracted from 22.5 to 9.5-fold. With this, the sensitivity of these sensors and assays to small changes in target concentration is improved. For example, for a receptor with useful dynamic range of just 4.5-fold (nH = 2.9), a change in target concentration from 5% below the midpoint of the binding curve to 5% above it changes the occupancy of the receptor –and the resulting output signal – by 7.2%, which is nearly three times greater than the 2.5% change that would be produced by a receptor that follows Langmuir isotherm binding (nH = 1, Fig. SI4†).The sensitivity enhancements we observe are less than would be expected from theory or previously reported model systems30,31 (Fig. SI5†). For example, at the highest depletant concentration we have employed (1.2 μM) theory predicts a pseudo-Hill coefficient of > 10 rather than the values of 2 to 2.9 that we observe. We suspect that this is due to the complexity of our systems, all of which entail surface-attached receptors (which, as previously reported, affects the binding properties of our receptor45) and two of which (the dipstick and ELISA) are complex, multistep assays. Irrespective, however, of how close to (or far from) ideal behaviour our systems are, the improved sensitivity we achieve will nevertheless prove of value in enhancing the precision of the bioanalytical techniques.
5. Conclusions
Many approaches for improving sensitivity of biomolecular receptors are complex. The rational introduction of cooperativity, for example, requires the design of coupled, binding-induced conformational changes to achieve the steepening of binding curves,81–84 and the final shape of the response curve will depend on the binding free energy landscape that such coupling produces.85 Sequestration, in contrast, appears easy to engineer into existing sensing platforms and assays as it only requires the addition of a high-affinity depletant. Moreover, the sensitivity associated with sequestration can be fine-tuned via the simple expedient of altering the concentration of the depletant. Given this relative simplicity, we believe that sequestration can be quickly and easily adapted in a diversity of scenarios to tune binding properties of detection assays. Consistent with this, here we have demonstrated the successful application of sequestration as a mechanism to induce steeper transitions in dose response curves for three different sensing techniques.Author contributions
A. C., C.·P., A. I., F. R. and K. P. designed research; A. C., C.·P.,G. O., A. I., J. G., performed experimental work. A. C., C.·P., A. I. and K. P. analyzed the data; A. C., C.·P., G. O.,F. R. and K. P. wrote the manuscript.Conflicts of interest
KWP serves on the advisory board of and owns equity in a company that is commercializing in vivo EAB sensors.Acknowledgements
This project has received funding from the National Institutes of Health (EB022015 Plaxco) as well as the European Union's Horizon 2020 research and innovation program under the Marie Sklodowska-Curie grant agreement No 799332 (SmartBioSense – Alejandro Chamorro) and No 101025241 (Entropic DNA Sensors – Andrea Idili). A. C. acknowledges support from Fondazione Umberto Veronesi for the post-doctoral fellowship 2022 (ID 0001420). Claudio Parolo (ISGlobal) acknowledges support from the Spanish Ministry of Science and Innovation and State Research Agency through the “Centro de Excelencia Severo Ochoa 2019–2023” Program (CEX2018-000806 S), and support from the Generalitat de Catalunya through the CERCA Program. Gabriel Ortega acknowledges the support from Ikerbasque, the Basque Foundation for Science. The authors acknowledge the use of the Biological Nanostructures Laboratory within the California NanoSystems Institute, supported by the University of California, Santa Barbara and the University of California, Office of the President.Notes and references
- A. P. F. Turner, Chem. Soc. Rev., 2013, 42, 3184–3196 RSC.
- G. Rosati, A. Idili, C. Parolo, C. Fuentes-Chust, E. Calucho, L. Hu, C. de C. Castro e Silva, L. Rivas, E. P. Nguyen, J. F. Bergua, R. Alvárez-Diduk, J. Muñoz, C. Junot, O. Penon, D. Monferrer, E. Delamarche and A. Merkoçi, ACS Nano, 2021, 15, 17137–17149 CrossRef CAS PubMed.
- J. Wang, Chem. Rev., 2008, 108, 814–825 CrossRef CAS PubMed.
- R. M. Lequin, Clin. Chem., 2005, 51, 2415–2418 CrossRef CAS PubMed.
- B. O'Farrell, Chapter 2.1 - Principles of Competitive and Immunometric Assays (Including ELISA), in The Immunoassay Handbook, ed. D. Wild, Elsevier, Oxford, 4th edn, 2013, pp. 89–107 Search PubMed.
- B. O'Farrell, Chapter 2.4 - Lateral Flow Immunoassay Systems: Evolution from the Current State of the Art to the Next Generation of Highly Sensitive, Quantitative Rapid Assays, in The Immunoassay Handbook, ed. D. Wild, Elsevier, Oxford, 4th edn, 2013, pp. 89–107 Search PubMed.
- Y. Xiao, R. Y. Lai and K. W. Plaxco, Nat. Protoc., 2007, 2, 2875–2880 CrossRef CAS PubMed.
- L. R. Schoukroun-Barnes, F. C. Macazo, B. Gutierrez, J. Lottermoser, J. Liu and R. J. White, Annu. Rev. Anal. Chem., 2016, 9, 163–181 CrossRef CAS PubMed.
- A. Idili, C. Parolo, G. Ortega and K. W. Plaxco, ACS Sens., 2019, 4, 3227–3233 CrossRef CAS PubMed.
- P. Dauphin-Ducharme, K. Yang, N. Arroyo-Currás, K. L. Ploense, Y. Zhang, J. Gerson, M. Kurnik, T. E. Kippin, M. N. Stojanovic and K. W. Plaxco, ACS Sens., 2019, 4, 2832–2837 CrossRef CAS PubMed.
- N. Arroyo-Currás, J. Somerson, P. A. Vieira, K. L. Ploense, T. E. Kippin and K. W. Plaxco, Proc. Natl. Acad. Sci., 2017, 114, 645–650 CrossRef PubMed.
- A. Idili, N. Arroyo-Currás, K. L. Ploense, A. T. Csordas, M. Kuwahara, T. E. Kippin and K. W. Plaxco, Chem. Sci., 2019, 10, 8164–8170 RSC.
- A. Idili, J. Gerson, T. Kippin and K. W. Plaxco, Anal. Chem., 2021, 93, 4023–4032 CrossRef CAS PubMed.
- R. M. Corn, Abstracts of papers of the American chemical society, Amer Chemical Soc,1155 16TH ST, NW, WASHINGTON, DC 20036 USA, 2005, 230, U330, U331 Search PubMed.
- F. Ricci, A. Vallée-Bélisle, A. J. Simon, A. Porchetta and K. W. Plaxco, Acc. Chem. Res., 2016, 49, 1884–1892 CrossRef CAS PubMed.
- F. W. Dahlquist, Methods Enzymol., 1978, 48, 270–299 CAS.
- M. Zacchia, M. L. Abategiovanni, S. Stratigis and G. Capasso, Kidney Dis., 2016, 2, 72–79 CrossRef PubMed.
- J. H. Howanitz and P. J. Howanitz, Am. J. Clin. Pathol., 2007, 127, 56–59 CrossRef CAS PubMed.
- H. C. Gerstein, Diabetes Care, 1999, 22, 659–660 CrossRef CAS PubMed.
- A. Paci, G. Veal, C. Bardin, D. Levêque, N. Widmer, J. Beijnen, A. Astier and E. Chatelut, Eur. J. Cancer, 2014, 50, 2010–2019 CrossRef CAS PubMed.
- M. Y. Fosso, Y. Li and S. Garneau-Tsodikova, Medchemcomm, 2014, 5, 1075–1091 RSC.
- J. Tamargo, J. Y. Le Heuzey and P. Mabo, Eur. J. Clin. Pharmacol., 2015, 71, 549–567 CrossRef CAS PubMed.
- D. Gonzalez, G. G. Rao, S. C. Bailey, K. L. R. Brouwer, Y. Cao, D. J. Crona, A. D. M. Kashuba, C. R. Lee, K. Morbitzer, J. H. Patterson, T. Wiltshire, J. Easter, S. W. Savage and J. R. Powell, Clin. Transl. Sci., 2017, 10, 443–454 CrossRef PubMed.
- M. Rybak, B. Lomaestro, J. C. Rotschafer, R. Moellering Jr., W. Craig, M. Billeter, J. R. Dalovisio and D. P. Levine, Am. J. Health Pharm., 2009, 66, 82–98 CrossRef CAS PubMed.
- Y. Fujisaka, A. Horiike, T. Shimizu, N. Yamamoto, Y. Yamada and T. Tamura, Jpn. J. Clin. Oncol., 2006, 36, 768–774 CrossRef PubMed.
- S. L. Berg JM and J. L. Tymoczko, Biochemistry, New York, 5th edn, 2002 Search PubMed.
- Q. Zhang, S. Bhattacharya and M. E. Andersen, Open Biol., 2013, 3, 130031 CrossRef PubMed.
- C. A. Hunter and H. L. Anderson, Angew. Chem., Int. Ed., 2009, 48, 7488–7499 CrossRef CAS PubMed.
- A. J. Simon, A. Vallée-Bélisle, F. Ricci and K. W. Plaxco, Proc. Natl. Acad. Sci., 2014, 111, 15048–15053 CrossRef CAS PubMed.
- F. Ricci, A. Vallée-Bélisle and K. W. Plaxco, PLoS Comput. Biol., 2011, 7, e1002171 CrossRef CAS PubMed.
- N. E. Buchler and F. R. Cross, Mol. Syst. Biol., 2009, 5, 272 CrossRef PubMed.
- S. Y. Kim and J. E. Ferrell, Cell, 2007, 128, 1133–1145 CrossRef CAS PubMed.
- N. V Valeyev, D. G. Bates, P. Heslop-Harrison, I. Postlethwaite and N. V Kotov, BMC Syst. Biol., 2008, 2, 48 CrossRef PubMed.
- J. E. Ferrell, Trends Biochem. Sci., 1996, 21, 460–466 CrossRef CAS PubMed.
- M. E. Crosby, Yale J. Biol. Med., 2007, 80, 141–142 Search PubMed.
- A. C. Notides, N. Lerner and D. E. Hamilton, Proc. Natl. Acad. Sci. U. S. A., 1981, 78, 4926–4930 CrossRef CAS PubMed.
- J. Tytgat and P. Hess, Nature, 1992, 359, 420–423 CrossRef CAS PubMed.
- K. Banerjee, B. Das and G. Gangopadhyay, J. Chem. Phys., 2013, 138, 165102 CrossRef PubMed.
- A. M. Erkine, S. F. Magrogan, E. A. Sekinger and D. S. Gross, Mol. Cell. Biol., 1999, 19, 1627–1639 CrossRef CAS PubMed.
- K. T. Hughes and K. Mathee, Annu. Rev. Microbiol., 1998, 52, 231–286 CrossRef CAS PubMed.
- F. Ricci, A. Vallée-Bélisle, A. J. Simon, A. Porchetta and K. W. Plaxco, Acc. Chem. Res., 2016, 49, 1884–1892 CrossRef CAS PubMed.
- A. Sena-Torralba, H. Torné-Morató, C. Parolo, S. Ranjbar, M. A. Farahmand Nejad, R. Álvarez-Diduk, A. Idili, M. R. Hormozi-Nezhad and A. Merkoçi, Adv. Mater. Technol., 2022, 2101450 CrossRef CAS.
- E. Delamarche, Y. Temiz, R. D. Lovchik, M. G. Christiansen and S. Schuerle, Angew. Chem., Int. Ed., 2021, 60, 17784–17796 CrossRef CAS PubMed.
- J. Wang and E. Katz, Anal. Bioanal. Chem., 2010, 398, 1591–1603 CrossRef CAS PubMed.
- C. Parolo, A. Idili, G. Ortega, A. Csordas, A. Hsu, N. Arroyo-Currás, Q. Yang, B. S. Ferguson, J. Wang and K. W. Plaxco, ACS Sens., 2020, 5, 1877–1881 CrossRef CAS PubMed.
- A. Chamorro-Garcia, G. Ortega, D. Mariottini, J. Green, F. Ricci and K. W. Plaxco, Chem. Commun., 2021, 57, 11693–11696 RSC.
- A. Chamorro-Garcia, A. dela Escosura-Muñiz, M. Espinosa-Castañeda, C. J. Rodriguez-Hernandez, C. de Torres and A. Merkoçi, Nanomedicine, 2016, 12, 53–61 CrossRef CAS PubMed.
- J. Turkevich, P. C. Stevenson and J. Hillier, Discuss. Faraday Soc., 1951, 11, 55–75 RSC.
- M.-C. Daniel and D. Astruc, Chem. Rev., 2004, 104, 293–346 CrossRef CAS PubMed.
- D. Kang, A. Vallée-Bélisle, A. Porchetta, K. W. Plaxco and F. Ricci, Angew. Chem., Int. Ed., 2012, 51, 6717–6721 CrossRef CAS PubMed.
- B. Wei, J. Zhang, X. Ou, X. Lou, F. Xia and A. Vallée-Bélisle, Anal. Chem., 2018, 90, 1506–1510 CrossRef CAS PubMed.
- S. S. Soni, D. Cruz, I. Bobek, C. Y. Chionh, F. Nalesso, P. Lentini, M. de Cal, V. Corradi, G. Virzi and C. Ronco, Int. Urol. Nephrol., 2010, 42, 141–150 CrossRef CAS PubMed.
- B. Yao, M.-C. Giel and Y. Hong, Mater. Chem. Front., 2021, 5, 2124–2142 RSC.
- P. Devarajan, Scand. J. Clin. Lab. Invest. Suppl., 2008, 241, 89–94 CrossRef PubMed.
- J. Mishra, C. Dent, R. Tarabishi, M. M. Mitsnefes, Q. Ma, C. Kelly, S. M. Ruff, K. Zahedi, M. Shao, J. Bean, K. Mori, J. Barasch and P. Devarajan, Lancet, 2005, 365, 1231–1238 CrossRef CAS.
- K. Damman, D. J. van Veldhuisen, G. Navis, A. A. Voors and H. L. Hillege, Eur. J. Heart Fail., 2008, 10, 997–1000 CrossRef CAS PubMed.
- H. Ding, Y. He, K. Li, J. Yang, X. Li, R. Lu and W. Gao, Clin. Immunol., 2007, 123, 227–234 CrossRef CAS PubMed.
- S. M. Tuladhar, V. O. Püntmann, M. Soni, P. P. Punjabi and R. G. Bogle, J. Cardiovasc. Pharmacol., 2009, 53, 261–266 CrossRef CAS PubMed.
- https://www.thermofisher.com/elisa/product/NGAL-Human-ELISA-Kit/KIT036, accessed October 2021.
- G. Lippi, R. Aloe, A. Storelli, G. Cervellin and T. Trenti, Clin. Chem. Lab. Med., 2012, 50, 1581–1584 CAS.
- L. Lei, J. Zhu, G. Xia, H. Feng, H. Zhang and Y. Han, Talanta, 2017, 162, 339–344 CrossRef CAS PubMed.
- C. H. Cho, J. H. Kim, D.-K. Song, T. J. Park and J. P. Park, Biosens. Bioelectron., 2019, 142, 111482 CrossRef CAS PubMed.
- J. Yukird, T. Wongtangprasert, R. Rangkupan, O. Chailapakul, T. Pisitkun and N. Rodthongkum, Biosens. Bioelectron., 2017, 87, 249–255 CrossRef CAS PubMed.
- J. Wang, J. Yu, Q. Yang, J. McDermott, A. Scott, M. Vukovich, R. Lagrois, Q. Gong, W. Greenleaf, M. Eisenstein, B. S. Ferguson and H. T. Soh, Angew. Chem., Int. Ed., 2017, 56, 744–747 CrossRef CAS PubMed.
- G. Rosati, M. Urban, L. Zhao, Q. Yang, C. de Carvalho Castro e Silva, S. Bonaldo, C. Parolo, E. P. Nguyen, G. Ortega, P. Fornasiero, A. Paccagnella and A. Merkoçi, Biosens. Bioelectron., 2022, 196, 113737 CrossRef CAS PubMed.
- R. J. White, A. A. Rowe and K. W. Plaxco, Analyst, 2010, 135, 589–594 RSC.
- B. R. Baker, R. Y. Lai, M. S. Wood, E. H. Doctor, A. J. Heeger and K. W. Plaxco, J. Am. Chem. Soc., 2006, 128, 3138–3139 CrossRef CAS PubMed.
- A. Idili, C. Parolo, R. Alvarez-Diduk and A. Merkoçi, ACS Sens., 2021, 6, 3093–3101 CrossRef CAS PubMed.
- H. Li, N. Arroyo-Currás, D. Kang, F. Ricci and K. W. Plaxco, J. Am. Chem. Soc., 2016, 138, 15809–15812 CrossRef CAS PubMed.
- H. Li, P. Dauphin-Ducharme, G. Ortega and K. W. Plaxco, J. Am. Chem. Soc., 2017, 139, 11207–11213 CrossRef CAS PubMed.
- A. Idili, N. Arroyo-Currás, K. L. Ploense, A. T. Csordas, M. Kuwahara, T. E. Kippin and K. W. Plaxco, Chem. Sci., 2019, 10, 8164–8170 RSC.
- P. Dauphin-Ducharme and K. W. Plaxco, Anal. Chem., 2016, 88, 11654–11662 CrossRef CAS PubMed.
- M. A. Pellitero, A. Shaver and N. Arroyo-Currás, J. Electrochem. Soc., 2020, 167, 037529 CrossRef CAS.
- A. Hill, J. Physiol., 1910, 40, i–vii Search PubMed.
- C. Parolo, A. Sena-Torralba, J. F. Bergua, E. Calucho, C. Fuentes-Chust, L. Hu, L. Rivas, R. Álvarez-Diduk, E. P. Nguyen, S. Cinti, D. Quesada-González and A. Merkoçi, Nat. Protoc., 2020, 15, 3788–3816 CrossRef CAS PubMed.
- J. He, Practical guide to ELISA development, in The Immunoassay Handbook, ed. D. Wild, Elsevier, Oxford, 4th edn, 2013, ch. 5.1, pp. 381–393 Search PubMed.
- R. de la Rica and M. M. Stevens, Nat. Protoc., 2013, 8, 1759–1764 CrossRef PubMed.
- A. Porchetta, A. Vallée-Bélisle, K. W. Plaxco and F. Ricci, J. Am. Chem. Soc., 2012, 134, 20601–20604 CrossRef CAS PubMed.
- S. P. Rafael, A. Vallée-Bélisle, E. Fabregas, K. Plaxco, G. Palleschi and F. Ricci, Anal. Chem., 2012, 84, 1076–1082 CrossRef CAS PubMed.
- C. A. Miller, J. M. Ho, S. E. Parks and M. R. Bennett, ACS Synth. Biol., 2021, 10, 258–264 CrossRef CAS PubMed.
- A. J. Simon, A. Vallée-Bélisle, F. Ricci, H. M. Watkins and K. W. Plaxco, Angew. Chem., Int. Ed., 2014, 53, 9471–9475 CrossRef CAS PubMed.
- H. Yu, J. Canoura, B. Guntupalli, X. Lou and Y. Xiao, Chem. Sci., 2017, 8, 131–141 RSC.
- A. J. Simon, A. Vallée-Bélisle, F. Ricci and K. W. Plaxco, Proc. Natl. Acad. Sci., 2014, 111, 15048–15053 CrossRef CAS PubMed.
- J. E. Dueber, E. A. Mirsky and W. A. Lim, Nat. Biotechnol., 2007, 25, 660–662 CrossRef CAS PubMed.
- G. Ortega, D. Mariottini, A. Troina, F. W. Dahlquist, F. Ricci and K. W. Plaxco, Proc. Natl. Acad. Sci., 2020, 117(32), 19136–19140 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc03901j |
| This journal is © The Royal Society of Chemistry 2022 |
