Open Access Article
Open Access ArticleDual targeting nanoparticles for epilepsy therapy†
Qinghong
Hou‡
abc,
Lulu
Wang‡
d,
Feng
Xiao
a,
Le
Wang
a,
Xiaoyan
Liu
a,
Lina
Zhu
 c,
Yi
Lu
*d,
Wenfu
Zheng
*b and
Xingyu
Jiang
c,
Yi
Lu
*d,
Wenfu
Zheng
*b and
Xingyu
Jiang
 *a
*a
aShenzhen Key Laboratory of Smart Healthcare Engineering, Guangdong Provincial Key Laboratory of Advanced Biomaterials, Department of Biomedical Engineering, Southern University of Science and Technology, Shenzhen 518055, P. R. China. E-mail: jiang@sustech.edu.cn
bCAS Key Laboratory for Biomedical Effects of Nanomaterials and Nanosafety, National Center for Nanoscience and Technology, Beijing 100190, P. R. China. E-mail: zhengwf@nanoctr.cn
cDepartment of Chemistry, School of Science, Tianjin University, Tianjin 300350, P. R. China
dCAS Key Laboratory of Brain Connectome and Manipulation, the Brain Cognition and Brain Disease Institute, Shenzhen Institute of Advanced Technology, Chinese Academy of Sciences, Shenzhen-Hong Kong Institute of Brain Science-Shenzhen Fundamental Research Institutions, Shenzhen 518055, P. R. China. E-mail: luyi@siat.ac.cn
First published on 19th October 2022
Abstract
For epilepsy therapy, one-third of the patients worldwide are resistant to antiepileptic drugs mainly due to the existence of the blood–brain barrier (BBB) that prevents the drugs from reaching the epileptic lesions. Here, we design a double targeting nanoparticle carrying lamotrigine (LTG) to cross the BBB and further concentrate at the neurons. We prepare the nanoparticles on a microfluidic chip by encapsulating LTG in poly(lactic-co-glycolic acid) (PLGA) to form a core (PL) and capping the core with a shell of lipids conjugated with the D-T7 peptide (targeting the BBB) and Tet1 peptide (targeting the neuron) to form D-T7/Tet1-lipids@PL nanoparticles (NPs). In vitro and in vivo experiments show that D-T7/Tet1-lipids@PL NPs have excellent neuron targeting, antiepileptic, and protecting effects. Our approach provides a new strategy for improving the therapeutic efficacy of existing antiepileptic drugs.
Introduction
Epilepsy is one of the most common neurological diseases. There are more than 70 million people worldwide suffering from epilepsy.1 Epilepsy is usually caused by abnormally increased neuronal excitability and abnormal activation of brain neuronal circuits due to the increase of extra neuronal excitatory neurotransmitters or the decrease of inhibitory neurotransmitters.2 Antiepileptic drugs (AEDs) are the main means of epilepsy treatment through various mechanisms.1,3 Although there are more than 25 AEDs,1 about one-third of epilepsy patients are resistant to existing drugs and cannot completely control seizures.4 The main cause of drug resistance is that most of the AEDs cannot pass through the blood–brain barrier (BBB).5 Moreover, even if the drugs enter the brain, it is difficult to specifically target the diseased neurons. Thus, it is necessary to explore a strategy to effectively deliver AEDs to the brain and target the neurons. Nano-delivery systems have been widely used for improving the utilization of drugs.6–14 The use of nanocarriers to improve the BBB-penetration efficiency of drugs has been extensively studied.15–19 However, there are few reported nanocarriers for delivering AEDs, and most of them are single-targeted nanocarriers, which still cause side effects. Recently, microfluidic technology has played important roles in many biomedical fields.20–22 In particular, microfluidic chips can easily synthesize NPs with uniform and small sizes,23–26 providing a platform for preparing nano-drugs.Here, we propose a dual-targeting nanocarrier system to deliver lamotrigine (LTG) to the diseased neurons to treat epilepsy. LTG is a first line AED in clinics.4,27 However, it has low solubility in water and is easily metabolized in the liver. Thus, high doses or repeated administration are required to achieve therapeutic concentrations,28,29 which can cause side effects such as nausea, headache, blurred vision, dizziness, and ataxia. To optimize the pharmacological action of LTG and minimize its side effects, the dual-targeting nanocarrier system had two components: (i) D-form T7 (D-T7) peptide, the retro-inverse sequence of the T7 peptide, which showed a high binding affinity with transferrin (Tf) receptor (a major component of the BBB) and was effective in guiding drug delivery to the central nervous system (CNS),30–35 was designed to target the BBB. (ii) Tet1 peptide, which can specifically bind to sphingomyelin and ganglioside (GT1b receptor) highly expressed on the surface of neurons.36–39 Although the ability of the T7 peptide, D-T7 peptide, and Tet1 peptide to target their respective targets and the combination of the T7 peptide and Tet1 peptide to treat Alzheimer's disease have been reported, the combined effects of the D-T7 peptide and Tet1 peptide to target the central nervous system (CNS) have not yet been attempted. The D-T7 peptide has a higher binding affinity to the Tf receptor than the T7 peptide for targeting the CNS, so the combined effects of the D-T7 peptide and Tet1 peptide to target the CNS need to be explored.32,33,38,39 We synthesized a dual-targeting delivery system on a two-step microfluidic chip which has been developed in the previous work.9 We encapsulated LTG in poly(lactic-co-glycolic acid, PLGA), a biodegradable and biocompatible polymer approved by the Food and Drug Administration (FDA), to form a core (PL) in the first step. We further coated a layer of lipids (DPPC, cholesterol, DSPE-PEG2k-D-T7, and DSPE-PEG2k-Tet1) as a shell to form a core–shell NP (D-T7/Tet1-lipids@PL). The co-modification of the D-T7 peptide and Tet1 peptide on the lipids@PL endowed the NPs with dual targeting capability to both the BBB and the neurons (Fig. 1B). We screened NPs with different D-T7/Tet1 concentration titrations. In vitro (BBB model) and in vivo (2 acute epilepsy models induced by kainic acid (KA) or pilocarpine and 1 chronic epilepsy model induced by KA, which are associated with human temporal lobe epilepsy.40,41) experiments showed that when the molar ratio of the D-T7 peptide to Tet1 peptide was 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, D-T7/Tet1-lipids@PL NPs showed the best performance in accumulating in the brain, inhibiting the seizures and protecting the neurons. To the best of our knowledge, this is the first work on dual targeting delivery systems with multiple types of in vivo epilepsy models, which has important implications for the clinical treatment of human temporal lobe epilepsy.
1, D-T7/Tet1-lipids@PL NPs showed the best performance in accumulating in the brain, inhibiting the seizures and protecting the neurons. To the best of our knowledge, this is the first work on dual targeting delivery systems with multiple types of in vivo epilepsy models, which has important implications for the clinical treatment of human temporal lobe epilepsy.
Results and discussion
To modify D-T7 and Tet1 peptides on the lipids@PL, the peptides were coupled with DSPE-PEG-Mal through a Michael addition reaction (Fig. S1†), and the coupling was confirmed by mass spectrometry (Fig. S2†). A microfluidic chip was used to synthesize D-T7/Tet1-lipids@PL with different D-T7/Tet1 ratios (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2, 1
2, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 2
1, and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (Fig. 1A). As controls, lipids@PL, D-T7-lipids@PL, and Tet1-lipids@PL were synthesized on the same chip. Transmission electron microscopy (TEM) and dynamic light scattering (DLS) showed that the D-T7/Tet1-lipids@PL NPs with various D-T7/Tet1 ratios had a typical core–shell structure and hydrodynamic diameters ranging from 58.89 ± 0.88 to 71.86 ± 0.31 nm (Fig. 1C–E, S3A and B†). We selected D-T7/Tet1-lipids@PL (2
1) (Fig. 1A). As controls, lipids@PL, D-T7-lipids@PL, and Tet1-lipids@PL were synthesized on the same chip. Transmission electron microscopy (TEM) and dynamic light scattering (DLS) showed that the D-T7/Tet1-lipids@PL NPs with various D-T7/Tet1 ratios had a typical core–shell structure and hydrodynamic diameters ranging from 58.89 ± 0.88 to 71.86 ± 0.31 nm (Fig. 1C–E, S3A and B†). We selected D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to characterize the physiochemical properties of the NPs. Compared to the size of NPs synthesized by a traditional method (124.30 ± 1.18 nm) (Fig. S3C and D†), the size of the NPs synthesized using the microfluidic chip was much smaller (68.93 ± 0.59 nm), and the synthesis speed was faster,33 demonstrating the advantage of the microfluidic chip. We tested the stability of various NPs, which are all stable at 4 °C for 10 days (Fig. S4†). D-T7/Tet1-lipids@PL (2
1) to characterize the physiochemical properties of the NPs. Compared to the size of NPs synthesized by a traditional method (124.30 ± 1.18 nm) (Fig. S3C and D†), the size of the NPs synthesized using the microfluidic chip was much smaller (68.93 ± 0.59 nm), and the synthesis speed was faster,33 demonstrating the advantage of the microfluidic chip. We tested the stability of various NPs, which are all stable at 4 °C for 10 days (Fig. S4†). D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) had a LTG encapsulation efficiency of approximately 85.85%, and a cumulative release rate of more than 50% within 72 h (Fig. S5†), indicating a high encapsulation capability and an appropriate drug release profile. UV-vis spectra characterization indicated that DSPE-PEG-D-T7 and DSPE-PEG-Tet1 encapsulation efficiency in D-T7/Tet1-lipids@PL (2
1) had a LTG encapsulation efficiency of approximately 85.85%, and a cumulative release rate of more than 50% within 72 h (Fig. S5†), indicating a high encapsulation capability and an appropriate drug release profile. UV-vis spectra characterization indicated that DSPE-PEG-D-T7 and DSPE-PEG-Tet1 encapsulation efficiency in D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was 83.33% and 71.34% respectively. The content of DSPE-PEG-D-T7 and DSPE-PEG-Tet1 in the D-T7/Tet1-lipids@PL (2
1) was 83.33% and 71.34% respectively. The content of DSPE-PEG-D-T7 and DSPE-PEG-Tet1 in the D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) solution was 57.33 and 43.05 μg mL−1 respectively (Fig. S6A and B†).
1) solution was 57.33 and 43.05 μg mL−1 respectively (Fig. S6A and B†).
We conducted experiments on the internalization of different DiI-labelled NPs in bEnd.3 cells which is a cell line from mice BBB. After 6 h of co-cultivation with the cells, the NPs displayed different intensities of fluorescence signals (Fig. S7A and C†). D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) showed higher uptake efficiency compared to other groups except Tet1-lipids@PL (Fig. S7B and D†).
1) showed higher uptake efficiency compared to other groups except Tet1-lipids@PL (Fig. S7B and D†).
We verified the internalization of various DiI-labelled NPs in hippocampal neurons and the human neuroblastoma (SY5Y) cell line. SY5Y cells were used as a control group to verify if the nanoparticles have a good targeting effect on neurons. According to the results of the confocal microscopy experiments, the NPs displayed different intensities of fluorescence signals. Except for Tet1-lipids@PL, the uptake efficiency of D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was higher than those of other groups (Fig. S8†). Quantitative data by flow cytometry revealed that among the experimental groups, D-T7/Tet1-lipids@PL (2
1) was higher than those of other groups (Fig. S8†). Quantitative data by flow cytometry revealed that among the experimental groups, D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) showed a higher level of internalization by the hippocampal neurons compared to other groups except Tet1-lipids@PL (Fig. S8, S9A and C†). This means that the Tet1 peptide plays a key role in targeting the neurons. Confocal microscopy images revealed that D-T7/Tet1-lipids@PL (2
1) showed a higher level of internalization by the hippocampal neurons compared to other groups except Tet1-lipids@PL (Fig. S8, S9A and C†). This means that the Tet1 peptide plays a key role in targeting the neurons. Confocal microscopy images revealed that D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) exhibited the highest fluorescence intensity in the hippocampal neurons among all the experimental groups, demonstrating its high internalization efficiency by neurons (Fig. S10A and B†). All the NPs showed a lower level of internalization in the SY5Y cells than those in the hippocampal neurons (Fig. S9B, D, S10B and D†), indicating the high specificity of the nanoparticles.
1) exhibited the highest fluorescence intensity in the hippocampal neurons among all the experimental groups, demonstrating its high internalization efficiency by neurons (Fig. S10A and B†). All the NPs showed a lower level of internalization in the SY5Y cells than those in the hippocampal neurons (Fig. S9B, D, S10B and D†), indicating the high specificity of the nanoparticles.
We verified the penetration ability of different NPs through an in vitro BBB model composed of a bEnd.3 monolayer and a hippocampal neuron layer (Fig. 2A). The bEnd3 cell is a mouse brain microvascular endothelial cell line which expresses the Tf receptor. D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was supposed to transcytosis the bEnd3 membranes by binding to the Tf receptor. Z-axis confocal imaging showed that different nanoparticles appeared in the bEnd.3 monolayer with different fluorescence intensities and penetration depth. Tet1-lipids@PL, although having the highest fluorescence inside the bEnd.3 monolayer, showed relatively low fluorescence intensity beneath the monolayer. This means that although Tet1-lipids@PL can bind to the bEnd.3 cells, they only have a weak chemotaxis tendency towards the hippocampal neurons beneath the bEnd.3 monolayer (Fig. 2B). D-T7/Tet1-lipids@PL (2
1) was supposed to transcytosis the bEnd3 membranes by binding to the Tf receptor. Z-axis confocal imaging showed that different nanoparticles appeared in the bEnd.3 monolayer with different fluorescence intensities and penetration depth. Tet1-lipids@PL, although having the highest fluorescence inside the bEnd.3 monolayer, showed relatively low fluorescence intensity beneath the monolayer. This means that although Tet1-lipids@PL can bind to the bEnd.3 cells, they only have a weak chemotaxis tendency towards the hippocampal neurons beneath the bEnd.3 monolayer (Fig. 2B). D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) had the largest penetration depth among all the experimental groups (Fig. 2B), which means that D-T7/Tet1-lipids@PL (2
1) had the largest penetration depth among all the experimental groups (Fig. 2B), which means that D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) had the most apparent chemotaxis tendency towards the neurons. Flow cytometry showed that all the NPs penetrated the BBB model and were internalized by the hippocampal neurons beneath the bEnd.3 monolayer (Fig. 2C and D). Among all the NP groups, D-T7/Tet1-lipids@PL (2
1) had the most apparent chemotaxis tendency towards the neurons. Flow cytometry showed that all the NPs penetrated the BBB model and were internalized by the hippocampal neurons beneath the bEnd.3 monolayer (Fig. 2C and D). Among all the NP groups, D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) showed the highest fluorescence intensity, demonstrating its excellent BBB penetration and neuron targeting performance (Fig. 2C and D).
1) showed the highest fluorescence intensity, demonstrating its excellent BBB penetration and neuron targeting performance (Fig. 2C and D).
To evaluate the BBB penetration ability of the nanoparticles, we used an in vivo imaging system (IVIS) to monitor the fluorescence distribution of DiR-labelled NPs in mice. Compared with other NPs, D-T7-lipids@PL and D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) showed a stronger fluorescence signal in the brain at 6- and 12-h time points (Fig. 3A, B, and S11A†). The presence of PEG in the nanoparticles can enhance the long-term circulation of the nanoparticles in mice, leading to increased fluorescence signals after 24 h. The mice were sacrificed and dissected after 24 h. The brains of the mice injected with D-T7/Tet1-lipids@PL (2
1) showed a stronger fluorescence signal in the brain at 6- and 12-h time points (Fig. 3A, B, and S11A†). The presence of PEG in the nanoparticles can enhance the long-term circulation of the nanoparticles in mice, leading to increased fluorescence signals after 24 h. The mice were sacrificed and dissected after 24 h. The brains of the mice injected with D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) showed the brightest fluorescent signal among all the experimental groups (Fig. 3C). We extracted the hippocampus from different experimental groups and found that the hippocampus of the mice treated with D-T7/Tet1-lipids@PL (2
1) showed the brightest fluorescent signal among all the experimental groups (Fig. 3C). We extracted the hippocampus from different experimental groups and found that the hippocampus of the mice treated with D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) had the strongest fluorescent signal, indicating its excellent performance to target the hippocampus region (Fig. 3D and S12†). To evaluate the neuron-targeting capability of D-T7/Tet1-lipids@PL (2
1) had the strongest fluorescent signal, indicating its excellent performance to target the hippocampus region (Fig. 3D and S12†). To evaluate the neuron-targeting capability of D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), the mouse brain tissues were dissected and NeuN (neuron marker) and GFAP (astrocyte marker) were stained specifically 6 h after the NP injection. Most of the D-T7/Tet1-lipids@PL (2
1), the mouse brain tissues were dissected and NeuN (neuron marker) and GFAP (astrocyte marker) were stained specifically 6 h after the NP injection. Most of the D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) NPs co-localized with the NeuN-labelled cells (Fig. 3E, S13 and S14†), indicating that D-T7/Tet1-lipids@PL (2
1) NPs co-localized with the NeuN-labelled cells (Fig. 3E, S13 and S14†), indicating that D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) can specifically target the neurons. A previous study reported that nanoparticles within 200 nm can also cross the BBB.5 The size of D-T7/Tet1-lipids@PL (2
1) can specifically target the neurons. A previous study reported that nanoparticles within 200 nm can also cross the BBB.5 The size of D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was less than 100 nm (Fig. 1D), so, it is easy for them to penetrate the BBB. Also, D-T7/Tet1-lipids@PL (2
1) was less than 100 nm (Fig. 1D), so, it is easy for them to penetrate the BBB. Also, D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) has dual targeting properties by binding to Tf receptors via the D-T7 peptide and binding to gangliosides on neurons via the Tet1 peptide. The Tf receptors have been reported to be expressed in the cerebral cortex and hippocampus, and are also widely expressed in the neurons.42 Thus, D-T7/Tet1-lipids@PL (2
1) has dual targeting properties by binding to Tf receptors via the D-T7 peptide and binding to gangliosides on neurons via the Tet1 peptide. The Tf receptors have been reported to be expressed in the cerebral cortex and hippocampus, and are also widely expressed in the neurons.42 Thus, D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) can not only target the BBB, but also accumulate in the neurons. Since both in vivo and in vitro experiments have proven that D-T7/Tet1-lipids@PL (2
1) can not only target the BBB, but also accumulate in the neurons. Since both in vivo and in vitro experiments have proven that D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) had the best BBB penetration and neuron targeting capability among the NPs, we selected it for further study. In addition, D-T7/Tet1-lipids@PL (2
1) had the best BBB penetration and neuron targeting capability among the NPs, we selected it for further study. In addition, D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was also present in the blood vessels of the brain (Fig. S15†). Apart from the brain, the liver and the spleen showed strong fluorescence, implying that the NPs tend to accumulate in the liver and can be metabolized quickly (Fig. S11B†).
1) was also present in the blood vessels of the brain (Fig. S15†). Apart from the brain, the liver and the spleen showed strong fluorescence, implying that the NPs tend to accumulate in the liver and can be metabolized quickly (Fig. S11B†).
We constructed two acute epilepsy models, the KA-initiated model (Fig. 4A) and the pilocarpine-initiated model (Fig. 4B), which were used for electrical signal monitoring and behavior analysis respectively. We verified the antiepileptic effect of D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on the KA epilepsy model. C57BL/6 mice were randomly divided into seven groups and injected with various NPs intravenously 2 h before the model construction (Fig. 4A). The PBS, LTG, lipids@PL, Tet1-lipids@PL, and D-T7-lipids@PL groups showed the onset of ictal seizures at about 30 min. However, ictal discharges start at about 1 h later (early ictal state, Fig. 4C, D, and S16†) in the D-T7/Tet1-lipids@PL (2
1) on the KA epilepsy model. C57BL/6 mice were randomly divided into seven groups and injected with various NPs intravenously 2 h before the model construction (Fig. 4A). The PBS, LTG, lipids@PL, Tet1-lipids@PL, and D-T7-lipids@PL groups showed the onset of ictal seizures at about 30 min. However, ictal discharges start at about 1 h later (early ictal state, Fig. 4C, D, and S16†) in the D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) group. About 2 h later (late ictal state), the local field potential (LFP) power (Fig. 4C) and amplitude (Fig. 4D) in the D-T7/Tet1-lipids@PL (2
1) group. About 2 h later (late ictal state), the local field potential (LFP) power (Fig. 4C) and amplitude (Fig. 4D) in the D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) group were obviously lower than those of other groups (Fig. 4C, D, and S16†). The D-T7-lipids@PL (2
1) group were obviously lower than those of other groups (Fig. 4C, D, and S16†). The D-T7-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) group, whether with a high (5 mg kg−1) or low (2 mg kg−1) LTG dose, exhibited apparent inhibitory effect on the seizures (Fig. 4C, D and S16†). Thus, D-T7/Tet1-lipids@PL (2
1) group, whether with a high (5 mg kg−1) or low (2 mg kg−1) LTG dose, exhibited apparent inhibitory effect on the seizures (Fig. 4C, D and S16†). Thus, D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) can delay and shorten the time of epileptic seizures and reduce the degree of epileptic seizures. In the experiments on the pilocarpine epilepsy model, 40 min after the injection of pilocarpine, the seizures occurred in the PBS (level 5), LTG (level 4), and lipids@PL (level 4) groups (Video S1†). By comparison, only mild seizures (level 1) occurred in the D-T7/Tet1-lipids@PL (2
1) can delay and shorten the time of epileptic seizures and reduce the degree of epileptic seizures. In the experiments on the pilocarpine epilepsy model, 40 min after the injection of pilocarpine, the seizures occurred in the PBS (level 5), LTG (level 4), and lipids@PL (level 4) groups (Video S1†). By comparison, only mild seizures (level 1) occurred in the D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) group after 1.5 h (Video S2†). 4 h later, the behavior of the D-T7/Tet1-lipids@PL (2
1) group after 1.5 h (Video S2†). 4 h later, the behavior of the D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) group returned to the normal state, while the mice in other groups were still epileptic (Video S3†). Thus, D-T7/Tet1-lipids@PL (2
1) group returned to the normal state, while the mice in other groups were still epileptic (Video S3†). Thus, D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) can delay the onset of epilepsy and reduce the degree of epileptic seizures. The electrophysiological signal monitoring and behavioral observation in the two acute epilepsy models confirmed that D-T7/Tet1-lipids@PL (2
1) can delay the onset of epilepsy and reduce the degree of epileptic seizures. The electrophysiological signal monitoring and behavioral observation in the two acute epilepsy models confirmed that D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) was excellent in the treatment of epilepsy.
1) was excellent in the treatment of epilepsy.
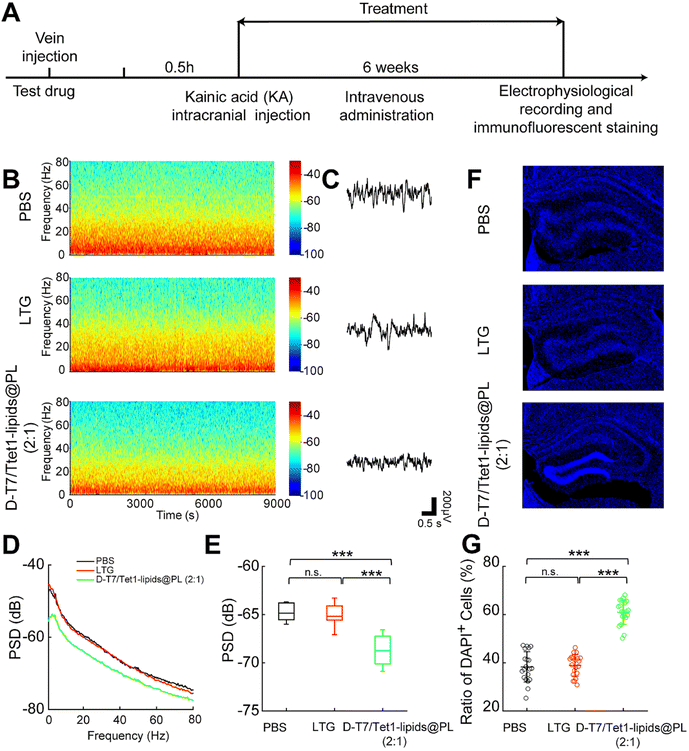
We further verified the antiepileptic effect of D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) on the KA-induced chronic epilepsy model (Fig. 5A). In the LFP results, spontaneous seizures in the dHPC of PBS and LTG groups were obviously observed, while the D-T7/Tet1-lipids@PL (2
1) on the KA-induced chronic epilepsy model (Fig. 5A). In the LFP results, spontaneous seizures in the dHPC of PBS and LTG groups were obviously observed, while the D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) group showed a relatively normal state (Fig. 5B and C). In addition, the LFP power of the D-T7/Tet1-lipids@PL (2
1) group showed a relatively normal state (Fig. 5B and C). In addition, the LFP power of the D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) group was significantly lower than those of other groups (Fig. 5D and E). Neuronal loss and dispersion of the granule cell layer of the dHPC are considered as the signs of chronic epilepsy.43,44 As shown in dHPC images, neuronal loss and dispersion of the granule cell layer of the dHPC in the mice treated with PBS or LTG were much more serious than those treated with D-T7/Tet1-lipids@PL (2
1) group was significantly lower than those of other groups (Fig. 5D and E). Neuronal loss and dispersion of the granule cell layer of the dHPC are considered as the signs of chronic epilepsy.43,44 As shown in dHPC images, neuronal loss and dispersion of the granule cell layer of the dHPC in the mice treated with PBS or LTG were much more serious than those treated with D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1), and the D-T7/Tet1-lipids@PL (2
1), and the D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) group had an intact hippocampal structure (Fig. 5F). The cell density of the granule cell layer in mice treated with PBS or LTG was significantly lower than that of the D-T7/Tet1-lipids@PL (2
1) group had an intact hippocampal structure (Fig. 5F). The cell density of the granule cell layer in mice treated with PBS or LTG was significantly lower than that of the D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) group (Fig. 5G). It is worth mentioning that D-T7/Tet1-lipids@PL (2
1) group (Fig. 5G). It is worth mentioning that D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) NPs were also located in cortex structures (Fig. 3E). As seizures usually propagate from the hippocampus to the cortex structures,45–47 our result suggested that D-T7/Tet1-lipids@PL (2
1) NPs were also located in cortex structures (Fig. 3E). As seizures usually propagate from the hippocampus to the cortex structures,45–47 our result suggested that D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) NPs may inhibit both the initiation and spreading of ictal discharges. The above data indicated that D-T7/Tet1-lipids@PL (2
1) NPs may inhibit both the initiation and spreading of ictal discharges. The above data indicated that D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) had an excellent therapeutic effect in treating epilepsy and had a long-term protective effect on the brain tissues.
1) had an excellent therapeutic effect in treating epilepsy and had a long-term protective effect on the brain tissues.
We evaluated the biocompatibility of D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) in vitro. Confocal microscopy showed that hippocampal neurons and bEnd.3 cells were viable after co-culturing with various NPs for 12 h (Fig. S17A and B†). CCK assays showed that after 24 h of incubation with an extremely high concentration of D-T7/Tet1-lipids@PL (2
1) in vitro. Confocal microscopy showed that hippocampal neurons and bEnd.3 cells were viable after co-culturing with various NPs for 12 h (Fig. S17A and B†). CCK assays showed that after 24 h of incubation with an extremely high concentration of D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) (1000 μg mL−1), more than 90% of the cells were alive (Fig. S17C and D†). Hemolysis tests indicated that even at an extremely high concentration (1000 μg mL−1), D-T7/Tet1-lipids@PL (2
1) (1000 μg mL−1), more than 90% of the cells were alive (Fig. S17C and D†). Hemolysis tests indicated that even at an extremely high concentration (1000 μg mL−1), D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) did not cause hemolysis (Fig. S18†). Hematological markers and biochemical parameters in mice blood showed that all the indexes of the D-T7/Tet1-lipids@PL (2
1) did not cause hemolysis (Fig. S18†). Hematological markers and biochemical parameters in mice blood showed that all the indexes of the D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)-treated mice were within the normal ranges in the acute (Fig. S19A†) and chronic (Fig. S20†) epilepsy models. Hematoxylin-eosin (H&E) staining showed no obvious pathological changes in the tissues of the mice treated with D-T7/Tet1-lipids@PL (2
1)-treated mice were within the normal ranges in the acute (Fig. S19A†) and chronic (Fig. S20†) epilepsy models. Hematoxylin-eosin (H&E) staining showed no obvious pathological changes in the tissues of the mice treated with D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) in the acute (Fig. S19B†) and chronic (Fig. S21†) epilepsy models. These results indicated that the series of lipids@PL NPs had good biocompatibility.
1) in the acute (Fig. S19B†) and chronic (Fig. S21†) epilepsy models. These results indicated that the series of lipids@PL NPs had good biocompatibility.
Conclusions
In this study, we used a microfluidic chip to synthesize a dual-targeting system (D-T7/Tet1-lipids@PL) with a core–shell structure to deliver lamotrigine for treating epileptic seizures. When the ratio of D-T7 to Tet1 was 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the D-T7/Tet1-lipids@PL NPs showed a maximum BBB penetration ability and the best targeting efficiency to the neurons both in vitro and in vivo. D-T7/Tet1-lipids@PL (2
1, the D-T7/Tet1-lipids@PL NPs showed a maximum BBB penetration ability and the best targeting efficiency to the neurons both in vitro and in vivo. D-T7/Tet1-lipids@PL (2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) realized excellent therapeutic effects on the seizures in both acute and chronic epilepsy models with high biosafety. Therefore, this nanocarrier may not only be used in treating epilepsy but its applications may also be extended to treating other disorders in the central nervous system such as glioma, Alzheimer's disease, and other psychiatric disorders.
1) realized excellent therapeutic effects on the seizures in both acute and chronic epilepsy models with high biosafety. Therefore, this nanocarrier may not only be used in treating epilepsy but its applications may also be extended to treating other disorders in the central nervous system such as glioma, Alzheimer's disease, and other psychiatric disorders.
Data availability
All other experimental data are available in the ESI.†Author contributions
Qinghong Hou, Wenfu. Zheng and Xingyu Jiang conceived the study; Qinghong Hou and Lulu Wang performed the experiments; Feng Xiao and Le Wang helped in the construction of the epileptic model; Xiaoyan Liu assisted in extracting hippocampal neurons. Qinghong Hou, Wenfu Zheng and Xingyu Jiang wrote the manuscript. All authors have given approval to the final version of the manuscript. Yi Lu, Lina Zhu, Wenfu Zheng, and Xingyu Jiang supervised the whole project.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
We thank the National Key R&D Program of China (2021YFF1200800, 2020YFA0908900, 2018YFA0902600, and 2021YFF1200100), National Natural Science Foundation of China (32071390 and 81730051), Guangdong Provincial Key Laboratory of Advanced Biomaterials (2022B1212010003), Shenzhen Key Laboratory of Smart Healthcare Engineering (ZDSYS20200811144003009), Shenzhen Science and Technology Program (KQTD20190929172743294), the Chinese Academy of Sciences (QYZDJ-SSW-SLH039), Shenzhen Bay Laboratory (SZBL2019062801004), Guangdong Innovative and Entrepreneurial Research Team Program (2019ZT08Y191), and Tencent Foundation through the XPLORER PRIZE for financial support. We would like to acknowledge the technical support from SUSTech CRF.Notes and references
- R. D. Thijs, R. Surges, T. J. O'Brien and J. W. Sander, Lancet, 2019, 393, 689–701 CrossRef.
- S. Shankar Raman, V. H. B. Narayanan and R. Durai, Assay Drug Dev. Technol., 2021, 19, 2–16 CrossRef CAS PubMed.
- M. A. Rogawski and W. Loscher, Nat. Rev. Neurosci., 2004, 5, 553–564 CrossRef CAS.
- O. Devinsky, A. Vezzani, T. J. O'Brien, N. Jette, I. E. Scheffer, M. de Curtis and P. Perucca, Nat. Rev. Dis. Prim., 2018, 4, 18024 CrossRef.
- Y. H. Tsou, X. Zhang, H. Zhu, S. Syed and X. Xu, Small, 2017, 13, 1701921 CrossRef PubMed.
- T. Liu, C. Wang, X. Gu, H. Gong, L. Cheng, X. Shi, L. Feng, B. Sun and Z. Liu, Adv. Mater., 2014, 26, 3433–3440 CrossRef CAS PubMed.
- L. Cheng, X. Wang, F. Gong, T. Liu and Z. Liu, Adv. Mater., 2020, 32, 1902333 CrossRef CAS.
- J. Wang, Y. Dong, Y. Li, W. Li, K. Cheng, Y. Qian, G. Xu, X. Zhang, L. Hu, P. Chen, W. Du, X. Feng, Y. Zhao, Z. Zhang and B.-F. Liu, Adv. Funct. Mater., 2018, 28, 1707360 CrossRef.
- J. Wang, W. Liu, Q. Tu, J. Wang, N. Song, Y. Zhang, N. Nie and J. Wang, Biomacromolecules, 2011, 12, 228 CrossRef CAS.
- X. Jiang, Y. Zheng, H. Chen, K. Leong, T. Wang and H. Mao, Adv. Mater., 2010, 22, 2556–2560 CrossRef CAS PubMed.
- X. Dong, R. K. Brahma, C. Fang and S. Yao, Chem. Sci., 2022, 13, 4239–4269 RSC.
- L. Chen, T. Zhao, M. Zhao, W. Wang, C. Sun, L. Liu, Q. Li, F. Zhang, D. Zhao and X. Li, Chem. Sci., 2020, 11, 2819–2827 RSC.
- H. Zhang, Z. Gao, X. Li, S. Ye and B. Tang, Chem. Sci., 2021, 12, 12429–12436 RSC.
- G. Yang, J. Tian, C. Chen, D. Jiang, Y. Xue, C. Wang, Y. Gao and W. Zhang, Chem. Sci., 2019, 10, 5766–5772 RSC.
- A. Babazadeh, F. Mohammadi Vahed and S. M. Jafari, J. Controlled Release, 2020, 321, 211–221 CrossRef CAS PubMed.
- K. R. Gajbhiye, A. Pawar, K. R. Mahadik and V. Gajbhiye, Colloids Surf., B, 2020, 187, 110770 CrossRef CAS.
- M. Agrawal, S. Saraf, S. Saraf, S. K. Dubey, A. Puri, R. J. Patel, Ajazuddin, V. Ravichandiran, U. S. Murty and A. Alexander, J. Controlled Release, 2020, 321, 372–415 CrossRef CAS PubMed.
- C. Diaz-Perlas, B. Oller-Salvia, M. Sanchez-Navarro, M. Teixido and E. Giralt, Chem. Sci., 2018, 9, 8409–8415 RSC.
- P. Zhao, Y. Wang, X. Kang, A. Wu, W. Yin, Y. Tang, J. Wang, M. Zhang, Y. Duan and Y. Huang, Chem. Sci., 2018, 9, 2674–2689 RSC.
- X. Li and X. Jiang, Adv. Drug Delivery, 2021, 128, 101 CrossRef PubMed.
- R. Dong, Y. Liu, L. Mou, J. Deng and X. Jiang, Adv. Mater., 2019, 31, 1805033 CrossRef CAS PubMed.
- Y. Liu and X. Jiang, Lab Chip, 2017, 17, 3960–3978 RSC.
- Q. Feng, L. Zhang, C. Liu, X. Li, G. Hu, J. Sun and X. Jiang, Biomicrofluidics, 2015, 9, 052604 CrossRef PubMed.
- L. Zhang, Q. Feng, J. Wang, J. Sun, X. Shi and X. Jiang, Angew. Chem., Int. Ed. Engl., 2015, 54, 3952–3956 CrossRef CAS PubMed.
- J. Sun, Y. Xianyu, M. Li, W. Liu, L. Zhang, D. Liu, C. Liu, G. Hu and X. Jiang, Nanoscale, 2013, 5, 5262–5265 RSC.
- X. Li, Q. Feng and X. Jiang, Adv. Healthcare Mater., 2019, 8, 1900672 CrossRef CAS PubMed.
- C. Zona, V. Tancredi, P. Longone, G. D'Arcangelo, M. D'Antuono, M. Manfredi and M. Avoli, Epilepsia, 2002, 43, 685–690 CrossRef CAS PubMed.
- H. Choi and M. J. Morrell, Expert Opin. Pharmacother., 2003, 2, 243–251 CrossRef.
- V. R. Yasam, S. L. Jakki, V. Senthil, M. Eswaramoorthy, S. Shanmuganathan, K. Arjunan and M. J. Nanjan, Expert Rev. Clin. Pharmacol., 2016, 9, 1533–1546 CrossRef CAS.
- J. Zhao, Z. Ye, J. Yang, Q. Zhang, W. Shan, X. Wang, Z. Wang, S. Ye, X. Zhou, Z. Shao and L. Ren, Biomaterials, 2020, 240, 119849 CrossRef CAS PubMed.
- L. Cai, C. Yang, W. Jia, Y. Liu, R. Xie, T. Lei, Z. Yang, X. He, R. Tong and H. Gao, Adv. Funct. Mater., 2020, 30, 1909999 CrossRef CAS.
- M. Yu, D. Su, Y. Yang, L. Qin, C. Hu, R. Liu, Y. Zhou, C. Yang, X. Yang, G. Wang and H. Gao, ACS Appl. Mater. Interfaces, 2019, 11, 176–186 CrossRef CAS PubMed.
- Q. Hou, L. Zhu, L. Wang, X. Liu, F. Xiao, Y. Xie, W. Zheng and X. Jiang, Nanoscale, 2022, 14, 3234–3241 RSC.
- X. Wei, C. Zhan, X. Chen, J. Hou, C. Xie and W. Lu, Mol. Pharm., 2014, 11, 3261–3268 CrossRef CAS.
- X. Wei, C. Zhan, Q. Shen, W. Fu, C. Xie, J. Gao, C. Peng, P. Zheng and W. Lu, Angew. Chem., Int. Ed. Engl., 2015, 54, 3023–3027 CrossRef CAS.
- J. Liu, Q. Teng, M. Garrity-Moses, T. Federici, D. Tanase, M. J. Imperiale and N. M. Boulis, Neurobiol. Dis., 2005, 19, 407–418 CrossRef CAS PubMed.
- P. Wang, X. Zheng, Q. Guo, Q. Yang, X. Pang, K. Qian, W. Lu, Q. Zhang and X. Jiang, J. Controlled Release, 2018, 279, 220–233 CrossRef CAS.
- D. S. H. Chu, J. G. Schellinger, M. J. Bocek, R. N. Johnson and S. H. Pun, Biomaterials, 2013, 34, 9632–9637 CrossRef CAS PubMed.
- A. Mathew, T. Fukuda, Y. Nagaoka, T. Hasumura, T. Morimoto, Y. Yoshida, T. Maekawa, K. Venugopal and D. S. Kumar, PLoS One, 2012, 7, e32616 CrossRef CAS.
- M. L'evesque and M. Avoli, Neurosci. Biobehav. Rev., 2013, 37, 2887–2899 CrossRef.
- M. L'evesque, G. Biagini, M. d. Curtis, V. Gnatkovsky, J. Pitsch, S. Wang and M. Avoli, Neurosci. Biobehav. Rev., 2021, 130, 274–291 CrossRef PubMed.
- T. Moos, J. Comp. Neurol., 1996, 375, 675–692 CrossRef CAS.
- V. Duveau, A. Madhusudan, M. Caleo, I. Knuesel and J. M. Fritschy, Hippocampus, 2011, 21, 935–944 CAS.
- V. Riban, V. Bouilleret, B. T. Pham-Lê, J. M. Fritschy, C. Marescaux and A. Depaulis, Neuroscience, 2002, 112, 101–111 CrossRef CAS.
- K. O. Cho, Z. R. Lybrand, N. Ito, R. Brulet, F. Tafacory, L. Zhang, L. Good, K. Ure, S. G. Kernie, S. G. Birnbaum, H. E. Scharfman, A. J. Eisch and J. Hsieh, Nat. Commun., 2015, 6, 6606 CrossRef CAS PubMed.
- J. Engel, Neuroscientist, 2001, 7, 340–352 CrossRef.
- Y. Lu, C. Zhong, L. Wang, P. Wei, W. He, K. Huang, Y. Zhang, Y. Zhan, G. Feng and L. Wang, Nat. Commun., 2016, 7, 10962 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2sc03298h |
| ‡ These authors contributed equally to this work |
| This journal is © The Royal Society of Chemistry 2022 |