Open Access Article
Open Access ArticleMultifunctional stimuli-responsive chemogenetic platform for conditional multicolor cell-selective labeling†
Pengfei
Chen
,
Rui
Wang
,
Ke
Wang
,
Jiao-Na
Han
,
Shi
Kuang
,
Zhou
Nie
 * and
Yan
Huang
* and
Yan
Huang
 *
*
State Key Laboratory of Chemo/Biosensing and Chemometrics, College of Chemistry and Chemical Engineering, Hunan Provincial Key Laboratory of Biomacromolecular Chemical Biology, Hunan University, Changsha, 410082, P. R. China. E-mail: niezhou.hnu@gmail.com; yanhuang@hnu.edu.cn
First published on 3rd October 2022
Abstract
Multicolor conditional labeling is a powerful tool that can simultaneously and selectively visualize multiple targets for bioimaging analysis of complex biological processes and cellular features. We herein report a multifunctional stimuli-responsive Fluorescence-Activating and absorption-Shifting Tag (srFAST) chemogenetic platform for multicolor cell-selective labeling. This platform comprises stimuli-responsive fluorogenic ligands and the organelle-localizable FAST. The physicochemical properties of the srFAST ligands can be tailored by modifying the optical-tunable hydroxyl group with diverse reactive groups, and their chemical decaging process caused by cell-specific stimuli induces a conditionally activatable fluorescent labeling upon binding with the FAST. Thus, the resulting switch-on srFASTs were designed for on-demand labeling of cells of interest by spatiotemporally precise photo-stimulation or unique cellular feature-dependent activation, including specific endogenous metabolites or enzyme profiles. Furthermore, diverse enzyme-activatable srFAST ligands with distinct colors were constructed and simultaneously exploited for multicolor cell-selective labeling, which allow discriminating and orthogonal labeling of three different cell types with the same protein tag. Our method provides a promising strategy for designing a stimuli-responsive chemogenetic labeling platform via facile molecular engineering of the synthetic ligands, which has great potential for conditional multicolor cell-selective labeling and cellular heterogeneity evaluation.
Introduction
Elucidating the localization and the activities of intracellular biomolecules for a deep understanding of biological processes in cells and organisms calls for effective imaging tools.1–4 Chemogenetic strategy, as a powerful toolkit, provides many options and great potential for visualizing biological events in real-time and in situ. In this strategy, genetically encoded activators (consisting of peptides or proteins) can specifically activate small synthetic ligands (fluorogens) to a fluorescent state by transient or covalent binding.5–7 Such hybrid approaches combine the excellent targeting-specificity of the genetically encoded protein tags with superior photophysical properties as well as tunability of the small synthetic ligands, which ensures a great deal of experimental versatility and designability.6 Accordingly, SNAP-tags,8,9 HaloTags,10 PYP-tags,11,12 fluorogen-activating proteins (FAPs)13 and fluorescence-activating and absorption-shifting Tags (FASTs)14–16 have been developed.As an emerging member of chemogenetic strategy, the FAST system contains a 14 kDa protein tag, which can non-covalently bind with a series of cell-permeant and nontoxic 4-hydroxybenzylidene rhodanine (HBR) derivatives and activate their fluorescence.14–16 The small size of the tag and its instantaneous and reversible binding with the ligands endow the FAST system with additional excellent properties such as (a) minimal functional perturbation to the target proteins, (b) excellent photophysical properties such as brightness and anti-photobleaching, and (c) much shorter maturation time than GFP-like fluorescent proteins.14 By changing the substituents on the 4-hydroxybenzyl moiety of the ligands, colours of the FAST:fluorogen complex have spanned from green-yellow to far-red, significantly improving its spectral tunability.15 Although the fluorogen promiscuity of the FAST system facilitates spectral flexibility, it is still challenging for FASTs to simultaneously label multiple targets with different colors because of indiscriminate recognition of structurally similar ligands by its protein tag. To address this problem, directed evolution of the FAST presented a feasible strategy to develop orthogonal labeling systems. Accordingly, two FAST variants that can selectively bind with different colored ligands (HMBR or HBR-3,5DOM) were recently developed, allowing for selective labeling of two targets with dual colors.17 However, further expanding orthogonal FAST tagging proteins requires laborious procedures to implement protein evolution and sophisticated multiple gene expressions. Hence, these challenges may present an opportunity for seeking additional solutions toward this problem.
Besides manipulation of FASTs, ligand engineering is an alternative approach with the potential to achieve multicolor selective labeling based on the identical FAST, which is yet to be demonstrated. Rationally designed ligands capable of responding to particular targets independently18–25 may offer an opportunity to realize not only conditional selective labeling, but also multicolor selective labeling. Given that complex biological samples include many diverse cell types, cellular heterogeneity in phenotypes and genotypes, a fundamental property of cellular ecosystems,26–28 might function as a key factor to enable conditional multicolor labeling of specific cell types of interest. The unique spatiotemporal distribution, metabolite profile, and endogenous enzymes of a cell of interest not only play a central relevance to its biological function, but are also highly useful as identification markers for discriminating diverse cell types.28–31 It is enlightening that these cellular characteristics may act as specific triggers for stimuli-responsive ligands with different colors to achieve heterogeneity-dependent cell-selective labeling, providing an additional approach for on-demand multicolor labeling of specific cell types without the requirement of diverse protein tags.
Herein, taking advantage of the cellular heterogeneity features and rational tunability of the ligands, we introduce a FAST-derived strategy to construct a multifunctional stimuli-responsive labeling platform, namely stimuli-responsive FAST (srFAST), for conditional multicolor cell-selective labeling. The srFAST consists of two parts: one is the FAST scaffold, which can activate the fluorescence of ligands exquisitely and provide an excellent subcellular location through genetic fusion with a localizable peptide or protein; the other is the rationally designed HBR-derived “caged” ligands, which are endowed with responsiveness to diverse stimuli such as light-inducible spatiotemporal activation, unique small-molecule metabolites, or the enzymatic expressions of specific cell types. These stimuli-activatable ligands can be conditionally activated by cell-specific physical or biochemical characteristics to enable on-demand fluorescent labeling of FAST-fused proteins, only in target cells. Meanwhile, using multicolor enzyme-responsive srFASTs, different cell types were selectively labeled with orthogonal multicolor fluorescence dependent on their independent endogenous enzymatic signatures. We envision that this proposed platform would enrich the applications of the FAST system in cell-specific multicolor labeling and provide a meaningful tool for cellular heterogeneous studies.
Results and discussion
Design principle of the srFAST platform
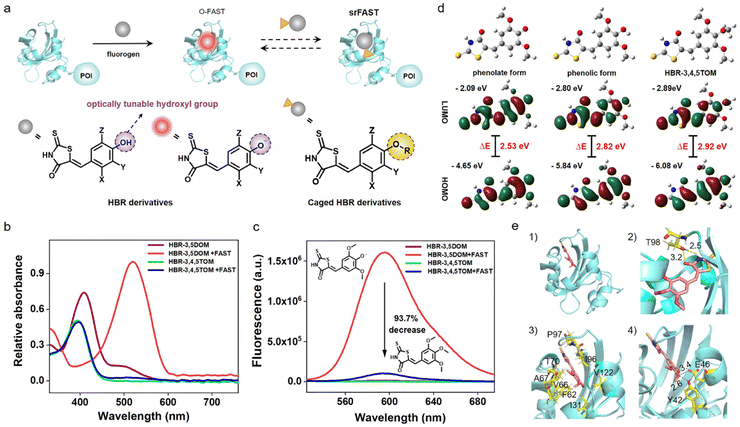
As an emerging fluorogenic chemogenetic protein tag, the FAST system features significant fluorescent enhancement and large absorption redshift of its HBR-derived ligands upon binding with the FAST, the protein (Fig. 1a and S1†).14,15 These noticeable spectroscopic changes depend on the inhibition of the intramolecular rotation of ligands by FAST binding and the protein microenvironment-caused deprotonation of the ligands' phenolic hydroxyl group. In addition to the key role in the spectroscopic variation, its characteristic hydroxyl group is likely to function as an optical tunable group ready for modification-mediated regulation, which has been widely exploited in various fluorescent dyes32–35 but still unexplored in the FAST system. Accordingly, we sought to investigate whether caging the hydroxyl group of the ligands can modulate the optical properties of the FAST system.Red-emissive ligand HBR-3,5DOM and its methoxy derivative HBR-3,4,5TOM were prepared for comparison and synthesized in one step by condensation of rhodanine to the corresponding substituted 4-hydroxy-benzaldehyde. We first investigated the photophysical properties of the compounds (Fig. 1b and c). The maximal absorption peak of HBR-3,4,5TOM is 393 nm, a slight blue shift to that of HBR-3,5DOM in the phenolic form (409 nm), and both compounds have little fluorescence in PBS buffer, with relative fluorescence quantum yields of 0.05% and 0.11%, respectively. In the presence of the FAST, deprotonated HBR-3,5DOM was stabilized and showed a large absorption redshift (409 nm to 518 nm) and a significant fluorescence increase at 600 nm. The FAST:HBR-3,5DOM complex (the original FAST system, O-FAST) exhibits a relative fluorescence quantum yield of 32% (Table S1†) comparable to that reported (31%) in the literature.15 However, HBR-3,4,5TOM exhibited a negligible absorption change, and only a weak fluorescence upon FAST binding with a relative fluorescence quantum yield of 4.4%, which was about 6.3% of that of the O-FAST at 600 nm. Time-dependent density functional theory (TDDFT) calculations revealed that the protonation and methylation of the oxygen atom increase the excitation energy gaps from 2.53 (HBR-3,5DOM in phenolate form) to 2.82 (HBR-3,5DOM in phenolic form) and 2.92 eV (HBR-3,4,5TOM), respectively, supporting the similarly blue-shifted absorption of the FAST:HBR-3,4,5TOM complex and free HBR-3,5DOM in comparison with the O-FAST. A close examination of the S1 excited state (Fig. S2†) showed that the electrons on the LUMO of HBR-3,4,5TOM and HBR-3,5DOM in the phenolic form are mainly located on the rhodanine framework, plausibly reasoning the decrease in the fluorescence efficiency of the FAST:HBR-3,4,5TOM complex due to intramolecular charge transfer (ICT).32,35 Further fluorescence competition assay (Fig. S3†) showed that HBR-3,4,5TOM could moderately interfere with the association between HBR-3,5DOM and the FAST when its concentration is significantly higher. The proposed Kd (28.2 μM) of HBR-3,4,5TOM upon FAST binding was much greater than that of HBR-3,5DOM (0.97 μM),15 indicating a decrease in affinity. Molecular docking studies (Fig. 1d) demonstrated that HBR-3,4,5TOM could enter the binding cavity of the FAST and be stabilized by the hydrogen bond with T98 as well as apolar interactions with I31, F62, V66, A67, T70, I96, P97 and V122. However, the hydrogen-bond network formed by the conserved residues Y42 and E46 to stabilize the phenolate anion36,37 in the O-FAST disappeared in the FAST:HBR-3,4,5TOM complex, potentially resulting in a decrease in affinity. Taken together, modifying the hydroxyl group of the ligand could diminish its binding with the FAST and efficiently reduce the complex fluorescence. Hence, the feature of the optically tunable hydroxyl group provides a good entrance to designing activatable ligands for a versatile srFAST platform.
Photo-srFAST for spatiotemporal cell-selective labeling of the nucleus
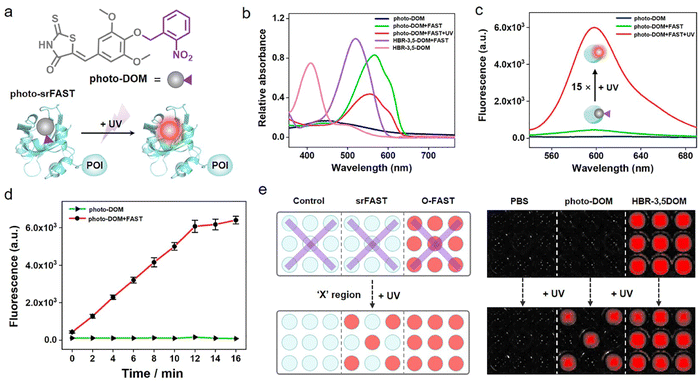
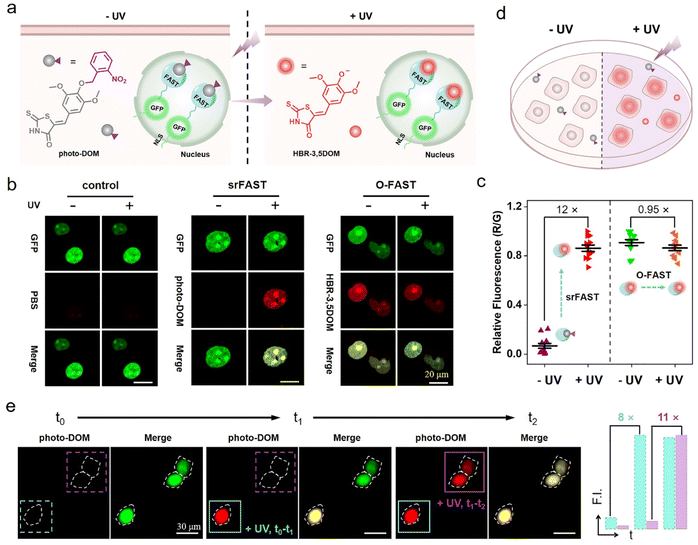
Light activation as an external regulation and control means has been widely used in spatiotemporal controlled imaging due to its quick, straightforward, and precisely controllable properties.38,39 Here, an optically controlled tagging system is explored as the preliminary attempt to achieve spatiotemporally selective labeling of cells. Here, the photoactivatable ligand photo-DOM containing a 2-nitrobenzyl group39,40 was successfully synthesized through a one-step nucleophilic reaction of HBR-3,5DOM and 1-(bromomethyl)-2-nitrobenzene (Fig. 2a). In comparison with HBR-3,5DOM, photo-DOM has a greater absorption enhancement with a large redshift from 445 nm to 567 nm after binding with the FAST (Fig. 2b), but shows weak fluorescence at 600 nm (Fig. 2c). Further competition assay indicated that 2-nitrobenzyl group modification of photo-DOM partially diminished the binding affinity with the FAST (Kd = 7.4 μM, Fig. S4†). Under UV irradiation (λ = 365 nm, 3 mW cm−2), the fluorescence intensity of the FAST:photo-DOM complex at 600 nm increased rapidly, and tended to be stable after 12 min with a significant enhancement about 15 times (Fig. 2c, d, and S5†), which are consistent with the uncaging dynamics of the same caging group reported in other literatures.40,41 These results proved the feasibility of our photo-responsive FAST system, named photo-srFAST. To demonstrate the ability of the photo-srFAST for spatiotemporally selective labeling, we performed patterned imaging in a 96-well plate. As shown in Fig. 2e, the photo-srFAST could pattern a specific “X” area with red fluorescence after UV irradiation. However for the O-FAST, red fluorescence was detected in all areas without selectivity. Thus, photo-decaging of the ligand's spectrum-tunable hydroxyl group is an efficient approach to develop srFASTs, and the constructed photo-srFAST can efficiently respond to photostimulation with an excellent S/N ratio.Then, we evaluated the feasibility of photo-srFAST in spatiotemporally selective labeling of target cells (Fig. 3a). The nucleus-localized FAST fusion protein (NLS-sfGFP-FAST) was transiently expressed in HEK293T cells, where the nuclear localization signal (NLS) and super-folded green fluorescent protein (sfGFP) were fused at FAST's N-terminal sequentially. As can be seen from Fig. 3b, the nucleus of the cells emitted the typical green fluorescence of sfGFP under the excitation of 488 nm, which confirmed the successful expression and localization of the FAST fusion protein. Being incubated with photo-DOM did not induce any detectable signal. After being illuminated with UV (λ = 365 nm, 3 mW cm−2) for 4 min, the cells appeared with red fluorescence, which was colocalized well with the green fluorescence, and the fluorescence intensity was significantly enhanced about 12 times after irradiation of 10 min (Fig. 3b, c and S6†), indicating the gradual formation of the photo-decaged fluorogenic complex. Flow cytometry analysis (Fig. S7†) also confirmed that photo-DOM could light up the target cell populations after UV illumination. Thus, photo-DOM is cell-permeant with a negligible fluorescence background, and the photo-srFAST is suitable for selective labeling of cells with a rapid and significant fluorescence response under UV irradiation. Comparatively, in the cells incubated with HBR-3,5DOM (the original FAST ligand) (Fig. 3b and c), both red and green fluorescence remained bright before and after UV irradiation, and the red fluorescence from the O-FAST was bleached by 15% after 10 min UV irradiation (Fig. S8†). In contrast, only the cells located at the selected area exposed to UV irradiation showed red fluorescence that merged as yellow in the presence of photo-DOM, while other cells out of the irradiated area remained green (Fig. 3d and S9†), suggesting spatially selective cell labeling by the photo-srFAST. At the same time, sequential labeling of the selected cells (Fig. 3e) was performed on a Leica DMi8 inverted microscope with an optical manipulative module, where the time and area of illumination could be finely controlled by its laser (λ = 405 nm, 40 mW cm−2). When the cells at specific locations were exposed to the laser in sequence, red fluorescence appeared instantaneously (5 s) and sequentially with 8 or 11-fold fluorescence enhancement, respectively, allowing for the near real-time implementation of the cell-specific labeling processes. Notably, even the adjacent cells in 50 μm can be manipulated precisely. CCK8 assay (Fig. S10†) showed that neither photo-DOM nor illumination had an obvious influence on the cell survival rate under the experimental conditions. Taken together, the photo-srFAST enables us to selectively and quickly label cells with high spatiotemporal imaging resolution through controlling UV irradiation, which has a potential application in spatiotemporal labeling and analysis.
H2O2-srFAST for metabolite-responsive cell-selective labeling of mitochondria
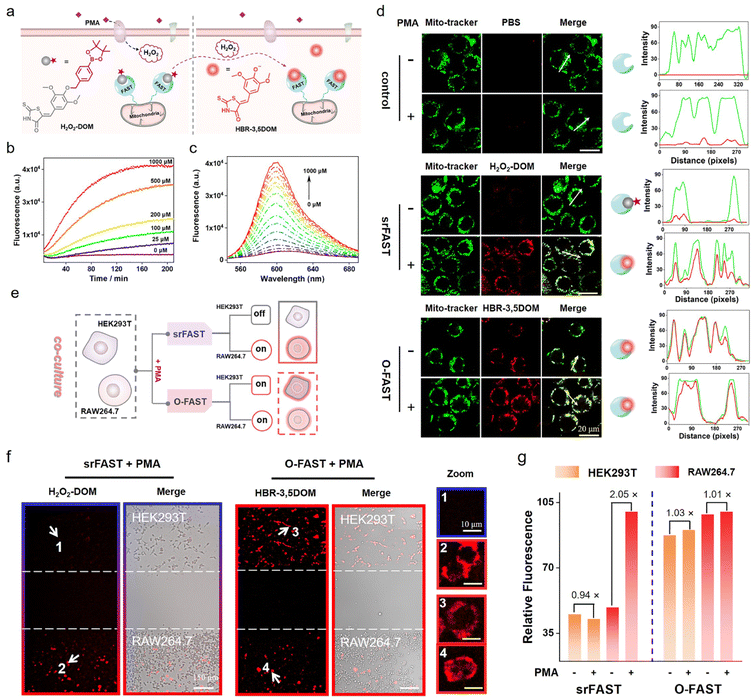
To demonstrate the versatility and expansibility of our srFAST platform, we sought to fabricate another stimuli-responsive ligand that can be specifically activated by a particular metabolite for cell-selective labeling. Generally, different cell types respond distinctively to the fluctuating environment and external stimuli and produce different metabolites under certain stress, leading to heterogeneous cellular responses.26,27 For example, respiratory burst is a rapid release process of reactive oxygen species (ROS) usually observed in immune cells, such as macrophages, as a pathogen-defending mechanism. Phorbol 12-myristate 13-acetate (PMA) is a potent stimulant of respiratory burst of macrophages, enabling activation of NADH oxidase and consequently massive generation of hydrogen peroxide and other ROS. Recently, mounting evidence supported that H2O2 produced by respiratory burst has a profound effect on macrophage signaling,42–44 implying that respiratory burst and its product H2O2 might function as the metabolic cue for selective identification and labeling of macrophages. Herein, we attempt to develop a H2O2-responsive FAST system, named H2O2-srFAST, for cell-selective labeling based on the heterogeneity of cellular response to a respiratory burst stimulant.Accordingly, we developed a H2O2-activatable ligand, H2O2-DOM, based on borate chemistry, an effective approach to build a turn-on probe of H2O2.42,43 H2O2-DOM underwent an absorption redshift from 540 nm to 575 nm upon FAST binding (Fig. S11†), and the resulting FAST:H2O2-DOM complex exhibited relative weak fluorescence at 600 nm in comparison with the O-FAST at an equimolecular concentration (Fig. S12†). Furthermore, the competition assay between H2O2-DOM and HBR-3,5DOM revealed a moderate affinity between H2O2-DOM and the FAST (Kd = 5.0 μM, Fig. S13†). Addition of H2O2 triggered a remarkable enhancement in fluorescence intensity due to H2O2-mediated boronate decaging of H2O2-DOM (Fig. 4b, c and S14†). A titration experiment presented that the emission intensity of the H2O2-srFAST at 600 nm exhibited a linear response toward H2O2 in the range of 5–250 μM with the detection limit of 1.5 μM (Fig. 4c and S14a†), which fits the endogenous H2O2 concentration in macrophages under PMA stimulation.42 Other compounds except for H2O2 generated negligible fluorescence in PBS buffer, illustrating the high selectivity of the H2O2-srFAST (Fig. S14b†).
Next, we investigated whether the H2O2-srFAST can be used for H2O2-responsive labeling of cells. HEK293T cells expressing the mitochondrial-targeted FAST protein (mito-FAST, the mitochondrial targeting sequence from subunit VIII of human cytochrome c oxidase was fused at FAST's N-terminal) were treated with H2O2-DOM and a commercialized mito-tracker (a specific dye for the localization of mitochondria in living cells). After the addition of 200 μM H2O2, bright red fluorescence was observed in cells, which overlapped with the green fluorescence from the mito-tracker (Fig. S15†). Thus, H2O2-DOM is cell-permeant with a low fluorescence background, and the H2O2-srFAST can respond to the micromolar level of H2O2 and label the mitochondria in living cells. Furthermore, RAW 264.7 macrophages which can produce endogenous H2O2 under PMA stimulation were used to test the potential of our H2O2-srFAST for target metabolite-responsive labeling of cells. As shown in Fig. 4d, in the absence of PMA, cells incubated with H2O2-DOM showed negligible red fluorescence. After PMA stimulation, bright red fluorescence was detected, which was perfectly colocalized with the green fluorescence from the mito-tracker. Flow cytometry analysis (Fig. S16†) also depicted that the red fluorescence intensity from the cell populations stimulated by PMA is about 4 times of that without PMA treatment. By contrast, the cells incubated with HBR-3,5DOM maintained red fluorescence before and after PMA stimulation. In addition, CCK8 assay suggested that H2O2-DOM had no obvious toxic or adverse effects on cells under experimental conditions (Fig. S17†). Thus, the H2O2-srFAST could be adapted for H2O2-responsive labeling of cells.
Further, the H2O2-srFAST is used for selective labeling of cells which can respond to PMA-stimulation and produce H2O2. Here, RAW 264.7 macrophages and HEK293T cells which respond differently under PMA stimulation are selected (Fig. S18†). Both cells expressing mito-FAST were co-cultured and stimulated with PMA, and then incubated with H2O2-DOM or HBR-3,5-DOM. For the O-FAST, it is impossible to distinguish the signals between the two different cell lines, since they were both labeled with similar red fluorescence (Fig. 4e–g). The merged images suggested the much higher transfection efficiency of HEK293T cells than RAW 264.7 cells. Even then we can easily distinguish the two co-cultured cell lines with the H2O2-srFAST. Only RAW 264.7 cells were labeled with strong red fluorescence due to the high level of respiratory burst-caused endogenous H2O2, with the signal intensity more than 2 times that from HEK293T cells (Fig. 4e–g and S19†). Therefore, the H2O2-srFAST can distinguish the cellular response of different cell types and selectively label the cell line of interest with the featured metabolite, demonstrating that our proposed srFAST platform has extensive potential in studying specific cell populations with metabolic heterogeneity.
Multicolor enzyme-srFASTs for protein-dependent orthogonal labeling of multiple cells
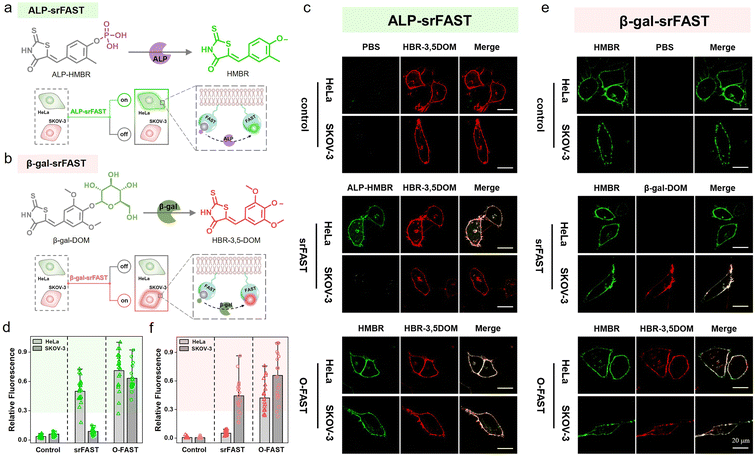
Encouraged by cell-selective labeling based on light-triggered spatiotemporal activation and specific metabolite-dependent chemical decaging, we further adapted the srFAST platform relying on cellular heterogeneity at the expression level of endogenous enzymes. Unique cell types have cell-specific endogenous enzyme expression and activity profiles to maintain diverse cellular functions, presenting a fundamental mechanism of cellular differentiation and heterogeneity. Moreover, aberrant expression of crucial enzymes in growth and metastasis regulation is highly associated with cancer development.28 For instance, alkaline phosphatase (ALP), a hydrolase in phosphate metabolism and associated with cell differentiation and viability, is overexpressed in several tumor cell lines including HeLa cells (a cell line of cervical cancer).44–46 β-Galactosidase (β-gal) removes galactose residues from substrates such as gangliosides, glycoproteins, and sphingolipids. It is known as an important biomarker for cell senescence and primary ovarian cancers, and is overexpressed in SKOV-3 cells (a cell line of ovarian cancer).47–49 Comparatively, both enzymes are expressed at a low level in normal cells, such as HEK293T cells.50,51 Thus, these cell-specific endogenous enzymes can be exploited to discriminate distinct cell types. We attempted to design two enzyme-activatable ligands with different colors, termed enzyme-srFASTs, for orthogonal multicolor selective labeling of cells with a distinct enzyme expression.For responding to ALP, a ligand ALP-HMBR was designed by blocking the hydroxyl group of the green-emissive ligand HMBR with a phosphate group (Fig. 5a). The FAST:ALP-HMBR complex (ALP-srFAST) displayed very weak fluorescence at 540 nm (Fig. S20 and S21†) and their interaction affinity was moderate (Kd = 5.4 μM, Fig. S22†). As expected, the phosphate group of ALP-HMBR could be specifically hydrolyzed to generate HMBR when ALP was presented (Fig. S23†),45,46,50 resulting in the recovery of green fluorescence of the decaged FAST:HMBR complex (Fig. S24†). The emission intensity of the ALP-srFAST at 540 nm is linearly proportional to the ALP concentration in the range of 2.0–50.0 U L−1 (Fig. S24†). Similarly, we developed the ligand β-gal-DOM by modifying HBR-3,5DOM with a galactose group and obtained a β-gal-srFAST (Fig. 5b). Unlike other activatable HBR-3,5DOM derivatives, β-gal-DOM has no detectable change in the absorption and fluorescence spectra before and after FAST addition (Fig. S26†). Ligand competition experiments revealed that the FAST was able to bind β-gal-DOM with low affinity (Kd = 16.3 μM, Fig. S27†). Addition of β-gal caused a remarkable fluorescence recovery of the FAST:β-gal-DOM complex at 600 nm owing to the hydrolyzation of the galactose group from β-gal-DOM, and there was an obvious linear relationship with the β-gal concentration from 0.01 to 0.12 U mL−1 (Fig. S28 and S29a†).47–49,51 Thanks to the excellent substrate specificity, both enzyme-srFASTs exhibited good selectivity toward their targets, including the modified ligands, respectively (Fig. S24d, S25 and S29b†). Collectively, srFASTs responding to two enzymes with distinct colors were developed in vitro.
For live-cell labeling, good biocompatibility of ALP-HMBR and β-gal-DOM was firstly confirmed for their negligible influence on cell viability in CCK8 assays (Fig. S30 and S31†). Next, the feasibility of the ALP-srFAST for efficient fluorogenic labeling of the cells with highly expressed endogenous ALP was confirmed. Both cell imaging and flow cytometry experiments exhibited that HeLa cells expressing the membrane-localized FAST (mem-FAST, the membrane localization signal GAP43 was fused at FAST's N-terminal) appeared with typical green fluorescence of the FAST:HMBR complex on their cell membrane when treated with ALP-HMBR, while there was no obvious fluorescence in the cells pre-treated with Na3VO4, a typical ALP inhibitor (Fig. S32†). Likewise, the cytomembrane of SKOV-3 cells overexpressing β-gal was labeled by the β-gal-srFAST with red fluorescence, confirmed by cell imaging and flow cytometric analysis (Fig. S33†). Together, these results proved that endogenous enzymes are potent markers for responsive labeling of target cells.
Further, the labeling selectivity of the enzyme-srFASTs was evaluated. Here, both HeLa and SKOV-3 cells transfected with the pcDNA3.1-mem-fast were separately incubated with the mixed ligands, where the uncaged ligands were devoted to verifying the successful expression and localization of mem-FAST. For the ALP-srFAST group (Fig. 5c and d), green fluorescence was only observed in ALP-positive HeLa cells, and the average fluorescence intensity was 6 times that of SKOV-3 cells, suggesting that the ALP-srFAST can optionally label HeLa cells. For the β-gal-srFAST group (Fig. 5e and f), only β-gal-positive SKOV-3 cells were stained by red fluorescence, with the average fluorescence intensity 8 times that of HeLa cells. However, for the corresponding O-FAST group, both cell lines incubated with the mixed original ligands were indiscriminately labeled with the same green or red fluorescence (Fig. 5c–f). In general, the two-color enzyme-srFASTs could respond to multiplex enzymes and label different cancer cells with distinct colors, opening novel prospects for multiple cell line discrimination and analysis.
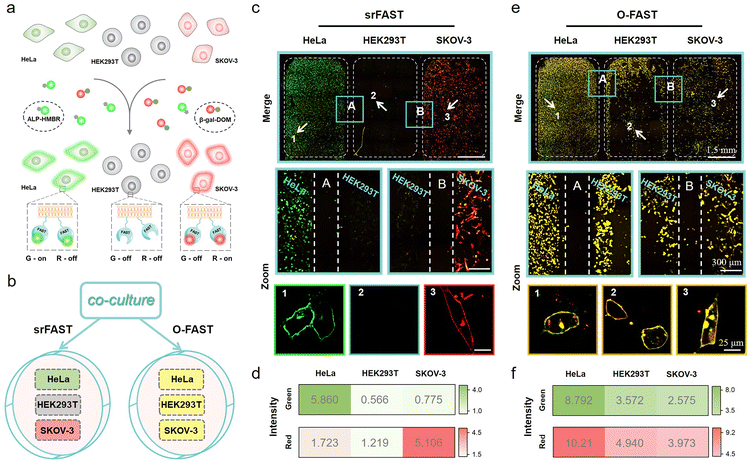
Finally, we sought to study the potential of multicolor enzyme-srFASTs for simultaneous orthogonal labeling of distinct cell lines in a complex multi-cell co-culture system (Fig. 6a). According to the different expression levels of the two enzymes, two cancer cell lines, HeLa (ALP+, β-gal−) and SKOV-3 (ALP−, β-gal+), and one normal cell line HEK293T (ALP−, β-gal−) were chosen. The three types of cells expressing the same mem-FAST were co-cultured, and then incubated with the mixture of ALP-HMBR and β-gal-DOM (srFAST) or HBR-3,5DOM and HMBR (O-FAST). For the srFAST group, it is quite easy to distinguish the three cell types, as HeLa cells were labeled with green fluorescence and SKOV-3 cells were labeled with red fluorescence, whereas no obvious fluorescence appeared in HEK293T cells (Fig. 6b–d and S34†). For the O-FAST group, comparatively, all cells were labeled with green and red fluorescence, merging as yellow equally (Fig. 6b, e, f and S34†). Therefore, multicolor enzyme-srFASTs are suitable for selectively labeling cells with orthogonal colors via the identical protein tag, and distinguishing different cell types. The ability to respond to multiple targets by using the same protein tag but different target-responding ligands with various spectral properties provides unprecedented experimental versatility and flexibility for multicolor imaging, without the need for reengineering. It is reasonable to envision that if there are cell lines co-overexpressing the two enzymes, they could be observed in yellow by employing multicolor enzyme-srFASTs. Specific labeling and distinguishing three or four cell lines with only one protein tag is a promising step toward overcoming the fluorogen promiscuity of the original FAST system, which also opens up exciting prospects for conditional multicolor cell-selective labeling and cellular heterogeneity monitoring at multiplex levels.
Conclusions
In summary, we developed a multifunctional stimuli-responsive chemogenetic platform (srFAST platform) for conditional cell-selective labeling via facile ligand engineering of the modular FAST system. The srFAST platform combines the subcellular localization of the genetically encoded FAST and the color and target tunability of synthetic ligands, providing excellent experimental versatility. Based on the srFAST platform, cells of interest were labeled on demand by spatiotemporally precise photo-activation or specific endogenous metabolites or enzyme profiles, demonstrated by proof-of-concept models, including photo-srFAST, H2O2-srFAST or enzyme-srFASTs, respectively. Further, multicolor enzyme-srFASTs were constructed for concurrent orthogonal labeling of three cell types with different colors, proving feasibility to achieve multiplex bioimaging with the same protein tag. The generality and modularity of this platform make it facile to enrich the srFAST platform via endowing an increasing number of spectrally tunable FAST fluorogens with additional responsiveness to diverse molecular features of cell heterogeneity. Moreover, the fast binding and reversible dynamics of the FAST system might enable further development of phase-specific enzyme-dependent srFAST systems to track diverse phases and progression of the cell cycle by simply changing the corresponding ligands. As a promising alternative approach for orthogonal multicolor labeling, we believe that the srFAST platform could expand the FAST toolbox and be of great potential in sophisticated imaging applications in multicellular systems.Data availability
Experimental procedures, compound characterization, NMR spectra, and additional data are located within the ESI.†Author contributions
P. Chen, N. Zhou and Y. Huang conceived the study, designed the experimental work, analysed the results and revised the manuscript. S. Kuang helped to revise the manuscript. P. Chen performed experimental work and wrote the manuscript. R. Wang, K. Wang and J. Han assisted in the synthesis of ligands, and also in plasmid construction, protein expression and purification.Conflicts of interest
The author(s) declare that they have no competing interests.Acknowledgements
This work was supported by the National Key R&D Program of China (2021YFA0910100), and the National Natural Science Foundation of China (No. 22074034, 22034002 and 21725503).Notes and references
- D. M. Chudakov, M. V. Matz, S. Lukyanov and K. A. Lukyanov, Physiol. Rev., 2010, 90, 1103–1163 CrossRef CAS PubMed.
- N. C. Shaner, P. A. Steinbach and R. Y. Tsien, Nat. Methods, 2005, 2, 905–909 CrossRef CAS PubMed.
- E. L. Snapp, Trends Cell Biol., 2009, 19, 649–655 CrossRef CAS PubMed.
- J. Liu and Z. Cui, Bioconjugate Chem., 2020, 31, 1587–1595 CrossRef CAS PubMed.
- C. A. Telmer, R. Verma, H. Teng, S. Andreko, L. Law and M. P. Bruchez, ACS Chem. Biol., 2015, 10, 1239–1246 CrossRef CAS PubMed.
- Z. Thiel, J. Nguyen and P. Rivera-Fuentes, Angew. Chem., Int. Ed., 2020, 59, 7669–7677 CrossRef CAS PubMed.
- C. Li, A. G. Tebo and A. Gautier, Int. J. Mol. Sci., 2017, 18, 1473 CrossRef PubMed.
- A. Keppler, S. Gendreizig, T. Gronemeyer, H. Pick, H. Vogel and K. Johnsson, Nat. Biotechnol., 2002, 21, 86–89 CrossRef PubMed.
- A. Gautier, A. Juillerat, C. Heinis, M. Kindermann, F. Beaufils and K. Johnsson, Chem. Biol., 2008, 15, 128–136 CrossRef CAS PubMed.
- G. V. Los, L. P. Encell, M. G. McDougall, D. D. Hartzell, N. Karassina, C. Zimprich, M. G. Wood, R. Learish, R. F. Ohana, M. Urh, D. Simpson, J. Mendez, K. Zimmerman, P. Otto, G. Vidugiris, J. Zhu, A. Darzins, D. H. Klaubert, R. F. Bulleit and K. V. Wood, ACS Chem. Biol., 2008, 3, 373–382 CrossRef CAS PubMed.
- M. Kumauchi, M. T. Hara, P. Stalcup, A. Xie and W. D. Hoff, Photochem. Photobiol., 2008, 84, 956–969 CrossRef CAS PubMed.
- Y. Hori, H. Ueno, S. Mizukami and K. Kikuchi, J. Am. Chem. Soc., 2009, 131, 16610–16611 CrossRef CAS PubMed.
- H. Ozhalici-Unal, C. L. Pow, S. A. Marks, L. D. Jesper, G. L. Silva, N. I. Shank, E. W. Jones, J. M. Burnette, P. B. Berget and B. A. Armitage, J. Am. Chem. Soc., 2008, 130, 12620–12621 CrossRef CAS PubMed.
- M. A. Plamont, E. Billon-Denis, S. Maurin, C. Gauron, F. M. Pimenta, C. G. Specht, J. Shi, J. Quérard, B. Pan, J. Rossignol, K. Moncoq, N. Morellet, M. Volovitch, E. Lescop, Y. Chen, A. Triller, S. Vriz, T. Le Saux, L. Jullien and A. Gautier, Proc. Natl. Acad. Sci. U. S. A., 2016, 113, 497–502 CrossRef CAS PubMed.
- C. G. Li, M. A. Plamont, H. L. Sladitschek, V. Rodrigues, I. Aujard, P. Neveu, T. Le Saux, L. Jullien and A. Gautier, Chem. Sci., 2017, 8, 5598–5605 RSC.
- C. Li, A. G. Tebo, M. Thauvin, M. A. Plamont, M. Volovitch, X. Morin, S. Vriz and A. Gautier, Angew. Chem., Int. Ed., 2020, 59, 17917–17923 CrossRef CAS PubMed.
- A. G. Tebo, B. Moeyaert, M. Thauvin, I. Carlon-Andres, D. Boken, M. Volovitch, S. Padilla-Parra, P. Dedecker, S. Vriz and A. Gautier, Nat. Chem. Biol., 2021, 17, 30–38 CrossRef CAS PubMed.
- T. Ueda, T. Tamura and I. Hamachi, ACS Sens., 2018, 3, 527–539 CrossRef CAS PubMed.
- M. Martineau, A. Somasundaram, J. B. Grimm, T. D. Gruber, D. Choquet, J. W. Taraska and L. D. Lavis, Nat. Commun., 2017, 8, 1412 CrossRef PubMed.
- D. Srikun, A. E. Albers, C. I. Nam, A. T. Iavarone and C. J. Chang, J. Am. Chem. Soc., 2010, 132, 4455–4465 CrossRef CAS PubMed.
- M. Abo, R. Minakami, K. Miyano, M. Kamiya, T. Nagano, Y. Urano and H. Sumimoto, Anal. Chem., 2014, 86, 5983–5990 CrossRef CAS PubMed.
- C. Deo, S. H. Sheu, J. Seo, D. E. Clapham and L. D. Lavis, J. Am. Chem. Soc., 2019, 141, 13734–13738 CrossRef CAS PubMed.
- P. E. Deal, P. Liu, S. H. Al-Abdullatif, V. R. Muller, K. Shamardani, H. Adesnik and E. W. Miller, J. Am. Chem. Soc., 2020, 142, 614–622 CrossRef CAS PubMed.
- H. H. Han, A. C. Sedgwick, Y. Shang, N. Li, T. T. Liu, B. H. Li, K. Q. Yu, Y. Zang, J. T. Brewster, M. L. Odyniec, M. Weber, S. D. Bull, J. Li, J. L. Sessler, T. D. James, X. P. He and H. Tian, Chem. Sci., 2020, 11, 1107–1113 RSC.
- S. Benson, F. De Moliner, W. Tipping and M. Vendrell, Angew. Chem., Int. Ed., 2022, e202204788 CAS.
- S. J. Altschuler and L. F. Wu, Cell, 2010, 141, 559–563 CrossRef CAS PubMed.
- A. Levchenko and I. Nemenman, Curr. Opin. Biotechnol., 2014, 28, 156–164 CrossRef CAS PubMed.
- C. Gnann, A. J. Cesnik and E. Lundberg, Trends Cancer, 2021, 7, 278–282 CrossRef CAS PubMed.
- D. J. Burgess, Nat. Rev. Genet., 2019, 20, 317 CrossRef CAS PubMed.
- Q. Tang, L. Liu, Y. Guo, X. Zhang, S. R. Zhang, Y. Jia, Y. F. Du, B. Cheng, L. Yang, Y. Y. Huang and X. Chen, Angew. Chem., Int. Ed., 2022, 61, e202113929 CAS.
- Y. D. Wang, W. T. Dai, Z. X. Liu, J. X. Liu, J. Cheng, Y. Y. Li, X. L. Li, J. Hu and J. H. Lv, Anal. Chem., 2021, 93, 671–676 CrossRef CAS PubMed.
- L. Yuan, W. Y. Lin, S. Zhao, W. S. Gao, B. Chen, L. W. He and S. S. Zhu, J. Am. Chem. Soc., 2012, 134, 13510–13523 CrossRef CAS PubMed.
- Z. R. Dai, G. B. Ge, L. Feng, J. Ning, L. H. Hu, Q. Jin, D. D. Wang, X. Lv, T. Y. Dou, J. N. Cui and L. Yang, J. Am. Chem. Soc., 2015, 137, 14488–14495 CrossRef CAS PubMed.
- W. Li, S. l. Yin, X. Y. Gong, W. Xu, R. H. Yang, Y. C. Wan, L. Yuan and X. B. Zhang, Chem. Commun., 2020, 56, 1349 RSC.
- C. Y. Zhang, L. Wei, C. Wei, J. Zhang, R. Y. Wang, Z. Xi and L. Yi, Chem. Commun., 2015, 51, 7505 RSC.
- K. S. Mineev, S. A. Goncharuk, M. V. Goncharuk, N. V. Povarova, A. I. Sokolov, N. S. Baleeva, A. Yu. Smirnov, I. N. Myasnyanko, D. A. Ruchkin, S. Bukhdruker, A. Remeeva, A. Mishin, V. Borshchevskiy, V. Gordeliy, A. S. Arseniev, D. A. Gorbachev, A. S. Gavrikov, A. S. Mishin and M. S. Baranov, Chem. Sci., 2021, 12, 6719 RSC.
- H. Benaissa, K. Ounoughi, I. Aujard, E. Fischer, R. Goïame, J. Nguyen, A. G. Tebo, C. Li, T. L. Saux, G. Bertolin, M. Tramier, L. Danglot, N. Pietrancosta, X. Morin, L. Jullien and A. Gautier, Nat. Commun., 2021, 12, 6989 CrossRef CAS PubMed.
- J. S. Zhang, S. H. D. Wong, X. Wu, H. Lei, M. Qin, P. Shi, W. Wang, L. M. Bian and Y. Cao, Adv. Mater., 2021, 33, 2105765 CrossRef CAS PubMed.
- W. H. Li and G. H. Zheng, Photochem. Photobiol. Sci., 2012, 11, 460–471 CrossRef CAS PubMed.
- S. Hauke, A. V. Appen, T. Quidwai, J. Ries and R. Wombacher, Chem. Sci., 2017, 8, 559–566 RSC.
- L. Y. Zhou, X. B. Zhang, Y. F. Lv, C. Yang, D. Q. Lu, Y. Wu, Z. Chen, Q. L. Liu and W. H. Tan, Anal. Chem., 2015, 87, 5626–5631 CrossRef CAS PubMed.
- B. C. Dickinson, C. Huynh and C. J. Chang, J. Am. Chem. Soc., 2010, 132, 5906–5915 CrossRef CAS PubMed.
- H. Iwashita, E. Castillo, M. S. Messina, R. A. Swanson and C. J. Chang, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2018513118 CrossRef CAS PubMed.
- C. J. Liang, Q. Z. Zheng, T. L. Luo, W. Q. Cai, L. Q. Mao and M. Wang, CCS Chem., 2022, 1–11 Search PubMed.
- H. W. Liu, K. Li, X. X. Hu, L. M. Zhu, Q. M. Rong, Y. C. Liu, X. B. Zhang, J. Hasserodt, F. L. Qu and W. H. Tan, Angew. Chem., Int. Ed., 2017, 56, 11788–11792 CrossRef CAS PubMed.
- Y. Li, S. J. Sun, L. Fan, S. F. Hu, Y. Huang, K. Zhang, Z. Nie and S. Z. Yao, Angew. Chem., Int. Ed., 2017, 56, 14888–14892 CrossRef CAS PubMed.
- Y. Gao, Y. L. Hu, Q. M. Liu, X. K. Li, X. M. Li, C. Y. Kim, T. D. James, J. Li, X. Chen and Y. Guo, Angew. Chem., Int. Ed., 2021, 60, 10756–10765 CrossRef CAS PubMed.
- G. Y. Jiang, G. J. Zeng, W. P. Zhu, Y. D. Li, X. B. Dong, G. X. Zhang, X. L. Fan, J. G. Wang, Y. Q. Wu and B. Z. Tang, Chem. Commun., 2017, 53, 4505 RSC.
- J. S. Huang, Y. Y. Jiang, J. C. Li, J. G. Huang and K. Y. Pu, Angew. Chem., Int. Ed., 2021, 60, 3999–4003 CrossRef CAS PubMed.
- K. Wang, L. Jiang, F. Zhang, Y. Q. Wei, K. Wang, H. S. Wang, Z. J. Qi and S. Q. Liu, Anal. Chem., 2018, 90, 14056–14062 CrossRef CAS PubMed.
- K. Z. Gu, Y. S. Xu, H. Li, Z. Q. Guo, S. J. Zhu, S. Q. Zhu, P. Shi, T. D. James, H. Tian and W. H. Zhu, J. Am. Chem. Soc., 2016, 138, 5334–5340 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc03100k |
| This journal is © The Royal Society of Chemistry 2022 |