Open Access Article
Open Access ArticleA combination therapy strategy for treating antibiotic resistant biofilm infection using a guanidinium derivative and nanoparticulate Ag(0) derived hybrid gel conjugate†
Ananta
Dey‡
 ac,
Manisha
Yadav‡
b,
Deepak
Kumar‡
b,
Anik Kumar
Dey
a,
Sweety
Samal
b,
Subhash
Tanwar
b,
Debrupa
Sarkar
b,
Sumit Kumar
Pramanik
ac,
Manisha
Yadav‡
b,
Deepak
Kumar‡
b,
Anik Kumar
Dey
a,
Sweety
Samal
b,
Subhash
Tanwar
b,
Debrupa
Sarkar
b,
Sumit Kumar
Pramanik
 *a,
Susmita
Chaudhuri
*b and
Amitava
Das
*a,
Susmita
Chaudhuri
*b and
Amitava
Das
 *c
*c
aCSIR – Central Salt and Marine Chemical Research Institute, Bhavnagar, Gujarat, India. E-mail: sumitpramanik@csmcri.res.in
bTranslational Health Science and Technology Institute (THSTI), Faridabad 121001, Haryana, India. E-mail: susmita@thsti.res.in
cIndian Institute of Science Education and Research Kolkata, Mohanpur, 741246, West Bengal, India. E-mail: amitava@iiserkol.ac.in
First published on 27th July 2022
Abstract
Bacteria organized in biofilms show significant tolerance to conventional antibiotics compared to their planktonic counterparts and form the basis for chronic infections. Biofilms are composites of different types of extracellular polymeric substances that help in resisting several host-defense measures, including phagocytosis. These are increasingly being recognized as a passive virulence factor that enables many infectious diseases to proliferate and an essential contributing facet to anti-microbial resistance. Thus, inhibition and dispersion of biofilms are linked to addressing the issues associated with therapeutic challenges imposed by biofilms. This report is to address this complex issue using a self-assembled guanidinium–Ag(0) nanoparticle (AD-L@Ag(0)) hybrid gel composite for executing a combination therapy strategy for six difficult to treat biofilm-forming and multidrug-resistant bacteria. Improved efficacy was achieved primarily through effective biofilm inhibition and dispersion by the cationic guanidinium ion derivative, while Ag(0) contributes to the subsequent bactericidal activity on planktonic bacteria. Minimum Inhibitory Concentration (MIC) of the AD-L@Ag(0) formulation was tested against Acinetobacter baumannii (25 μg mL−1), Pseudomonas aeruginosa (0.78 μg mL−1), Staphylococcus aureus (0.19 μg mL−1), Klebsiella pneumoniae (0.78 μg mL−1), Escherichia coli (clinical isolate (6.25 μg mL−1)), Klebsiella pneumoniae (clinical isolate (50 μg mL−1)), Shigella flexneri (clinical isolate (0.39 μg mL−1)) and Streptococcus pneumoniae (6.25 μg mL−1). Minimum bactericidal concentration, and MBIC50 and MBIC90 (Minimum Biofilm Inhibitory Concentration at 50% and 90% reduction, respectively) were evaluated for these pathogens. All these results confirmed the efficacy of the formulation AD-L@Ag(0). Minimum Biofilm Eradication Concentration (MBEC) for the respective pathogens was examined by following the exopolysaccharide quantification method to establish its potency in inhibition of biofilm formation, as well as eradication of mature biofilms. These effects were attributed to the bactericidal effect of AD-L@Ag(0) on biofilm mass-associated bacteria. The observed efficacy of this non-cytotoxic therapeutic combination (AD-L@Ag(0)) was found to be better than that reported in the existing literature for treating extremely drug-resistant bacterial strains, as well as for reducing the bacterial infection load at a surgical site in a small animal BALB/c model. Thus, AD-L@Ag(0) could be a promising candidate for anti-microbial coatings on surgical instruments, wound dressing, tissue engineering, and medical implants.
Introduction
Most bacteria exist in sessile communities covered with biofilms, and biofilm formation enables single-cell organisms to assume a temporary multicellular lifestyle, which allows these bacteria to subvert innate immune defenses.1–3 The widespread distribution of biofilms in various diseases and their resilience to numerous conventional anti-microbial treatments and increasing anti-microbial resistance have contributed towards an alternate approach, “biofilm inhibition and dispersal,” as a major interventional option and a future strategy.4 Typically, biofilm development passes through three distinct stages: attachment, maturation (active sessile cells), and release. Within a biofilm, one or more types of bacteria and/or fungi share nutrients and DNA and undergo changes to evade the immune system.5,6 The structural integrity of a biofilm, self-synthesized by microorganisms, primarily relies upon the extracellular polymeric matrix, such as extracellular DNA (eDNA), polysaccharides, proteins, amyloid fibers, lipids, and bacteriophages. Biofilms protect bacteria from host defense, anti-microbial chemotherapies, and extracellular enzymatic activity, which leads to the manifestation of chronic, non-healing infections.7,8 A high-density bacterial colony, protected by a biofilm, also triggers small molecule signals through quorum sensing to induce virulence and drug resistance mechanisms. Additionally, biofilms form a physical barrier that prevents most immune cells from detecting pathogenic bacteria.9 Literature reports tend to suggest that the biofilm-associated anti-microbial resistance seems to be multifactorial, which may vary from organism to organism and pose a serious challenge to the conventional clinical therapeutic options for treating microbial infections.7,8,10,11 Despite their significance, effective therapeutic options for inhibiting biofilm-associated microbial infections remain limited, and the phenomenon of biofilm resistance remains a subject that has many unaddressed issues.6 There are two distinct phases of the bacterial life cycle: unicellular (planktonic) and multicellular (biofilms or sessile cells).12,13 The alternation between the two phases requires the transition from planktonic cells to sessile cells to initiate biofilm formation and from sessile cells to detached cells to allow a return to the planktonic mode of growth.14 The challenges to conventional mono-therapeutic approaches posed by biofilms have given way to combinatorial therapies that inhibit biofilm formation and/or dispersion of established biofilms, as well as act as anti-microbial chemotherapy.15–19 A recent report reveals that the use of certain synthetic guanidinium derivatives could effectively induce biofilm disassembly in the developmental cycle of Bacillus subtilis.20–22 It is proposed that the guanidine functionalities participate in hydrogen bond formation in the extracellular polymeric matrix weakening its mechanical stability which induces dispersion. Guanidine is a strong base (pKa is 13.6) and exists almost exclusively as guanidinium in water. Presumably, a guanidinium derivative with pyridine moieties (AD-L) as the additional hydrogen bond-forming functionalities could be efficient in inhibiting the formation of biofilms and their dispersion.23 Importantly, AD-L is found to form a hydrogel, which could be utilized for topical formulations.Guanidine is known to be a versatile ligand that can form stable complexes with various metal ions.24 However, its use for the reduction of Ag(I) to Ag(0) is not reported in the literature. In contrast, hydrazine and its derivatives, including hydrazide, are known to produce Ag(0) from Ag(I).25,26 Inhibitory and bactericidal effects of nanoparticulate Ag(0) (AgNPs) on Gram-positive, Gram-negative, and fungal pathogens are well established.27–30 Despite such advantages, the applications of AgNPs inadvertently suffer from various limitations. AgNPs are prone to oxidation, which causes the loss of antibacterial activity, apart from contributing to health and environmental issues induced by Ag+.31–35 Furthermore, the synthesis and functionalization processes for the generation and stabilization of AgNPs often raise health issues.36–38 Literature reports also reveal that AgNPs smaller than 10 nm could directly modify the penetrability of the bacterial cells and induce cell lysis.39–41 However, biofilm formation hinders the transport of AgNPs, and the bacterial pathogens get eventual protection from the bactericidal effects of AgNPs. This gives a distinct scope for using AgNPs along with a therapeutic agent that exhibits biofilm dispersion/inhibitory influence. More recently, hydrogels with high water content and biocompatibility have been widely used in biomedical applications such as tissue engineering and also as controlled release materials.42–48 A hydrogel could be used for the sustained release of AgNPs to induce bactericidal influence on planktonic bacteria with a prolonged inhibition time.29,30,42,49–52 We have exploited the coordination as well as the reducing property of the guanidinium-hydrazide derivative, AD-L, for in situ reduction of Ag(I) to nanoparticulate Ag(0) and stabilize such AgNPs within the gel matrix of AD-L. A recent report has also demonstrated that the in situ synthesized AgNPs can be effectively stabilized in hydrogel networks without using any external capping agent for developing a well-ordered AgNP-hybrid hydrogel as a soft composite.53 Typically, Ag(I)–Ag(0) redox potential is ∼0.22 V (vs. NHE), and being a mild oxidant, it is expected to oxidize hydrazine to ammonia and its derivatives to the corresponding amine forms.25,54 We intend to pursue a small molecule-based hybrid gel for inhibition of biofilm formation and dispersion of established biofilms, with the subsequent use of a bactericidal agent for eradication of the planktonic cells—a proof of concept for a combination therapy approach.55 In this study, an improved efficacy is demonstrated by using a cationic guanidinium core for biofilm inhibition and dispersion of preformed biofilms, along with subsequent bactericidal activity towards planktonic bacteria by using in situ generated Ag(0) through reduction of Ag(I) by the hydrazide derivative of the guanidinium ion. Minimum Inhibitory Concentrations (MICs) of the AD-L@Ag(0) formulation for certain extremely drug-resistant biofilm-forming bacterial pathogens are assessed, and most importantly, the evaluated MIC for the respective pathogens is superior to that in the available literature reports. Further, the efficacy of this hybrid gel formulation is also demonstrated in the reduction of bacterial infection load at a surgical site in a small animal BALB/c mouse model.
Materials and methods
Synthesis of AD-L
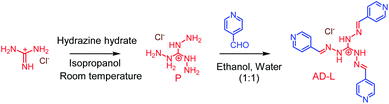
The AD-L ligand was synthesized following a literature procedure (Scheme 1).56 To a water solution of triaminoguanidinium chloride (0.25 g, 2.38 mmol), 4-pyridinecarboxaldehyde (0.675 mL, 7.17 mmol) dissolved in ethanol was added and stirred for 6 h at 85 °C. Then the obtained reaction mixture was kept at ambient temperature to yield a yellow precipitate. The obtained precipitate was washed using ethanol–water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) and dried under a vacuum. Yield 1.5 g (82%). 1H NMR (600 MHz, DMSO-d6) δ (ppm) 8.76 (6H, d, J = 6.3 Hz), 8.68 (3H s), 7.99 (6H, d, J = 5.8 Hz). MS (ESI): m/z calculated for [M–Cl−]+ (C19H18N9)+: 372.1836; found: 372.1696. Elemental analysis calculated for C19H18ClN9: C, 55.58; H, 4.92; N, 30.76. Found C, 55.55; H, 4.90; N, 30.75. (FT-IR) (ν cm−1): –C
1, v/v) and dried under a vacuum. Yield 1.5 g (82%). 1H NMR (600 MHz, DMSO-d6) δ (ppm) 8.76 (6H, d, J = 6.3 Hz), 8.68 (3H s), 7.99 (6H, d, J = 5.8 Hz). MS (ESI): m/z calculated for [M–Cl−]+ (C19H18N9)+: 372.1836; found: 372.1696. Elemental analysis calculated for C19H18ClN9: C, 55.58; H, 4.92; N, 30.76. Found C, 55.55; H, 4.90; N, 30.75. (FT-IR) (ν cm−1): –C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N str. frequency (∼1595.92 cm−1).
N str. frequency (∼1595.92 cm−1).
Characterization techniques
X-ray photoelectron spectroscopic (XPS) studies were performed on xerogel samples of AD-L and AD-L@Ag(0) using a Thermo Scientific instrument (Model: ESCALAB 250 Xi XPS) equipped with a monochromatic AlKα X-ray excitation source having an X-ray spot size of 650 × 650 μm2. Pass energies of 150 eV for survey scans and 20 eV for narrow scans with the C1s SP3 hydrocarbon peak set at a BE of 284.8 eV were used. To overcome the issue of sample charging, a low-energy electron flood gun was used. Data were processed using a Thermo Scientific™ Avantage data system and CasaXps processing software. Peak areas were measured typically after satellite subtraction and background subtraction, either with a linear background or by following the methods of Shirley. The deconvolution of the core-level spectra was performed by choosing a Shirley or spline Tougaard background with the GL(30) line shape (70% Gaussian, 30% Lorentzian) to determine the chemical species of the xerogel. Scanning electron microscopy (SEM) images were recorded by utilizing a high-resolution scanning electron microscope (SEM; JEOL JSM 7100F, Japan) having an accelerating voltage of 15 kV. Samples were prepared on a silicon wafer substrate/HPAN support/alumina support, and a thin (≈4 nm) gold coating was sputtered deposited (EM ACE200, Leica Microsystems) to avoid sample charging under the electron beam. A transmission electron microscope (TEM; JEOL JEM 2100, Japan) operated at 200 kV was utilized for recording the images while studying the morphology of the xerogel transferred onto copper grids. Zeta Cad streaming current and a zeta potential meter were used to measure the surface charge of the xerogel. UV-visible absorption spectra were recorded using a Shimadzu UV-2600 spectrophotometer in the range of 200–800 nm. Dynamic rheological measurements were performed on a rheometer (TA instruments AR2000; New Castle Delaware 19720). Viscosity at varying shear rates was measured using a cone plate geometry (diameter 40 mm, 52 μm gap).Determination of minimum inhibitory concentration (MIC) and minimum bactericidal concentration (MBC)
The minimum inhibitory concentration (MIC) was estimated as the lowest concentration of the anti-microbial agent that will suppress the visible growth of microorganisms after a certain incubation period. As per the Clinical and Laboratory Standards Institute (CLSI) guidelines, the minimum bactericidal concentration (MBC) determination57–60 (MBC) assay was used for evaluating this new anti-microbial agent/drug for predicting its efficacy towards eradication (killing of 99.9% of a specific bacterial species) of a particular type of bacterium. The MIC and MBC were estimated for each isolate that we used in the present study, i.e., Acinetobacter baumannii (ATCC BAA-2800), Pseudomonas aeruginosa (ATCC 27853), Staphylococcus aureus (ATCC 33592), Klebsiella pneumoniae (ATCC 19050), Escherichia coli (clinical isolate P10192), Klebsiella pneumoniae (clinical isolate 16280), Shigella flexneri (clinical isolate 1A) and Streptococcus pneumoniae (ATCC 49619), by following standard protocols elaborated in S2 (ESI†).Time kill assay
Time kill assay was used to study the time kinetics of the activity of AD-L@Ag(0) against a bacterial strain for determining the bacteriostatic and/or bactericidal activity(ies). Time kill assays were performed for Klebsiella pneumoniae (clinical isolate, 16280) and Staphylococcus aureus (ATCC 33592) by following a literature procedure. Concentrations equal to half of MIC, MIC, twice the MIC, and four times the MIC of the AD-L@Ag(0) nano-formulation were tested. The detailed procedure is described in S2 (ESI†).Crystal violet (CV) for MBIC (minimum biofilm inhibitory concentration) determination
Crystal violet (CV) is a basic dye that binds non-specifically to negatively charged surface molecules such as polysaccharides and eDNA in the extracellular matrix and is generally used to evaluate biofilm biomass. Previously mentioned isolates of Acinetobacter baumannii, Pseudomonas aeruginosa, Staphylococcus aureus, Klebsiella pneumoniae, Escherichia coli, Klebsiella pneumoniae, Shigella flexneri and Streptococcus pneumoniae were used for this assay. For crystal violet staining, a 48 hour old biofilm was used after removing planktonic cells. Then the biofilm was stained with an equal volume of 0.5% CV and incubated for 45 minutes at room temperature. Excess CV was then removed by washing twice with sterile PBS. The biofilm was then detached using 95% ethanol and quantified on the basis of turbidity. Turbidity was measured by taking OD595 in a microplate ELISA reader.EPS quantification for MBEC (minimum biofilm eradication concentration) determination
Exopolysaccharide (EPS) quantification was carried out to measure the biofilm eradication effect of AD-L@Ag(0) in Acinetobacter baumannii, Pseudomonas aeruginosa, Staphylococcus aureus, Klebsiella pneumoniae, Escherichia coli, Klebsiella pneumoniae, Shigella flexneri and Streptococcus pneumoniae. Sulfuric acid (ACS reagent, 95.0–98.0%, Cat. No. 258105, Sigma), phenol (Sigma-Aldrich), and a 96-well flat-bottomed (polystyrene) microplate (Corning, Cat. No. 3595) were used in the phenol–sulfuric acid assay.61 For EPS quantification, 2× MBIC, MBIC, and 0.5× MBIC concentrations of AD-L@Ag(0) were used in triplicate along with no-treatment control. Details of the EPS quantification method are elaborated in S2 (ESI†).Microscopic visualization of biofilms
For a better understanding of the effect of AD-L@Ag(0) on biofilms, biofilms with or without AD-L@Ag(0) treatment were observed under a confocal microscope. Initially, a mature biofilm that formed on glass coverslips was washed thrice with sterile PBS (0.1 M, 7.4 pH) to remove any planktonic cells. Then it was fixed using a fixative (4% paraformaldehyde, 2.5% glutaraldehyde, and 0.1 M phosphate buffer having pH 7.2) and stained with the extracellular polysaccharide dye Filmtracer Sypro Ruby (Thermo, Waltham, Massachusetts, United States). The nuclei of all cells were stained with DAPI stain. The image was captured in the confocal microscope FV300 Olympus.Cytotoxicity analysis of AD-L@Ag(0)
Cytotoxicity of the AD-L@Ag(0) formulation was evaluated against the commonly used epithelial cell line (Vero-E6, Vero, ATCC CCL-81) using the methyl thiazolyl tetrazolium (MTT) method. The MTT method was performed as per the previously published standard protocol by ATCC (S2 in the ESI†).62In vivo proof-of-concept for AD-L@Ag(0)
All experiments were performed in compliance with the national and institutional guidelines regarding biosafety (The Department of Biotechnology Biosafety Guidelines, Govt. of India) and animal ethics (Committee for the Purpose of Control and Supervision of Experiments on Animals – CPCSEA). The protocols were approved by the Institutional Biosafety Committee (certificate no. IBSC/107/2019) and Institutional Animal Ethics Committee (certificate no. IEAC/THSTI/63). Further, to demonstrate in vivo evidence of reduction of surgical site infection by AD-L@Ag(0), experiments with a BALB/c mouse were planned. Prior to the experiment, all 6–8 week old mice were anaesthetized using ketamine/xylazine mouse cocktail at 0.1 ml per 20 g mouse weight. A surgical wound was created on the right-hand side of their backbone with a sterile scalpel by making a longitudinal incision of 1 cm in length. The wound was infected by the insertion of a segment of silk suture through the skin with a suturing needle and was secured by knotting. 5 μL bacterial suspensions of Klebsiella pneumoniae (clinical), Staphylococcus aureus and Pseudomonas aeruginosa were added onto the suture with a pipette (1 × 103 CFU per mL). The control group was maintained as such. The treatment group was treated with the AD-L@Ag(0) suspension gel at a concentration of 100 μg ml−1 for Klebsiella pneumoniae and 5 μg ml−1 for Staphylococcus aureus and Pseudomonas aeruginosa. Each mouse was housed separately in a ventilated cage to protect them from further injuries with proper bedding and diet. AD-L@Ag(0) application to each treatment group mouse was carried out every 24 hours for 3 days, and skin swab sampling was performed until day 4 post-infection. All the mice were checked twice daily throughout the experiment. The swab sample was cultured on a MHB agar plate by the spread plate method. At the end of the experiment, the mice were euthanized humanely using an overdose of inhalant CO2.Statistical analysis
The overall plan of analysis of the anti-biofilm and bactericidal effect of the formulation is described in Fig. 1. Two independent sets of each experiment were performed in triplicate for all biological assays. The difference in proliferation levels of treatment and control groups was compared by using GraphPad Prism version 9.0.0 for Windows (GraphPad Software, San Diego, California USA). Statistical analysis was performed by using a non-parametric test as follows – for MBIC and cytotoxicity analysis, two-way ANOVA followed by Dunnett's multiple comparison test; for EPS, one-way ANOVA followed by Dunnett's multiple comparison test; and for animal study, two-way ANOVA corrected by Tukey's multiple comparison test.Results and discussion
Synthesis and characterization of AD-L@Ag(0)
The guanidinium derivative AD-L was synthesized by following a previously reported literature procedure.56 The gelation ability of the pure ligand (AD-L), as well as AD-L in the presence of silver nitrate, was examined in a MeOH–H2O mixture solvent. The gel was achieved at a solvent ratio of MeOH–H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v). The synthesized ligand was dispersed initially in 1 ml of MeOH–H2O (1
1, v/v). The synthesized ligand was dispersed initially in 1 ml of MeOH–H2O (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) in a glass vial and was thoroughly dissolved through sonication of the dispersion in the glass vial. After that, the solution was kept at room temperature without any further disturbance. The solution containing pure AD-L took 4 hours to form a mechanically stable gel, while a mechanically stable gel was found to form instantly for the other solution containing equimolar amounts of AD-L and AgNO3. The critical gelation concentration (cgc) was evaluated as 6.2 wt% in the case of pure AD-L, and the stability of the gels was examined through the vial inversion technique. Importantly, in the presence of AgNO3, the critical gelation concentration was found to be lower and was evaluated as 3.4 wt%. The pure ligand AD-L produced a yellow colour gel, whereas the metallogel (AD-L@Ag(0)) showed a light orange colour. Recent studies revealed that the pKa value of the guanidinium derivative varies between 12 and 13.5.63–65 The pKa of the pyridine moiety with extended conjugation at the 4-position is ∼4.5.66,67 Thus, AD-L is expected to present solely as a mono-cationic species in neutral or physiological pH. Guanidine is known to be a versatile ligand that can form stable complexes with various metal ions.19 Nandibewoor and his co-workers reported oxidation of guanidinium ions to urea by Ag(I) species.68 Also, hydrazide derivatives are known to act as reducing agents for Ag(I) species and generate nanoparticulate Ag(0) following a two-electron redox process.24,68–70 Silver is more efficient in exhibiting plasmon excitation than gold and copper, which show surface plasmon resonances (SPR) in the visible region of the spectrum. AgNPs contain localized plasmons, and their interaction with an electromagnetic wave leads to localized plasmon resonance. Importantly, the SPR of AgNPs can be tuned to a wavelength in the visible spectrum as a function of their size, shape, and morphology.71,72 Spherical AgNPs with a diameter of ∼2–5 nm show typically a surface plasmon resonance (SPR) band around 400 nm, and this accounts for the spectral change and the visually detectable colour change from light yellow to light orange for the gel.72,73 Further, methanol is not known to reduce Ag(I) to Ag(0) under the normal experimental conditions that we have followed in this study.74,75 Various alcohols can only reduce Ag(I) to Ag(0) under strongly alkaline conditions,76 which is not the case for the present study. Thus, in the present situation, guanidinium and hydrazide could both contribute to the effective reduction of Ag(I) to Ag(0). Importantly, both urea (oxidized form of AD-L) and guanidinium (unreacted) derivatives are known to form stable gels (vide infra) and induce dispersion of biofilms.20–22,77,78 The presence of Ag(0) in the xerogel of AD-L@Ag(0) was proved from the data obtained from an XPS experiment (vide infra). It is important to note that the solvent composition has a significant role in the stabilization of the metallogel (AD-L@Ag) and the gel from the pure ligand (AD-L). No gelation was noticed in pure methanol or pure water. Deviation of solvent composition from methanol–water (1
1, v/v) in a glass vial and was thoroughly dissolved through sonication of the dispersion in the glass vial. After that, the solution was kept at room temperature without any further disturbance. The solution containing pure AD-L took 4 hours to form a mechanically stable gel, while a mechanically stable gel was found to form instantly for the other solution containing equimolar amounts of AD-L and AgNO3. The critical gelation concentration (cgc) was evaluated as 6.2 wt% in the case of pure AD-L, and the stability of the gels was examined through the vial inversion technique. Importantly, in the presence of AgNO3, the critical gelation concentration was found to be lower and was evaluated as 3.4 wt%. The pure ligand AD-L produced a yellow colour gel, whereas the metallogel (AD-L@Ag(0)) showed a light orange colour. Recent studies revealed that the pKa value of the guanidinium derivative varies between 12 and 13.5.63–65 The pKa of the pyridine moiety with extended conjugation at the 4-position is ∼4.5.66,67 Thus, AD-L is expected to present solely as a mono-cationic species in neutral or physiological pH. Guanidine is known to be a versatile ligand that can form stable complexes with various metal ions.19 Nandibewoor and his co-workers reported oxidation of guanidinium ions to urea by Ag(I) species.68 Also, hydrazide derivatives are known to act as reducing agents for Ag(I) species and generate nanoparticulate Ag(0) following a two-electron redox process.24,68–70 Silver is more efficient in exhibiting plasmon excitation than gold and copper, which show surface plasmon resonances (SPR) in the visible region of the spectrum. AgNPs contain localized plasmons, and their interaction with an electromagnetic wave leads to localized plasmon resonance. Importantly, the SPR of AgNPs can be tuned to a wavelength in the visible spectrum as a function of their size, shape, and morphology.71,72 Spherical AgNPs with a diameter of ∼2–5 nm show typically a surface plasmon resonance (SPR) band around 400 nm, and this accounts for the spectral change and the visually detectable colour change from light yellow to light orange for the gel.72,73 Further, methanol is not known to reduce Ag(I) to Ag(0) under the normal experimental conditions that we have followed in this study.74,75 Various alcohols can only reduce Ag(I) to Ag(0) under strongly alkaline conditions,76 which is not the case for the present study. Thus, in the present situation, guanidinium and hydrazide could both contribute to the effective reduction of Ag(I) to Ag(0). Importantly, both urea (oxidized form of AD-L) and guanidinium (unreacted) derivatives are known to form stable gels (vide infra) and induce dispersion of biofilms.20–22,77,78 The presence of Ag(0) in the xerogel of AD-L@Ag(0) was proved from the data obtained from an XPS experiment (vide infra). It is important to note that the solvent composition has a significant role in the stabilization of the metallogel (AD-L@Ag) and the gel from the pure ligand (AD-L). No gelation was noticed in pure methanol or pure water. Deviation of solvent composition from methanol–water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) resulted in precipitation. However, a stable gel was formed in the methanol–water (1
1, v/v) resulted in precipitation. However, a stable gel was formed in the methanol–water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) medium without any significant change in the rheological properties (vide infra).
1, v/v) medium without any significant change in the rheological properties (vide infra).
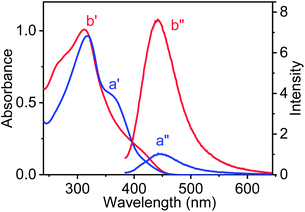
UV-vis spectroscopic studies were performed for AD-L and AD-L@Ag(0) in their respective gel state in a methanol–water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) solvent mixture. The UV-visible spectra of AD-L are shown in Fig. 2 (a′ – blue trace). It shows an absorbance maximum at 317 nm and a hump at ∼359 nm. The UV-visible spectra of the as-synthesized metallogel (AD-L@Ag(0)) show a new hump in the region ∼410 nm with a maximum at 311 nm. Thus, the absorbance of the 340–393 nm band for AD-L was found to decrease, and an increase in absorbance at around ∼410 nm was observed, apart from the shift in the absorption maximum to 311 nm for AD-L@Ag. This new hump at ∼410 nm matches well with the characteristic spectral band of the spherical Ag(0) nanoparticles having diameters in the range of 2–5 nm.72,79,80 It was observed that the nanoparticles were stable for more than 6 months when stored in the gel matrix at ∼10 °C. Importantly, characteristic photoluminescence of the nanoclusters of Ag(0) nanoparticles was observed in the solution. The emission spectra of the respective gel (methanol–water (1
1, v/v) solvent mixture. The UV-visible spectra of AD-L are shown in Fig. 2 (a′ – blue trace). It shows an absorbance maximum at 317 nm and a hump at ∼359 nm. The UV-visible spectra of the as-synthesized metallogel (AD-L@Ag(0)) show a new hump in the region ∼410 nm with a maximum at 311 nm. Thus, the absorbance of the 340–393 nm band for AD-L was found to decrease, and an increase in absorbance at around ∼410 nm was observed, apart from the shift in the absorption maximum to 311 nm for AD-L@Ag. This new hump at ∼410 nm matches well with the characteristic spectral band of the spherical Ag(0) nanoparticles having diameters in the range of 2–5 nm.72,79,80 It was observed that the nanoparticles were stable for more than 6 months when stored in the gel matrix at ∼10 °C. Importantly, characteristic photoluminescence of the nanoclusters of Ag(0) nanoparticles was observed in the solution. The emission spectra of the respective gel (methanol–water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, v/v) medium) were also recorded and are shown in Fig. 2a′′ and b′′. Fig. 2a′′ shows the emission spectrum of AD-L@Ag(0), having a maximum at 447 nm (λExt = 410 nm). However, only a weak luminescence was observed for the precursor ligand, i.e. AD-L, following excitation at 359.81 The observed blue emission strongly supports the formation of silver nanoparticles through the reduction of Ag(I) to Ag(0).81 The ligand stabilized Ag nanoparticles were found to be stable under ambient conditions.
1, v/v) medium) were also recorded and are shown in Fig. 2a′′ and b′′. Fig. 2a′′ shows the emission spectrum of AD-L@Ag(0), having a maximum at 447 nm (λExt = 410 nm). However, only a weak luminescence was observed for the precursor ligand, i.e. AD-L, following excitation at 359.81 The observed blue emission strongly supports the formation of silver nanoparticles through the reduction of Ag(I) to Ag(0).81 The ligand stabilized Ag nanoparticles were found to be stable under ambient conditions.
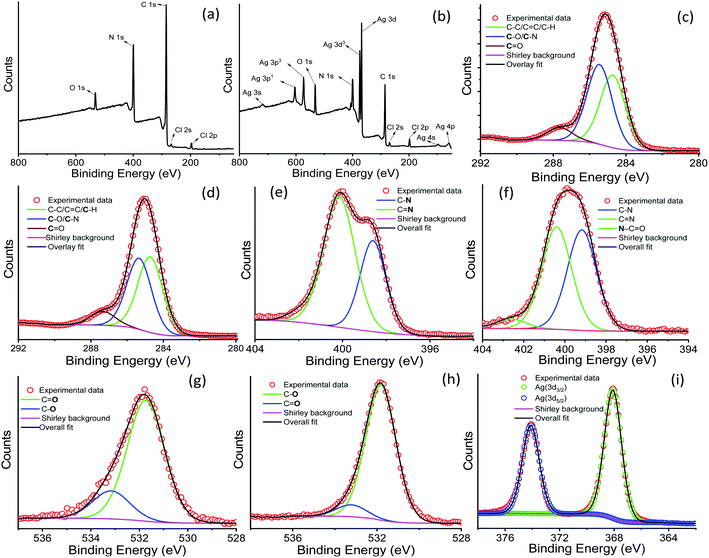
X-ray photoelectron spectroscopic studies were performed using xerogel samples of AD-L and AD-L@Ag(0) to understand the chemical composition and valence state of Ag, C, and N-atoms. The XPS spectrum of pure ligand AD-L exhibited four peaks at binding energies of 196.99 eV, 284.85 eV, 399.13 eV, and 532.01 eV, which were ascribed to Cl 2p, C 1s, N 1s, and O 1s elements, respectively (Fig. 3a). The O 1s peak of ligand AD-L may arise due to the presence of a small amount of carbonate counter anions. Fig. 3b shows the survey spectra of the silver metal-containing AD-L@Ag(0) gel. The core-level XPS spectrum of AD-L@Ag(0) showed the corresponding peaks at binding energies of 197.5 eV (Cl2p), 284.8 eV, 398.85 eV (N1s), and 531.09 eV (O1s). The Ag 3d spectrum in Fig. 3b-(v) is found to split into 3d5/2 and 3d3/2 with a spin–orbit splitting of ∼6 eV, which corresponds to unperturbed metallic silver and is typically anticipated for Ag(0) atoms in the nanoparticle core.82,83 The fine splitting of O1s and N1s confirms the formation of urea functionalities in AD-L@Ag, which is evident from the N 1s spectrum, as shown in Fig. 3. These results confirm the presence of O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N at 402.87 eV and the formation of urea functionalities as the oxidation product of AD-L by AgNO3. Further, the enhancement of the peak intensity at 287.66 eV of carbonyl functionalities in the O 1s spectrum also confirms such a proposition (Table 1).
C–N at 402.87 eV and the formation of urea functionalities as the oxidation product of AD-L by AgNO3. Further, the enhancement of the peak intensity at 287.66 eV of carbonyl functionalities in the O 1s spectrum also confirms such a proposition (Table 1).
| Fine spectra | Energy (eV) | Species | AD-L (at%) | AD-L@Ag (at%) |
|---|---|---|---|---|
| C1s | 284.5 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C–C, C–H C, C–C, C–H |
26.34 | 31.57 |
| 286.1 | C–N | 57.12 | 46.94 | |
| 287.8 | O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O (COOO) O (COOO) |
15.64 | 23.41 | |
| N1s | 398.7 | C–N | 31.93 | 39.56 |
| 401.7 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N N |
58.33 | 51.56 | |
| 403.3 | O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–N C–N |
— | 9.74 | |
| O1s | 531.6 | C–O | 68.56 | 77.12 |
| 532.8 | C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
29.56 | 22.79 | |
| Ag 3d | 368.24 | Ag (3d3/2) | — | 61.32 |
| 376.19 | Ag (3d5/2) | — | 28.68 |
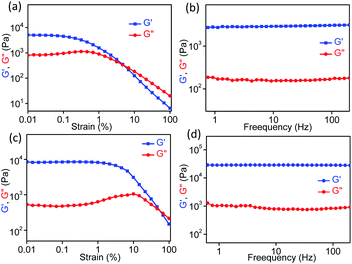
Rheology is the most important defining feature of supramolecular gels and is used to analyze the mechanical properties of bulk gel samples.84 Several factors can influence the rheological properties, e.g., fiber morphology, fiber length and strength, the number and type of crosslink, and the distribution of the fibers in space. However, the situation is rather complex for a gel that is formed from two different gelator molecules either through a self-sorting or a co-assembly process, which could be the process for AD-L@Ag(0). Rheological measurements (strain and frequency sweeps) were carried out to gain insight into the mechanical properties of the synthesized ligand gel and metallogel in terms of elastic storage modulus (G′, the contribution of elastic, i.e., solid-like) and elastic loss modulus (G′′, liquid-like) (Fig. 3). For rheological experiments, these gels were obtained by adding an aqueous solution of silver salt to a solution of the ligand in methanol to attain the final methanol–H2O ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
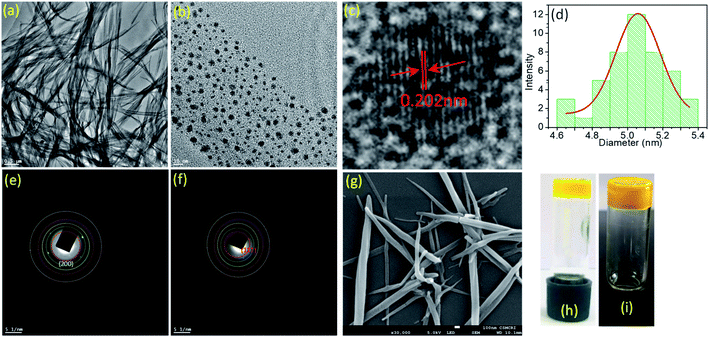
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v). For both ligand AD-L and metallogel AD-L@Ag(0), the strain sweep experiments displayed linear viscoelastic behavior when varying the strain amplitude below a certain level known as the critical strain to a fixed frequency (f = 1 Hz) (Fig. 4a and c). However, above the critical strain, the G′ values were found to decline rapidly and a cross-over between G′ and G′′ suggested that the gel network had collapsed and converted to a sol (Fig. 4a and c). The strain sweep experiment further exhibited that the metallogel undergoes a transition into a sol above 10% strain, whereas the ligand gel displayed a continuous decline even below 10% strain (Fig. 4a and c). This result proposes that the metallogel is more organized than the ligand gel. In frequency sweep experiments with a small amplitude of strain (0.1%), the storage modulus (G′) was observed to be invariant with the variation of frequency for both the gels. Further, G′ was higher than G′′ in both cases, which suggests the elastic nature of the gels (Fig. 4b and d). While the general behaviors of the G′ and G′′ were analogous for all the gels, the G′ of the metallogel (AD-L@Ag(0)) was found to be higher in magnitude than the G′ of the ligand gel (AD-L). This result indicated the higher rigidity of the metallogel than that of the ligand gel. To investigate the morphological properties of the metallogel, transmission electron microscopy (TEM) and field-emission scanning electron microscopy (FE-SEM) were employed. The TEM images of the as-synthesized nanoparticles at different magnifications are presented in Fig. 5a and b. The HRTEM image (Fig. 5c) shows the lattice fringes which are symbolic of plasmonic Ag(0) nanoparticles. The most interesting observation from the TEM images is that the metallogel has an entangled fibril morphology, and all the Ag nanoparticles are uniformly distributed on the fibril network. We successfully detected the SAED patterns of the silver nanoparticles (Fig. 5e and f). The SAED pattern reveals that the nanoparticles consist of (111) and (200) planes. The HRTEM images reveal that the silver nanoparticles are crystalline, consisting of good quality lattice fringes with significant d-spacing values. Careful analysis of the HRTEM images reveals two groups of lattice fringes; the lattice distances are 0.20 nm and 0.23 nm, which are the indicators of the (200) and (111) planes of Ag(0), respectively.37 From the statistical analysis (Fig. 5d) of the TEM images, we can conclude that the average particle size of the Ag nanoparticles is 5 nm. Fig. 5g shows the FE-SEM images of the metallogel, which has a fibrillar morphology.
1 (v/v). For both ligand AD-L and metallogel AD-L@Ag(0), the strain sweep experiments displayed linear viscoelastic behavior when varying the strain amplitude below a certain level known as the critical strain to a fixed frequency (f = 1 Hz) (Fig. 4a and c). However, above the critical strain, the G′ values were found to decline rapidly and a cross-over between G′ and G′′ suggested that the gel network had collapsed and converted to a sol (Fig. 4a and c). The strain sweep experiment further exhibited that the metallogel undergoes a transition into a sol above 10% strain, whereas the ligand gel displayed a continuous decline even below 10% strain (Fig. 4a and c). This result proposes that the metallogel is more organized than the ligand gel. In frequency sweep experiments with a small amplitude of strain (0.1%), the storage modulus (G′) was observed to be invariant with the variation of frequency for both the gels. Further, G′ was higher than G′′ in both cases, which suggests the elastic nature of the gels (Fig. 4b and d). While the general behaviors of the G′ and G′′ were analogous for all the gels, the G′ of the metallogel (AD-L@Ag(0)) was found to be higher in magnitude than the G′ of the ligand gel (AD-L). This result indicated the higher rigidity of the metallogel than that of the ligand gel. To investigate the morphological properties of the metallogel, transmission electron microscopy (TEM) and field-emission scanning electron microscopy (FE-SEM) were employed. The TEM images of the as-synthesized nanoparticles at different magnifications are presented in Fig. 5a and b. The HRTEM image (Fig. 5c) shows the lattice fringes which are symbolic of plasmonic Ag(0) nanoparticles. The most interesting observation from the TEM images is that the metallogel has an entangled fibril morphology, and all the Ag nanoparticles are uniformly distributed on the fibril network. We successfully detected the SAED patterns of the silver nanoparticles (Fig. 5e and f). The SAED pattern reveals that the nanoparticles consist of (111) and (200) planes. The HRTEM images reveal that the silver nanoparticles are crystalline, consisting of good quality lattice fringes with significant d-spacing values. Careful analysis of the HRTEM images reveals two groups of lattice fringes; the lattice distances are 0.20 nm and 0.23 nm, which are the indicators of the (200) and (111) planes of Ag(0), respectively.37 From the statistical analysis (Fig. 5d) of the TEM images, we can conclude that the average particle size of the Ag nanoparticles is 5 nm. Fig. 5g shows the FE-SEM images of the metallogel, which has a fibrillar morphology.
Literature reports reveal that Ag(0) nanoparticles with positive surface charge typically show the best bactericidal activity against various bacterial pathogens, followed by the Ag(0) NPs that do not possess any surface charge.85–88 This is true for both Gram-positive and Gram-negative bacteria. Ag(0) NPs release Ag+-ions following a slow oxidation process.89 The zeta potential (ζ) evaluated for Ag(0) in the AD-L@Ag(0) gel carries a positive charge (+30 mV), while that for AD-L is +13.4 mV. This confirms an overall positive surface potential for the Ag(0) NPs that are produced in situ. Literature reports also reveal that hydrogel matrices as self-assembled networks protect the NPs from aggregation or cluster formation.88,90 In the present study, the presence of the pyridine functionalities of the cationic guanidinium ions or the corresponding urea functionalities in their oxidized form would further help in stabilizing the Ag(0) NPs from agglomerate formation, and indeed Ag(0) NPs were not found to form bigger aggregates when examined during one month.
One would expect that cationic guanidinium derivatives would be attracted towards the negatively charged cell membrane of bacteria and interact with the biofilm components such as exopolysaccharides, nucleic acids, lipids, and proteins via electrostatic, hydrophobic, H-bonding, van der Waals forces and ionic interactions, respectively. However, such reports are rather scarce in the contemporary literature.91,92 Banerjee and his co-workers have shown that guanidinium-based Ionic Covalent Organic Nanosheets (iCONs) possess a bactericidal effect against both Gram +ve (S. aureus) and Gram −ve (E. coli) bacteria. Electrostatic interactions between the positively charged covalent organic nanosheets (CONs) derived from guanidinium ions and the negatively charged phospholipid bilayer of the bacterial membrane were speculated to be the reason for the bactericidal effect of guanidinium-based CONs.22 A recent report also tends to suggest that polyhexamethylene guanidine hydrochloride (PHMG-Cl) could adversely affect the biofilm-associated antibiotic-resistant eDNA release by Klebsiella strain—a key factor for a pan-drug-resistant Klebsiella strain.93 These results have helped us in developing Ag(0) NPs (with positive surface charges) stabilized in a hydrogel matrix of cationic guanidinium ion derivatives.
Antibacterial activity of AD-L@Ag(0)
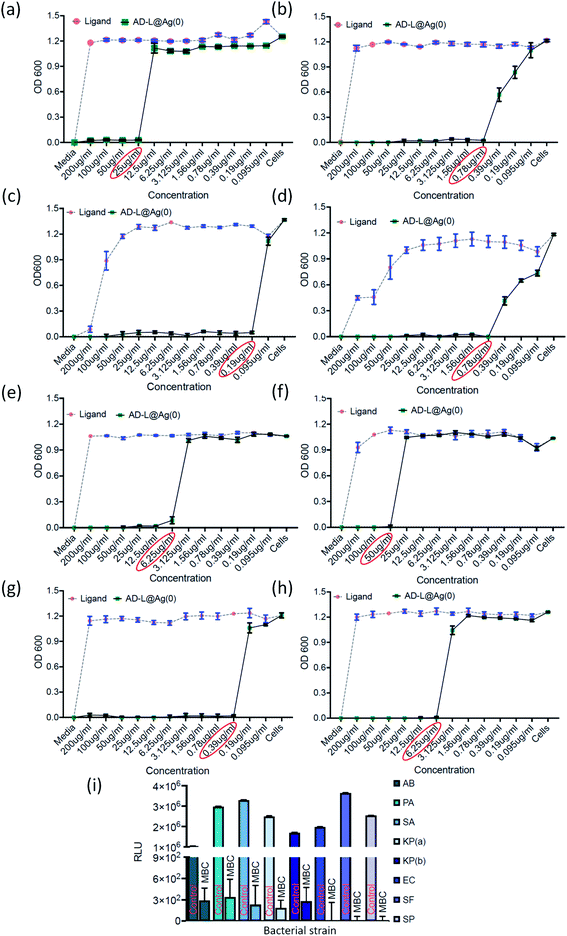
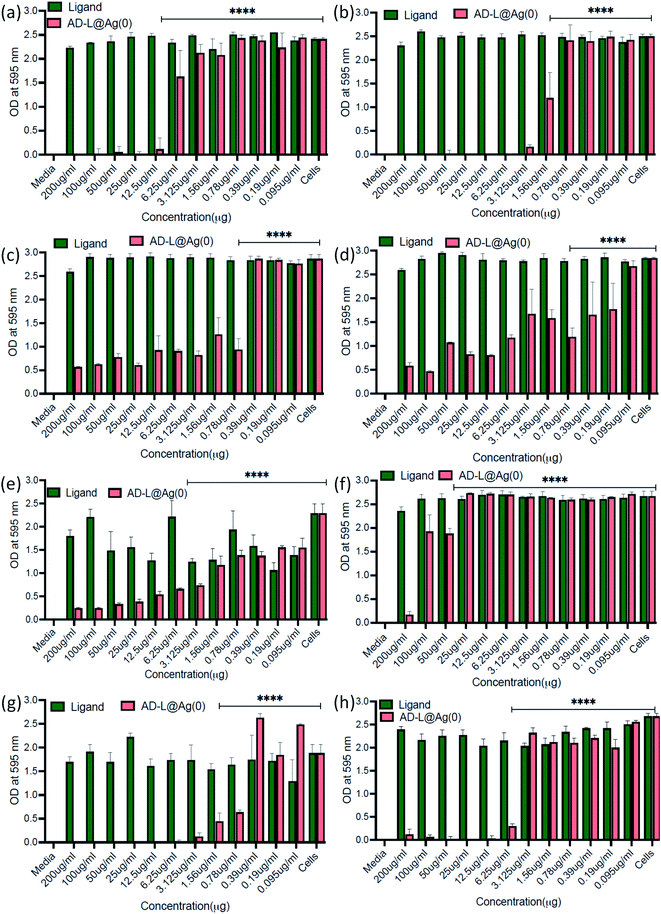
The antibacterial activity of the biosynthesized AD-L@Ag(0) was evaluated against various antibiotic-resistant bacteria in nutrient broth through turbidity measurement. The MIC values of AD-L@Ag(0) for Acinetobacter baumannii, Pseudomonas aeruginosa, Staphylococcus aureus, Klebsiella pneumoniae, Escherichia coli, Klebsiella pneumoniae (clinical), Shigella flexneri and Streptococcus pneumoniae were found to be 25 μg mL−1, 0.78 μg mL−1, 0.19 μg mL−1, 0.78 μg mL−1, 6.25 μg mL−1, 50 μg mL−1, 0.39 μg mL−1, and 6.25 μg mL−1 respectively (Fig. 6a–h). The zero CFU and lower ATP reading suggest that the MBC remains the same as the MIC concentration except for Acinetobacter baumannii, Staphylococcus aureus and Klebsiella pneumoniae (clinical) and changed to 50 μg mL−1, 100 μg ml−1, and 0.39 μg mL−1, respectively (Fig. 6i). Although there is no significant difference between the MICs of Gram-positive vs. Gram-negative and ATCC vs. clinical strains, the MIC of ATCC Gram-positive strains (0.19 μg ml−1) is much lower than the average ATCC MIC (8.5 μg ml−1), and in the case of clinical isolates also, the MIC of Gram-positive strains (6.25 μg ml−1) is much lower than that of Gram-negative strains (18.89 μg ml−1). A similar trend was observed in MBC values. All the clinical isolates used in this study are multi drug-resistant and AD-L@Ag(0) was able to kill all bacterial isolates at less than 10 μg ml−1 concentration, except two isolates Acinetobacter baumannii (50 μg ml−1) and Klebsiella pneumoniae (100 μg ml−1). The respective MIC and MBC values along with the MBIC and MBEC values for all isolates are provided in Table 2.| S. No. | Bacterial isolate | MIC | MBC | MBIC (50/90) | MBEC |
|---|---|---|---|---|---|
| 1 | Acinetobacter baumannii | 25 μg ml−1 | 50 μg ml−1 | 6.25/12.5 μg ml−1 | 25 μg ml−1 |
| 2 | Pseudomonas aeruginosa | 0.78 μg ml−1 | 0.78 μg ml−1 | 3.125/1.56 μg ml−1 | — |
| 3 | Staphylococcus aureus | 0.19 μg ml−1 | 0.39 μg ml−1 | 0.78/— μg ml−1 | 0.39 μg ml−1 |
| 4 | Klebsiella pneumoniae | 0.78 μg ml−1 | 0.78 μg ml−1 | 0.78/— μg ml−1 | 0.39 μg ml−1 |
| 5 | Escherichia coli | 6.25 μg ml−1 | 6.25 μg ml−1 | 3.125/50 μg ml−1 | — |
| 6 | Klebsiella pneumoniae (clinical) | 50 μg ml−1 | 100 μg ml−1 | 50/200 μg ml−1 | 500 μg ml−1 |
| 7 | Shigella flexneri | 0.39 μg ml−1 | 0.39 μg ml−1 | 0.78/3.125 μg ml−1 | 0.39 μg ml−1 |
| 8 | Streptococcus pneumoniae | 6.25 μg ml−1 | 6.25 μg ml−1 | 6.25/12.5 μg ml−1 | — |
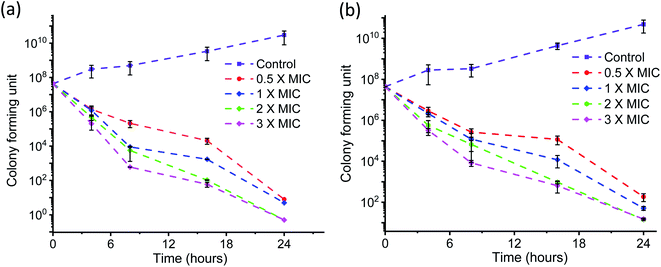
The time kinetics for killing the bacteria by AD-L@Ag(0) at different concentrations (above MIC, MIC and half of MIC) was also tested by the conventional plate count method. The time–kill curves for AD-L@Ag(0) were made using the highly anti-microbial resistant clinical strains of Klebsiella pneumoniae (Fig. 7a) and Staphylococcus aureus (Fig. 7b).
Rapid killing during the first 2 h of the assay was observed for concentrations above 2× MIC, and MIC and half of MIC showed little effect from zero to 4 h but the growth rate then decreased eventually. 3× MIC concentration exhibits the best bactericidal activity time–kill curve for Staphylococcus aureus (Fig. 7b) during the first 4 hours of the assay. But for the rest of the MIC concentration, the curve shows a delayed killing pattern.
Anti-biofilm activity of AD-L@Ag(0)
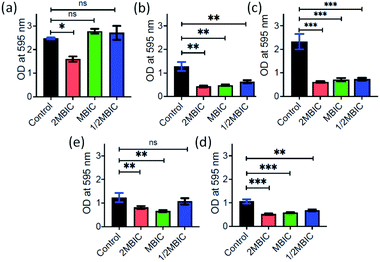
Inspired by the antibacterial property and good dispersibility of AD-L@Ag(0) we attempted to assess the biofilm inhibition and eradication properties of the formulation. During evolution, bacteria develop different mechanisms to get rid of antibiotics i.e., via efflux pump, changing the conformation of the drug target, enzymatic degradation of the antibacterial drug, changing the membrane permeability of the drug, biofilm production etc.94. Among all these strategies, biofilms have received significant attention for the discovery of new therapeutics. Biofilms are complex structures having bacterial colonies inside, and these are embedded inside extracellular polymeric substances, a matrix which is generally composed of eDNA, proteins, and polysaccharides, leading to high resistance to antibiotics. In this study, the MBIC50 and MBIC90 (Minimum Biofilm Inhibitory Concentration at 50% and 90% reduction, respectively) were measured for the AD-L@Ag(0) formulation and only the guanidine ligand by crystal violet assay for different bacterial strains including both Gram positive and Gram negative bacteria.The MBIC50 values for Acinetobacter baumannii, Pseudomonas aeruginosa, Staphylococcus aureus, Klebsiella pneumoniae, Escherichia coli, Klebsiella pneumoniae (clinical), Shigella flexneri and Streptococcus pneumoniae were found to be 6.25 μg mL−1, 3.125 μg mL−1, 0.78 μg mL−1, 0.78 μg mL−1, 3.125 μg mL−1, 50 μg mL−1, 0.78 μg mL−1, and 6.25 μg mL−1, respectively (Fig. 8). Furthermore, the MBIC90 values were 12.5 μg mL−1, 1.56 μg mL−1, 50 μg mL−1, 200 μg mL−1, 3.125 μg mL−1, and 12.5 μg mL−1 for Acinetobacter baumannii, Pseudomonas aeruginosa, Escherichia coli, Klebsiella pneumoniae (clinical), Shigella flexneri and Streptococcus pneumoniae, respectively. This MBIC concentration of the AD-L@Ag(0) formulation is considered based on statistically significant inhibition levels (p < 0.0001) compared to no treatment control. Furthermore, the exopolysaccharide quantification of biofilms was performed to check the biofilm eradication property of AD-L@Ag (0). The MBEC was determined with the treatment of AD-L@Ag(0) at 2× MBIC50, MBIC50 concentration and half of the MBIC50. Significant reductions in exopolysaccharides were observed even in half of the MBIC in most of the bacteria (Fig. 9). The MBEC values for Staphylococcus aureus, Klebsiella pneumoniae, and Shigella flexneri were 0.39 μg mL−1 (51.22% reduction), 0.39 μg mL−1 (68.64%), and 0.78 μg mL−1 (35.59%), respectively. The clinical isolate of Klebsiella pneumoniae (clinical) showed significant reduction of biofilms at the MBIC (1.56 μg mL−1), that is 46.53%. However, for Acinetobacter baumannii, upon treatment with AD-L@Ag(0) at the MBIC and half of the MBIC, no significant eradication of biofilms was observed; however, it does have a significant impact (35.51%) on biofilm integrity at the 2× MBIC concentration. On the other hand, non-significant eradication was observed in Escherichia coli, Pseudomonas aeruginosa, and Streptococcus pneumoniae even at 2× MBIC. From these results, we can hypothesize that AD-L@Ag(0) has biofilm eradication activity at a higher concentration and it can disintegrate or disperse the EPS component of mature biofilms resulting in planktonic cells exposed to the bactericidal effect of the formulation.
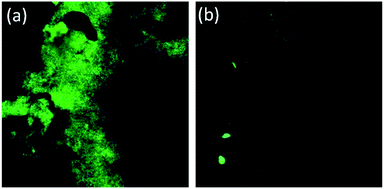
Additionally, to understand the alterations in the ultrastructural and physicochemical properties of biofilms after AD-L@Ag(0) treatment, confocal microscopy images were obtained at settings for DAPI – laser transmission – 0.8% and PMT voltage – 468, and for SYPRO – laser transmission – 0.6% and PMT voltage – 538. The extensive damage of the biofilm membrane was reflected through a dramatic reduction of DAPI emission (Fig. 10). A decrease in fluorescence of DAPI following treatment with colistin, an anti-microbial peptide, was also reported by Doktycz et al., respectively.95 Thus, confocal microscopy images demonstrated alteration in the ultrastructural morphology of the biofilm surface and provided an important insight into the anti-microbial mechanisms of AD-L@Ag(0).
Biocompatibility and in vivo proof-of-concept
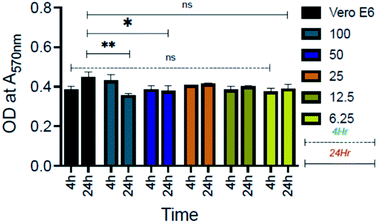
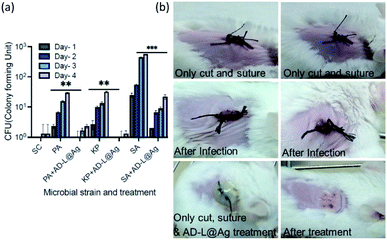
Furthermore, biocompatibility studies using 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyl tetrazolium bromide (MTT) assay performed on an epithelial cell line (Vero, ATCC CCL-81) (Fig. 11) indicate that AD-L@Ag(0) is non-cytotoxic and biocompatible thereby making it an excellent candidate for biomedical applications. To investigate further the application of AD-L@Ag(0) to treat multi-drug-resistant infections in vivo, we employed a mouse surgical site infection model to examine the efficacy of AD-L@Ag(0). The surgical wound was created on the right-hand side of their backbone with a sterile scalpel by making a longitudinal incision of 1 cm in length. The wound was infected by the insertion of a segment of silk suture through the skin using a suturing needle with 5 μL bacterial suspensions of Klebsiella pneumoniae (clinical), S. aureus (ATCC) and Pseudomonas aeruginosa (ATCC). The mice treated with the AD-L@Ag(0) suspension gel at the MIC concentration showed significant inhibition of bacterial growth in vivo (Fig. 12, Table S1†). The local bacterial counts in wound areas in control groups are much higher than those in the AD-L@Ag(0) treated group, which is consistent with wound sizes. Thus, AD-L@Ag(0) possesses outstanding anti-microbial activity and has potential in clinical multidrug resistant pathogen-induced wound treatment. This pilot experiment validates the efficacy of the current formulation in clearing bacterial infection at surgical sites and opens up prospects for detailed biocompatibility and safety studies in small animals.AD-L@Ag(0) as a combination therapy: bactericidal, biofilm-inhibitory, biofilm-eradicatory
Antibiotics that are highly active against Gram-negative bacteria are usually found to be less toxic to Gram-positive bacteria.96 In this study, we have shown that the AD-L@Ag(0) gel has similar antibacterial and anti-biofilm properties against Gram-positive and Gram-negative isolates. The zeta potential evaluated for Ag(0) in the AD-L@Ag(0) gel carries positive charge (+30.2 mV). Literature reports have established that cationic guanidinium derivatives and Ag+ are found to be attracted to the negatively charged membrane of bacteria and interact with the biofilm components such as exopolysaccharides, nucleic acids, lipids, and proteins via electrostatic, hydrophobic, H-bonding, van der Waals forces and ionic interactions, respectively.91,92 PHMG-Cl might form a DNA–PHMG-Cl complex, bind to AT base pairs by molecular docking, and inactivate biofilm-associated antibiotic-resistant eDNA release by Klebsiella strain.93Guanidinium-based Ionic Covalent Organic Nanosheets (iCONs) were earlier shown to have a bactericidal effect against both Gram +ve (S. aureus) and Gram −ve (E. coli) bacteria. Electrostatic interactions between positively charged iCONs and the negatively charged phospholipid bilayer of the bacterial membrane were speculated to be the reason for the bactericidal effect of guanidinium-based iCONs.22 Presumably, the positively charged surface of the AD-L@Ag(0) hydrogel in the present study favored an electrostatic interaction with the bacteria and biofilm components, which are majorly negatively charged.31 TEM studies show that the average particle size of Ag nanoparticles is ∼5 nm (Fig. 5g). Zhang et al. (2016) showed that the smaller-sized Ag nanoparticles were found to be more toxic to bacteria than large-sized Ag nanoparticles.97 Previous studies showed that the antibacterial efficacy improved for AgNPs having a size <10 nm, while those with ∼5 nm are most efficient.31,98 Certain reports reveal that AgNPs having a size of ∼5 nm could infuse through biofilm pores,99 and the larger particles are reported to be removed by efflux activity.100 The guanidinium derivative alone has been shown to have a minimal antibacterial effect even at 120 μg ml−1 and is prone to cause regrowth within 6–8 h of treatment.101 Our formulation has an augmented bactericidal effect integrating the functionalities of Ag(0) and the cationic guanidinium ions in AD-L@Ag(0). Data from the MIC, MBC, MBIC, MBEC, and time–kill assays in the current study corroborate these characteristics and demonstrate that the present formulation showed significant bactericidal activity at 50 μg ml−1 in as soon as 2–4 hours even for XDR clinical isolates.
In addition to the bactericidal effect, AD-L@Ag(0) has demonstrated a biofilm inhibitory as well as a biofilm dispersal effect at higher concentrations. It has also been reported that the acidic environment of the biofilm favors the disassembly of ligand–metal coordinations, as well as the supramolecular assembly.102 These factors presumably would favor access of AgNPs and guanidinium ions to the bacterial membrane and biofilm components to induce an enhanced bactericidal activity. For most of the physical or chemical agents with biofilm dispersal activity, there is a risk of mobilization of planktonic cells to nearby tissues and the bloodstream causing bacteremia.103 The present formulation with an efficient bactericidal activity can potentially address this challenge by exterminating the detached planktonic cells.
Conclusions
In this study, we have successfully demonstrated the efficacy of a unique organic–inorganic hybrid gel, derived from a guanidinium derivative and in situ generated Ag(0) nanoparticles stabilized in a soft gel matrix, as a therapeutic combination reagent for biofilm inhibition, dispersion and eradication of certain extremely drug-resistant bacterial pathogens that have clinical relevance. Experimental results confirm that the hydrazido derivative of the guanidinium ion is primarily effective in the inhibition and dispersion of biofilms observed in different multidrug resistant clinical isolates. Whereas, the AgNPs generated through in situ reduction of Ag(I) species and stabilized in the gel matrix are effective chiefly in inducing the bactericidal property of this hybrid gel through effective killing of planktonic bacteria. Importantly, this gel-hybrid and the Ag-nanoparticle bound nano-fibers were found to be non-cytotoxic. The formulation showed a significant bactericidal effect on laboratory strains as well as clinical isolates of clinically relevant extremely drug-resistant bacterial species, including both Gram positive and Gram negative bacteria. Further, in vivo proof-of-concept study showed that the formulation was effective in reducing the bacterial infection load at the surgical site in a small animal model. These results demonstrate that the current formulation harbors a significant promise for treating surgical wounds, as well as for use as a coating for surgical accessories, biomedical implants and engineered human tissue. To the best of our knowledge, the efficacy achieved by this hybrid gel formulation as a combination therapeutic agent in biofilm inhibition, dispersion, eradication and bacterial killing of the above-mentioned pathogens is superior to that reported in the presently available literature. The nano-formulation was observed to be a membrane disruptor, showing a reduction in membrane integrity and up-regulation of membrane associated proteins.74,78 Detailed cellular and molecular studies for elucidating the mechanism of action of the formulation are being conducted and will be part of our future report. This study opens up a new paradigm for developing anti-microbial agents with hybrid functionalities for effectively controlling extremely drug-resistant pathogens.Author contributions
AD, SC and SKP has conceptualised, executed, analysed data and drafted the manuscript. AD, MY, DK, ST, SS and DS have performed the experiments. All authors have contributed in refining the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
A. Das acknowledges SERB (India) Grants (CRG/2020/000492 & JCB/2017/000004). AD and SKP acknowledge a DBT (India) grant (BT/PR22251/NNT/28/1274/2017) for partially supporting this research. A. Dey acknowledges the CSIR for a senior research fellowship. M. Yadav acknowledges a UGC fellowship. D. Kumar acknowledges a DBT research fellowship. S. Chaudhuri acknowledges the DBT-THSTI for fund support. We also thank the Analytical & Environment Science Division & Centralized Instrument Facility of the CSIR-CSMCRI, Bhavnagar, and Small Animal Facility, THSTI. This manuscript bears a CSIR-CSMCRI PRIS No. 153/2022.Notes and references
- M. Kostakioti, M. Hadjifrangiskou and S. J. Hultgren, Cold Spring Harbor Perspect. Med., 2013, 3, a010306 CrossRef PubMed.
- D. Lebeaux, J. M. Ghigo and C. Beloin, Microbiol. Mol. Biol. Rev., 2014, 78, 510–543 CrossRef PubMed.
- A. S. Lee, H. de Lencastre, J. Garau, J. Kluytmans, S. Malhotra-Kumar, A. Peschel and S. Harbarth, Nat. Rev. Dis. Primers, 2018, 4, 18033 CrossRef PubMed.
- N. Rabin, Y. Zheng, C. Opoku-Temeng, Y. Du, E. Bonsu and H. O. Sintim, Future Med. Chem., 2015, 7, 493–512 CrossRef CAS PubMed.
- R. David, Nat. Rev. Microbiol., 2010, 8, 613 CrossRef PubMed.
- D. Davies, Nat. Rev. Drug Discovery, 2003, 2, 114–122 CrossRef CAS PubMed.
- G. Hughes and M. A. Webber, Br. J. Pharmacol., 2017, 174, 2237–2246 CrossRef CAS PubMed.
- R. J. Melander and C. Melander, ACS Infect. Dis., 2017, 3, 559–563 CrossRef CAS PubMed.
- D. H. Limoli, C. J. Jones and D. J. Wozniak, Microbiol. Spectrum, 2015, 3, 1–19 Search PubMed.
- Y. Jiao, G. D. Cody, A. K. Harding, P. Wilmes, M. Schrenk, K. E. Wheeler, J. F. Banfield and M. P. Thelen, Appl. Environ. Microbiol., 2010, 76, 2916–2922 CrossRef CAS PubMed.
- L. Hall-Stoodley, J. W. Costerton and P. Stoodley, Nat. Rev. Microbiol., 2004, 2, 95–108 CrossRef CAS PubMed.
- A. Penesyan, I. T. Paulsen, S. Kjelleberg and M. R. Gillings, npj Biofilms Microbiomes, 2021, 7, 80 CrossRef CAS PubMed.
- I. Guzmán-Soto, C. McTiernan, M. Gonzalez-Gomez, A. Ross, K. Gupta, E. J. Suuronen, T.-F. Mah, M. Griffith and E. I. Alarcon, iScience, 2021, 24, 102443 CrossRef PubMed.
- K. Lee Calvin, J. de Anda, E. Baker Amy, R. Bennett Rachel, Y. Luo, Y. Lee Ernest, A. Keefe Joshua, S. Helali Joshua, J. Ma, K. Zhao, R. Golestanian, A. O'Toole George and C. L. Wong Gerard, Proc. Natl. Acad. Sci., 2018, 115, 4471–4476 CrossRef CAS PubMed.
- J. V. d. O. Santos, A. L. F. Porto and I. M. F. Cavalcanti, Antibiotics, 2021, 10, 520 CrossRef PubMed.
- N. Goel, S. W. Fatima, S. Kumar, R. Sinha and S. K. Khare, Biotechnol. Rep., 2021, 30, e00613 CrossRef CAS PubMed.
- M. R. Kiedrowski and A. R. Horswill, Ann. N. Y. Acad. Sci., 2011, 1241, 104–121 CrossRef CAS PubMed.
- H. Koo, R. N. Allan, R. P. Howlin, P. Stoodley and L. Hall-Stoodley, Nat. Rev. Microbiol., 2017, 15, 740–755 CrossRef CAS PubMed.
- N. Kandoth, S. Barman, A. Chatterjee, S. Sarkar, A. K. Dey, S. K. Pramanik and A. Das, Adv. Funct. Mater., 2021, 31, 2104480 CrossRef CAS.
- I. Kolodkin-Gal, S. Cao, L. Chai, T. Böttcher, R. Kolter, J. Clardy and R. Losick, Cell, 2012, 149, 684–692 CrossRef CAS PubMed.
- T. Böttcher, I. Kolodkin-Gal, R. Kolter, R. Losick and J. Clardy, J. Am. Chem. Soc., 2013, 135, 2927–2930 CrossRef PubMed.
- S. Mitra, S. Kandambeth, B. P. Biswal, A. Khayum M, C. K. Choudhury, M. Mehta, G. Kaur, S. Banerjee, A. Prabhune, S. Verma, S. Roy, U. K. Kharul and R. Banerjee, J. Am. Chem. Soc., 2016, 138, 2823–2828 CrossRef CAS PubMed.
- I. M. Garcia, S. B. Rodrigues, M. E. Rodrigues Gama, V. C. Branco Leitune, M. A. Melo and F. M. Collares, Bioact. Mater., 2020, 5, 228–232 CrossRef.
- M. R. E. Santos, S. M. Ferreira, P. V. Mendonça, F. De Bon, A. C. Serra and J. F. J. Coelho, Polym. Chem., 2019, 10, 4944–4953 RSC.
- R. Banerjee, K. Das, A. Das and S. Dasgupta, Inorg. Chem., 1989, 28, 585–588 CrossRef CAS.
- U. Nickel, A. zu Castell, K. Pöppl and S. Schneider, Langmuir, 2000, 16, 9087–9091 CrossRef CAS.
- J. López-Esparza, L. F. Espinosa-Cristóbal, A. Donohue-Cornejo and S. Y. Reyes-López, Ind. Eng. Chem. Res., 2016, 55, 12532–12538 CrossRef.
- T. C. Dakal, A. Kumar, R. S. Majumdar and V. Yadav, Front. Microbiol., 2016, 7, 1831 Search PubMed.
- J. R. Morones, J. L. Elechiguerra, A. Camacho, K. Holt, J. B. Kouri, J. T. Ramírez and M. J. Yacaman, Nanotechnology, 2005, 16, 2346–2353 CrossRef CAS PubMed.
- R. P. Allaker, J. Dent. Res., 2010, 89, 1175–1186 CrossRef CAS PubMed.
- S. Anees Ahmad, S. Sachi Das, A. Khatoon, M. Tahir Ansari, M. Afzal, M. Saquib Hasnain and A. Kumar Nayak, Mater. Sci. Energy Technol., 2020, 3, 756–769 CAS.
- E. Urnukhsaikhan, B.-E. Bold, A. Gunbileg, N. Sukhbaatar and T. Mishig-Ochir, Sci. Rep., 2021, 11, 21047 CrossRef PubMed.
- J. Li, L. Kang, B. Wang, K. Chen, X. Tian, Z. Ge, J. Zeng, J. Xu and W. Gao, ACS Sustainable Chem. Eng., 2019, 7, 1146–1158 CrossRef CAS.
- X. Fei, M. Jia, X. Du, Y. Yang, R. Zhang, Z. Shao, X. Zhao and X. Chen, Biomacromolecules, 2013, 14, 4483–4488 CrossRef CAS PubMed.
- B. Banerji, S. K. Pramanik, P. Sanphui, S. Nikhar and S. C. Biswas, Chem. Biol. Drug Des., 2013, 82, 401–409 CrossRef CAS PubMed.
- S. Hiroto, Y. Miyake and H. Shinokubo, Chem. Rev., 2017, 117, 2910–3043 CrossRef CAS PubMed.
- R. Bandichhor, in Hazardous Reagent Substitution: A Pharmaceutical Perspective, The Royal Society of Chemistry, 2018, 10.1039/9781782623847-00001, pp. 1–17.
- J. Pinto, D. Magrì, P. Valentini, F. Palazon, J. A. Heredia-Guerrero, S. Lauciello, S. Barroso-Solares, L. Ceseracciu, P. P. Pompa, A. Athanassiou and D. Fragouli, ACS Appl. Mater. Interfaces, 2018, 10, 16095–16104 CrossRef CAS PubMed.
- Y. Cao, R. Jin and C. A. Mirkin, J. Am. Chem. Soc., 2001, 123, 7961–7962 CrossRef CAS PubMed.
- J. B. Wright, K. Lam, D. Hansen and R. E. Burrell, Am. J. Infect. Control, 1999, 27, 344–350 CrossRef CAS PubMed.
- D. M. Eby, N. M. Schaeublin, K. E. Farrington, S. M. Hussain and G. R. Johnson, ACS Nano, 2009, 3, 984–994 CrossRef CAS PubMed.
- S. H. Aswathy, U. Narendrakumar and I. Manjubala, Heliyon, 2020, 6, e03719 CrossRef CAS PubMed.
- H. Y. Lee, H. K. Park, Y. M. Lee, K. Kim and S. B. Park, Chem. Commun., 2007, 2959–2961, 10.1039/B703034G.
- J. Song, H. Kang, C. Lee, S. H. Hwang and J. Jang, ACS Appl. Mater. Interfaces, 2012, 4, 460–465 CrossRef CAS PubMed.
- R. Tiwari, S. Banerjee, D. Tyde, K. D. Saha, A. Ethirajan, N. Mukherjee, S. Chattopadhy, S. K. Pramanik and A. Das, Bioconjugate Chem., 2021, 32, 245–253 CrossRef CAS PubMed.
- S. K. Pramanik and A. Das, Chem. Commun., 2021, 57, 12058–12073 RSC.
- S. K. Pramanik, S. Seneca, M. Peters, L. D'Olieslaeger, G. Reekmans, D. Vanderzande, P. Adriaensens and A. Ethirajan, RSC Adv., 2018, 8, 36869–36878 RSC.
- R. Tiwari, P. S. Shinde, S. Sreedharan, A. K. Dey, K. A. Vallis, S. B. Mhaske, S. K. Pramanik and A. Das, Chem. Sci., 2021, 12, 2667–2673 RSC.
- F. Xu, H. Padhy, M. Al-Dossary, G. Zhang, A. R. Behzad, U. Stingl and A. Rothenberger, J. Mater. Chem. B, 2014, 2, 6406–6411 RSC.
- Z. Xu, M. Li, X. Li, X. Liu, F. Ma, S. Wu, K. W. K. Yeung, Y. Han and P. K. Chu, ACS Appl. Mater. Interfaces, 2016, 8, 16584–16594 CrossRef CAS PubMed.
- J. Hopf, M. Waters, V. Kalwajtys, K. E. Carothers, R. K. Roeder, J. D. Shrout, S. W. Lee and P. D. Nallathamby, Nanoscale Adv., 2019, 1, 4812–4826 RSC.
- J. Yang, Y. Chen, L. Zhao, Z. Feng, K. Peng, A. Wei, Y. Wang, Z. Tong and B. Cheng, Composites, Part B, 2020, 197, 108139 CrossRef CAS.
- P. Rajamalli, P. Malakar, S. Atta and E. Prasad, Chem. Commun., 2014, 50, 11023–11025 RSC.
- J. W. Cahn and R. E. Powell, J. Am. Chem. Soc., 1954, 76, 2568–2572 CrossRef CAS.
- T. Tolker-Nielsen, APMIS, Suppl., 2014, 1–51, DOI:10.1111/apm.12335.
- A. Maity, M. Gangopadhyay, A. Basu, S. Aute, S. S. Babu and A. Das, J. Am. Chem. Soc., 2016, 138, 11113–11116 CrossRef CAS PubMed.
- C. Rodríguez-Melcón, C. Alonso-Calleja, C. García-Fernández, J. Carballo and R. Capita, Biology, 2022, 11, 46 CrossRef PubMed.
- D. M. Hacek, D. C. Dressel and L. R. Peterson, J. Clin. Microbiol., 1999, 37, 1881–1884 CrossRef CAS PubMed.
- H. Karballaei Mirzahosseini, M. Hadadi-Fishani, K. Morshedi and A. Khaledi, Microb. Drug Resist., 2020, 26, 815–824 CrossRef CAS PubMed.
- N. C. d. S. Santos, R. B. d. L. Scodro, E. G. Sampiron, A. L. Ieque, H. C. d. Carvalho, T. d. S. Santos, L. D. Ghiraldi Lopes, P. A. Z. Campanerut-Sá, V. L. D. Siqueira, K. R. Caleffi-Ferracioli, J. J. V. Teixeira and R. F. Cardoso, Microb. Drug Resist., 2020, 26, 752–765 CrossRef CAS PubMed.
- M. DuBois, K. A. Gilles, J. K. Hamilton, P. A. Rebers and F. Smith, Anal. Chem., 1956, 28, 350–356 CrossRef CAS.
- T. L. Riss and A. L. Niles, Eli Lilly & Company and the National Center for Advancing Translational Sciences, 2013, https://www.ncbi.nlm.nih.gov/books/NBK144065/ Search PubMed.
- P. V. Bharatam, D. S. Patel and P. Iqbal, J. Med. Chem., 2005, 48, 7615–7622 CrossRef CAS PubMed.
- B. Xu, M. I. Jacobs, O. Kostko and M. Ahmed, ChemPhysChem, 2017, 18, 1503–1506 CrossRef CAS PubMed.
- P. E. Mason, G. W. Neilson, J. E. Enderby, M.-L. Saboungi, C. E. Dempsey, A. D. MacKerell and J. W. Brady, J. Am. Chem. Soc., 2004, 126, 11462–11470 CrossRef CAS PubMed.
- Y. Zuo, C. Chen, X. Li, Z. Deng, Y. Chen, J. Behler, G. Csányi, A. V. Shapeev, A. P. Thompson, M. A. Wood and S. P. Ong, J. Phys. Chem. A, 2020, 124, 731–745 CrossRef CAS PubMed.
- A. Güven, Int. J. Mol. Sci., 2005, 6, 257–275 CrossRef.
- J. C. Abbar, S. D. Lamani and S. T. Nandibewoor, Ind. Eng. Chem. Res., 2009, 48, 7550–7560 CrossRef CAS.
- R. Banerjee, A. Das and S. Dasgupta, J. Chem. Soc., Dalton Trans., 1990, 2271–2275, 10.1039/DT9900002271.
- A. Furst, R. C. Berlo and S. Hooton, Chem. Rev., 1965, 65, 51–68 CrossRef CAS.
- D. D. Evanoff Jr and G. Chumanov, Chemphyschem, 2005, 6, 1221–1231 CrossRef PubMed.
- T. Huang and X.-H. N. Xu, J. Mater. Chem., 2010, 20, 9867–9876 RSC.
- A. Chhatre, P. Solasa, S. Sakle, R. Thaokar and A. Mehra, Colloids Surf., A, 2012, 404, 83–92 CrossRef CAS.
- L. Bar, F. Perissinotto, L. Redondo-Morata, M. I. Giannotti, J. Goole and P. Losada-Pérez, Colloids Surf., B, 2022, 210, 112239 CrossRef CAS PubMed.
- R. Banerjee, A. Das and S. Dasgupta, J. Chem. Soc., Dalton Trans., 1990, 1207–1212, 10.1039/DT9900001207.
- J. F. Gomes, A. C. Garcia, E. B. Ferreira, C. Pires, V. L. Oliveira, G. Tremiliosi-Filho and L. H. S. Gasparotto, Phys. Chem. Chem. Phys., 2015, 17, 21683–21693 RSC.
- B. Mishra, R. M. Golla, K. Lau, T. Lushnikova and G. Wang, ACS Med. Chem. Lett., 2016, 7, 117–121 CrossRef CAS PubMed.
- R. C. Van Lehn, M. Ricci, P. H. J. Silva, P. Andreozzi, J. Reguera, K. Voïtchovsky, F. Stellacci and A. Alexander-Katz, Nat. Commun., 2014, 5, 4482 CrossRef CAS PubMed.
- D. Lin, H. Wu, R. Zhang and W. Pan, Chem. Mater., 2009, 21, 3479–3484 CrossRef CAS.
- R. Jin, C. Zeng, M. Zhou and Y. Chen, Chem. Rev., 2016, 116, 10346–10413 CrossRef CAS PubMed.
- K. Jia, P. Wang, L. Yuan, X. Zhou, W. Chen and X. Liu, J. Mater. Chem. C, 2015, 3, 3522–3529 RSC.
- T. C. Kaspar, T. Droubay, S. A. Chambers and P. S. Bagus, J. Phys. Chem. C, 2010, 114, 21562–21571 CrossRef CAS.
- N. J. Firet, M. A. Blommaert, T. Burdyny, A. Venugopal, D. Bohra, A. Longo and W. A. Smith, J. Mater. Chem. A, 2019, 7, 2597–2607 RSC.
- E. R. Draper and D. J. Adams, Chem. Soc. Rev., 2018, 47, 3395–3405 RSC.
- A. Abbaszadegan, Y. Ghahramani, A. Gholami, B. Hemmateenejad, S. Dorostkar, M. Nabavizadeh and H. Sharghi, J. Nanomater., 2015, 2015, 720654 Search PubMed.
- C. Liao, Y. Li and S. C. Tjong, Int. J. Mol. Sci., 2019, 20, 449 CrossRef PubMed.
- L. Wang, C. Hu and L. Shao, Int. J. Nanomed., 2017, 12, 1227–1249 CrossRef CAS PubMed.
- S. Chernousova and M. Epple, Angew. Chem., Int. Ed. Engl., 2013, 52, 1636–1653 CrossRef CAS PubMed.
- S. Kittler, C. Greulich, J. Diendorf, M. Köller and M. Epple, Chem. Mater., 2010, 22, 4548–4554 CrossRef CAS.
- Y. Murali Mohan, K. Vimala, V. Thomas, K. Varaprasad, B. Sreedhar, S. K. Bajpai and K. Mohana Raju, J. Colloid Interface Sci., 2010, 342, 73–82 CrossRef CAS PubMed.
- G. McDonnell and A. D. Russell, Clin. Microbiol. Rev., 1999, 12, 147–179 CrossRef CAS PubMed.
- A. S. Joshi, P. Singh and I. Mijakovic, Int. J. Mol. Sci., 2020, 21, 7658 CrossRef CAS PubMed.
- O. V. Moshynets, T. P. Baranovskyi, O. S. Iungin, N. P. Kysil, L. O. Metelytsia, I. Pokholenko, V. V. Potochilova, G. Potters, K. L. Rudnieva, S. Y. Rymar, I. V. Semenyuta, A. J. Spiers, O. P. Tarasyuk and S. P. Rogalsky, Int. J. Mol. Sci., 2022, 23, 731 CrossRef CAS PubMed.
- L. A. Dever and T. S. Dermody, Arch. Intern. Med., 1991, 151, 886–895 CrossRef CAS PubMed.
- S. Hasim, D. P. Allison, B. Mendez, A. T. Farmer, D. A. Pelletier, S. T. Retterer, S. R. Campagna, T. B. Reynolds and M. J. Doktycz, Front. Microbiol., 2018, 9, 219 CrossRef PubMed.
- E. Escamilla-García, A. G. Alcázar-Pizaña, J. C. Segoviano-Ramírez, C. Del Angel-Mosqueda, A. P. López-Lozano, E. Cárdenas-Estrada, M. A. De La Garza-Ramos, C. E. Medina-De La Garza and M. Márquez, Int. J. Microbiol., 2017, 2017, 5924717 Search PubMed.
- X.-F. Zhang, Z.-G. Liu, W. Shen and S. Gurunathan, Int. J. Mol. Sci., 2016, 17, 1–34 Search PubMed.
- S. Agnihotri, S. Mukherji and S. Mukherji, RSC Adv., 2014, 4, 3974–3983 RSC.
- T.-O. Peulen and K. J. Wilkinson, Environ. Sci. Technol., 2011, 45, 3367–3373 CrossRef CAS PubMed.
- P. D. Nallathamby, K. J. Lee, T. Desai and X.-H. N. Xu, Biochemistry, 2010, 49, 5942–5953 CrossRef CAS PubMed.
- H. Han, J. Zhu, D.-Q. Wu, F.-X. Li, X.-L. Wang, J.-Y. Yu and X.-H. Qin, Adv. Funct. Mater., 2019, 29, 1806594 CrossRef.
- J. Wu, F. Li, X. Hu, J. Lu, X. Sun, J. Gao and D. Ling, ACS Cent. Sci., 2019, 5, 1366–1376 CrossRef CAS PubMed.
- J. Wille and T. Coenye, Biofilm, 2020, 2, 100027 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc02980d |
| ‡ All authors equally contributed to this work. |
| This journal is © The Royal Society of Chemistry 2022 |