Open Access Article
Open Access ArticleIntact living-cell electrolaunching ionization mass spectrometry for single-cell metabolomics†
Yunlong
Shao
a,
Yingyan
Zhou
a,
Yuanxing
Liu
a,
Wenmei
Zhang
a,
Guizhen
Zhu
a,
Yaoyao
Zhao
*a,
Qi
Zhang
a,
Huan
Yao
b,
Hansen
Zhao
b,
Guangsheng
Guo
ac,
Sichun
Zhang
 b,
Xinrong
Zhang
b,
Xinrong
Zhang
 b and
Xiayan
Wang
b and
Xiayan
Wang
 *a
*a
aDepartment of Chemistry and Biology, Center of Excellence for Environmental Safety and Biological Effects, Beijing Key Laboratory for Green Catalysis and Separation, Beijing University of Technology, Beijing 100124, P. R. China. E-mail: zhaoyaoyao@bjut.edu.cn; xiayanwang@bjut.edu.cn
bDepartment of Chemistry, Tsinghua University, Beijing 100084, P. R. China
cMinzu University of China, Beijing 100081, P. R. China
First published on 17th June 2022
Abstract
While single-cell mass spectrometry can reveal cellular heterogeneity and the molecular mechanisms of intracellular biochemical reactions, its application is limited by the insufficient detection sensitivity resulting from matrix interference and sample dilution. Herein, we propose an intact living-cell electrolaunching ionization mass spectrometry (ILCEI-MS) method. A capillary emitter with a narrow-bore, constant-inner-diameter ensures that the entire living cell enters the MS ion-transfer tube. Inlet ionization improves sample utilization, and no solvent is required, preventing sample dilution and matrix interference. Based on these features, the detection sensitivity is greatly improved, and the average signal-to-noise (S/N) ratio is about 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 of single-cell peaks in the TIC of ILCEI-MS. A high detection throughput of 51 cells per min was achieved by ILCEI-MS for the single-cell metabolic profiling of multiple cell lines, and 368 cellular metabolites were identified. Further, more than 4000 primary single cells digested from the fresh multi-organ tissues of mice were detected by ILCEI-MS, demonstrating its applicability and reliability.
1 of single-cell peaks in the TIC of ILCEI-MS. A high detection throughput of 51 cells per min was achieved by ILCEI-MS for the single-cell metabolic profiling of multiple cell lines, and 368 cellular metabolites were identified. Further, more than 4000 primary single cells digested from the fresh multi-organ tissues of mice were detected by ILCEI-MS, demonstrating its applicability and reliability.
Introduction
Single-cell metabolite analysis is a promising method to reveal cellular heterogeneity at the most basic and subtle small molecule level,1–5 especially living single-cell metabolite analysis,6 enabling the gathering of the most veritable bio-chemical information and reflecting the cell status with higher fidelity, including cell differentiation and division,7–9 communication,1,10 interaction with the environment,11–13 and stress response.14 This diverse information is of great significance for the development of cell biology and medicine and facilitates cancer diagnosis, prognosis, and treatment.15 Mass spectrometry (MS) is the principal method for single-cell metabolite analysis because of its ability to afford rapid, wide-spectrum detection along with excellent qualitative structure determination.12 Different MS-based methods have been developed to profile single-cell metabolites.16–20 Secondary-ion MS (SIMS)16 and matrix-assisted laser desorption/ionization MS (MALDI-MS)18,19 require a high-vacuum environment and complex sample preparation, preventing the analysis of living cells. Cytometry by time of flight (CyTOF)20 can be used to detect metal-labeled proteins in living cells; however, it is incapable of measuring small-molecule metabolites that are difficult to label. Therefore, electrospray ionization MS (ESI-MS), which does not require labeling, is more suitable for living single-cell metabolite analysis.11,21–26 In the detection of living cells, cell samples are usually intact cells or a part of the native components of living cells. Therefore, due to the complex biological structure of cells, the liquid system is no longer a homogeneous solution. However, current studies on single-cell ESI-MS still use the electrospray mode developed for homogeneous solutions, resulting in some insurmountable problems related to the ionization of heterogeneous single-cell samples. The use of a large amount of sheath fluid and sheath gas in the electrospray process results in the severe dilution of samples. The use of nanoliters of solvent for dissolution or extraction can result in thousand-fold sample dilution, which is lethal for trace components in single cells with picoliter volumes. In addition, because the ionization process occurs before the sample enters the MS ion inlet, the sample is subject to diffusion, dilution, and loss in the atmosphere; this results in problems such as a low sample-acquisition ratio in the MS ion inlet, thereby reducing the utilization of single-cell samples.27 These problems limit the sensitivity of single-cell MS detection.Although many bioelectrospray techniques based on high-throughput single-cell manipulation have been developed for tissue engineering and regenerative medicine,28–34 most are not suitable for MS. Therefore, to achieve high-sensitivity and high-throughput single-cell MS, a novel ionization technique that can be used for complex living-cell samples is needed.
Herein, we propose an intact living-cell electrolaunching ionization MS (ILCEI-MS) method using a narrow-bore capillary [the inner diameter (I.D.) is slightly smaller than the average cell diameter] to achieve efficient single-cell separation and transport while avoiding sample dilution by the sheath fluid. The novel ILCEI technique can realize undiluted living single-cell flow MS. In the ILCEI method, the constant-I.D., thin-walled tip of a quartz tube serves as the outlet, which reduces the volume of droplets formed by the combined action of the applied electric field and the surface tension on the port. Based on observation with an online ILCEI-MS visualization platform, the emitted single-cell droplet contains only a small amount of thin-layer buffer, wherein one cell acts as one droplet. The cell components are almost undiluted. Moreover, the cells remain intact and alive during flight. Ions are generated after a charged single-cell droplet forms in the heated transfer tube of the mass spectrometer, eliminating the loss of sample ions during transport through the atmospheric environment and mass spectrometer ion entrance, which is not possible with ESI. Therefore, the utilization of single-cell samples is significantly improved. Because of the abovementioned characteristics, the single-cell MS total ion chromatogram (TIC) of B104 cells obtained with ILCEI-MS has an average S/N ratio of ∼20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; thus, the detection sensitivity is greatly improved. Moreover, a throughput of approximately 51 cells per min was obtained for single-cell MS. Using the ILCEI-MS method, we analyzed more than 482 A549 cells and identified 368 cellular metabolites in one experiment. Additionally, more than 2800 in vitro-cultured living cells were discriminated with exact cell-type attribution. Hence, the ILCEI-MS method shows satisfactory single-cell detection sensitivity and stability. The high-throughput detection of 4072 single cells from multiple mouse organs in a tumor-bearing mouse model of non-small lung cancer (NSCLC) demonstrated the general applicability of this method in practical complex biological samples. ILCEI-MS is an easy-to-use method with a simple structure and stable operation. Moreover, ILCEI-MS is highly compatible with commercial MS ion source equipment and has low operating requirements.
1; thus, the detection sensitivity is greatly improved. Moreover, a throughput of approximately 51 cells per min was obtained for single-cell MS. Using the ILCEI-MS method, we analyzed more than 482 A549 cells and identified 368 cellular metabolites in one experiment. Additionally, more than 2800 in vitro-cultured living cells were discriminated with exact cell-type attribution. Hence, the ILCEI-MS method shows satisfactory single-cell detection sensitivity and stability. The high-throughput detection of 4072 single cells from multiple mouse organs in a tumor-bearing mouse model of non-small lung cancer (NSCLC) demonstrated the general applicability of this method in practical complex biological samples. ILCEI-MS is an easy-to-use method with a simple structure and stable operation. Moreover, ILCEI-MS is highly compatible with commercial MS ion source equipment and has low operating requirements.
Results and discussion
Configuration and visual characterization of the ILCEI-MS system
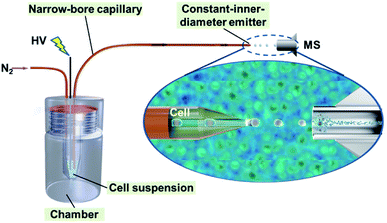
Based on our previous work on narrow-bore capillary chromatography,35–41 we developed a single-cell MS analysis method using a novel ILCEI technique. This analysis method aims to realize high-throughput single-cell online dispersion, dilution-free MS injection, and high-sensitivity MS detection. As shown in Fig. 1, the method is simple and requires only a narrow capillary and an in-house-built pressurized poly (methyl methacrylate) chamber. The capillary has a constant-I.D. throughout its entirety and a thin-walled tip. A freshly prepared cell suspension is loaded into a solution vial inside the pressurized chamber and introduced into the capillary driven by nitrogen gas for the injection of cells. The cells arrange themselves along the capillary axis to form a single-cell flow and are launched from the constant-I.D., thin-walled tip into the MS inlet for detection. A high voltage is applied directly via a stainless-steel needle to the cell suspension to launch single cells.The necessity of each key strategy in the method was verified through a series of comparative experiments. A narrow capillary with an I.D. similar to the diameter of a cell is a prerequisite for separating single cells (Fig. S1†). A capillary wall thinned by etching is needed to generate a single-cell MS detection signal from a constant-I.D. capillary emitter (Fig. S2†). The tip with a constant-I.D. has a low risk of fluid clogging, enabling a stable flow rate and a long operation life, which are important factors for the MS detection of complex biological samples (Fig. S3†).
To objectively and comprehensively investigate the operation of the system, an online ILCEI-MS visualization platform was designed and built (Fig. S4†). The operation of the ILCEI-MS system was visually recorded using this platform. A single A549 cell moving in a narrow capillary propelled by a mobile phase was observed in real time and recorded at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 frames per second (fps), as shown in Video S1.† The cell was slightly squeezed from its original spherical shape into a nearly cylindrical shape owing to the confinement effect as the cell passed through the narrow capillary (the I.D. was slightly smaller than the cell diameter; Fig. 2, stage I). Additional video footage of a single cell being transported inside the capillary is provided in Video S2.† In this video, the single cell appears similar to a plug and is pushed along the capillary by the mobile phase. Once a single cell enters the capillary and begins its transport, its distance from the cell in front of it remains constant because the liquid is almost incompressible. Consequently, the single cells move in sequence. This observation is consistent with the expected behavior of cells in narrow-bore capillaries. Thus, the use of such capillaries is a simple and reliable method for single-cell dispersion and transport. Notably, even though the capillary was clogged with a few very large cells (due to cell heterogeneity), it could be flushed by applying a hydraulic pressure as low as 70–200 bar using a liquid chromatography system or simply a chromatography pump.
000 frames per second (fps), as shown in Video S1.† The cell was slightly squeezed from its original spherical shape into a nearly cylindrical shape owing to the confinement effect as the cell passed through the narrow capillary (the I.D. was slightly smaller than the cell diameter; Fig. 2, stage I). Additional video footage of a single cell being transported inside the capillary is provided in Video S2.† In this video, the single cell appears similar to a plug and is pushed along the capillary by the mobile phase. Once a single cell enters the capillary and begins its transport, its distance from the cell in front of it remains constant because the liquid is almost incompressible. Consequently, the single cells move in sequence. This observation is consistent with the expected behavior of cells in narrow-bore capillaries. Thus, the use of such capillaries is a simple and reliable method for single-cell dispersion and transport. Notably, even though the capillary was clogged with a few very large cells (due to cell heterogeneity), it could be flushed by applying a hydraulic pressure as low as 70–200 bar using a liquid chromatography system or simply a chromatography pump.
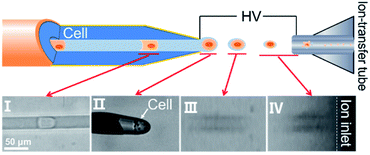
The ejection of an entire single cell from the constant-I.D.-emitter was recorded in real time at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fps (Video S3†). An intact individual cell was ejected from the capillary tip and carried a tiny amount of solvent to form the initial droplet that flew into the MS inlet. We termed this initial droplet the “single-cell droplet” (Fig. 2, stage II) and this process the “electrolaunching” (Fig. 2, stage II–IV). This unique launching phenomenon occurred at a certain voltage at the constant-I.D., thin-walled tip of the narrow capillary, which is key to resolving the dilution problem. We expected to achieve the same result when such a tip was used as an emitter. The results confirm that the ILCEI-MS method achieved single-cell injection rather than the injection of multiple cells simultaneously.
000 fps (Video S3†). An intact individual cell was ejected from the capillary tip and carried a tiny amount of solvent to form the initial droplet that flew into the MS inlet. We termed this initial droplet the “single-cell droplet” (Fig. 2, stage II) and this process the “electrolaunching” (Fig. 2, stage II–IV). This unique launching phenomenon occurred at a certain voltage at the constant-I.D., thin-walled tip of the narrow capillary, which is key to resolving the dilution problem. We expected to achieve the same result when such a tip was used as an emitter. The results confirm that the ILCEI-MS method achieved single-cell injection rather than the injection of multiple cells simultaneously.
After synchronizing the high-speed microphotography with the single-cell MS data acquisition, three cells were successively electrolaunched and recorded using a high-speed camera at 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 fps (at this shooting speed, the camera could record up to 5 s); the corresponding mass spectra were collected simultaneously (Video S4†). The time interval of the electrolaunching of the three cells was consistent with the retention time of the MS peaks (Fig. S5 and Video S5†). This observation demonstrates that each signal peak in the TIC obtained by the ILCEI-MS system corresponds to a single cell; the peaks do not arise from the superposition of multiple cell signals. Further, we compared the ILCEI-MS results of the cell membrane debris sample and the intact living cell sample and found obvious differences in the MS peak shape and ion intensity between the two samples (Fig. S6†). Three samples were chosen to evaluate the signal stability of ILCEI-MS under complex solvent conditions: 40 mmol L−1 ammonium formate aqueous solution (mobile phase); the cell lysate of lung cancer cells (the supernatant collected after centrifugation at 17
000 fps (at this shooting speed, the camera could record up to 5 s); the corresponding mass spectra were collected simultaneously (Video S4†). The time interval of the electrolaunching of the three cells was consistent with the retention time of the MS peaks (Fig. S5 and Video S5†). This observation demonstrates that each signal peak in the TIC obtained by the ILCEI-MS system corresponds to a single cell; the peaks do not arise from the superposition of multiple cell signals. Further, we compared the ILCEI-MS results of the cell membrane debris sample and the intact living cell sample and found obvious differences in the MS peak shape and ion intensity between the two samples (Fig. S6†). Three samples were chosen to evaluate the signal stability of ILCEI-MS under complex solvent conditions: 40 mmol L−1 ammonium formate aqueous solution (mobile phase); the cell lysate of lung cancer cells (the supernatant collected after centrifugation at 17![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 g to simulate the massive leakage of cellular components into the solution); and a liposome suspension with an average diameter of 200 nm (to simulate solutions containing a large number of cell membrane fragments). We collected 1000 scans, and the average relative standard deviation was less than 7%. The experimental results demonstrate that ILCEI-MS has good signal stability (Fig. S7 and S8†). The concern that cell debris or background noise may cause false-positive results in single-cell assays was ruled out. The visual characterization of the ILCEI-MS analysis system is shown in Fig. 2.
000 g to simulate the massive leakage of cellular components into the solution); and a liposome suspension with an average diameter of 200 nm (to simulate solutions containing a large number of cell membrane fragments). We collected 1000 scans, and the average relative standard deviation was less than 7%. The experimental results demonstrate that ILCEI-MS has good signal stability (Fig. S7 and S8†). The concern that cell debris or background noise may cause false-positive results in single-cell assays was ruled out. The visual characterization of the ILCEI-MS analysis system is shown in Fig. 2.
To further study the changes after the single-cell droplet was formed, we shifted the microscope observation field to the atmospheric section (Fig. 2, stage III) between the emitter and the MS inlet (Fig. 2, stage IV). The combined videos (Video S6†) show that the single-cell droplet did not undergo clear droplet splitting during its flight before it entered the ion-transfer tube. To further confirm that the cells remained intact before entering the MS inlet, we used a cover glass to obstruct and receive the single-cell droplet ejected by the emitter. Bright-field and fluorescence imaging revealed a distinct stereoscopic cell profile along with good single-cell dispersion and the absence of luminous cell debris, demonstrating that the cell entered the MS inlet in an intact state (Fig. S9†). To further verify that the ILCEI-MS system is a true living-cell MS detection system, we evaluated the viability of cells about to enter the mass spectrometer using CCK8 assay. The experimental details are provided in the Experimental section and Fig. S10.† Before entering the mass spectrometer, 87.22% of the cells were still alive, demonstrating that ILCEI-MS is truly a living-cell analysis method.
The formation of single-cell droplets eliminated the dilution of single-cell samples during electrolaunching. Most of the space in single-cell droplets is occupied by a cell, and the solvent is present only as an ultrathin film on the droplet surface, which reduces the dilution factor by hundreds or thousands of times compared to traditional methods. Furthermore, less solvent means less interference from impurities and less charge competition.42 The above two points are key to improving the sensitivity of single-cell metabolite analysis.
Formation mechanism of the single-cell droplet
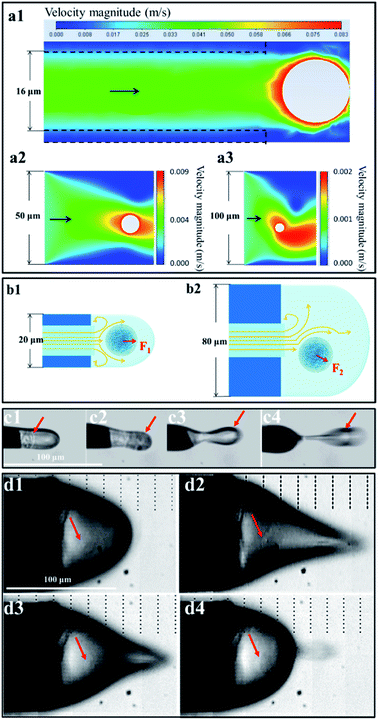
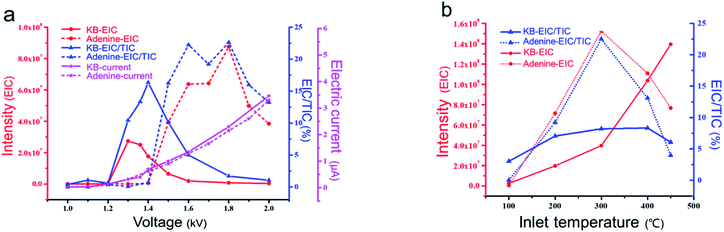
We speculated that the formation of the single-cell droplet could be attributed primarily to the flow field and electric field created by the narrow-bore, constant-I.D., thin-walled emitter. The following experimental investigation was carried out based on this speculation.To clearly characterize the experimental results, we defined the extracted ion chromatogram (EIC) as the total intensity of the extracted ion signals during a period (area integration of peaks in the extracted ion chromatogram) and the TIC as the intensity of the total ion signal during a period (integration of peaks in the TIC). The detection sensitivities of the samples in the same period were evaluated based on the EIC/TIC. The experimental results showed that the relationship between the voltage and the MS signal differed between the KB cell suspension and adenine aqueous solution, although the two samples had nearly overlapping current curves. The highest mass spectral signal intensity and sample detection sensitivity were obtained at 1.3–1.4 kV for the KB cell suspension; however, for the adenine aqueous solution, the mass spectrometry signal intensity began to increase when the spray voltage exceeded 1.4 kV, and the optimum signal was obtained at 1.6–1.8 kV. According to other studies on electrospray theory,43–47 the voltage required for an adenine aqueous solution spray system to form a steady jet should be 1.6–1.8 kV. The KB cell suspension had the same mobile phase as the adenine aqueous solution and produced almost the same ionization current; however, it produced a completely different mass spectrum response signal, demonstrating that ILCEI is different from ESI. This phenomenon is interesting as it reveals a disadvantage of the electrospray mode in the steady jet state: the conventional ESI technique was developed based on a homogeneous solution model and thus cannot handle the characteristics of heterogeneous systems such as cell suspensions. In contrast, the ILCEI technique was developed specifically for heterogeneous systems.
Preliminary study on the ionization mechanism of ILCEI-MS
Based on observation with a high-speed camera, the volume of the initial droplets produced by ILCEI remained almost constant before entering the mass spectrometer (Fig. 2; also see Video S6† for details). This is completely different from the continuous splitting of the initial droplets into secondary droplets in ESI. To further show the difference between ILCEI and ESI, we compared the cell suspension and homogeneous solution during mass spectrometry injection (Fig. S11 and Video S8†). The A549 cell suspension exhibited less droplet splitting (Fig. S11c2†) because of the presence of whole cells in the initial droplets of the cell suspension under electrolaunching, which hindered droplet shrinkage. A large surface area was maintained, and the Rayleigh limit was not reached,48 preventing the droplets from splitting. Because droplet splitting is accompanied by a random distribution of net charge, decreased droplet splitting results in more charge remaining on the surface of the mother droplet. In addition, a very small volume of the mobile phase means that very few impurity molecules are present in the mother liquid droplets. Thus, the cell components retained in the mother droplets will have a greater chance of gaining charge, thereby enhancing the ionization of the single-cell sample. The cells intercepted at the entrance of the mass spectrometer retained their structures and remained alive (see Fig. S9 and S10†). Thus, all cell components entered the mass spectrometer, avoiding sample loss in the atmospheric path and improving the sample collection rate. Hence, we believe that the ionization of single-cell components should occur in the ion-transfer tube of the mass spectrometer. Referring to similar studies on “inlet ionization”,27,49 we propose the following hypotheses: (1) after entering the MS inlet, the single-cell droplet experiences rapid boiling and explosion in a low-pressure, high-temperature environment; and (2) the sample molecules rapidly move to the gas phase and acquire random charges during the explosion. According to these assumptions, the proportion of sample molecules converted to gas-phase ions will increase as the intensity of the explosion increases.The ionization processes that occurred after a single-cell droplet entered the ion-transfer tube could not be characterized visually using a high-speed camera. Therefore, we conducted a preliminary experimental verification. We increased the temperature of the ion-transfer tube from 100 °C to 450 °C in stages (the normal adjustable parameter range of a mass spectrometer) and recorded the changes in the MS signal intensity (EIC) and the MS sensitivity (EIC/TIC) for a KB cell suspension and a 14 μmol L−1 adenine standard solution (Fig. 4b). For both samples (i.e., the KB cell suspension and the adenine standard solution), the EIC/TIC value gradually increased with increasing temperature below 300 °C; when the temperature exceeded 300 °C, the EIC/TIC value of the KB cell suspension continued to increase, whereas that of the adenine standard solution gradually decreased. Below 300 °C, the signal intensity of both samples increased with increasing temperature; this is because a reasonable increase in the temperature of the ion-transfer tube promoted the desolation of the sample and thus increased the intensity of the signal arising from the sample, regardless of whether ionization occurred in or out of the MS inlet. For a homogeneous solution, it is easy to reach the temperature required for complete evaporation. When the temperature continued to increase, the signal intensity decreased under the combined effects of background ion interference and the space-charge effect. However, single-cell droplets with larger volumes had more complex constituents than the initial droplets produced by homogeneous solution electrospray; thus, higher temperatures were required to complete the desolvation process quickly. These results corroborate our hypothesis that the cells are rapidly gasified in the ion-transfer tube to achieve ionization. As shown in Fig. 4b, the single-cell MS signal intensity obtained with our system can be further improved by increasing the temperature of the mass spectrometer ion-transfer tube to ≥400 °C. The main feature of inlet ionization is that it avoids the diffusion and annihilation of sample ions in the atmosphere, which greatly improves the detection sensitivity and sample utilization.
Performance evaluation of ILCEI-MS for single-cell detection
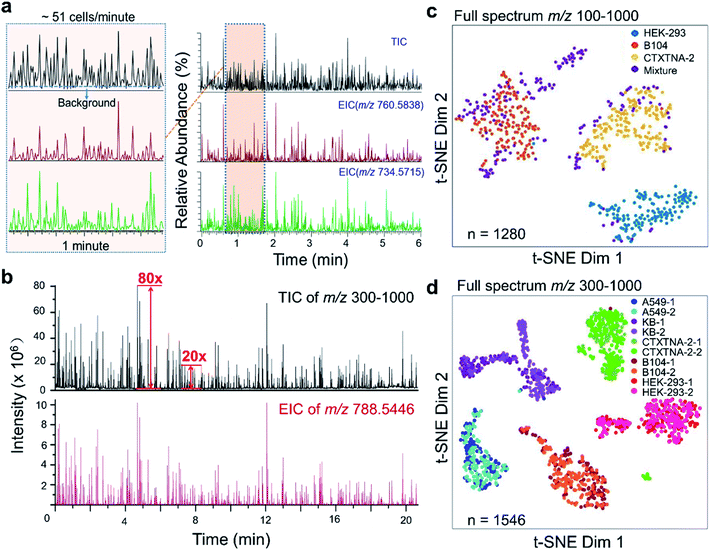
The detection throughput and sensitivity of the ILCEI-MS system were tested using B104 cells as an example. A throughput of approximately 51 cells per min was obtained for single-cell MS (Fig. 5a), and the average S/N ratio of single-cell signal peaks in TIC (baseline intensity = 1 × 106) was approximately 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and the highest S/N ratio reached 80
1, and the highest S/N ratio reached 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in the scan range of m/z 300–1000 (Fig. 5b). We analyzed over 482 A549 cells and obtained approximately 5200 peaks in the m/z range of 100–1000. Among them, 368 cellular metabolites were assigned (Table S2†). On average, 800 ions and 745 ions (Fig. S12†) related to cellular metabolites in the positive and negative ion modes were detected from living single cells, respectively. The expanded coverage of single-cell metabolites demonstrates that ILCEI-MS has great potential for in-depth studies of cellular heterogeneity at the metabolic level. We also evaluated the cell discrimination capability and repeatability of ILCEI-MS. First, MS analysis was performed on HEK-293, B104, and CTXTNA-2 cells and a mixture of these three cell types. Fig. 5c shows that these three cell types were clearly discriminated, and the cell mixture was well resolved into these three cell types. Next, A549, KB, CTXTNA-2, B104, and HEK-293 cells were repeatedly detected by the ILCEI-MS system at different times of one day to evaluate the repeatability (Fig. 5d); the single-cell mass spectra of these five types of cell are shown in Fig. S13.† The cells of the same type were clustered together in the t-distributed stochastic neighbor embedding (t-SNE) map, demonstrating the good repeatability and weak batch effect of the ILCEI-MS system.
1 in the scan range of m/z 300–1000 (Fig. 5b). We analyzed over 482 A549 cells and obtained approximately 5200 peaks in the m/z range of 100–1000. Among them, 368 cellular metabolites were assigned (Table S2†). On average, 800 ions and 745 ions (Fig. S12†) related to cellular metabolites in the positive and negative ion modes were detected from living single cells, respectively. The expanded coverage of single-cell metabolites demonstrates that ILCEI-MS has great potential for in-depth studies of cellular heterogeneity at the metabolic level. We also evaluated the cell discrimination capability and repeatability of ILCEI-MS. First, MS analysis was performed on HEK-293, B104, and CTXTNA-2 cells and a mixture of these three cell types. Fig. 5c shows that these three cell types were clearly discriminated, and the cell mixture was well resolved into these three cell types. Next, A549, KB, CTXTNA-2, B104, and HEK-293 cells were repeatedly detected by the ILCEI-MS system at different times of one day to evaluate the repeatability (Fig. 5d); the single-cell mass spectra of these five types of cell are shown in Fig. S13.† The cells of the same type were clustered together in the t-distributed stochastic neighbor embedding (t-SNE) map, demonstrating the good repeatability and weak batch effect of the ILCEI-MS system.
Primary single-cell analysis
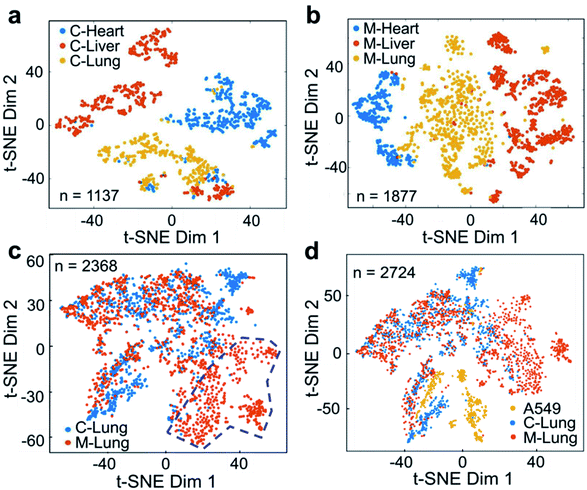
The cell suspension obtained from the digestion of fresh tissue is usually a mixture of different types of cell, such as epithelial cells, glial cells, and muscle cells. These cells are quite different in size, structure, physicochemical characteristics, etc., presenting challenges in terms of the universal applicability of analytical methods. Therefore, we further studied the metabolic profiles of primary cells digested from fresh tissue. A549 cells were used to establish a tumor-bearing mouse model of NSCLC. As shown in Fig. S14,† the lung mass in the lung cancer mouse was obvious, and the liver was enlarged and hard, indicating successful cancer cell transplantation. ILCEI-MS was used successfully to detect 4072 single cells from the single-cell suspension (Fig. S15 and S16†) digested from tissues of hearts, livers and lungs. The original MS data are shown in Fig. S17–S22.† On average, 773 ions and 787 ions (Fig. S23†) related to cellular metabolites were detected in the positive ion mode from 2368 individual single cells from lung tissues of the lung cancer model mice and control mice, respectively. To visualize the heterogeneity of cells in multiple organs, we performed a t-SNE analysis based on the single-cell metabolite profiles acquired by ILCEI-MS. Fig. 6a and b show that the differences in the heart, liver, and lung cells of the lung cancer model mice were more apparent than those in the control mice. Remarkably, the lung tissue cells from the lung cancer model mice exhibited greater metabolic differences (Fig. 6b), indicating more cell subpopulations, consistent with the literature.50 Further comparison of the single cells from the lung tissues of the lung cancer model mice and control mice showed partial overlap and partial difference in the cell community. Interestingly, a specific subset of cells (marked by the blue dashed circle in Fig. 6c) appeared in the lung tissue cells of the lung cancer model mice. The metabolomic information contained in this cell subset may provide valuable information for further studies on biomarkers for NSCLC. The primary lung cells in the control and lung cancer model mice were clearly distinguished from cultured A549 cells in vitro, which were used to establish a lung cancer model (Fig. 6d). These results suggest that the human lung cancer cell line A549 underwent metabolic reprogramming after entering the mice to adapt to the microenvironment in vivo and meet the needs of their own growth.51 These results effectively demonstrate the reliability and universal applicability of the ILCEI-MS method.Conclusion
Single-cell sequencing has contributed to great progress in the fields of single-cell genomics52 and transcriptomics.53 However, metabolites cannot be amplified like DNA/RNA. Therefore, the development of single-cell metabolomics relies on high-sensitivity detection methods. At present, single-cell metabolite MS methods suffer from several problems. First, the detection sensitivity and detection coverage of existing methods are insufficient, restricting the investigation of cell heterogeneity. Second, many single-cell MS detection methods involve complicated systems, require difficult operation procedures, and have high manufacturing costs and poor stability. These problems arise because sample dilution, waste, and matrix interference in the electrospray process have not been well resolved. In addition, these problems become more prominent when heterogeneous cell samples (e.g., living single cells) are ionized.In this study, we developed a new ionization method that continuously introduces live single cells into a mass spectrometer. With a cell detection throughput of 51 cells per min, ILCEI-MS achieved high sensitivity for single-cell detection, and the average S/N ratio of single-cell signal peaks is about 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in the TIC. The average information coverage of a single cell is greater than 700 metabolite-related ions. These promising results can be attributed to the following two characteristics: first, the narrow-bore capillary exerts a significant spatial confinement effect on the cell, resulting in reliable single-cell high-throughput separation along with orderly long-distance cell transport while avoiding the dilution of cell samples by sheath solution and sheath gas. Second, a novel intact living-cell electrolaunching ionization process was developed based on a constant-I.D., thin-walled emitter at the end of a narrow capillary. This approach can realize single-cell MS injection with almost no mobile phase dilution, little matrix interference, and inlet ionization, resulting in greatly improved single-cell sample utilization. A series of single-cell detection experiments with a high sampling volume were tested, demonstrating that the ILCEI-MS system has good single-cell recognition ability and stability. The universal applicability of ILCEI-MS for actual biological samples was demonstrated by detecting 4072 primary single cells digested from fresh mouse heart, liver, and lung tissues.
1 in the TIC. The average information coverage of a single cell is greater than 700 metabolite-related ions. These promising results can be attributed to the following two characteristics: first, the narrow-bore capillary exerts a significant spatial confinement effect on the cell, resulting in reliable single-cell high-throughput separation along with orderly long-distance cell transport while avoiding the dilution of cell samples by sheath solution and sheath gas. Second, a novel intact living-cell electrolaunching ionization process was developed based on a constant-I.D., thin-walled emitter at the end of a narrow capillary. This approach can realize single-cell MS injection with almost no mobile phase dilution, little matrix interference, and inlet ionization, resulting in greatly improved single-cell sample utilization. A series of single-cell detection experiments with a high sampling volume were tested, demonstrating that the ILCEI-MS system has good single-cell recognition ability and stability. The universal applicability of ILCEI-MS for actual biological samples was demonstrated by detecting 4072 primary single cells digested from fresh mouse heart, liver, and lung tissues.
Although the mechanism of the ILCEI-MS requires further study, it does not hinder its application in fields closely related to cell heterogeneity. For example, ILCEI-MS can be used in developmental biology to study the metabolic molecular mechanism of cell differentiation in the early embryo, related research has emerged in the field of genomics.54 We expect that this method can also be applied to high-throughput single-organelle metabolite analysis in, for example, the nucleus and mitochondria.
Data availability
The data supporting the findings of this study are available within the article and in the ESI.† The raw data are available from the authors upon reasonable request.Author contributions
X. Y. W. supervised the project. X. Y. W. and Y. L. S. designed the experiments. Y. L. S. designed and modified the instrument under the guidance of X. Y. W. and Yaoyao Z. Q. Z., Y. L. S., and Yingyan Z. performed the high-speed photography and the corresponding data analysis. Y. L. S., W. M. Z., Yaoyao Z., Y. X. L., and G. Z. Z. prepared the materials and performed the MS detection. Yaoyao Z., G. S. G., S. C. Z., Y. X. L., X. R. Z., W. M. Z., Y. L. S., H. Y., and H. S. Z. performed the data analysis. Y. L. S. designed and performed the simulations under the guidance of X. Y. W. The manuscript was written by Y. L. S., Yaoyao Z., Yingyan Z., W. M. Z., and X. Y. W. All authors discussed the results and commented on the manuscript. All authors approved the final manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was funded by the National Natural Science Foundation of China (No. 22127805, 21625501, and 21936001), the Beijing Outstanding Young Scientist Program (BJJWZYJH01201910005017), and the Analysis Center of Chemical Performance of Beijing University of Technology. We thank Xianfa Yang of Guangzhou Regenerative Medicine and Health Guangdong Laboratory for helpful discussions and Hanyu Yuan of the Beijing Computing Center for help with the violin plots.Notes and references
- F. J. Hartmann, D. Mrdjen, E. McCaffrey, D. R. Glass, N. F. Greenwald, A. Bharadwaj, Z. Khair, S. G. S. Verberk, A. Baranski, R. Baskar, W. Graf, D. Van Valen, J. Van den Bossche, M. Angelo and S. C. Bendall, Nat. Biotechnol., 2021, 39, 186–197 CrossRef CAS PubMed.
- M. Fessenden, Nature, 2016, 540, 153–155 CrossRef CAS PubMed.
- V. Pareek, H. Tian, N. Winograd, S. J. Benkovic and H. Tian, Science, 2020, 368, 283–290 CrossRef CAS PubMed.
- Q. S. Huang, S. F. Mao, M. Khan, W. W. Li, Q. Zhang and J. M. Lin, Chem. Sci., 2020, 11, 253–256 RSC.
- C. Seydel, Nat. Methods, 2021, 18, 1452–1456 CrossRef CAS PubMed.
- M. H. Zhuang, Z. H. Hou, P. Y. Chen, G. L. Liang and G. M. Huang, Chem. Sci., 2020, 11, 7308–7312 RSC.
- X. Xu, J. Wang, L. Wu, J. Guo, Y. Song, T. Tian, W. Wang, Z. Zhu and C. Yang, Small, 2020, 16, e1903905 CrossRef PubMed.
- M. Oginuma, Y. Harima, O. A. Tarazona, M. Diaz-Cuadros, A. Michaut, T. Ishitani, F. Xiong and O. Pourquie, Nature, 2020, 584, 98–101 CrossRef CAS PubMed.
- S. Trefely and K. E. Wellen, Science, 2018, 360, 603–604 CrossRef CAS PubMed.
- M. N. Artyomov and J. Van den Bossche, Cell Metab., 2020, 32, 710–725 CrossRef CAS PubMed.
- H. Zhu, N. Wang, L. Yao, Q. Chen, R. Zhang, J. Qian, Y. Hou, W. Guo, S. Fan, S. Liu, Q. Zhao, F. Du, X. Zuo, Y. Guo, Y. Xu, J. Li, T. Xue, K. Zhong, X. Song, G. Huang and W. Xiong, Cell, 2018, 173, 1716–1727 CrossRef CAS PubMed.
- L. Zhang and A. Vertes, Angew. Chem., Int. Ed., 2018, 57, 4466–4477 CrossRef CAS PubMed.
- M. Xue, W. Wei, Y. Su, J. Kim, Y. S. Shin, W. X. Mai, D. A. Nathanson and J. R. Heath, J. Am. Chem. Soc., 2015, 137, 4066–4069 CrossRef CAS PubMed.
- P. J. Hines, Science, 2016, 354, 843 CrossRef PubMed.
- R. Zenobi, Science, 2013, 342, 1243259 CrossRef CAS PubMed.
- Z. Y. Yuan, Q. M. Zhou, L. S. Cai, L. Pan, W. L. Sun, S. W. Qumu, S. Yu, J. X. Feng, H. S. Zhao, Y. C. Zheng, M. L. Shi, S. Li, Y. Chen, X. R. Zhang and M. Q. Zhang, Nat. Methods, 2021, 18, 1223–1232 CrossRef CAS PubMed.
- L. Rappez, M. Stadler, S. Triana, R. M. Gathungu, K. Ovchinnikova, P. Phapale, M. Heikenwalder and T. Alexandrov, Nat. Methods, 2021, 18, 799–805 CrossRef CAS PubMed.
- K. Scupakova, F. Dewez, A. K. Walch, R. M. A. Heeren and B. Balluff, Angew. Chem., Int. Ed., 2020, 59, 17447–17450 CrossRef CAS PubMed.
- E. K. Neumann, T. J. Comi, S. S. Rubakhin and J. V. Sweedler, Angew. Chem., Int. Ed., 2019, 58, 5910–5914 CrossRef CAS PubMed.
- Q. L. Liu, W. J. Ge, T. T. Wang, J. Y. Lan, S. Martinez-Jarquin, C. Wolfrum, M. Stoffel and R. Zenobi, Angew. Chem., Int. Ed., 2021, 60, 24534–24542 CrossRef CAS PubMed.
- W. F. Zhang, N. Li, L. Lin, Q. S. Huang, K. Uchiyama and J. M. Lin, Small, 2020, 16, 1903402 CrossRef CAS PubMed.
- H. Yao, H. S. Zhao, X. Zhao, X. Y. Pan, J. X. Feng, F. J. Xu, S. C. Zhang and X. R. Zhang, Anal. Chem., 2019, 91, 9777–9783 CrossRef CAS PubMed.
- J. F. Cahill, J. Riba and V. Kertesz, Anal. Chem., 2019, 91, 6118–6126 CrossRef CAS PubMed.
- S. Xu, M. Liu, Y. Bai and H. Liu, Angew. Chem., Int. Ed., 2021, 60, 1806–1812 CrossRef CAS PubMed.
- Z. Z. Shen, H. S. Zhao, H. Yao, X. Y. Pan, J. L. Yang, S. C. Zhang, G. J. Han and X. R. Zhang, Chem. Sci., 2022, 13, 1641–1647 RSC.
- L. Song, K. Chingin, M. Wang, D. Zhong, H. Chen and J. Xu, Anal. Chem., 2022, 94, 4175–4182 CrossRef CAS PubMed.
- V. S. Pagnotti, N. D. Chubatyi and C. N. McEwen, Anal. Chem., 2011, 83, 3981–3985 CrossRef CAS PubMed.
- S. N. Jayasinghe, P. A. M. Eagles and A. N. Qureshi, Biotechnol. J., 2006, 1, 86–94 CrossRef CAS PubMed.
- S. N. Jayasinghe, A. N. Qureshi and P. A. M. Eagles, Small, 2006, 2, 216–219 CrossRef CAS PubMed.
- P. A. M. Eagles, A. N. Qureshi and S. N. Jayasinghe, J. Biol. Chem., 2006, 394, 375–378 CAS.
- P. K. Odenwälder, S. Irvine, J. R. McEwan and S. N. Jayasinghe, Biotechnol. J., 2007, 2, 622–630 CrossRef PubMed.
- S. Sahoo, W. C. Lee, J. C. H. Goh and S. L. Toh, Biotechnol. Bioeng., 2010, 106, 690–698 CrossRef CAS PubMed.
- D. I. Braghirolli, F. Zamboni, P. C. Chagastelles, D. J. Moura, J. Saffi, J. A. P. Henriques, D. A. Pilger and P. Pranke, Biomicrofluidics, 2013, 7, 044130 CrossRef CAS PubMed.
- H. J. Chun, C. H. Park, I. K. Kwon, G. Khang, N. Maurmann, L. E. Sperling and P. Pranke, in Cutting-edge enabling technologies for regenerative medicine, Springer Press, Singapore, 2018, vol. 1078, pp. 79–100 Search PubMed.
- Y. L. Shao, Y. Y. Zhou, Y. Y. Wu, Q. Zhang, Z. H. Yu, G. S. Guo and X. Y. Wang, Talanta, 2020, 218, 121096 CrossRef CAS PubMed.
- X. Y. Wang, V. Veerappan, C. Cheng, X. Jiang, R. D. Allen, P. K. Dasgupta and S. R. Liu, J. Am. Chem. Soc., 2010, 132, 40–41 CrossRef CAS PubMed.
- X. Y. Wang, L. Liu, Q. S. Pu, Z. F. Zhu, G. S. Guo, H. Zhong and S. R. Liu, J. Am. Chem. Soc., 2012, 134, 7400–7405 CrossRef CAS PubMed.
- L. Liu, V. Veerappan, Q. R. Pu, C. Cheng, X. Y. Wang, L. P. Lu, R. D. Allen and G. S. Guo, Anal. Chem., 2014, 86, 729–736 CrossRef CAS PubMed.
- L. Liu, V. Veerappan, Y. Z. Bian, G. S. Guo and X. Y. Wang, Sci. China: Chem., 2015, 58, 1605–1611 CrossRef CAS.
- R. N. Li, Y. L. Shao, Y. M. Yu, X. Y. Wang and G. S. Guo, Chem. Commun., 2017, 53, 4104–4107 RSC.
- W. M. Zhang, L. Liu, Q. Zhang, D. T. Zhang, Q. Hu, Y. N. Wang, X. Y. Wang, Q. S. Pu and G. S. Guo, Chem. Commun., 2020, 56, 2423–2426 RSC.
- A. C. Susa, Z. J. Xia and E. R. Williams, Anal. Chem., 2017, 89, 3116–3122 CrossRef CAS PubMed.
- G. I. Taylor, Proc. R. Soc. London, Ser. A, 1966, 291, 145–158 Search PubMed.
- M. Cloupeau and B. Prunet-Foch, J. Aerosol Sci., 1994, 25, 10211036 Search PubMed.
- I. Hayati, A. I. Bailey and T. F. Tadros, Nature, 1986, 319, 41–43 CrossRef.
- A. Jaworek and A. Krupa, J. Aerosol Sci., 1999, 30, 873–893 CrossRef CAS.
- P. Nemes, I. Marginean and A. Vertes, Anal. Chem., 2007, 79, 3105–3116 CrossRef CAS PubMed.
- L. Rayleigh, Philos. Mag., 1882, 14, 184–186 Search PubMed.
- C. N. McEwen, V. S. Pagnotti, E. D. Inutan and S. Trimpin, Anal. Chem., 2010, 82, 9164–9168 CrossRef CAS PubMed.
- A. Prat, O. Karginova, J. S. Parker, C. Fan, X. P. He, L. Bixby, J. C. Harrell, E. Roman, B. Adamo, M. Troester and C. M. Perou, Breast Cancer Res. Treat., 2013, 142, 237–255 CrossRef CAS PubMed.
- D. F. Quail and J. A. Joyce, Nat. Med., 2013, 19, 1423–1437 CrossRef CAS PubMed.
- C. A. Lareau, L. S. Ludwig, C. Muus, S. H. Gohil, T. Zhao, Z. Chiang, K. Pelka, J. M. Verboon, W. Luo, E. Christian, D. Rosebrock, G. Getz, G. M. Boland, F. Chen, J. D. Buenrostro, N. Hacohen, C. J. Wu, M. J. Aryee, A. Regev and V. G. Sankaran, Nat. Biotechnol., 2021, 39, 451–461 CrossRef CAS PubMed.
- A. H. Rizvi, P. G. Camara, E. K. Kandror, T. J. Roberts, I. Schieren, T. Maniatis and R. Rabadan, Nat. Biotechnol., 2017, 35, 551–560 CrossRef CAS PubMed.
- G. Peng, S. Suo, G. Cui, F. Yu, R. Wang, J. Chen, S. Chen, Z. Liu, G. Chen, Y. Qian, P. P. L. Tam, J. D. J. Han and N. Jing, Nature, 2019, 572, 528–532 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc02569h |
| This journal is © The Royal Society of Chemistry 2022 |