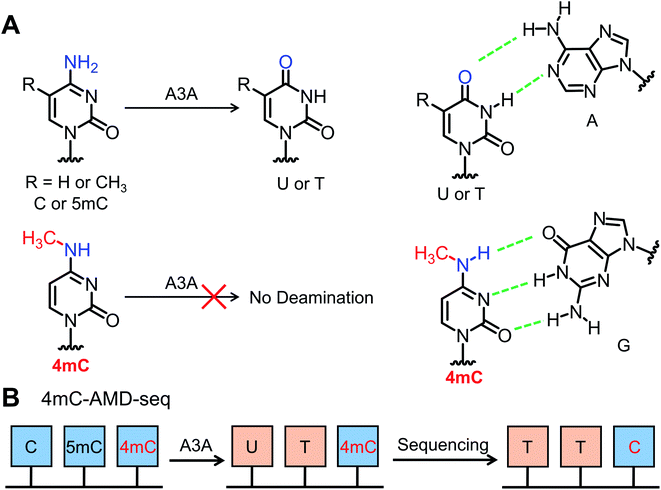
Open Access Article
Open Access ArticleGenome-wide mapping of N4-methylcytosine at single-base resolution by APOBEC3A-mediated deamination sequencing†
Jun
Xiong‡
abc,
Ping
Wang‡
de,
Wen-Xuan
Shao
ab,
Gaojie
Li
de,
Jiang-Hui
Ding
ab,
Neng-Bin
Xie
ab,
Min
Wang
ab,
Qing-Yun
Cheng
a,
Conghua
Xie
 ac,
Yu-Qi
Feng
ac,
Yu-Qi
Feng
 ab,
Weimin
Ci
*de and
Bi-Feng
Yuan
ab,
Weimin
Ci
*de and
Bi-Feng
Yuan
 *abcf
*abcf
aDepartment of Radiation and Medical Oncology, Zhongnan Hospital of Wuhan University, School of Public Health, Wuhan University, Wuhan 430071, China. E-mail: bfyuan@whu.edu.cn
bSauvage Center for Molecular Sciences, Department of Chemistry, Wuhan University, Wuhan 430072, China
cCancer Precision Diagnosis and Treatment and Translational Medicine Hubei Engineering Research Center, Zhongnan Hospital of Wuhan University, Wuhan, China
dKey Laboratory of Genomics and Precision Medicine, China National Center for Bioinformation, Beijing Institute of Genomics, Chinese Academy of Sciences, Beijing 100101, China. E-mail: ciwm@big.ac.cn
eUniversity of Chinese Academy of Sciences, Beijing 100049, China
fWuhan Research Center for Infectious Diseases and Cancer, Chinese Academy of Medical Sciences, China
First published on 11th August 2022
Abstract
N 4-methylcytosine (4mC) is a natural DNA modification occurring in thermophiles and plays important roles in restriction-modification (R-M) systems in bacterial genomes. However, the precise location and sequence context of 4mC in the whole genome are limited. In this study, we developed an APOBEC3A-mediated deamination sequencing (4mC-AMD-seq) method for genome-wide mapping of 4mC at single-base resolution. In the 4mC-AMD-seq method, cytosine and 5-methylcytosine (5mC) are deaminated by APOBEC3A (A3A) protein to generate uracil and thymine, both of which are read as thymine in sequencing, while 4mC is resistant to deamination and therefore read as cytosine. Thus, the readouts of cytosines from sequencing could manifest the original 4mC sites in genomes. With the 4mC-AMD-seq method, we achieved the genome-wide mapping of 4mC in Deinococcus radiodurans (D. radiodurans). In addition, we confirmed that 4mC, but not 5mC, was the major modification in the D. radiodurans genome. We identified 1586 4mC sites in the genome of D. radiodurans, among which 564 sites were located in the CCGCGG motif. The average methylation levels in the CCGCGG motif and non-CCGCGG sequence were 70.0% and 22.8%, respectively. We envision that the 4mC-AMD-seq method will facilitate the investigation of 4mC functions, including the 4mC-involved R-M systems, in uncharacterized but potentially useful strains.
Introduction
DNA molecules employ four canonical nucleobases of adenine (A), thymine (T), cytosine (C), and guanine (G) to encode genetic information in living organisms. In addition to the four-base hereditary information, a variety of naturally modified nucleobases have been identified in the genomes of living organisms.1–6N6-Methyladenine (6mA) and 5-methylcytosine (5mC) are the two most extensively characterized modifications in bacterial DNA.7–11 In addition to 6mA and 5mC, N4-methylcytosine (4mC) was found to be present in some bacterial genomes in the 1980s.12 Similar to 6mA and 5mC, 4mC also participates in the bacterial restriction-modification systems (R-M systems) that protect bacteria against bacteriophages and other invasive foreign DNA.13 The traditional view of the functions of these DNA modifications in R-M systems has been progressively broadened to additional roles, such as the epigenetic regulation of gene expression.13,14 It has been proposed that the methyl group of 6mA, 5mC, and 4mC protrudes from the double helix, which may alter the interaction between DNA binding proteins and their recognition sites.15–174mC is mostly found in the genomes of some thermophilic bacteria.18 It has been recently reported that 4mC also exists in the genome of D. radiodurans, a bacterium with an exceptional DNA repair system.19D. radiodurans is well known for its capability of resistance to γ irradiation, UV light, oxidizing agents and chemical mutagens.20,21 However, knowledge about 4mC-specific R-M systems, such as 4mC motifs and the corresponding methyltransferases and restriction endonucleases, is much less than that for 6mA and 5mC,22 which is largely due to a lack of efficient analytical methods for the detection of 4mC in genomes. Revealing the functions of 4mC in genomes relies on the sensitive detection, accurate quantification, and mapping of 4mC in genomes. Mass spectrometry has been established as an effective technique for qualitative and quantitative analysis of nuclei acid modifications.23–28 However, it is unable to achieve genome-wide mapping of DNA modifications.
Bisulfite sequencing enables researchers to identify 5mC sites in genomes at single-base resolution.29 However, traditional bisulfite sequencing cannot distinguish 4mC from 5mC because 5mC resists deamination and 4mC is partially resistant to deamination,30 which will lead to complicated sequencing results and is not suitable to accurately differentiate 4mC from 5mC. A 4mC-Tet-assisted-bisulfite-sequencing (4mC-TAB-seq) approach was recently established for mapping 4mC.31 In 4mC-TAB-seq, Tet protein was used to oxidize 5mC to 5-carboxylcytosine (5caC), which is read as thymine in bisulfite sequencing. Therefore, both cytosine and 5mC are read as thymine in 4mC-TAB-seq, whereas 4mC sites are partially read as cytosine. This approach was coupled with the high-throughput sequencing platform, making it possible to map 4mC in the Caldicellulosiruptor kristjanssonii genome. However, the harsh conditions for chemical deamination in bisulfite-based strategies could lead to as much as 99.9% degradation of input DNA.32,33
Single-molecule real-time (SMRT) sequencing is capable of detecting modified bases.34 However, compared with the commonly used high-throughput sequencing technologies, SMRT sequencing is more costly and frequently suffers from a high false-positive rate and systematically overestimates 4mC. Greer et al. reported that SMRT sequencing analysis identified 6.0% of the cytosines as 4mC in E. coli.35 However, the LC-MS/MS quantification of E. coli demonstrated that 4mC was below the limit of detection (<0.00005%).35 This large discrepancy between 4mC values suggests that SMRT sequencing incorrectly calls 4mC peaks.35 Hence, the development of a 4mC-specific mapping method compatible with high-throughput sequencing platforms will be particularly important for deciphering its functions.
It has been reported that the apolipoprotein B mRNA-editing catalytic polypeptide-like (APOBEC) family enzymes can convert cytosine to uracil in single-stranded DNA (ssDNA) and mediate critical functions in viral infection and carcinogenesis.36 Recently, it was reported that APOBEC3A (A3A) protein exhibited high deamination activity towards both cytosine and 5mC in DNA.37–40 The cytosine and 5mC could be readily deaminated regardless of sequence context.41,42 Considering that 4mC contains a methyl group at the N4 position and was partially resistant to bisulfite-mediated chemical deamination, we reasoned that 4mC might also be resistant to deamination by A3A protein (Fig. 1). Along this line, we developed an A3A-mediated deamination sequencing (4mC-AMD-seq) method for mapping 4mC in DNA at single-base resolution. In addition, the 4mC-AMD-seq method was successfully applied to map 4mC sites in D. radiodurans DNA.
Materials and methods
Chemicals and reagents
2′-Deoxycytidine (dC), 2′-deoxyguanosine (dG), 2′-deoxyadenosine (dA), thymidine (T), 2′-deoxynucleoside 5′-triphosphates (dATP, dCTP, dGTP, TTP) and phosphodiesterase I were purchased from Sigma-Aldrich (St. Louis, MO, USA). 5-Methyl-2′-deoxycytidine (5mdC) was purchased from Berry & Associates (Dexter, MI, USA). N4-Methyl-2′-deoxycytidine-5′-triphosphate (4mdCTP) and 5-methyl-2′-deoxycytidine-5′-triphosphate (5mdCTP) were purchased from TriLink BioTechnologies (San Diego, CA, USA). 2′-Deoxyurdine (dU) was purchased from Meryer Chemical Technology Co., Ltd (Shanghai, China). 3-Methyl-2′-deoxycytidine (3mdC) was synthesized and the details can be found in the ESI.† S1 nuclease, DNase I, and alkaline phosphatase were purchased from Takara Biotechnology Co. Ltd (Dalian, China).Bacteria culture and DNA isolation
D. radiodurans strain R1 was purchased from China General Microbiological Culture Collection Center (CGMCC 1.3828). D. radiodurans cells were grown in tryptone glucose yeast extract (TGY) liquid media or on agar plates (0.5% tryptone, 0.1% glucose, and 0.3% yeast extract) at 30 °C. As for the stable isotope labelling, D. radiodurans cells were cultured in TGY liquid medium supplemented with 50 or 500 μg mL−1 of D3-methionine to metabolically label the methyl group in genomic DNA with the CD3 group. The cells were collected after culturing for 12 h and then DNA was extracted. Bacterial DNA was isolated using an Ezup Column Bacteria Genomic DNA Purification kit (Sangon, Shanghai, China) according to the manufacturer's instructions.Preparation of DNA with 4mC or 5mC modification
Three kinds of 215-bp double-stranded DNA (dsDNA) substrates (DNA-C, DNA-4mC, and DNA-5mC) were prepared as the standards for the method development. DNA-C, DNA-4mC, and DNA-5mC share the same sequence context and the detailed sequence information can be found in ESI Table S1.† As for the synthesis of DNA-C, 0.5 ng of pUC19 DNA was used as the template for PCR amplification. PCR amplification was carried out in a 50 μL reaction solution including 2.5 U of Taq DNA polymerase (Takara Biomedical Technology), 5 μL of 10× reaction buffer, 1 μL of dATP, dGTP, TTP, and dCTP (2.5 mM for each), 2 μL of 10 μM forward primer (5′-GAGTGAGTGAGGGAGGAAG-3′), and 2 μL of 10 μM reverse primer (5′-CCACTCACAATTCCACACAACATAC-3′). DNA-4mC and DNA-5mC were prepared by PCR amplification using 4mdCTP and 5mdCTP instead of dCTP, respectively. In DNA-4mC and DNA-5mC DNA substrates, all the cytosines were replaced by 4mC or 5mC respectively (except for the cytosines in PCR primers). PCR conditions were 95 °C for 2 min, 30 cycles of 95 °C for 1 min, 52 °C for 2 min, 72 °C for 3 min, followed by 72 °C for 7 min. The PCR products were purified by agarose gel electrophoresis and recovered using a Gel Extraction kit (Omega Bio-Tek Inc., Norcross, GA, USA).Expression and purification of recombinant A3A protein
The plasmid for A3A protein expression (pET-41a-A3A) was purchased from TsingKe Co., Ltd. pET-41a-A3A plasmid contains the full-length coding sequence of A3A in the pET-41a vector, which carries the glutathione S-transferase (GST) tag and the human rhinovirus 3C protease (HRV 3C) site. A3A protein was expressed and purified according to the previously described procedure,41 and the details can be found in the ESI.†Deamination assay
Three kinds of 215-bp dsDNA (DNA-C, DNA-4mC, and DNA-5mC) were used as the substrates to evaluate the deaminase activity of A3A protein on cytosine, 4mC and 5mC. The dsDNA substrates were first denatured at 95 °C for 10 min and then cooled at 0 °C for 2 min. Then A3A protein was added into the denatured DNA. The deamination reaction was carried out at 37 °C for 2 h in a 20 μL solution of 20 mM MES (pH 6.5), 0.1% Triton X-100, 2 μL of DMSO, and 1 pmol of DNA substrate (DNA-C, DNA-4mC or DNA-5mC). Varied amounts of A3A protein were used for the deamination reaction in the steady-state kinetics study. The deamination reaction was terminated by incubating the solution at 95 °C for 10 min. The deamination rate was determined by LC-MS/MS analysis or Sanger sequencing.Enzymatic digestion of DNA
Enzymatic digestion of DNA was carried out according to a previously described method.43,44 The detailed procedure can be found in the ESI.†LC-MS/MS analysis
LC-MS/MS analysis of nucleosides was performed on an LC-ESI-MS/MS system consisting of a Shimadzu LC-30AD UPLC (Tokyo, Japan) and a Shimadzu LC/MS-8045 mass spectrometer. Data acquisition and processing were performed using LabSolution Software (Shimadzu). The LC separation was performed on a Shimadzu Shim-pack GIST C18 column (2.1 i.d. × 100 mm, 2 μm) with a flow rate of 0.3 mL min−1 at 40 °C. Water with 0.05% formic acid (solvent A) and methanol (solvent B) were employed as mobile phases. A gradient of 0–1.5 min 5% B, 1.5–3 min 5–40% B, 3–6.5 min 40% B, 6.5–7 min 40–5% B, and 7–12 min 5% B was used.Tandem MS/MS was performed under positive electrospray ionization (ESI) mode. The ESI parameters were set as follows: interface voltage at 3.0 kV, nebulization gas flow rate at 3.0 L min−1, drying gas flow rate at 10.0 L min−1, desolvation line (DL) temperature at 250 °C, and heat block temperature at 400 °C. The MS fragments of 4mdC and 5mdC were obtained under product ion scan mode. The parent ion of [M + H]+ (m/z 242.2) was fragmented under collision-induced dissociation (CID) with a collision energy of 10 V or 40 V. The mass transitions of dG (268.2 → 152.1), dA (252.2 → 136.1), dC (228.2 → 112.1), T (243.2 → 127.0), 4mdC/5mdC (242.2 → 126.1), 4mdC (242.2 → 95.1), and dU (251.2 → 135.1) were used for monitoring under multiple reaction monitoring (MRM) mode. The MRM parameters of all nucleosides were optimized and the detailed mass spectrometer parameters are listed in ESI Table S2.†
4mC in D. radiodurans DNA was also examined on an LTQ Orbitrap Elite mass spectrometer (Thermo Fisher Scientific, USA) equipped with a Dionex Ultimate 3000 UPLC system (Thermo Fisher Scientific, USA). The LC separation conditions were the same as those for the Shimadzu LC/MS-8045 mass spectrometer system. The MS analysis was performed under positive ion mode with full scan detection (MS1, m/z 200–300) at a resolution of 60![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000. The MS2 fragmentation was based on the precursor ion of [M + H]+ (m/z 242.11) under CID with a collision energy of 15 eV. The most abundant product ion (m/z 126.06) in MS2 spectra was selected for the MS3 analysis under CID mode with a collision energy of 28 eV. The data analysis was carried out using Xcalibur v3.0.63 (Thermo Fisher Scientific, USA).
000. The MS2 fragmentation was based on the precursor ion of [M + H]+ (m/z 242.11) under CID with a collision energy of 15 eV. The most abundant product ion (m/z 126.06) in MS2 spectra was selected for the MS3 analysis under CID mode with a collision energy of 28 eV. The data analysis was carried out using Xcalibur v3.0.63 (Thermo Fisher Scientific, USA).
Steady-state kinetics study
The deaminase activity of A3A protein toward cytosine, 4mC and 5mC on DNA was evaluated by a steady-state kinetics study. A3A protein concentration and time interval for each dsDNA substrate (DNA-C, DNA-4mC, and DNA-5mC) were optimized with the deamination rate within 20% at 37 °C (ESI Table S3†). The concentration of the dsDNA substrate (DNA-C, DNA-4mC, and DNA-5mC) was initially 100 nM and A3A concentrations were 2.4 nM for DNA-C and DNA-5mC, and 240 nM for DNA-4mC. Increasing dsDNA concentrations (0.1–1 μM) were used to construct the Michaelis–Menten equation curve. Quantitative measurement of nucleosides was conducted by LC-MS/MS analysis, and the standard curves of dC, 4mdC, and 5mdC were constructed to calculate the amounts of the nucleosides. Deamination rate was calculated by quantifying dC, 4mdC, and 5mdC of A3A-treated and untreated samples with the following formula: deamination rate = ([C0] – [C])/t. The [C0] value represents the initial amount of dC, 4mdC or 5mdC without A3A treatments, and the [C] value represents the amount of dC, 4mdC or 5mdC after A3A treatment, and the t value represents the incubation time. kcat and KM were calculated with nonlinear regression fitting to the Michaelis–Menten equation: deamination rate = (kcat)([S])/(KM + [S]). kcat and KM values represent the maximum rate of reaction and the Michaelis constant respectively, and [S] is the concentration of the DNA substrate.Single-base resolution analysis of 4mC in DNA by Sanger sequencing
A3A protein-treated and untreated dsDNA substrates (DNA-C, DNA-4mC, and DNA-5mC) were amplified by PCR using EpiMark® Hot Start Taq DNA polymerase (New England Biolabs). PCR amplification was carried out with initial denaturation at 95 °C for 3 min, and 35 cycles of 95 °C for 30 s and 65 °C for 75 s, followed by 5 min of elongation at 65 °C. The forward primer (5′-GAGTGAGTGAGGGAGGAAG-3′) and reverse primer (5′-CCACTCACAATTCCACACAACATAC-3′) were used for the PCR amplification. The resulting PCR products were subjected to Sanger sequencing (TsingKe).Quantitative analysis of the level of 4mC at individual sites in DNA by colony sequencing
To quantitatively measure the level of 4mC at individual sites in DNA, different ratios of DNA-C and DNA-4mC were mixed and subjected to colony sequencing. Briefly, the DNA-C and DNA-4mC were mixed with percentages of DNA-4mC/(DNA-4mC + DNA-C) being 0%, 33%, 66%, and 100%. These samples were then treated with A3A protein at 37 °C for 2 h followed by PCR amplification and colony sequencing. Forty clones for each sample were picked up and subjected to Sanger sequencing (TsingKe).Preparation of spike-in DNA
The PCR amplificated 215 bp dsDNA (DNA-4mC) was used as the 4mC spike-in DNA. The 304 bp unmodified C spike-in and 237 bp 5mC modified spike-in were produced by PCR amplification from the pUC19 plasmid with dCTP and 5mdCTP, respectively. Detailed sequences of the primers and the spike-ins can be found in ESI Table S1.†Sequencing library preparation for 4mC-AMD-seq
The 4mC-AMD-seq libraries for D. radiodurans were prepared as follows. D. radiodurans DNA (100 ng in 400 μL of water) was fragmented to an average size of 300–400 bp using an ultrasonic homogenizer (Scientz Biotechnology Co., Ltd, China). The sheared DNA was lyophilized to dryness and then reconstituted in water. The resulting DNA, spiked with 0.1% of C spike-in, 5mC spike-in, and 4mC spike-in, was end-repaired and adenylated at 30 °C for 20 min and immediately at 72 °C for 20 min using a Hieff NGS® Ultima Endprep Mix (Yeasen Biotechnology Co., Ltd, Shanghai). Adaptor-OH and adaptor-P (ESI Table S4†) were mixed at a final concentration of 20 μM each in 1× annealing buffer (Beyotime Biotechnology, Shanghai) and then incubated at 95 °C for 5 min followed by cooling gradually to 25 °C. Adaptor ligation was performed at 20 °C for 15 min using a Hieff NGS® Ultima DNA Ligation Module (Yeasen) followed by DNA purification using KAPA Pure beads (Roche). The adaptor ligation was confirmed by 20% native PAGE. The resulting DNA (16 μL) was added with 2 μL of DMSO and then denatured at 95 °C for 10 min and immediately cooled at 0 °C. The deamination reaction by A3A protein was carried out at 37 °C for 2 h at a final concentration of 20 mM MES (pH 6.5), 0.1% Triton X-100 and 6 μM A3A protein. The resulting A3A-deaminated DNA was amplified by PCR with 10 cycles using pre-P5 primer, pre-P7 primer (ESI Table S4†), and Q5U® Hot Start High-Fidelity DNA polymerase (New England Biolabs). After purification with KAPA Pure beads, the DNA products were amplified by PCR with 5 cycles using P5-universal primer, P7-index primer (ESI Table S4†), and Q5® Hot Start High-Fidelity 2× Master Mix (New England Biolabs). The final PCR products were purified with KAPA Pure beads and 1.5% agarose gel electrophoresis and examined using an Agilent 2100 before sequencing on the Illumina NovaSeq 6000 platform (Novogene Co., Ltd, China).Data processing
FastQC (v0.11.8) software was used to perform quality control on the raw data (fastq format) obtained by sequencing. Then, the raw reads were trimmed to remove low-quality bases and adaptor sequences with Trim Galore (v0.6.7) and Cutadapt (v3.5 with Python 3.9.7). Trimmed reads were mapped to the reference genome of D. radiodurans (NCBI accession: ASM832978v1) by Bisamrk (v0.23.1). Then, Bismark (v0.23.1) was used to remove PCR duplication and extract methylation sites. For each site, the number of ‘‘C’’ bases was counted as 4mC sites (denoted NC) and the number of ‘‘T’’ bases was counted as not 4mC sites (denoted NT). The sequencing depth and coverage of samples were calculated using samtools (v1.9) software. For screening of 4mC sites, parameters of the sequencing depth ≥ 100 and methylation level (defined as methylation level = NC/(NC + NT)) ≥ 0.1 were used based on statistical results generated from the Bismark methylation extractor. The 6 bp contexts of upstream and downstream of 4mC sites were extracted and used for the motif searching and identification with STREME (v5.4.1).Results
Differential deamination of cytosine, 4mC and 5mC by A3A protein
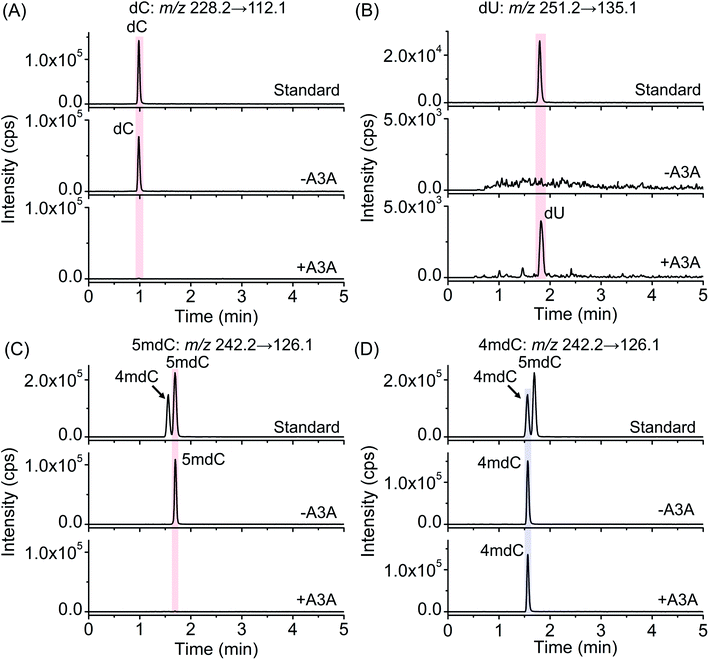
The previous report demonstrated that A3A protein could efficiently remove the NH2 group at the N4 position of both cytosine and 5mC in DNA to form uracil and thymine, respectively (Fig. 1A).41 Different from cytosine and 5mC, 4mC carries a methyl group that replaces one hydrogen atom of the NH2 group at the N4 position of cytosine. We speculated that the methyl group might compromise the deamination of 4mC by A3A protein (Fig. 1A). In this respect, the differential deamination between cytosine/5mC and 4mC by A3A protein may offer the opportunity to develop a single-base resolution method to map 4mC in DNA. In this proposed strategy, A3A protein converts cytosine and 5mC to form uracil and thymine, respectively, both of which will pair with adenine (Fig. 1A). If 4mC is resistant to the deamination by A3A protein, 4mC still pairs with guanine (Fig. 1A). Therefore, the differential coding properties of cytosine/5mC and 4mC after conversion by A3A protein can be recorded by sequencing, which can achieve the single-base resolution mapping of 4mC in DNA (Fig. 1B).We first expressed and purified the recombinant A3A protein (ESI Fig. S1†). Three synthesized 215-bp dsDNA (DNA-C, DNA-4mC, and DNA-5mC) were employed to evaluate the deamination of cytosine, 4mC and 5mC by A3A protein (ESI Table S1 and Fig. S2†). The dsDNA substrates were first denatured to single-stranded DNA (ssDNA) and then treated with A3A protein. The resulting products were enzymatically digested and the released nucleosides were analyzed by LC-MS/MS (ESI Table S2†). The results showed that the peak of dC from DNA-C was almost undetectable and dU was observed from the deamination of cytosine (Fig. 2A and B). In addition, the peak of 5mdC from DNA-5mC was also undetectable after A3A treatment (Fig. 2C). However, the peak intensity of 4mdC from DNA-4mC remained the same after A3A treatment (Fig. 2D). These results demonstrated that, as per our speculation, A3A protein could efficiently deaminate cytosine/5mC, but not 4mC in DNA.
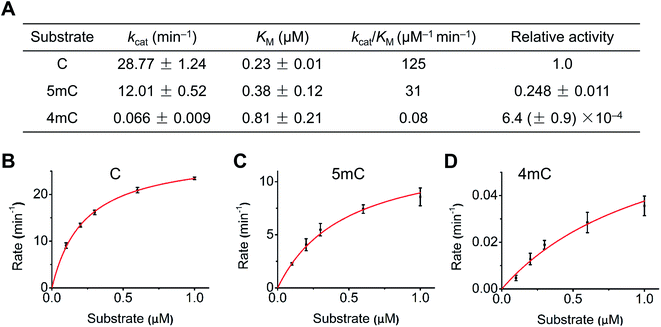
We next quantitatively evaluated the deamination efficiency of A3A protein toward cytosine, 4mC and 5mC by carrying out the steady-state kinetics study (detailed reaction conditions can be found in ESI Table S3†). The results showed that A3A exhibited high deamination activity toward cytosine (kcat/KM = 125 μM−1 min−1) and 5mC (kcat/KM = 31 μM−1 min−1) (Fig. 3A). However, A3A showed a dramatic diminution of deamination activity toward 4mC (kcat/KM = 0.08 μM−1 min−1, Fig. 3A). The steady-state kinetics analysis revealed a 1562-fold and 387-fold decrease of 4mC deamination rate compared to that of cytosine and 5mC, respectively (Fig. 3A–D). Collectively, the quantitative evaluation by steady-state kinetics analysis demonstrated that A3A exhibited differential deamination activity toward cytosine, 4mC and 5mC.
Single-base resolution analysis of 4mC in DNA by Sanger sequencing
The differential deamination of cytosine, 4mC and 5mC by A3A protein offers the opportunity to map 4mC sites in DNA at single-base resolution with the A3A-mediated deamination sequencing (4mC-AMD-seq). In this respect, we first employed Sanger sequencing to examine the nucleobase conversion in DNA-C, DNA-4mC and DNA-5mC after A3A treatment. The results showed that all the cytosines in DNA-C and all the 5mC in DNA-5mC were read as thymine upon A3A treatment (Fig. 4A and B). In contrast, all the 4mC sites in DNA-4mC were still read as cytosine after A3A treatment (Fig. 4C). These results displayed that cytosine/5mC were read as thymine, while 4mC was read as cytosine with the 4mC-AMD-seq method, indicating that the 4mC-AMD-seq was capable of the single-base resolution analysis of 4mC in DNA.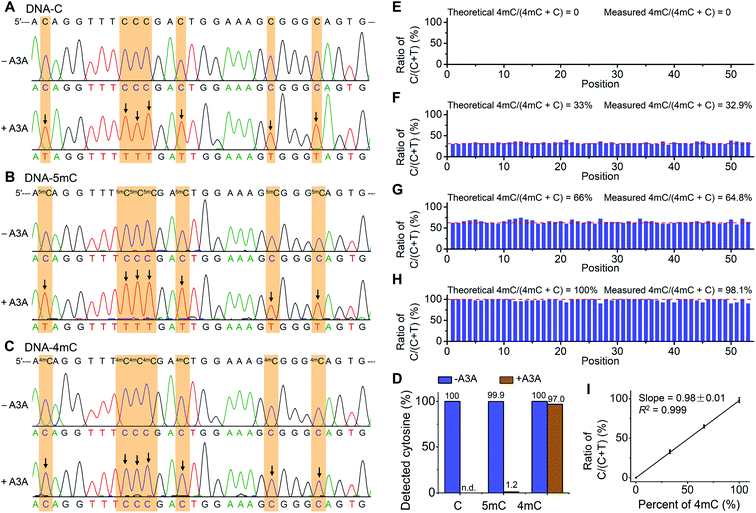 | ||
| Fig. 4 Single-base resolution analysis of 4mC in DNA by Sanger sequencing. (A–C) 215-bp dsDNA substrates (DNA-C, DNA-5mC, and DNA-4mC) were employed for the development of the 4mC-AMD-seq method by Sanger sequencing. Both C and 5mC were deaminated upon A3A treatment and all the cytosines and 5mC sites were read as thymine. 4mC was resistant to deamination by A3A protein and was still read as cytosine. Arrows denote deamination events (C-to-T conversion). (D) Evaluation of the C-to-T conversion upon A3A treatment by colony sequencing. (E–H) Quantitative evaluation of the level of 4mC at individual sites by 4mC-AMD-seq with colony sequencing. Various ratios of DNA-C and DNA-4mC were mixed and used as the DNA template for 4mC-AMD-seq. The X-axis indicates the position of cytosines (53 cytosines per strand). Detailed sequences are shown in ESI Table S1.† (I) Linear regression analysis by plotting the measured ratios of C/(C + T) with colony sequencing to the theoretical percentages of 4mC/(4mC + C). | ||
We next quantitatively measured the C-to-T conversion rates at each cytosine, 4mC and 5mC site. The deaminated DNA-C, DNA-4mC, and DNA-5mC were amplified by PCR and then subjected to colony sequencing. The results showed that all the 2120 cytosine sites in DNA-C from 40 clones were read as thymine after A3A treatment (Fig. 4D, and ESI Fig. S3 and S4†), indicating that cytosines were completely deaminated by A3A protein. As for DNA-5mC, 2094 5mC sites from 40 clones (totally 2120 cytosines) were read as thymine after A3A treatment with the overall 5mC-to-T conversion rate being 98.8% and only 1.2% residual cytosines being detected (Fig. 4D and ESI Fig. S5 and S6†). In contrast, 2056 cytosine sites were detected from 40 clones in DNA-4mC after A3A treatment with the overall 4mC-to-T conversion rate being only 3.0%, and 97.0% 4mC sites are kept (Fig. 4D and ESI Fig. S7 and S8†). Collectively, the colony sequencing results demonstrated the distinctly different coding properties between cytosine/5mC and 4mC in DNA upon A3A treatment, suggesting that the 4mC-AMD-seq method is capable of mapping 4mC in DNA at single-base resolution.
Quantitative analysis of the level of 4mC at individual sites in DNA
We next assessed the possibility of the quantitative measurement of the 4mC level at individual sites in DNA by the 4mC-AMD-seq method. To this end, we prepared a variety of mixtures of DNA-C and DNA-4mC with the molar percentages of DNA-4mC/(DNA-C + DNA-4mC) being 0%, 33%, 66% and 100%. The mixture was subjected to A3A treatment, PCR amplification, and colony sequencing. The results showed that all the cytosines from 40 clones were converted to thymine from the sample of 0% DNA-4mC (Fig. 4E). In the samples of 33%, 66%, and 100% of DNA-4mC, we observed the mean measured ratios of C/(C + T) from original cytosine/4mC sites to be 32.9% (28–40%), 64.8% (58–74%), and 98.1% (90–100%), respectively (Fig. 4F–4H). The linear regression analysis showed that the measured levels of 4mC were proportional to the theoretical percentages of 4mC (slope = 0.98 ± 0.01, R2 = 0.999) (Fig. 4I). Taken together, the results demonstrated that the quantitative analysis of the 4mC level at individual sites in DNA could be achieved by the 4mC-AMD-seq method.Determination of 4mC in D. radiodurans DNA
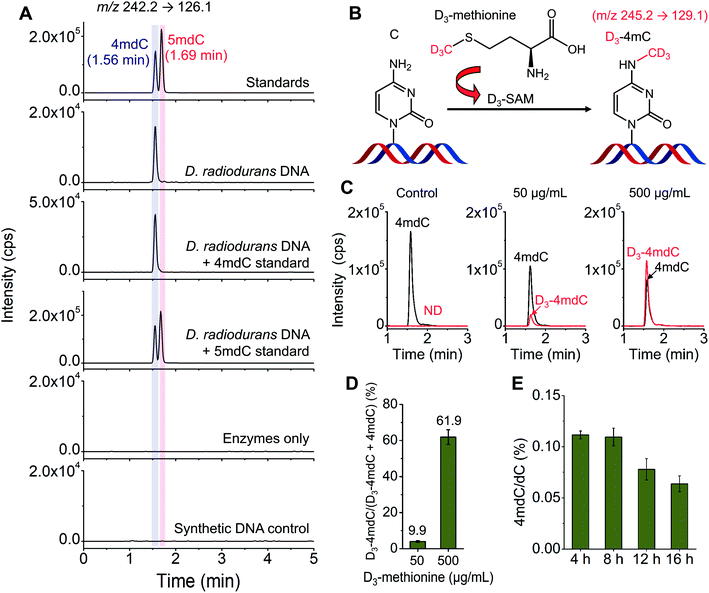
D. radiodurans is a kind of bacterium with extreme resistance to the lethal damage of DNA.20 DNA modifications may play roles in the resistance capability of D. radiodurans to damaging reagents. However, there were discrepant reports on the existence of methylated cytosine in D. radiodurans DNA.45,46D. radiodurans DNA was previously reported to contain 5mC detected by immunoblotting.45 In contrast, a recent study by the enzyme-linked apta-sorbent assay revealed the absence of 5mC in D. radiodurans DNA.46Herein, we employed LC-MS/MS to determine the methylated cytosines in D. radiodurans DNA. Under the optimized LC conditions, 4mC and 5mC can be well separated with the retention times of 4mC and 5mC being 1.56 min and 1.69 min, respectively (Fig. 5A). The LC-MS/MS analysis showed that 4mC was clearly detected; however, 5mC was not detectable in D. radiodurans DNA (Fig. 5A). We also added the 4mC or 5mC standard to the digested D. radiodurans DNA. The results further confirmed that the detected peak from D. radiodurans DNA was 4mC, but not 5mC (Fig. 5A). Moreover, no 4mC signal was detected from the enzymes-only control or the synthetic DNA control (Fig. 5A), excluding the potential source of 4mC from enzymes used for DNA digestion.
The stable isotope tracing monitored by mass spectrometry further confirmed the existence of 4mC in D. radiodurans DNA (Fig. 5B). We treated D. radiodurans cells with stable isotope-labelled methionine (D3-methionine). Methionine could be converted to S-adenosyl-L-methionine (SAM), which was a universal methyl donor for DNA methylation.47 The result showed that with the increased concentration of D3-methionine from 50 to 500 μg mL−1, the detected percentage of D3-4mdC was increased from 9.9% to 61.9% (Fig. 5C and D and ESI Fig. S9†), suggesting the presence of 4mC in D. radiodurans DNA, and the methyl group of 4mC originates from SAM.
The tandem MS/MS analysis showed that both 4mC and 5mC produced the fragment ion m/z 126 under a collision energy of 10 V (ESI Fig. S10†). With a higher collision energy of 40 V, 4mC produces a characteristic ion m/z 95 in comparison with 5mC (ESI Fig. S10†). The product-ion spectra of 4mC and 5mC standards indicated that the ion of m/z 95 should arise from the loss of methylamine (31 Da) (ESI Fig. S10†), which is consistent with the presence of a methyl group on a nitrogen atom. Therefore, the ion of m/z 95 is a characteristic fragment of 4mC compared with 5mC. D. radiodurans DNA showed similar fragment ions (including the characteristic ion of m/z 95) to the 4mC standard (ESI Fig. S10†), indicating the presence of 4mC in D. radiodurans DNA. In addition, high-resolution mass spectrometry further verified that the characteristic ion of m/z 95.0238 was produced by the 4mC standard and D. radiodurans DNA (ESI Fig. S11 and S12†). Collectively, these results suggested that 4mC, but not 5mC, existed in D. radiodurans DNA. Our results also showed that other DNA modifications, such as 3-methylcytosine (3mC), 5hmC, 5fC, and 5caC, were absent in D. radiodurans (ESI Fig. S13 and S14†).
With the confirmed presence of 4mC in D. radiodurans DNA, we further quantified the content of 4mC in D. radiodurans DNA. The LC-MS/MS analysis showed that the global content of 4mC in D. radiodurans DNA with 4, 8, 12 and 16 h of culturing was 0.111%, 0.110%, 0.078% and 0.064% (4mdC/dC), respectively (Fig. 5E and ESI Fig. S15†). The measured levels of 4mC are comparable to a previous report that there is approximately 0.13% of 4mC (4mC/C) in the genome of D. radiodurans.19 It should be noted that the overall content of 4mC in D. radiodurans DNA does not indicate the methylation level at individual loci.
Genome-wide mapping of 4mC in D. radiodurans DNA by 4mC-AMD-seq
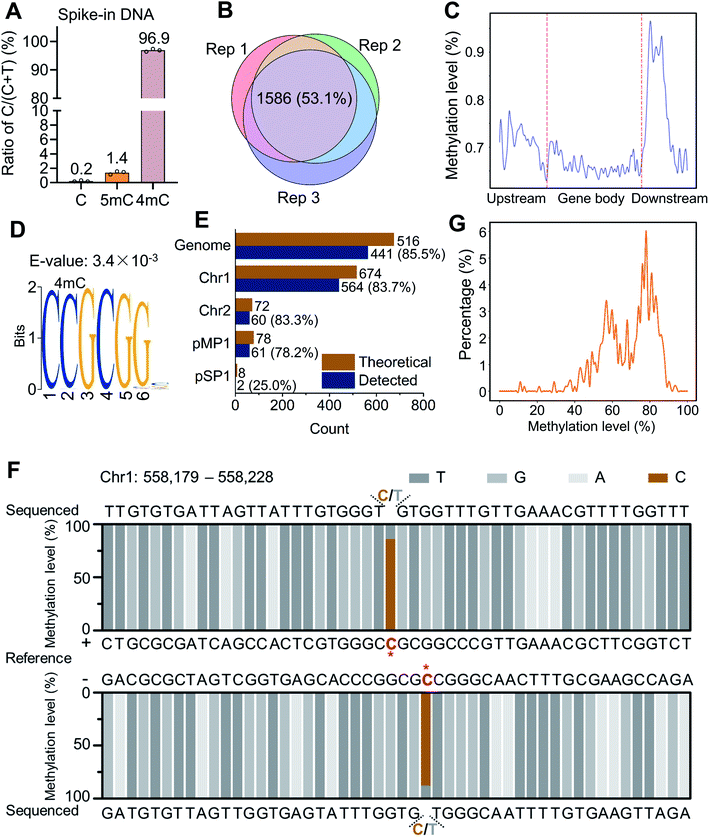
We next employed the 4mC-AMD-seq method to map 4mC sites in D. radiodurans DNA (ESI Fig. S16 and Table S5†). We validated the deamination rates of A3A towards C, 5mC, and 4mC with spike-in controls by colony sequencing (ESI Table S6†). In accordance with the method development results, only 0.2% of cytosine sites on unmodified C spike-in DNA were read as C after A3A deamination (Fig. 6A), which is comparable with the previously reported deamination rate of A3A to cytosines in DNA (0.2–0.3% remaining residual cytosines).39,48 0.2% of non-deaminated cytosines indicated a low false positive rate for 4mC-AMD-seq. We also observed an average non-deamination rate of 5mC (1.4%) and 4mC (96.9%) on 5mC and 4mC spike-in DNA (Fig. 6A). Although 0.2% of cytosines in spike-in DNA are not deaminated, the non-deaminated cytosines were not at the same sites of DNA and they were randomly distributed in DNA, which would not affect the identification of 4mC in high-throughput sequencing with high sequencing depth (average 616× sequencing depth, ESI Table S5†).We first analyzed the raw reads produced by 4mC-AMD-seq. Three biological replicates were measured and 63.1–79.4% of the raw reads from each replicate could be uniquely mapped to the D. radiodurans R1 reference genome, yielding an average read coverage of 99.3% and average sequencing depth of 616× of genome (ESI Table S5†). To accurately identify 4mC sites, a threshold of sequencing depth (≥100) and methylation level (≥10%) was employed for 4mC site calling. We identified 2067, 2148, and 2345 4mC sites in D. radiodurans DNA in three biological replicates (Rep 1, Rep 2, and Rep 3), respectively (ESI Table S5†). The 4mC sites were highly correlated among the three replicates (Pearson correlation coefficients > 0.98, ESI Fig. S17†). Among the three replicates, 1586 4mC sites (53.1%) were overlapped (Fig. 6B). The annotation analysis showed that 4mC sites were mainly enriched in the gene downstream region (Fig. 6C and ESI Fig. S18†).
The distribution analysis of 4mC sites showed that a highly conserved motif of C(4mC)GCGG was observed (E-value = 3.4 × 10−3) (Fig. 6D). Among the overlapped 4mC sites from the three replicates, 35.6% of 4mC sites (564 sites) were present in the C(4mC)GCGG context, which accounted for 83.7% of the total number of CCGCGG in D. radiodurans DNA (Fig. 6E). 4mC in C(4mC)GCGG context occurred on all the two chromosomes and two plasmids of D. radiodurans (Fig. 6E). From a representative region that contains the C(4mC)GCGG motif obtained by 4mC-AMD-seq, it can be seen that the second cytosine was highly methylated on both of the + and − strands (86.1% and 88.0%, respectively) (Fig. 6F). In addition, the statistical analysis demonstrated that the overall methylation level of the second cytosine of the CCGCGG motif in different replicates (Rep 1, Rep 2, and Rep 3) was mainly between 50% and 90% (Fig. 6G and ESI Fig. S19†).
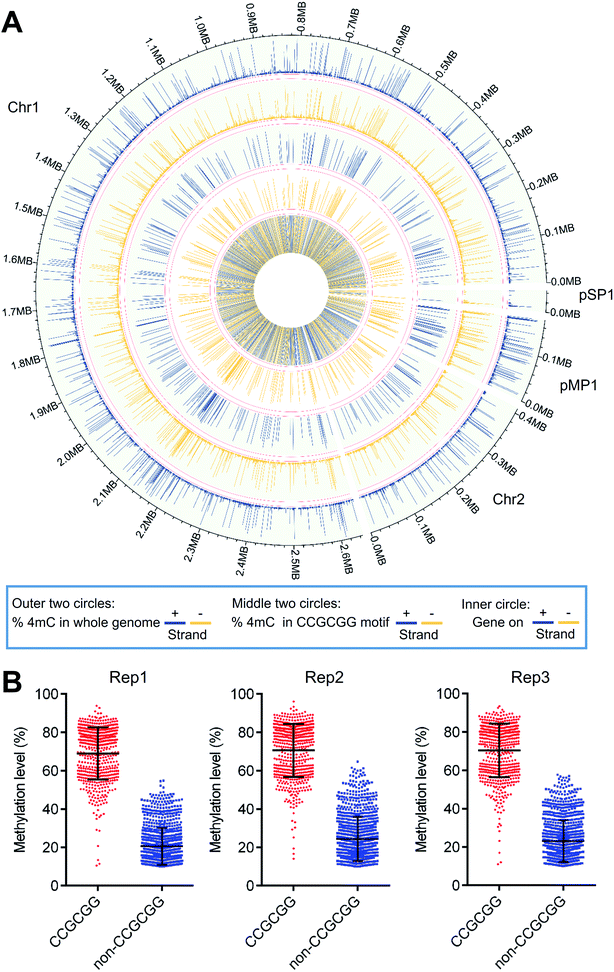
A Circos plot was generated to illustrate the overall distribution and level of 4mC sites (1586 sites) that were commonly identified in all three replicates (Fig. 7A). The results showed that 4mC sites were generally symmetrically distributed in both the + and − strands of D. radiodurans DNA in terms of their location and level (Fig. 7A, outer two circles). The overall 4mC level in D. radiodurans DNA detected by 4mC-AMD-seq (0.30 ± 0.11%, ESI Table S6†) was comparable with the overall 4mC level detected by LC-MS/MS (0.1%) (Fig. 5E). Compared to the methylation level of all the 4mC sites in the whole genome, the methylation level of 4mC in the C(4mC)GCGG motif was much higher (Fig. 7A, middle two circles; ESI Fig. S17†). The average methylation levels in the CCGCGG motif and the non-CCGCGG sequence were 70.0% and 22.8%, respectively (Fig. 7B).
Having obtained the 4mC motif in D. radiodurans DNA by the 4mC-AMD-seq method, we further verified 4mC modification in the CCGCGG motif in seven specific regions using Sanger sequencing. After deamination with A3A protein, D. radiodurans DNA was amplified by locus-specific primers (ESI Table S7†), and the PCR products were subjected to Sanger sequencing. The result indicated that all seven CCGCGG loci were N4-methylated at the second cytosine (ESI Fig. S20†), indicating the good accuracy of the 4mC-AMD-seq method in profiling 4mC in genomes.
Discussion
Both 6mA and 5mC have long been known to exist in bacterial DNA and function in R-M systems. Similar to 6mA and 5mC, 4mC also exists in some thermophilic bacteria. However, relatively little is known about the function of 4mC beyond its role in the R-M systems.Studies on the biological functions of 4mC heavily rely on the methods used for 4mC analysis. In general, analysis of 4mC includes the qualitative and quantitative detection of 4mC and the mapping analysis of 4mC in genomes. Since three isomers of methylated cytosines, 3mC, 4mC and 5mC, could be potentially present in genomes of living organisms, effective separation and distinct detection of these methylated cytosines are essential for their functional studies. Due to the lack of an effective detection method, there were discrepant reports on the existence of methylated cytosine in D. radiodurans DNA.45,46 With the optimized composition of the mobile phases in LC-MS/MS analysis, we achieved the unambiguous detection of 3mC, 4mC and 5mC from DNA, which offers a valuable platform to clarify some previous conflicting findings. It will also be feasible to confirm modifications already identified, or to identify methylated cytosines in uncharacterized species by our developed LC-MS/MS method.
The crystal structure of A3A bound to single-stranded DNA indicated that the carbonyl oxygen atoms of W98 and S99 formed a bifurcated hydrogen bond to NH2, which favors cytosine positioning.49 However, after the replacement of one hydrogen atom by the methyl group in 4mC, only one hydrogen bond could be formed with one of the carbonyl oxygen atoms of W98 or S99. In addition, the interaction between 4mC and W98 or S99 may be affected by the steric hindrance of the CH3 group at the N4 position of 4mC, which could compromise the deamination activity toward 4mC. This observation and speculation encouraged us to characterize the differential deamination activity of A3A protein toward cytosine/5mC and 4mC, with which we eventually develop the bisulfite-free 4mC-AMD-seq method for single-base resolution mapping of 4mC in D. radiodurans DNA. This idea of selective conversion of nucleobases or modified nucleobases in DNA by enzymes that enables the subsequent altered base pairing of nucleobases and modified nucleobases may also be utilized to map other nucleic acid modifications in the future. In case no wild-type enzymes meet the requirements, protein evolution technology could be employed to screen appropriate enzymes.
Unlike the harsh chemical conditions typically used in bisulfite-based sequencing approaches, the deamination reaction by A3A protein was performed under mild and non-destructive conditions in the 4mC-AMD-seq method, which could allow the low initial DNA input. This is particularly useful for those mapping studies of DNA modifications with limited biological or clinical samples. Since A3A could effectively deaminate cytosine and 5mC, but not 4mC in DNA, the 4mC-AMD-seq method is also capable of precisely mapping 4mC under the circumstance where both 4mC and 5mC exist in genomes. In addition, the non-destructive 4mC-AMD-seq method can be potentially coupled with long-read sequencing technologies, such as nanopore sequencing, in future studies.
We identified 1586 4mC sites in the D. radiodurans genome and 564 sites were located in the C(4mC)GCGG motif, which is consistent with previously reported results obtained by SMRT sequencing.19 In addition, it has been demonstrated that the M.DraR1 methyltransferase is responsible for methylating the second cytosine in the CCGCGG motif.19 Knockout of M.DraR1 in D. radiodurans showed a 9-fold increase of spontaneous rifampin mutation frequency and >100-fold increase in transformation frequency,19 indicating that 4mC in genomes might be involved in minimizing spontaneous mutagenesis and recombination. Thus, the presence of 4mC in the D. radiodurans genome may contribute to its resistance to stress conditions. We envision that the 4mC-AMD-seq method will facilitate the investigation of 4mC functions, including the 4mC-involved R-M systems, in uncharacterized but potentially useful strains.
Conclusions
In summary, we characterized that A3A protein was capable of readily deaminating cytosine and 5mC in DNA to form U and T, while 4mC could resist the deamination by A3A protein. With this unique property of A3A protein, we developed the 4mC-AMD-seq method for mapping 4mC in DNA at single-base resolution. The developed 4mC-AMD-seq method also allowed the quantitative evaluation of the level of 4mC at individual sites in DNA. With the developed method, we achieved the genome-wide mapping of 4mC in D. radiodurans. We detected 1586 4mC sites in the genome of D. radiodurans, of which 564 sites were located in the C(4mC)GCGG motif. 4mC sites in D. radiodurans were mainly enriched in the downstream region of the genes. Collectively, these results support that the developed 4mC-AMD-seq method can precisely map 4mC sites at single-base resolution as well as providing the quantitative level of 4mC at individual sites throughout the whole genomes of living organisms, which may facilitate uncovering the uncharacterized functions of 4mC in the future.Data availability
The sequencing data supporting the findings of this study have been deposited into the NCBI Gene Expression Omnibus (GEO) database with accession number GSE189965.Author contributions
J. X. and B. F. Y. designed the experiments and interpreted the data. J. X., J. H. D., N. B. X., M. W. and Q. Y. C performed the expression and purification of A3A protein, and evaluation of A3A deamination. J. X. and W. X. S. cultured the D. radiodurans and extracted genomic DNA. J. X performed the library preparation and high-throughput sequencing. G. L., P. W., W. C. and C. X. analyzed the sequencing data. J. X. and Y. Q. F. analyzed the mass spectrometry data. J. X. and B. F. Y. wrote the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The work is supported by the National Natural Science Foundation of China (22074110 and 21721005).References
- B. Mark and J. F. McGouran, Nat. Rev. Chem., 2018, 2, 332–348 CrossRef.
- A. Hofer, Z. J. Liu and S. Balasubramanian, J. Am. Chem. Soc., 2019, 141, 6420–6429 CrossRef CAS PubMed.
- L. Y. Zhao, J. Song, Y. Liu, C. X. Song and C. Yi, Protein Cell, 2020, 11, 792–808 CrossRef CAS PubMed.
- M. Nappi, A. Hofer, S. Balasubramanian and M. J. Gaunt, J. Am. Chem. Soc., 2020, 142, 21484–21492 CrossRef CAS PubMed.
- C. J. Ma, L. Li, W. X. Shao, J. H. Ding, X. L. Cai, Z. R. Lun, B. F. Yuan and Y. Q. Feng, Chem. Sci., 2021, 12, 14126–14132 RSC.
- F. Tang, J. Yuan, B. F. Yuan and Y. Wang, J. Am. Chem. Soc., 2022, 144, 454–462 CrossRef CAS PubMed.
- G. Z. Luo and C. He, Nat. Struct. Mol. Biol., 2017, 24, 503–506 CrossRef CAS PubMed.
- C. Luo, P. Hajkova and J. R. Ecker, Science, 2018, 361, 1336–1340 CrossRef CAS PubMed.
- Y. Feng, J. J. Chen, N. B. Xie, J. H. Ding, X. J. You, W. B. Tao, X. Zhang, C. Yi, X. Zhou, B. F. Yuan and Y. Q. Feng, Chem. Sci., 2021, 12, 11322–11329 RSC.
- Y. Feng, N. B. Xie, W. B. Tao, J. H. Ding, X. J. You, C. J. Ma, X. Zhang, C. Yi, X. Zhou, B. F. Yuan and Y. Q. Feng, CCS Chem., 2021, 2, 994–1008 CrossRef.
- Q. Wang, J. H. Ding, J. Xiong, Y. Feng, B. F. Yuan and Y. Q. Feng, Chin. Chem. Lett., 2021, 32, 3426–3430 CrossRef CAS.
- A. Janulaitis, S. Klimasauskas, M. Petrusyte and V. Butkus, FEBS Lett., 1983, 161, 131–134 CrossRef CAS PubMed.
- K. Vasu and V. Nagaraja, Microbiol. Mol. Biol. Rev., 2013, 77, 53–72 CrossRef CAS PubMed.
- R. A. Gaultney, A. T. Vincent, C. Lorioux, J. Y. Coppee, O. Sismeiro, H. Varet, R. Legendre, C. A. Cockram, F. J. Veyrier and M. Picardeau, Nucleic Acids Res., 2020, 48, 12102–12115 CrossRef CAS PubMed.
- A. Jeltsch, Chembiochem, 2002, 3, 274–293 CrossRef CAS PubMed.
- M. A. Sanchez-Romero, I. Cota and J. Casadesus, Curr. Opin. Microbiol., 2015, 25, 9–16 CrossRef CAS PubMed.
- L. Liu, Y. Zhang, M. Liu, W. Wei, C. Yi and J. Peng, J. Mol. Biol., 2020, 432, 1035–1047 CrossRef CAS PubMed.
- M. Ehrlich, M. A. Gama-Sosa, L. H. Carreira, L. G. Ljungdahl, K. C. Kuo and C. W. Gehrke, Nucleic Acids Res., 1985, 13, 1399–1412 CrossRef CAS PubMed.
- S. Li, J. Cai, H. Lu, S. Mao, S. Dai, J. Hu, L. Wang, X. Hua, H. Xu, B. Tian, Y. Zhao and Y. Hua, Front. Microbiol., 2019, 10, 1905 CrossRef PubMed.
- Y. Kawaguchi, M. Shibuya, I. Kinoshita, J. Yatabe, I. Narumi, H. Shibata, R. Hayashi, D. Fujiwara, Y. Murano, H. Hashimoto, E. Imai, S. Kodaira, Y. Uchihori, K. Nakagawa, H. Mita, S. I. Yokobori and A. Yamagishi, Front. Microbiol., 2020, 11, 2050 CrossRef PubMed.
- J. R. Battista, Annu. Rev. Microbiol., 1997, 51, 203–224 CrossRef CAS PubMed.
- D. Chung, J. Farkas, J. R. Huddleston, E. Olivar and J. Westpheling, PLoS One, 2012, 7, e43844 CrossRef CAS PubMed.
- S. Liu and Y. Wang, Chem. Soc. Rev., 2015, 44, 7829–7854 RSC.
- Y. Dai, B. F. Yuan and Y. Q. Feng, RSC Chem. Biol., 2021, 2, 1096–1114 RSC.
- W. Y. Lai, J. Z. Mo, J. F. Yin, C. Lyu and H. L. Wang, TrAC, Trends Anal. Chem., 2019, 110, 173–182 CrossRef CAS.
- K. D. Clark, C. Lee, R. Gillette and J. V. Sweedler, ACS Cent. Sci., 2021, 7, 1183–1190 CrossRef CAS PubMed.
- M. Y. Cheng, X. J. You, J. H. Ding, Y. Dai, M. Y. Chen, B. F. Yuan and Y. Q. Feng, Chem. Sci., 2021, 12, 8149–8156 RSC.
- Q. Y. Cheng, J. Xiong, C. J. Ma, Y. Dai, J. H. Ding, F. L. Liu, B. F. Yuan and Y. Q. Feng, Chem. Sci., 2020, 11, 1878–1891 RSC.
- M. Berney and J. F. McGouran, Nat. Rev. Chem., 2018, 2, 332–348 CrossRef CAS.
- G. Vilkaitis and S. Klimasauskas, Anal. Biochem., 1999, 271, 116–119 CrossRef CAS PubMed.
- M. Yu, L. Ji, D. A. Neumann, D. H. Chung, J. Groom, J. Westpheling, C. He and R. J. Schmitz, Nucleic Acids Res., 2015, 43, e148 CrossRef PubMed.
- K. Tanaka and A. Okamoto, Bioorg. Med. Chem. Lett., 2007, 17, 1912–1915 CrossRef CAS PubMed.
- Y. Liu, Z. Hu, J. Cheng, P. Siejka-Zielinska, J. Chen, M. Inoue, A. A. Ahmed and C. X. Song, Nat. Commun., 2021, 12, 618 CrossRef PubMed.
- S. Ardui, A. Ameur, J. R. Vermeesch and M. S. Hestand, Nucleic Acids Res., 2018, 46, 2159–2168 CrossRef CAS PubMed.
- Z. K. O'Brown, K. Boulias, J. Wang, S. Y. Wang, N. M. O'Brown, Z. Hao, H. Shibuya, P. E. Fady, Y. Shi, C. He, S. G. Megason, T. Liu and E. L. Greer, BMC Genomics, 2019, 20, 445 CrossRef PubMed.
- S. U. Siriwardena, K. Chen and A. S. Bhagwat, Chem. Rev., 2016, 116, 12688–12710 CrossRef CAS PubMed.
- M. A. Carpenter, M. Li, A. Rathore, L. Lackey, E. K. Law, A. M. Land, B. Leonard, S. M. Shandilya, M. F. Bohn, C. A. Schiffer, W. L. Brown and R. S. Harris, J. Biol. Chem., 2012, 287, 34801–34808 CrossRef CAS PubMed.
- P. Wijesinghe and A. S. Bhagwat, Nucleic Acids Res., 2012, 40, 9206–9217 CrossRef CAS PubMed.
- E. K. Schutsky, J. E. DeNizio, P. Hu, M. Y. Liu, C. S. Nabel, E. B. Fabyanic, Y. Hwang, F. D. Bushman, H. Wu and R. M. Kohli, Nat. Biotechnol., 2018, 36, 1083–1090 CrossRef CAS PubMed.
- N. B. Xie, M. Wang, T. T. Ji, X. Guo, J. H. Ding, B. F. Yuan and Y. Q. Feng, Chem. Sci., 2022, 13, 7046–7056 RSC.
- E. K. Schutsky, C. S. Nabel, A. K. F. Davis, J. E. DeNizio and R. M. Kohli, Nucleic Acids Res., 2017, 45, 7655–7665 CrossRef CAS PubMed.
- Q. Y. Li, N. B. Xie, J. Xiong, B. F. Yuan and Y. Q. Feng, Anal. Chem., 2018, 90, 14622–14628 CrossRef CAS PubMed.
- M. Y. Chen, C. B. Qi, X. M. Tang, J. H. Ding, B. F. Yuan and Y. Q. Feng, Chin. Chem. Lett., 2022, 33, 3772–3776 CrossRef.
- M. Y. Chen, Z. Gui, K. K. Chen, J. H. Ding, J. G. He, J. Xiong, J. L. Li, J. Wang, B. F. Yuan and Y. Q. Feng, Chin. Chem. Lett., 2022, 33, 2086–2090 CrossRef CAS.
- N. A. Patil, B. Basu, D. D. Deobagkar, S. K. Apte and D. N. Deobagkar, Biochim. Biophys. Acta, Gen. Subj., 2017, 1861, 593–602 CrossRef CAS PubMed.
- A. Ferrandi, F. Castani, M. Pitaro, S. Tagliaferri, C. B. de la Tour, R. Alduina, S. Sommer, M. Fasano, P. Barbieri, M. Mancini and I. M. Bonapace, Biochim. Biophys. Acta, Gen. Subj., 2019, 1863, 118–129 CrossRef CAS PubMed.
- P. K. Chiang, R. K. Gordon, J. Tal, G. C. Zeng, B. P. Doctor, K. Pardhasaradhi and P. P. McCann, FASEB J., 1996, 10, 471–480 CrossRef CAS PubMed.
- Z. Y. Sun, R. Vaisvila, L. M. Hussong, B. Yan, C. Baum, L. Saleh, M. Samaranayake, S. X. Guan, N. Dai, I. R. Correa, S. Pradhan, T. B. Davis, T. C. Evans and L. M. Ettwiller, Genome Res., 2021, 31, 291–300 CrossRef PubMed.
- T. Kouno, T. V. Silvas, B. J. Hilbert, S. M. D. Shandilya, M. F. Bohn, B. A. Kelch, W. E. Royer, M. Somasundaran, N. Kurt Yilmaz, H. Matsuo and C. A. Schiffer, Nat. Commun., 2017, 8, 15024 CrossRef PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: Synthesis of 3-methyl-2′-deoxycytidine (3mdC); expression and purification of recombinant A3A protein; enzymatic digestion of DNA; confirmation of 4mC modification in D. radiodurans DNA by tandem mass spectrometry analysis and high-resolution mass spectrometry analysis; analysis of 4mC in specific loci by 4mC-AMD-seq in D. radiodurans DNA; distribution of 4mC in the gene body and the upstream and downstream of genes; analysis of the methylation level distribution of 4mC sites; Tables S1–S7; Fig. S1–S20. See https://doi.org/10.1039/d2sc02446b |
| ‡ These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |