Open Access Article
Open Access ArticlePharmacomodulation of a ligand targeting the HBV capsid hydrophobic pocket†
Mathilde
Briday
a,
François
Hallé
b,
Lauriane
Lecoq
*a,
Sylvie
Radix
b,
Juliette
Martin
a,
Roland
Montserret
a,
Marie
Dujardin
a,
Marie-Laure
Fogeron
a,
Michael
Nassal
 c,
Beat H.
Meier
d,
Thierry
Lomberget
c,
Beat H.
Meier
d,
Thierry
Lomberget
 *b and
Anja
Böckmann
*b and
Anja
Böckmann
 *a
*a
aMolecular Microbiology and Structural Biochemistry (MMSB) UMR 5086 CNRS/Université de Lyon, Labex Ecofect, 7 Passage du Vercors, 69367 Lyon, France. E-mail: lauriane.lecoq@ibcp.fr; anja.bockmann@ibcp.fr
bUniversité de Lyon, Université Lyon 1, CNRS UMR 5246 Institut de Chimie et Biochimie Moléculaires et Supramoléculaires (ICBMS), Faculté de Pharmacie-ISPB, 8 Avenue Rockefeller, FR-69373 Lyon Cedex 08, France. E-mail: thierry.lomberget@univ-lyon1.fr
cDepartment of Medicine II/Molecular Biology, University Hospital Freiburg, Medical Center, University of Freiburg, Freiburg 79106, Germany
dPhysical Chemistry, ETH Zürich, 8093 Zürich, Switzerland
First published on 7th July 2022
Abstract
Hepatitis B virus (HBV) is a small enveloped retrotranscribing DNA virus and an important human pathogen. Its capsid-forming core protein (Cp) features a hydrophobic pocket proposed to be central notably in capsid envelopment. Indeed, mutations in and around this pocket can profoundly modulate, and even abolish, secretion of enveloped virions. We have recently shown that Triton X-100, a detergent used during Cp purification, binds to the hydrophobic pocket with micromolar affinity. We here performed pharmacomodulation of pocket binders through systematic modifications of the three distinct chemical moieties composing the Triton X-100 molecule. Using NMR and ITC, we found that the flat aromatic moiety is essential for binding, while the number of atoms of the aliphatic chain modulates binding affinity. The hydrophilic tail, in contrast, is highly tolerant to changes in both length and type. Our data provide essential information for designing a new class of HBV antivirals targeting capsid–envelope interactions.
Introduction
Despite the availability of an effective prophylactic vaccine, chronic infection with hepatitis B virus (HBV) causes close to a million deaths each year, with currently only limited means to cure infection by immunomodulation with type-I interferon and nucleos(t)ide analogs blocking reverse transcription.1 The development of new antivirals is thus a pressing but unmet need. During the viral life cycle of HBV (recently reviewed in2), the virion envelope proteins first interact through low affinity interactions with the surface of the hepatocyte, and then through specific interactions with the cellular receptor, NTCP. This allows for endocytosis, and release of the nucleocapsid in the cytoplasm. Capsids are transported to the nucleus, with subsequent release and repair of the viral DNA into an episomal minichromosome from which viral mRNAs are transcribed; then proteins are synthesized by the ribosome, to form nucleocapsids containing pregenomic (pg) RNA and the viral polymerase. After maturation, capsids are either re-imported to the nucleus, or they are enveloped for virion formation and secretion.The capsid-forming core protein (Cp) is the building block of nucleocapsids and is involved in many crucial steps of the HBV life cycle; it is therefore emerging as a new antiviral target.3,4 Cp comprises 183 amino acids, of which the first 140 constitute the assembly domain required to form ordered capsid shells, connected via a short linker to the arginine-rich C-terminal domain (CTD) involved notably in nucleic acid binding.2,5–7 The X-ray crystallographic structure of truncated CTD-less Cp149![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 8 is shown in Fig. 1A. The assembly domain features five αhelices of which α3 and α4 from two monomers associate into a four-helix bundle that protrudes as a spike from the capsid surface. 120 of the so-formed Cp dimers then assemble into the icosahedral T = 4 capsid (Fig. 1B),20 or into a minor class of T = 3 capsids which comprise only 90 dimers. The interdimer contacts are mediated by the last 30 residues of the assembly domain, including helix α5. The currently most advanced anti-Cp drugs are the Capsid Assembly Modulators (CAMs),9 all targeting the interdimer interface in the capsid and interfering with the assembly of proper, genome containing nucleocapsids (as recently reviewed in2,10,11). A new and unexplored option to interfere with nucleocapsid dynamics in general, and nucleocapsid envelopment in particular, is offered by the hydrophobic pocket formed in the Cp dimer at the base of the spike (Fig. 1C), which we have recently shown by solid-state NMR to bind the detergent Triton X-100 (TX100),12 followed by the resolution of cryo-EM structure of the complex.13 Binding causes well-detectable conformational alterations in the capsid structure which essentially localize to a region implicated in HBV nucleocapsid envelopment, i.e. virion formation.
8 is shown in Fig. 1A. The assembly domain features five αhelices of which α3 and α4 from two monomers associate into a four-helix bundle that protrudes as a spike from the capsid surface. 120 of the so-formed Cp dimers then assemble into the icosahedral T = 4 capsid (Fig. 1B),20 or into a minor class of T = 3 capsids which comprise only 90 dimers. The interdimer contacts are mediated by the last 30 residues of the assembly domain, including helix α5. The currently most advanced anti-Cp drugs are the Capsid Assembly Modulators (CAMs),9 all targeting the interdimer interface in the capsid and interfering with the assembly of proper, genome containing nucleocapsids (as recently reviewed in2,10,11). A new and unexplored option to interfere with nucleocapsid dynamics in general, and nucleocapsid envelopment in particular, is offered by the hydrophobic pocket formed in the Cp dimer at the base of the spike (Fig. 1C), which we have recently shown by solid-state NMR to bind the detergent Triton X-100 (TX100),12 followed by the resolution of cryo-EM structure of the complex.13 Binding causes well-detectable conformational alterations in the capsid structure which essentially localize to a region implicated in HBV nucleocapsid envelopment, i.e. virion formation.
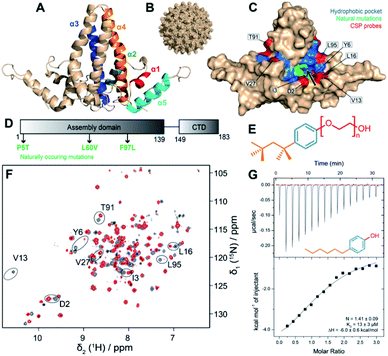 | ||
| Fig. 1 Workflow to assess binding to the hydrophobic pocket. (A) Organization of the five helices of Cp149 dimer (PDB 1QGT8) coloured on one monomer. (B) Structure of the HBV capsid, assembled from 120 copies of Cp149 dimer.8 (C) Structure of the Cp149 dimer in surface representation. The residues of the hydrophobic pocket impacted by the interaction of TX10012 are highlighted in blue. The eight residues whose NMR signals were used to quantify CSPs are highlighted in red. The pocket entrance is indicated by an arrow. Naturally occurring mutations are plotted in green. (D) Sequence of the full-length core protein (Cp183) with mutations highlighted. (E) Structure of the TX100 molecule (compound 1a), with the hydrophilic tail in red, the aromatic moiety in blue, and the alkyl part in para-position in orange. (F) Solution-state NMR spectra of the 2H13C15N-Cp149 apo dimer in grey, and with four equivalents TX100 in red ([Cp149monomer] = 80 μM and [TX100] 320 μM). Peaks used to assess CSPs are circled and labelled with their amino-acid type and position in the sequence. The assigned apo spectrum is shown in Fig. S2,† and the full set of CSPs between the two spectra are shown in Fig. S3.† (G) ITC raw data and binding isotherms displaying the titration of 4-hexylphenol (compound 2a) against Cp149 reassembled capsids at pH 7.5 and 298 K (KD = 13 ± 3 μM). For an overview of ITC runs see Fig. S4.† | ||
Envelopment occurs once the initially encapsidated viral pregenomic (pg) RNA is reverse transcribed, inside the capsid, into mature, double-stranded DNA, signalling readiness of the capsid for interaction with the envelope proteins. The mechanism of this regulation is still undetermined, although several hypotheses have been proposed.12,14–16 Indeed, envelopment is modulated by several naturally-occurring Cp mutations,17,18 many of them being located in and around Cp's hydrophobic pocket, such as F/I97L, P5T and L60V (Fig. 1C and D). Mutations in other envelopment-disabling positions also localize to the hydrophobic pocket, namely at L95 and K96.19 Interestingly, uncoupling of envelopment from genome maturation by the F/I97L mutation can be reverted by the A119F mutation in the preS envelope protein domain.20 In all cases, the most plausible interpretation for the mutationally altered envelopment phenotypes is a modification of the core-envelope interaction. In addition to the hydrophobic pocket in the Cp spike base, the spike tip has also been identified as interacting domain, as indicated by inhibition of envelopment by spike-binding peptides.21,22 The detailed binding site(s) of the envelope proteins on the core protein, as well as a molecular view of the interaction, remain to be determined.
We have recently found that TX100 binds to the hydrophobic pocket in Cp.12 The residues impacted by TX10012 binding are highlighted in blue in Fig. 1C, and colocalize with the residues described to alter envelopment phenotypes (F/I97L, P5T and L60V). Our finding thus opens a new avenue to target the capsid–envelope interaction, by taking this molecule as a lead compound for the development of new antivirals being capable of blocking capsid envelopment. Having determined TX100′s affinity to Cp,12 we here investigated the pharmacophore of Cp hydrophobic pocket binders to establish how the three chemical moieties of the molecule contribute to capsid binding. The variations we explored concern, as shown in Fig. 1E, firstly the TX100 highly hydrophobic branched aliphatic chain carrying five methyl groups (in orange); secondly, the aromatic ring (in blue); and thirdly the (–CH2–CH2-O-)n hydrophilic tail (in red), connected to the aromatic ring in para position to the aliphatic chain.
Results
We tested in this work the capsid's hydrophobic pocket binding capacity for compounds with variations in the three TX100 moieties. To explore the chemical space of the pocket, we used 28 de novo synthesized compounds and 12 commercially available molecules (for a complete overview see Fig. S1; for chemical syntheses see ESI†).To assess binding, we first used solution-state NMR on the truncated Cp dimer (Cp149), which allowed to speed up analysis of the large number of compounds compared to the use of solid-state NMR on assembled capsids.23,24 As shown in the example spectra in Fig. 1F, chemical shift perturbations (CSPs) are a clear indicator for TX100 binding, and represent, when the exchange rate is in slow regime, the chemical shifts of the bound form. This is for example the case for TX100 and 4-tert-octylphenol (OP) (compounds 1a and 1c) as can be seen in the spectra in Fig. S5 and S6† where upon titration only peaks of the free and the bound forms can be observed. For compounds with lower affinity (higher KD) resulting in a fast exchange regime, peaks can be found at intermediate positions. This is the case for 4-hexylphenol (4HF, compound 2a) for which a titration is shown in Fig. S7.† In this case, if one considers that the compounds have similar binding modes and contacts, the extent of the CSPs induced by the different compounds can be correlated with the strength of binding.
Where binding constants were in an adequate range, we complemented the NMR data by isothermal titration calorimetry (ITC) measurements to quantify binding affinities with Cp149 reassembled capsids, as shown in the example of 4HF in Fig. 1G. ITC reveals that only compounds 1a and 1c have a KD below 10 μM, while all other compounds tested have lower affinities (see Table 1) and are therefore, as shown for 4HF (Fig. S7†), in fast exchange. Thus, one can consider that CSPs are representative of the binding strength at same molar ratios (4 molar equivalents).25
| Ligand | K D (μM) | ΔH (kcal mol−1) |
|---|---|---|
| 1a | 8.3 ± 0.5 | −4.59 ± 0.05 |
| 1c | 7.5 ± 0.5 | −4.79 ± 0.14 |
| 2a | 13 ± 3 | −6.0 ± 0.6 |
| 4a | 18 ± 7 | −5 ± 3 |
| 4b | 14 ± 7 | −7 ± 4 |
| 4c | 30 ± 4 | −12.7 ± 1.2 |
| 6g | 68 ± 7 | −3.41 ± 0.09 |
Chemical-shift assignments of the Cp149 dimer at pH 7.5, used as reference for CSP calculations, are shown in Fig. S2† and were deposited at the BMRB under accession number 51294. The assigned reference spectrum is shown in Fig. S2A.† Unlike in solid-state NMR analysis, where we mainly used 13C detection on protonated samples,12 in solution NMR not all amino acids of Cp are visible due to incomplete back exchange of several NH in the hydrophobic core of the Cp dimer after expression in deuterated medium. While this prevented the use of some residues localized directly in the hydrophobic pocket, we selected eight amino acids located near the entry of the pocket (in red in Fig. 1C), which can be detected to measure CSPs (as shown on the sequence in Fig. S2B†). As exemplified in Fig.S3† for TX100, CSPs can then be derived upon addition of each ligand.
The aromatic moiety is central for pocket binding
First, to investigate the importance of the aromatic moiety for binding, we tested the interaction of reduced TX100 (compound 1b), shown in Fig. 2A, where the aromatic ring is replaced by a cyclohexane moiety. The solution NMR spectrum recorded on Cp149 with compound 1b only showed very small CSPs (Fig. 2B) (for full spectra, see Fig. S8–S13†), and ITC did not yield meaningful information. This suggests the central importance of the aromatic moiety to yield a productive interaction with the hydrophobic pocket. We thus considered in the following only compounds conserving the aromatic ring of TX100.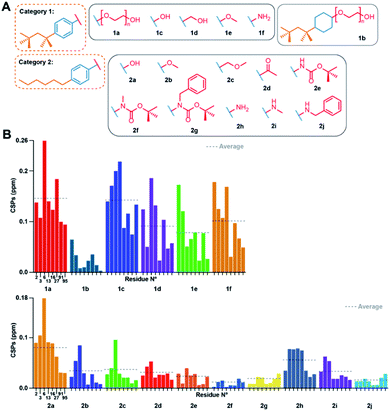 | ||
| Fig. 2 Effects of modulations of the TX100 aromatic ring and hydrophilic tail. (A) Chemical structures of compounds tested from categories 1 and 2. (B) NMR CSPs induced by the compounds for the eight selected capsid resonances (for full spectra see Fig. S8 and S9†). CSP averages are indicated by dotted lines. Residue numbers of the representative peaks are given below the CSPs; all CSPs throughout the manuscript are shown at the same scale. | ||
The hydrophilic tail is dispensable for pocket binding, and can be replaced by –OH, and to a lesser extent, by CH2–OH or –NH2
We next determined whether the hydrophilic chain of TX100 is important for binding to the hydrophobic pocket. Fig. 2A shows the compounds used grouped in two categories, one carrying a tert-octyl group as hydrophobic moiety (category 1), the other a linear hexyl chain (category 2), which resulted in slightly higher water solubility. The NMR spectra of the Cp149 dimer in the presence of all listed compounds (Fig. S8 and S9†) resulted in the CSPs shown in Fig. 2B, with those from categories 1 and 2 shown in separate plots. Category 1 compounds (except 1b) showed high CSPs values, with averages (indicated by dotted lines) higher than 0.05 ppm when compared to the apo state. In particular, compound 1c, namely OP, gave CSPs as high as those derived for TX100. Category 2 compounds induced consistently smaller CSPs, indicating that the n-hexyl moiety binds less strongly than the tert-octyl moiety. Intra-category comparisons revealed that replacement of the polyethylene glycol (PEG) chain by –OH and –NH2 impacts binding, but only to a small extent, a shared result for both categories. However, replacing the PEG chain by bulkier groups (compounds 2e, 2f, 2g, 2j) weakened the interaction. The presence of a possible hydrogen-bonding partner could thus reveal another element of the pharmacophore. The binding parameters of the different compounds with the capsid determined by ITC are reported in Table 1. Individual curves are shown in Fig. S4.† In general, the data showed that the enthalpies for all compounds are negative and in the same range, indicating that the interaction is exothermic. The results confirmed compounds 1c and 2a, both bearing a hydroxyl group instead of the PEG tail, as best alternative binders to TX100. Hence, while the hydrophilic moiety as a whole plays a minor role in binding, the position of the first oxygen in the chain is important. H-bond donor is required, as methyl ether (2b), methoxymethyl (2c) and ketone (2d) resulted in rather small CSPs, and no measurable interaction by ITC. The positioning of bulky groups, like those in compounds 2e, 2f, 2g and 2j, resulted in the smallest CSPs, i.e. less efficient binding.Fully hydrophobic aliphatic, aromatic and functionalized chains can successfully replace the tert-octyl group
We next assessed variations in the hydrophobic element. We investigated compounds featuring different chain lengths and branching, and insertions of oxygen/nitrogen atoms, assigned to categories 3 and 4 (Fig. 3A). We started from both –OH and –NH2 groups instead of the hydrophilic tail, based on their previously assessed roughly similar binding affinities. We investigated different branched hydrocarbon chains, but also amide, ester, and ether groups to establish whether their polarity would favour binding to the accessible main chain amide or carboxyl groups in the capsid's hydrophobic pocket. Fig. 3B displays the CSPs measured for categories 3 and 4 (for full spectra, see Fig. S10 and S11†).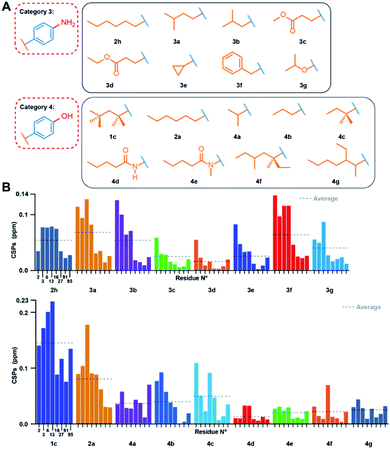 | ||
| Fig. 3 Effects of modulations of the TX100 hydrophobic moiety. (A) Compounds tested from categories 3 and 4. (B) NMR chemical-shift perturbations induced by compounds on the eight selected capsid resonances (for full spectra see Fig. S10 and S11†). CSPs averages are indicated by dotted lines. | ||
The CSPs indicate an optimal atom number of four to eight carbons, as indicated by the similar CSPs induced by compounds 1c, 2a, 2h, 3a, 3b and 4c (average CSP between 0.046 and 0.142 ppm) (see Table S1† for solution NMR parameters and Table S2† for the list of all averaged CSPs). Linear or branched three-carbon chains reduced CSPs to below 0.050 ppm, as observed for compounds 4a and 4b. When long branched alkyl chains were introduced para to the phenolic OH group, the interaction also weakened, as seen for compounds 4f and 4g. The CSPs also revealed that investigated groups with heteroatoms did not establish new favourable interactions, as demonstrated by the small CSPs of compounds 3c, 3d, 3e, 4d, and 4e (average CSP < 0.029 ppm). Surprisingly, while a cyclopropyl moiety (3e) did not bind to the pocket, a benzyl moiety (3f) displayed substantial binding, as indicated by a CSP of 0.071 ppm. ITC measurements showed comparable affinities (between 14 and 30 μM) for compounds 4a, 4b, and 4c, and no measurable interactions for 4f and 4g, consistent with the NMR data.
A single aromatic ring is essential for binding, and hydrophilic/hydrophobic groups need to be in para position
We further investigated the impact of increasing the size of the aromatic moiety. In the respective category 5 compounds (Fig. 4A), the aromatic part is replaced by a naphthalene (5a and b) or a benzofuran (5c) ring. The CSPs for these molecules are very small (Fig. 4B upper panel; for full spectra see Fig. S12†), showing that fused aromatic ring systems block the interaction, disabling binding of compounds 5a, 5b and 5c into the hydrophobic pocket of Cp.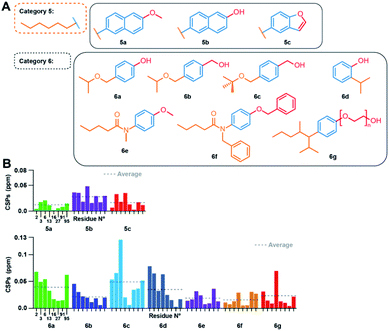 | ||
| Fig. 4 Effect of modulations of the TX100 aromatic ring, and concurrently on the hydrophilic and hydrophobic groups. (A) Compounds tested from categories 5 and 6. (B) NMR CSPs induced by the compounds on the eight selected capsid resonances (for full spectra see Fig. S12 and S13†). CSPs averages are indicated by dotted lines. | ||
Subsequently, we also established a variety of simultaneous modifications of the hydrophobic and hydrophilic groups (category 6). The only compound of this category displaying a significant interaction with the capsid was 6c, with an average CSP of 0.048 ppm (for full spectra, see Fig. S13†). Analysis of CSPs induced by 6b and 6c bearing both a benzyl alcohol on the hydrophilic part shows that a terminal tert-butyl group on the hydrophobic chain seems to favor the interaction, since the CSPs for 6c are more than twice as high as for 6b. Yet, CSPs for 6c still remain lower than for 1d, confirming the even higher impact of the 4-tert-butyl moiety on binding. Both compounds 6d and 6g were tested by ITC (Table 1), the latter displaying weak affinity, and the former no measurable data. The lack of affinity of 6d, compared to its para isomer 4a is revealed by ITC (Fig. S4†), which highlights that the relative position of the hydrophilic and hydrophobic groups is central for the interaction. NMR however report small, but, within the error bars, same CSPs for 4a and 6d.
Docking of TX100 and OP on the core protein dimer
We used the information derived from the binding study to dock the 4-tert-octylphenol (OP) and TX100 molecules to the Cp hydrophobic pocket. For this purpose, we defined a search box comprising residues which showed significant CSPs on binding of TX100.12 We computed multiple binding models and, based on our NMR and ITC results, selected those where the hydrophilic moiety is positioned outside the hydrophobic pocket. Fig. 5A and B show the best models obtained by docking respectively molecule 1c (OP) and 1a (TX100). In both models, the tert-octyl chains share a similar position inside the pocket. However, the aromatic rings are oriented differently, with the PEG tail from TX100 protruding outside the pocket near I59-L60 and the N-terminus of the protein (M1 to Y6) (Fig. 5A), while the phenol ring of OP is rather directed towards Q99, V13, L16 and S17 (Fig. 5B). Comparison of both docked models with the recent cryo-EM structure of the HBV capsid in presence of TX100 (PDB 7PZK)13 is shown in Fig. 5C and D. In the cryo-EM structure, the hydrophobic moiety of TX100 can insert deeper in the pocket as permitted by the rotation of F97 aromatic ring identified in ref. 13 The docking with OP (Fig. 5D) likely represents a more realistic orientation since it is found closer to the cryo-EM structure than docking with TX100 (Fig. 5C). Yet, even if the docking did not predict the F97 sidechain rotation, it correctly modelled the reorientation of K96 sidechain in the case of TX100, where it aligns well with the cryo-EM structure (Fig. 5C).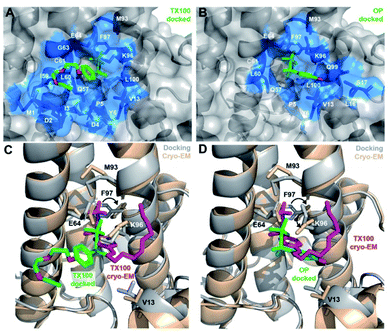 | ||
| Fig. 5 Computed models of OP and TX100 bound in the hydrophobic pocket of the Cp149 dimer. Docked model of (A) OP (compound 1c) and (B) TX100 (1a) in the hydrophobic pocket of Cp149 dimer. Residues located in the direct proximity of ligands (<5 Å) are represented in blue sticks. (C and D) Comparison of bound TX100 in the cryo-EM structure (in magenta, protein in wheat, PDB 7PZK)13 with the docked models (in green, protein in grey) for (C) TX100 and (D) OP. Neighbouring residues V13, E64, M93, K96 and F97 are shown as sticks. In both cryo-EM structure13 and docked models, TX100 contains n = 3 ethylene oxide units in the PEG tail. | ||
Solid-state NMR CSPs can distinguish the conformational changes caused by the different chemical entities
The CSPs of the bound state reveal the impact of the binder on the protein structure, especially when 13C CSPs are considered, which mostly reflect changes in backbone dihedral angles. In a first approximation, small carbon CSPs reflect small changes, and large carbon CSPs substantial impact on the structure. Amide proton and nitrogen chemical shifts, in contrast, are more sensitive to differences in hydrogen bonding and chemical environment variations. We thus established the structural changes using 13C CSPs for compounds 1c and 2a, and compared them to those induced by TX100. Extracts from 13C–13C-dipolar assisted rotational resonance (DARR) spectra for P5, A58 and R98 are shown in Fig. 6A, and 13C-CSPs for these and other residues of interest are shown in Fig. 6B for the three compounds. Full aliphatic regions of DARR spectra are shown in Fig. S14A and more† extracts in Fig. S14B.†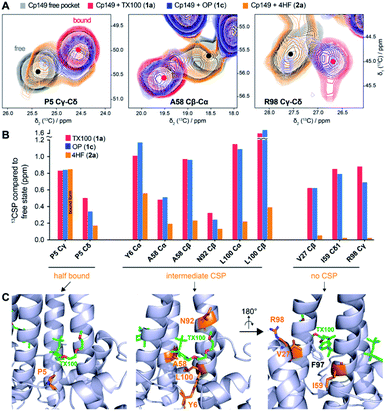 | ||
| Fig. 6 Solid-state NMR can probe the impact of the ligand's hydrophilic tail on the hydrophobic pocket. (A) Comparison of 2D extracts from 13C–13C-DARR solid-state NMR spectra of Cp149 capsids reassembled without compounds (empty pocket, grey), and in presence of TX100 (bound pocket, red), OP (blue), or 4HF (orange). Full aliphatic regions of DARR spectra are shown in Fig. S14.† (B) Comparison of 13C-CSPs induced by the three compounds. Three groups could be identified based on their behaviour when bound to 4HF: P5 at the entrance of the pocket showing a mixture of free/bound pocket; the residues showing intermediate CSPs; and the residues showing no CSPs with 4HF. (C) Structure of Cp183-TX100 (PDB 7PZK)13 with residues involved in the three CSP classes shown in orange. The right panel is rotated by 180° to display the bottom of the hydrophobic pocket and the proximity with F97 sidechain. | ||
Interestingly, OP induces CSPs highly similar to binding of TX100, with spectra almost perfectly overlapping for most residues. This confirms that the PEG tail is not involved in the interaction, and that replacement by a hydroxyl group has almost no impact, even on residues surrounding the pocket. On the contrary, the binding with 4HF induces smaller carbon CSPs for residues within the hydrophobic pocket. In particular, the P5 Cγ-Cδ cross peak in presence of 4HF reveals the presence of two species, respectively corresponding to the bound and free hydrophobic pockets with each about 50% occupancy, as based on peak intensities, suggesting that only half of the pockets are occupied. P5 Cγ is located in the direct vicinity of the ligand, at around 4 Å from the aromatic ring according to the cryo-EM structure13 and docked models (Fig. 5). As such, it is a good indicator of binding and can be used to estimate the percentage of occupancy. However, for the other residues in the vicinity of the ligand such as Y6, A58, N92 and L100, we detect a broad peak, at an intermediate chemical shift between the free and the bound state, as illustrated for A58 (Fig. 6A-B). Finally, a third group could be identified, represented by residues V27, I59 and R98, which show almost no CSPs upon 4HF binding, but high CSPs upon binding of TX100 and OP (Fig. 6B). Interestingly, these residues are located at the bottom of the hydrophobic pocket, behind F97 (Fig. 6C). Hence, the lack of significant CSPs for these residues, as illustrated for R98, is likely due to an absence of a larger conformational rearrangement in presence of 4HF, in contrast to the F97 side-chain rotation observed with TX100.13 We therefore conclude that, compared to the branched tert-octyl moiety, the less sterically hindered n-hexyl group might not be able to induce the rotation of the F97 side chain.13
Discussion
The exploration of 40 different TX100 analogues allowed us to identify the structural and chemical requirements for micromolar binding of TX100 to the capsid-forming HBV Cp. Results from NMR and ITC were complementary, with NMR allowing to characterize also weaker binding events, and ITC able to provide a KD for notably the strongest binders. To summarize (Fig. 7), we established that, with respect to binding, (i) the TX100 hydrophilic tail can be reduced to a single hydroxyl function without significant loss; (ii) its replacement by –NH2, –OCH3 and –CH2OH is possible with minor losses; (iii) in contrast, replacement of the aromatic ring by a cyclohexane ring has a deleterious effect, either due to steric hindrance, loss of π interactions or the different relative orientation of the hydrophobic/hydrophilic moiety; (vi) a hydrophobic moiety of 4–8 atoms is needed to establish significant binding. The para positioning on the aromatic ring proved to be essential, and exceedingly bulky groups on either side hindered binding. Further studies will now attempt to improve the potency of TX100 derivatives by combining hydrophobic, aromatic and hydrophilic moieties. Perspectives include (i) increase lipophilicity of the hydrophobic chain using fluorine bearing carbons (CF2/CF3), thereby decreasing solvation and strengthening affinity to the targeted hydrophobic pocket, and (ii) link the hydrophilic moiety to a functionalized molecule, to induce multivalent interactions with the Cp dimer, for instance targeting the capsid-assembly modulator (CAM) binding pocket.3,26A recent cryo-EM structure13 allowed to determine the position of the previously identified TX100 molecule12 in the hydrophobic pocket; while electron density was observed in the pocket already in previous EM studies,27,28 the nature of the molecule had not been determined, likely due to the lack of resolution. Indeed, while the EM density allows positioning of TX100 mainly based on the aromatic ring and the terminal –C(CH3)3 group, chemical composition and hydrophilic tail were not accessible from the electron densities.
A docking study based on NMR chemical shifts, coupled with the reasonable assumption that the hydrophobic part, rather than the hydrophilic part, fits into the hydrophobic pocket, results in a bundle of structures, many of which show an orientation very similar to that obtained by cryo-EM.13 Both structural models position the oxygen of the phenolic ring in a position where it can interact with residue K96 at the pocket entry.
In contrast to NMR,12 cryo-EM reported only limited structural changes13 in the presence of TX100, predominantly repositioning of the F97 aromatic ring. NMR indeed detects numerous and beyond-the-pocket-extending CSPs between the apo and bound states. This is likely due to the higher sensitivity of NMR which can identify, with atomic resolution, changes in local dihedral angle orientations, but also differences in the local magnetic field as induced by different positioning of aromatic side chains. When considering that the largest change described in the cryo-EM study13 is the movement of the aromatic F97 ring, one can deduce that cryo-EM can detect changes which give rise to 13C CSPs > 1.5 ppm, which is 5 times higher than the limit under which we consider the CSP insignificant (Fig. 3A in ref. 12). Thus, while cryo-EM has the clear advantage to position the binders inside the pocket, solid-state NMR can identify the residues impacted by binding, including those which react in an allosteric manner.
One should mention that TX100 is known to cause endocrine disruption29 and, besides a variety of other phenols, particularly 4-tert-octylphenol is considered as a major environmental pollutant,30 as it is toxic to many organisms. In order to consider TX100 as a starting point for further compound development, this toxicity must be addressed. Alternatives to TX100 are currently under development, as for instance the new detergent Nereid, an environmentally-friendly replacement candidate for TX100 based on substitution of the phenol with a benzylic alcohol.31 Interestingly, we here show that the benzylic alcohol version (compound 1d) retains strong binding capacity to Cp, which points out a direction towards less endocrine-disrupting Cp pocket binders.
Conclusions
By presenting a detailed analysis of the binding properties of the hydrophobic pocket that plays a central role in envelopment, our work opens a new avenue to target both the capsid–envelope interaction, and probably the dynamics of the HBV capsid in the viral life cycle in general.2 The pharmacomodulation through the experimental identification of the important parts of the ligands binding to the hydrophobic pocket is a first step towards the development of a potential binder that would not exhibit endocrine disrupting activity. It opens possibilities to design amino substituted compounds, which could lead to an antiviral treatment.Data availability
The 1H, 13C, 15N backbone chemical shifts of the Cp149 core protein dimer of hepatitis B virus at pH 7.5 and a temperature of 295 K have been deposited in the BioMagResBank (BMRB) (http://www.bmrb.wisc.edu/) under accession number 51294.Author contributions
MB prepared Cp samples, together with MD and MLF, measured ITC, together with RM, and NMR spectra, together with LL; TL, FH and SR designed compounds, FH and SR synthesized compounds; JM made docking experiments; MB and LL analyzed data; AB, TL, and LL designed the study, together with MN and BHM. MB, LL and AB prepared the initial manuscript, together with MN and BHM, and input from all authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by the French Agence Nationale de Recherches sur le Sida et les hépatites virales (ANRS, ECTZ10048, ECTZ71388), the CNRS (CNRS-Momentum 2018), and a CNRS Prematuration Project, and the LABEX ECOFECT (ANR-11-LABX-0048) within the Université de Lyon program Investissements d’Avenir (ANR11-IDEX-0007). Financial support from the IR-RMN-THC Fr3050 CNRS for conducting the research is gratefully acknowledged.References
- G. C. Fanning, F. Zoulim, J. Hou and A. Bertoletti, Nat. Rev. Drug Discovery, 2019, 18, 827–844 CrossRef CAS PubMed.
- M. Niklasch, P. Zimmermann and M. Nassal, Biomedicines, 2021, 9, 1577 CrossRef CAS PubMed.
- U. Viswanathan, N. Mani, Z. Hu, H. Ban, Y. Du, J. Hu, J. Chang and J.-T. Guo, Antiviral Res., 2020, 182, 104917 CrossRef CAS PubMed.
- A. Diab, A. Foca, F. Zoulim, D. Durantel and O. Andrisani, Antiviral Res., 2018, 149, 211–220 CrossRef CAS PubMed.
- M. Nassal, Virus Res., 2008, 134, 235–249 CrossRef CAS PubMed.
- M. Nassal, Gut, 2015, 64, 1972–1984 CrossRef CAS PubMed.
- C. Seeger and W. S. Mason, Virology, 2015, 479–480, 672–686 CrossRef CAS PubMed.
- S. A. Wynne, R. A. Crowther and A. G. W. Leslie, Mol. Cell, 1999, 3, 771–780 CrossRef CAS PubMed.
- T. M. Block, K.-M. Chang and J.-T. Guo, Annu. Rev. Virol., 2021, 8, 437–458 CrossRef PubMed.
- C. J. Schlicksup and A. Zlotnick, Curr. Opin. Virol., 2020, 45, 43–50 CrossRef CAS PubMed.
- S. Fung, H. S. J. Choi, A. Gehring and H. L. A. Janssen, Hepatology, 2022, 76, 233–250 CrossRef CAS PubMed.
- L. Lecoq, S. Wang, M. Dujardin, P. Zimmermann, L. Schuster, M.-L. Fogeron, M. Briday, M. Schledorn, T. Wiegand, L. Cole, R. Montserret, S. Bressanelli, B. H. Meier, M. Nassal and A. Böckmann, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2022464118 CrossRef CAS PubMed.
- C. Makbul, C. Kraft, M. Grießmann, T. Rasmussen, K. Katzenberger, M. Lappe, P. Pfarr, C. Stoffer, M. Stöhr, A.-M. Wandinger and B. Böttcher, Viruses, 2021, 13, 2115 CrossRef CAS PubMed.
- J. Summers and W. S. Mason, Cell, 1982, 29, 403–415 CrossRef CAS PubMed.
- X. Ning, D. Nguyen, L. Mentzer, C. Adams, H. Lee, R. Ashley, S. Hafenstein and J. Hu, PLoS Pathog., 2011, 7, e1002255 CrossRef CAS PubMed.
- J. Xi, H. Liu and J. Hu, J. Virol., 2021, JVI.01305–JVI.01321 Search PubMed.
- T. T. Yuan, G. K. Sahu, W. E. Whitehead, R. Greenberg and C. Shih, J. Virol., 1999, 73, 5731–5740 CrossRef CAS PubMed.
- K. Li, F. Zoulim, C. Pichoud, K. Kwei, S. Villet, J. Wands, J. Li and S. Tong, J. Virol., 2007, 81, 9202–9215 CrossRef CAS PubMed.
- D. Ponsel and V. Bruss, J. Virol., 2003, 77, 416–422 CrossRef CAS PubMed.
- S. Le Pogam, T. T. Yuan, G. K. Sahu, S. Chatterjee and C. Shih, J. Virol., 2000, 74, 9099–9105 CrossRef CAS PubMed.
- B. Bottcher, N. Tsuji, H. Takahashi, M. R. Dyson, S. Zhao, R. A. Crowther and K. Murray, EMBO J., 1998, 17, 6839–6845 CrossRef CAS PubMed.
- C. Makbul, V. Khayenko, H. M. Maric and B. Böttcher, Microorganisms, 2021, 9, 956 CrossRef CAS PubMed.
- L. Lecoq, S. Wang, T. Wiegand, S. Bressanelli, M. Nassal, B. H. Meier and A. Böckmann, Biomol. NMR Assignments, 2018, 12, 205–214 CrossRef CAS PubMed.
- L. Lecoq, M.-L. Fogeron, B. H. Meier, M. Nassal and A. Böckmann, Viruses, 2020, 12, 1069 CrossRef CAS PubMed.
- M. P. Williamson, Prog. Nucl. Magn. Reson. Spectrosc., 2013, 73, 1–16 CrossRef CAS PubMed.
- C. R. Bourne, M. G. Finn and A. Zlotnick, J. Virol., 2006, 80, 11055–11061 CrossRef CAS PubMed.
- B. Böttcher, in Macromolecular protein complexes III: Structure and function, Springer International Publishing, Cham, 2020, pp. 451–470 Search PubMed.
- B. Böttcher and M. Nassal, J. Mol. Biol., 2018, 430, 4941–4954 CrossRef PubMed.
- A. C. Nimrod and W. H. Benson, Crit. Rev. Toxicol., 1996, 26, 335–364 CrossRef CAS PubMed.
- L. W. B. Olaniyan, O. O. Okoh, N. T. Mkwetshana and A. I. Okoh, in Reviews of environmental contamination and toxicology, 248, Springer International Publishing, Cham, 2018, pp. 81–109 Search PubMed.
- J. Farcet, J. Kindermann, M. Karbiener, R. Scheinecker, O. Kostner and T. R. Kreil, J. Med. Virol., 2021, 93, 3880–3889 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc02420a |
| This journal is © The Royal Society of Chemistry 2022 |