Open Access Article
Open Access ArticleCryo-EM structure of acylpeptide hydrolase reveals substrate selection by multimerization and a multi-state serine-protease triad†
Anna J.
Kiss-Szemán
 a,
Pál
Stráner
a,
Pál
Stráner
 b,
Imre
Jákli
b,
Imre
Jákli
 ab,
Naoki
Hosogi
ab,
Naoki
Hosogi
 c,
Veronika
Harmat
c,
Veronika
Harmat
 ab,
Dóra K.
Menyhárd
ab,
Dóra K.
Menyhárd
 *ab and
András
Perczel
*ab and
András
Perczel
 *ab
*ab
aLaboratory of Structural Chemistry and Biology, Institute of Chemistry, Eötvös Loránd University, Budapest – 1117, Hungary. E-mail: dora.k.menyhard@ttk.elte.hu; perczel.andras@ttk.elte.hu
bMTA-ELTE Protein Modelling Research Group, Eötvös Loránd Research Network, Budapest – 1117, Hungary
cEM Application Department, EM Business Unit, JEOL Ltd, Tokyo 196-8556, Japan
First published on 18th May 2022
Abstract
The first structure of tetrameric mammalian acylaminoacyl peptidase, an enzyme that functions as an upstream regulator of the proteasome through the removal of terminal N-acetylated residues from its protein substrates, was determined by cryo-EM and further elucidated by MD simulations. Self-association results in a toroid-shaped quaternary structure, guided by an amyloidogenic β-edge and unique inserts. With a Pro introduced into its central β-sheet, sufficient conformational freedom is awarded to the segment containing the catalytic Ser587 that the serine protease catalytic triad alternates between active and latent states. Active site flexibility suggests that the dual function of catalysis and substrate selection are fulfilled by a novel mechanism: substrate entrance is regulated by flexible loops creating a double-gated channel system, while binding of the substrate to the active site is required for stabilization of the catalytic apparatus – as a second filter before hydrolysis. The structure not only underlines that within the family of S9 proteases homo-multimerization acts as a crucial tool for substrate selection, but it will also allow drug design targeting of the ubiquitin-proteasome system.
Introduction
The mode and purpose of protein multimerization are still under debate. A great number of proteins function as homodimers and a higher degree of homo- and hetero-association is also common, while the active form of only approximately 1/4 of human enzymes is monomeric.1,2 Multimerization carries numerous advantages derived partly from the decrease of surface over volume ratio; it may restrain denaturation and promiscuous interactions, increase stability of the active sites by reducing internal motion and reinforcing their spatial protection, or decrease aggregation propensity by shielding – or utilizing – interaction-prone hot-spots, as well as allowing for cooperativity and providing a vehicle for introducing allosteric regulation.3–5The homo-multimer state is a special case of assembly, one that is reached overcoming the entropic disadvantage coupled to self-association of the monomeric units containing interaction prone surfaces, over association with any other protein (or non-protein) partners.6,7 This requires homomeric contact surfaces to achieve a high degree of selectivity which seems to be fine-tuned and modulated by intrinsic dynamics,8 insertions and deletions9 and also by regions remote from the interaction surfaces2 to achieve or preserve functionality. However, examples of different homo-multimerization forms fulfilling the same function can also be found and in these cases association surfaces seem not to be conserved or specified, shifting along the outer surface of the monomers.5 Thus, it is still to be clarified whether the complexity of multimeric forms arose to create specific functions or simply through neutral or adaptive evolutionary processes.
Here we considered acylaminoacyl peptidase (AAP), a member of the S9 serine protease – or oligopeptidase – family, which is tetrameric in its mammalian form.10–12 The functional form of S9 proteases ranges from monomeric through dimeric, tetrameric to hexameric. The central locus of multimerization is preserved, but not the sequence of the association motifs or the relative orientation of the interacting monomers. The most important interaction center is a β-edge at the surface of the catalytic domain – the outermost member of an 8-stranded β-sheet forming the core of this region. This β-edge carries varying amyloidogenic sequence motifs, and thus it remains highly interaction-prone in all S9 proteases.
At the core of the physiological functions of AAP lies its ability to cleave the N-terminal acetylated (N-Ac) amino-acid from proteins and peptides.13 Removal of the N-Ac tail of proteins leads to dramatic changes in their physicochemical properties such as charge, fold, localization, and protein–protein interactions, but perhaps most importantly, it affects ubiquitination and – through it – the degradation pathway proteins will enter.14,15 Although the regulation of protein lifespan through the addition or removal of the terminal N-Ac is far from clarified, AAP has been shown to play a significant role in protein maturation and degradation processes,16–19 as well as being a key upstream modulator of proteasomal function.20,21 Downregulation of AAP leads to the accumulation of misfolded proteins through disruption of the ubiquitin-proteasome system (UPS),22–24 a crucial sustainer of healthy homeostasis. Inhibiting components of the UPS is already in use as an anti-cancer therapeutic strategy and potential further targets are intensively researched – a field where thus AAP could also be considered.
AAP is a serine-protease enzyme, however, its unique structure and multimerization state seem to create such a seclusion of its active site that potentiates interactions very much unlike those of regular serine-proteases, the most striking example of which is its interaction with and inhibition by carbapenem antibiotics.25–28 Furthermore, AAP was suggested to be able to catalyze the cleavage of amyloid oligomers formed in the progress of Alzheimer's disease.29 It was also shown to be a key protein in cellular response to DNA-damage30 and oxidative stress,31,32 and was proposed to be a potential tumor suppressor.33–35 AAP seems to contribute to the membrane localization of major oncogenic protein K-Ras which may allow K-Ras induced cancers to be targeted through AAP inhibition.36
A detailed understanding of AAP function and catalysis has so far been hindered by the lack of its structure. In spite of reports on its production, and preliminary results of crystallographic studies,13,37–40 the structure of mammalian AAP could not be determined over four decades that passed since its first characterization. Structures of archeal and bacterial AAPs are unfit for model building, since they not only share low sequence identity with the mammalian enzyme (13–20%, Table S1†), but also mostly assume different oligomeric forms.41
Here we present the cryo-EM determined structure of AAP isolated from porcine liver (pAAP). The tetrameric structure shows a gated and channeled intricate interior. Unique inserts that appear in only the mammalian variants participate in forming the main interaction surfaces of the multimer, therefore defining flexibility and architecture of the complex. Our MD simulations show that these large insertions function as shutters for the substrate access channel system, providing the means for substrate screening and selection. Thus, in the case of pAAP, tetramerization seems to be a prerequisite for reaching the catalytically competent form. We found the classical serine protease active site of pAAP in an alternating state via the unprecedented destabilization of the active Ser region, shifting between an active and a latent conformer, as confirmed by both the experimentally determined cryo-EM map and MD simulations. The determined structure will allow, for the first time, structure-based modeling, drug design and detailed analysis of the catalysis of this many faceted enzyme.
Results and discussion
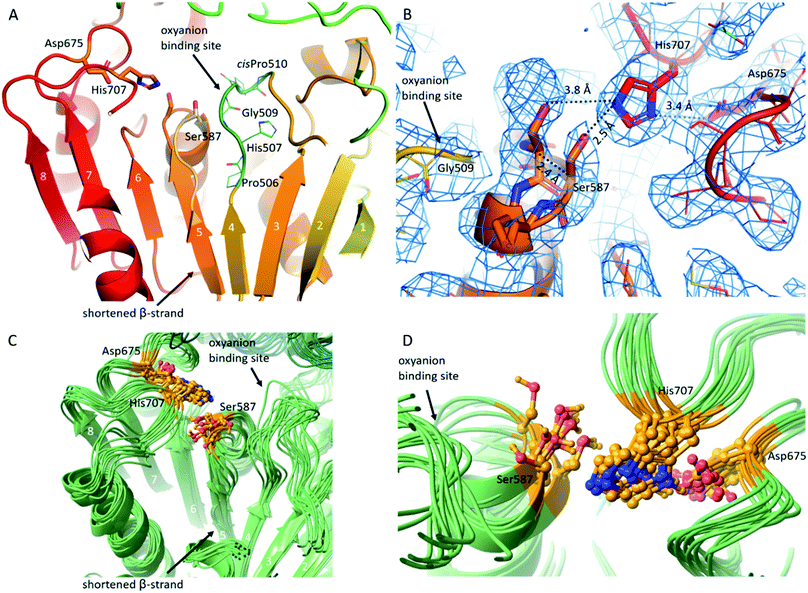
Using cryo-EM, the 3.27 Å resolution structure of the pAAP tetramer was determined (Fig. 1 and S1†). The overall structure is toroidal shaped, showing D2 point group symmetry, with two large openings on the outer surface and an intricate antechamber formed on the inside, from where the active sites of the monomeric units can be reached. To characterize the robustness and flexibility of the determined structure and to model those segments that could not be resolved in the cryo-EM map, MD simulations were also carried out, using the experimentally determined structure as a starting model.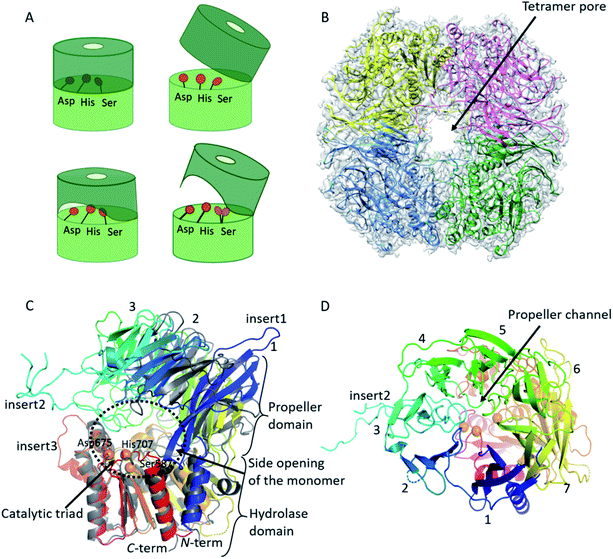 | ||
| Fig. 1 Cryo-EM structure of tetrameric AAP from porcine liver. (A) Schematic representation of the monomeric building blocks thus far detected among S9 serine proteases (with the hydrolase domain (light green) containing the catalytic triad (orange circles), and the propeller domain (deep green) that shields the active site): closed monomer with the catalytic triad in active conformation, accessible only through the propeller channel (upper left), open structure where the catalytic triad is inactivated, but readily accessible (upper right), a semi-closed structure with permanent side opening, with the catalytic triad in active form (lower left) and an open structure with a large side opening obtained in this study, with the catalytic Ser alternating between active and latent conformations (lower right).41 (B) Cryo-EM determined structure of the toroidal pAAP tetramer. Cryo-EM map surface (gray) is shown at a 0.010 threshold level (UCSF Chimera42 chain A – yellow, chain B – red, chain C – green, chain D – cyan). (C) and (D) The monomeric unit of pAAP (from N- to C-termini colored with rainbow colors) viewed from the side with the main insertions labeled (C) and from the direction of the propeller domain (D). The open conformer of ApAAP (PDB id: 3o4g, grey) is overlaid in (C) showing their similar extent of opening. The three types of substrate entrances are labeled: side opening of the monomer, tetramer pore and propeller channel. | ||
pAAP monomers present an open structure with a conformationally destabilized catalytic apparatus
AAP is an oligopeptidase belonging to the S9 family of serine proteases. The monomeric unit of AAPs – and of other members of the S9 family such as prolyl-oligopeptidase (POP), dipeptidyl-peptidase 4 (DPP4) or oligopeptidase B (OPB) – is built from two domains: the better conserved hydrolase domain and the highly variable propeller domain (usually composed of 7 blades, each formed by four antiparallel β-strands) (Fig. 1, S2 and Table S2†). The active site with the catalytic Ser/His/Asp triad is buried between the two.41The pAAP monomers contain a 7-bladed propeller domain (residues 23–457) that is tilted away from the hydrolase (residues 1–22 and 458–732) by approximately 40°, creating a large opening near the active site beneath blade2 (Fig. 1C) that can be accessed from the central cavity of the tetramer. The opening created by domain orientation is similar in extent to that seen in the case of the open form of Aeropyrum pernix AAP (ApAAP).43 In contrast to ApAAP, where the resting state of the enzyme was shown to be a mixture of inactivated open and catalytically competent closed conformers, in case of pAAP interdomain contacts within the tetramer suggest that the domain-organization is rigid. The entrance of the pAAP monomer is further shaped by the loops of blades 3–5, downscaling the large opening to approximately 20–25 Å diameter. These segments are not present in the open form of ApAAP, which thus presents a decidedly wider entrance inside its monomeric unit.
Comparing the structure of the monomer of pAAP to those of the S9 family members determined previously reveals three unique insertions on its surface (Table S2 and Fig. S2, S3†). All four strands of blade1 are elongated, protruding from the outer surface of the propeller domain (Fig. 1: insert1: residues 33–46, 52–61 and 69–80). Blade3 also contains an unusually long insert (Fig. 1: insert2: residues 168–222), forming a long, crossed loop that reaches the surface and contributes to the shaping of the tetramer entrance. A long insertion, containing mainly hydrophobic residues, can also be found in the hydrolase domain (Fig. 1, insert3: residues 636–644), facing blade4 of the propeller. Catalytic Ser587 is placed on a tight turn between a β-strand and an α-helix (Fig. 2) – the conformational strain imposed on the backbone contributes to its activation.
In pAAP, a crucial substitution was found in the middle of the β-strand running parallel and adjacent to the strand leading up to the catalytic Ser587. In place of the Val, Ile, Ala or Gly residues found in most of the S9 proteases in this spot, in pAAP (and also in the human variant) Pro506 appears (Fig. 2A and S4†). The presence of the Pro breaks the β-strand and abolishes the stabilizing interaction between the neighboring strands, allowing greater flexibility for the segment containing the catalytic Ser587. Accordingly, we detected conformational heterogeneity of the 584–591 segment – hosting Ser587 – in the cryo-EM map, which could be described with two conformers of this segment: one can be considered as active, while the other as a latent state of the catalytic apparatus based on their conformation and the corresponding H-bonding network (Fig. 2B and S5†). In the latent state, Cα of the catalytic Ser587 is shifted by 2.4 Å away from the catalytic His707, which places the Ser-Oγ at 3.8 Å from the Nε2 of His (while they are at 2.5 Å from each other in the active conformer). The conformational heterogeneity of the 584–591 segment appears to be localized to the catalytic sites which are distant from each other within the tetramer (the closer pairs are separated by more than 30 Å (measured at Ser587 Cαs)) and show no signs of being in (direct) structural communication with each other, and thus it is reasonable to suppose that the different conformational states of the Ser-loop are evenly distributed among the subunits (both in time and space). Since these regions are also buried deep within the tetrameric assembly, they resulted in no global structural differences that could have been detected during data processing – at least not at the 4.12 Å resolution of the unsymmetrized C1 map – and thus the final models were built using the D2 averaged map of significantly better resolution (3.27 Å). This procedure, however, does not allow differentiation of conformational patterns within the tetramer (resulting in a single, averaged picture of the monomers).
On the other hand, MD simulation of the pAAP tetramer (using a model based on the experimentally determined structure) also sampled both conformers seen in the cryo-EM results, in addition to a number of intermediate states (Fig. 2C and D). While the distance of His707 Nδ1 and the carboxylate oxygens of Asp675 remained within H-bonding distance in over 97% of the snapshots, in only 16.3% of the snapshots was the Ser587–His707 H-bond intact. Inactivation of the catalytic triad was previously reported for the open conformers of monomeric or dimeric S9 proteases43,44 (and for the truncated version of Deinococcus radiodurans carboxypeptidase, DrCP45), but in all cases this was achieved by the destabilization of the His-loop (containing the catalytic His). A serine protease containing multiple conformers of the Ser-loop is unprecedented, to the best of our knowledge. The cryo-EM and simulation results thus both suggest that the active site of pAAP is conformationally heterogeneous and probably requires the docking of the substrate to reach a fully catalytically competent state.
The primary substrate specificity pocket is hydrophobic, similarly to that of other AAPs (Table S3†). However, compared to archeal AAPs, it is narrower with a Trp sidechain lining its lower region (Trp628; Fig. S6†) possibly playing a role in shifting S1 specificity of pAAP towards smaller P1 residues (Val, Ala37). AAPs are exopeptidases, but were also reported to have endopeptidase properties.13,46,47 The substrate binding groove of ApAAP and PhAAP is widened near the surface so they can accommodate substrates, where the site of hydrolysis is not the peptide bond following the first (N-acetylated) residue of the chain, but further along the sequence.48,49 In contrast, this region in the pAAP structure is restricted by Phe274 and Cys275 (of an insert to blade4) forming a barrier locked in place by hydrogen bonds with His325 (Fig. S6†). This also forces the conserved Arg677 into a latent conformation forming a salt bridge with Asp624, from which however, Arg677 may be released to form a hydrogen bond with the P2 residue of larger substrates. These features indicate that pAAP and its human variant might be less effective endopeptidases than ApAAP or PhAAP.
Another notable feature of AAPs is that a cisPro also appears at the active site ensuring the conformation of its AAP-specific HGGP motif that forms the oxyanion site.50 The corresponding cisPro510 of pAAP restricts the pliability of the 507–519 loop placing the backbone N-H of Gly509 in ideal position for coordinating the tetrahedral transition state of the catalytic reaction. Based on the structure, this loop – as an important interaction hub – can be effected by a pathogenic mutation of human AAP, the Thr541Met mutation that causes 50–60% decrease in activity.51 Thr541 makes contacts with loop 507–519, and the Thr → Met switch may lead to loop restructuring, and through that, distortion of the oxyanion site (Fig. S7 and S8).†52
The model of the monomeric unit of human AAP (hAAP) can be found in the AlphaFold Database.53 The model is remarkably close to the experimentally determined structure of pAAP (backbone RMSD for the propeller and hydrolase domains is 1.29 Å and 1.28 Å, respectively, while along the full sequence, 1.48 Å) with a few distinct and functionally important differences. The domain opening was slightly underestimated (Fig. S9†), the cis conformer of Pro510 was not predicted, neither was the liberation of the Ser-loop – or the mode of tetramerization.
Highly interconnected tetrameric assembly with a double-gated shutter system
Formation of the tetrameric structure of mammalian pAAP is guided by two main interaction surfaces and is fine-tuned by the long blade3 insertions (insert2) that wrap around the outer pore of the toroid, creating a highly interconnected structure (Fig. 3). All three unique inserts shown in Fig. 1 participate in forming the key interaction surfaces of the tetramer. The primary interaction surface (A/B and C/D interactions, Fig. 3B) contains, among others, the outermost β-strand of the core β-sheet of the hydrolase domain, the unique (mammalian-specific) insert2 of the propeller and insert3 of the hydrolase domain. Since the segments that participate in forming the interaction surface come from both domains, and numerous propeller-hydrolase contacts are also formed, tetramerization fixes the inter-domain opening of the monomers. The secondary, orthogonal interaction surface (A/D, B/C, Fig. 3B) is formed by insert1 – the extra elongation of blade1 and blade2. The topology of this interaction is unusual – there is an opening between blade1 and blade2 of each propeller domain creating a deep crevice – this is where blade1 of the neighboring monomer is docked in a domain-swapping interaction. And since the blade1 extensions only appear in pAAP (and hAAP, according to the sequence fit), this type of interunit interaction has thus far been unseen in the S9 family. A third, small interaction surface was also detected between monomers of A/C and B/D between blade2 of one monomer and insert2 of blade3 of the opposing monomer (Fig. 3B) completing the network of interconnections between all monomers. Simply put, this means that insert2 of blade3 is long enough not only to reach the outer surface of the tetramer and wind along the hydrolase domain of the neighboring unit, but also to progress along the pore all the way to the monomer at the opposing end.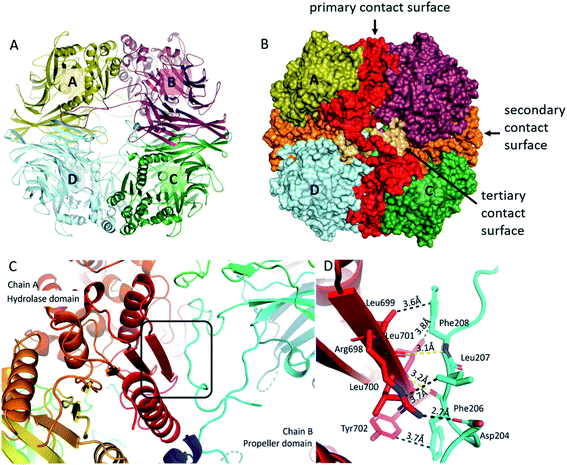 | ||
| Fig. 3 Tetrameric assembly of pAAP. (A) and (B) The primary (red), the secondary (orange) and the tertiary (light orange) interaction surfaces of the tetrameric assembly. For representing pAAP the cryo-EM structure was used, with missing segments (residues 1–9, 39, 110–115, 183–198, 496–497) completed based on the results of the MD simulations. Interaction energies and complex formation significance scores (CSS) obtained using PISA54 are as follows: primary surface: −37.9 kcal mol−1, CSS = 1.0; secondary surface: −11.0 kcal mol−1, CSS = 1.0; third surface: −5.7 kcal mol−1, CSS = 0.08. (C) The central 8-stranded β-sheet of the hydrolase domain with the aggregation-prone β-edge (red, chain A) is covered by the a perpendicularly running segment (residues 204–208) of insert2 (light blue) of the neighbouring monomer (chain B). (D) Details of the interaction: several hydrophobic contacts, a backbone hydrogen bond (yellow) and a salt bridge formed at the contact surface (distances averaged over the four monomers are shown). | ||
The presence of two orthogonal interaction surfaces that both have a significant contribution to the stability of the multimer means that pAAP is not merely a loose association of dimers but a genuine tetramer. Since tetramerization is responsible for the emergence of the double-gated shutter system that seems to be able to screen potential substrates of the enzyme, the inserts present that make this specific mode of multimerization possible are of great significance. The only other member of the S9 family that is known to form a tetrameric structure is DrCP.45 However, in that case only one type of interunit interaction contributes to the stability of the multimer, one that is similar to the primary interaction surface of pAAP, because insert1 is missing and insert2 is not long enough to reach the surface of the tetramer. Thus, the resultant tetramer is characteristically different from that of mammalian AAP.
Being an interconnected tetramer, pAAP possesses all the possible substrate routes previously seen in oligopeptidase structures: leading through the propeller channel or through the tetramer pore and monomer side opening (Fig. 1). The propeller channel is quite wide here compared to other oligopeptidases,55 and therefore it may serve as a potential alternative entry-route towards the catalytic cavity (Fig. S10†) for smaller substrates. In fact, using the FTMap server56 small-ligand binding sites were located along the propeller channel, indicating its accessibility (Fig. S10†). To explore the possible roles of all different pores and channels in the substrate entrance, we analyzed the MD trajectories from this respect also. The segments that could not be resolved in the cryo-EM map (residues 1–8, 39, 110–115, 183–198, 496–497) proved also to be the most flexible in the MD simulations (Fig. S11†). Two especially mobile regions were found. One is the outer segment of insert2 that forms the shape and size of the entrance pores on the two opposing surfaces of the tetramer and the other is the 110–117 moiety of blade2 that guards the entrance to the monomeric units in the inner cavity, where the corresponding segments of all four monomers are placed into proximity by tetramerization (Fig. 4). Both may allow or block the further advance of the substrates, and neither would be present in different multimerization states.
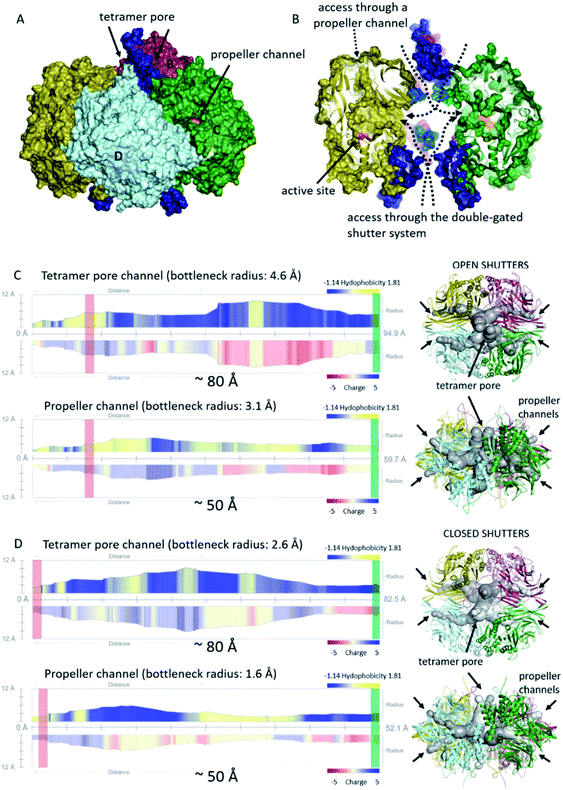 | ||
| Fig. 4 Cavities and channel systems of the pAAP tetramer. (A) and (B) The molecular surface of the pAAP tetramer with the ‘shutters’ of the double gated channel system colored in dark blue (outer pore of the toroid) and light blue (monomer side openings). The entrance of the propeller channel is also labeled. (B) The cross-section shows inner antechambers and the monomer side openings leading to the active sites (orange). (C) and (D) Analysis of the accessibility through the two main routes to the active site with the channel profiles calculated by MoleONline.55 Representative structures of molecule dynamics simulations are shown with (C) both shutters open and with (D) both shutters closed. Red rectangle shows the place of the catalytic triad, and the green rectangle marks the outer surface of the tetramer on the channel profiles (the distance between the two is indicated). | ||
We carried out clustering of the conformations accessed by these most mobile regions during the MD simulation and selected extreme conformations of both. These were added to the cryo-EM determined core structure. Channel analysis55 of the resultant (hybrid) structural models indicates that the gating loops fluctuate between endpoints of an open and a closed structure: a state that provides an approximately 80 Å long channel through the tetramer pore and the central ante-chamber to the monomer side openings and active sites, and another where the catalytic triad can only be accessed by the significantly narrower (∼60% in diameter) channel (Fig. 4). It is important to note that the width of the long channel from the outer pore to the active site is just wide enough for allowing unstructured – or unfolded – chains or shorter oligopeptides to reach the catalytic apparatus. Opening of the outer pore is a frequent phenomenon during the simulation – 25.3% of the structures belong to this class (while 24.9% can be categorized as closed, with 49.8% of the snapshots carrying the outer loop in intermediate conformation) and was also identified by PCA analysis as the most significant fluctuation of the structure (Fig. S11†). On the other hand, the opening of the inner gate at the monomer side is a rare event (4.2% of the snapshots). The results indicate that the two most flexible regions of pAAP together form a double-gated shutter system that larger substrates (i.e. oligopeptides, protein segments) must encounter with while reaching the active sites: firstly at either of the two outer openings of the tetramer toroid, secondly within the interior chamber at the monomer openings.
The outer gatekeeping loops contain a large number of charged and polar amino acids, in distinct patches: a KKRK and a TSDDE motif followed by a RKK motif. Entering substrates must pass between two such gating-loops. We propose that interacting with these highly charged segments might be sufficient to promote the unfolding of the substrates, by decoupling salt-bridges and H-bonds, destabilizing their secondary and tertiary structure. The flexibility of the long loops allows interaction with a variety of targets. Curiously, Ser187 of human AAP (also present in pAAP) – located in the central negatively charged motif of insert2 – was identified as a phosphorylation site.57 Phosphorylation at this spot could influence interaction with the substrates and between the two gatekeepers themselves, offering a possible mode of regulating AAP function.
The interaction hot-spot: β-edge of the hydrolase domain
Within the S9 protease family, the histidine residue of the catalytic triad is placed on a loop (His-loop) that is connected to the terminal β-strand of the 8-membered central β-sheet, at the surface of the core region of the hydrolase domain. This terminal β-strand – key member of the primary interaction surface of pAAP – is situated as an “ideal” aggregation primer (“sticky β-edge”). The corresponding sequences in various oligopeptidases were recognized by five different predictors as being (at least partially) aggregation-prone (Table S4†). In the case of monomeric oligopeptidases, the sticky β-strand is covered by long N-terminal extensions, while in the case of dimeric, tetrameric and hexameric oligopeptidases, where these unprotected interaction-prone β-strands run along the outermost surface of the monomers, they are hidden by the process of multimerization41,42 (Fig. 3C and S12†).A similar scenario was recently outlined in connection with steroid hormone receptors. The large, hydrophobic surface segment of monomeric ketosteroid receptors is covered by a C-terminal extension, while it is hidden by dimerization in estrogen receptors that do not carry the C-terminal tail. The authors conclusively demonstrated that these mechanisms provide structural stabilization (by covering aggregation-prone surfaces), but do not affect function. A model was proposed, where the evolutionary transition in the composition of the vulnerable interaction hot spots – making their protection more efficient while simultaneously destabilizing their unprotected forms – “entrenches” these regions, trapping the achieved multimerization state and interaction topology.58 Intriguingly, among S9 proteases, it is only the central locus of interaction that persevered – the outermost sticky β-edge of the hydrolase domain – however, the mode of its protection greatly varies. And even more importantly, we believe that, among these enzymes, multimerization is a key component of the substrate selection and preparation apparatus.
Conclusion
Oligopeptidases pre-screen their substrates based on size. Some members of the S9 family were proposed to provide access to the active site through the narrow channel dissecting the propeller domain,59 but in the case of most S9 proteases, two different substrate selection scenarios could be deciphered based on the determined structures. In one, the loops containing the His and Asp residues of the triad (His-loop and Asp-loop) are stabilized by inter-domain interactions, which provides the means for disassembly of the catalytic apparatus where the domains shifted away from one another. This was shown to be the case for POP, OPB and ApAAP, where the monomers open up in a clamshell-like motion, inactivating the active site, but also making it readily accessible. Only those substrates will be cleaved by these enzymes that – besides being compatible with the binding surface and substrate specificity pockets of the enzyme interior – also allow the reclosing of the domains, thus resulting in the restoration of the inter-domain interactions that lock the His- and Asp-loops in a conformation that reinstates the functional triad. This requires a build-up that does not restrict domain movement. Accordingly, POP and OPB are monomeric, while ApAAP forms dimers in a way that only involves hydrolase–hydrolase type of interactions, allowing the free movement of the propeller domains. Another strategy for pre-screening of substrates applied by S9 oligopeptidases is providing a permanent entrance to the active site but shielding it by multimerization (Fig. 1A). Dimeric DPP4 and hexameric PhAAP are examples for this latter.41Mammalian AAP seems to apply a hybrid of the two strategies observed so far: it possesses an open monomer, the entrance of which is however narrowed by the inner loops of blades 3–5, and its closing is prohibited by the hydrolase–propeller interactions with the neighboring monomers. Thus, in this case, activation and inactivation of the catalytic triad have been decoupled from the opening and closing of the monomers. Instead, with a Pro inserted into the middle of the fourth strand of the central β-sheet, sufficient conformational freedom is awarded to the 584–591 segment containing the catalytic Ser587 that the active site alternates between the active and inactive conformation even in the absence of domain movements. This is truly a twist on the classical serine protease setup – to the best of our knowledge, no such liberation of the catalytic triad has been previously detected. It is a necessary addition to the S9 oligopeptidase build-up too, since beside functioning as an oligopeptidase – assisting the degradation of smaller peptides – mammalian AAP also removes terminal N-acetylated amino acids from intact proteins,17,18 processing a considerably greater variety of substrates than the other family members do. The highly charged clusters of the flexible gatekeeper loops, lining the outer pore together with the shielded interior of the tetramer, might add a chaperon-like function60 to the entrance: enabling it to first strip the substrate protein segments of their solvent shell and then stabilize the exposed hydrophobic residues, promoting the unfolding of the chain that will allow the substrate to reach the buried and gated active sites.
The structure of pAAP is a splendidly fine-tuned system: self-association guided by the interaction prone β-edge and unique inserts leads to self-compartmentalization that equips the enzyme with its “channels-and-shutters” system guaranteeing that only selected substrates can reach the active site. The His-loop sequentially follows the hot-spot of self-association, and the segments responsible for shaping the outer pore of the tetramer are directly connected to loops that shape the entrance of the monomeric units – also linking self-association and catalysis. Therefore, in the case of mammalian AAP, multimerization is a prerequisite for controlled catalytic function.
The pAAP structure lends further support to the previous hypothesis that among the closely related enzymes of the S9 protease family, the different modes and extents of homo-multimerization equip the “all-purpose” serine protease catalytic machinery embedded in all with unique substrate selection mechanisms.
The determined structure of pAAP thus not only provides a sufficient model of the human enzyme (Fig. S13 and S14†) that will allow drug design efforts, but also contributes to our understanding of the significance and mechanisms of protein multimerization.
Materials and methods
Cryo-EM sample preparation and data collection
The preparation and purification of the mammalian AAP sample (from porcine liver) were based on a previous method (Fig. S15†)39 with an additional size exclusion chromatographic step using a Superose 6 30/100 column on an AKTA FPLC system (GE Healthcare, 20 mM TRIS, pH = 8, 0.15 M NaCl, 1 mM EDTA, 1 mM DTT). Tetrameric composition was verified by size exclusion chromatography. To monitor that the catalytically competent form of the enzyme was preserved during the purification process, concentrated samples of pAAP were incubated with N-acetyl-alanine p-nitroanilide (AANA, eNovation Chemicals LLC) as a substrate (1.6 μM in 5% DMF/water) in buffer (50 mM phosphate, pH = 8, 0.3 M NaCl, 1 mM EDTA, 5 mM mercaptoethanol) at 37 °C (reaction mixture: 10 μl of AANA solution, 985 μl buffer, 5 μl protein sample). The formation of p-nitroaniline was measured spectrophotometrically by monitoring the increase in absorbance at 410 nm.The purified protein sample (3 μl) of 6![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) mg
mg![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ml−1 concentration in 10 mM TRIS (pH = 7.5) buffer was placed on a Quantifoil R1.2/1.3 grid (GIG, 1.0
ml−1 concentration in 10 mM TRIS (pH = 7.5) buffer was placed on a Quantifoil R1.2/1.3 grid (GIG, 1.0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) μm hole size, 200 mesh) and was vitrified. After 2 and 4
μm hole size, 200 mesh) and was vitrified. After 2 and 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) s of blotting time, the grid (4
s of blotting time, the grid (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) °C, 90% humidity) was plunge-frozen in liquid ethane (Leica EMGP). Cryo-EM single particle data collection was performed using a CRYO ARM 300 microscope operated at 300
°C, 90% humidity) was plunge-frozen in liquid ethane (Leica EMGP). Cryo-EM single particle data collection was performed using a CRYO ARM 300 microscope operated at 300 ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) kV equipped with a K3 camera (Gatan). Images were recorded at 80
kV equipped with a K3 camera (Gatan). Images were recorded at 80![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000-fold magnification corresponding to 0.95
000-fold magnification corresponding to 0.95![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Å per pixel using a 20 eV energy filter (Omega Filter) with an exposure time of 4 s and a total electron dose of 37.5
Å per pixel using a 20 eV energy filter (Omega Filter) with an exposure time of 4 s and a total electron dose of 37.5 ![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) e Å−2. The spherical aberration coefficient (Cs) was 2.7 mm and the defocus range was 0.5–2.5 μm. A total of 1157 micrographs were collected from a single grid (ESI – Detailed methods, Fig. S1 and Table S5†).
e Å−2. The spherical aberration coefficient (Cs) was 2.7 mm and the defocus range was 0.5–2.5 μm. A total of 1157 micrographs were collected from a single grid (ESI – Detailed methods, Fig. S1 and Table S5†).
Data availability
Cryo-EM structures and atomic models have been deposited in the Electron Microscopy Data Bank (EMDB) and PDB, respectively, with the following accession codes: EMD-13691 and PDB: 7PX8.Author contributions
A. J. K.-Sz, V. H., D. K. M. and A. P. conceived the project, A. J. K.-Sz, P. S., V. H. and D. K. M. designed the experiments. P. C. provided funding and conceptual input. A. J. K.-Sz, V. H., and D. K. M. analysed the data, prepared figures and tables and co-wrote the manuscript. A. J. K.-Sz. isolated and purified the protein with help of P. S. N. H. and conducted the cryo-EM experiment. I. J. helped with data transition and processing.Conflicts of interest
The authors declare that they have no conflict of interest.Acknowledgements
This work was supported by project no. 2018-1.2.1-NKP-2018-00005 implemented with the support provided from the National Research, Development and Innovation Fund of Hungary, financed under the 2018-1.2.1-NKP funding scheme; no. VEKOP-2.3.2-16-2017-00014 and VEKOP-2.3.3-15-2017-00018 of the European Union and the State of Hungary, co-financed by the European Regional Development Fund; by MedInProt Grants from the Hungarian Academy of Sciences; and within the framework of the ELTE Thematic Excellence Programme 2020, National Challenges Subprogramme – TKP2020-NKA-06, supported by the Hungarian Ministry for Innovation and Technology; as well as by the Hungarian Scientific Research Fund (NKFIH-OTKA) grant K116305. We also thank K. Okayama, R. Tatsumi (EM Application Department, EM Business Unit, JEOL Ltd, Tokyo) and G. Oszkó (JEOL (EUROPE)SAS) for the essential role in our cooperation.References
- A. Chang, et al., BRENDA, the ELIXIR core data resource in 2021: new developments and updates, Nucleic Acids Res., 2021, 49(D1), D498–D508 CrossRef CAS PubMed.
- G. K. A. Hochberg, et al., Structural principles that enable oligomeric small heat-shock protein paralogs to evolve distinct functions, Science, 2018, 359(6378), 930–935 CrossRef CAS.
- N. J. Marianayagam, M. Sunde and J. M. Matthews, The power of two: protein dimerization in biology, Trends Biochem. Sci., 2004, 29, 618–625 CrossRef CAS PubMed.
- R. E. A. Gwyther, D. D. Jones and H. L. Worthy, Better together: building protein oligomers naturally and by design, Biochem. Soc. Trans., 2019, 47(6), 1773–1780 CrossRef CAS PubMed.
- M. Lynch, Evolutionary diversification of the multimeric states of proteins, Proc. Natl. Acad. Sci. U. S. A., 2013, 110(30), E2821–E2828 CrossRef CAS PubMed.
- I. Ispolatov, et al., Binding properties and evolution of homodimers in protein-protein interaction networks, Nucleic Acids Res., 2005, 33, 3629–3635 CrossRef CAS PubMed.
- K. Hashimoto, et al., Caught in self-interaction: evolutionary and functional mechanisms of protein homooligomerization, Phys. Biol., 2011, 8, 035007 CrossRef PubMed.
- T. Perica, et al., Evolution of oligomeric state through allosteric pathways that mimic ligand binding, Science, 2014, 346(6216), 1254346 CrossRef PubMed.
- K. Hashimoto and A. R. Panchenko, Mechanisms of protein oligomerization, the critical role of insertions and deletions in maintaining different oligomeric states, Proc. Natl. Acad. Sci. U. S. A., 2010, 107(47), 20352–20357 CrossRef CAS PubMed.
- S. Tsunasawa, K. Narita and K. Ogata, Purification and properties of acylamino acid-releasing enzyme from rat liver, J. Biochem., 1975, 77, 89–102 CAS.
- K. Kobayashi and J. A. Smith, Acyl-peptide hydrolase from rat liver. Characterization of enzyme reaction, J. Biol. Chem., 1987, 262(24), 11435–11445 CrossRef CAS PubMed.
- M. Mitta, et al., The primary structure of porcine liver acylamino acid-releasing enzyme deduced from cDNA sequences, J. Biochem., 1989, 106(4), 548–551 CrossRef CAS PubMed.
- R. G. Krishna and F. Wold, Specificity determinants of acylaminoacyl-peptide hydrolase, Protein Sci., 1992, 1(5), 582–589 CrossRef CAS PubMed.
- H. Aksnes, A. Drazic, M. Marie and T. Arnesen, First things first: vital protein marks by N-terminal acetyltransferases, Trends Biochem. Sci., 2016, 41, 746–760 CrossRef CAS PubMed.
- C. S. Hwang, A. Shemorry and A. N. Varshavsky, N-terminal acetylation of cellular proteins creates specific degradation signals, Science, 2010, 327, 973–977 CrossRef CAS PubMed.
- J. Perrier, A. Durand, T. Giardina and A. Puigserver, Catabolism of intracellular N-terminal acetylated proteins: involvement of acylpeptide hydrolase and acylase, Biochimie, 2005, 87(8), 673–685 CrossRef CAS PubMed.
- T. Arnesen, Towards a functional understanding of protein N-terminal acetylation, PLoS Biol., 2011, 9(5), e1001074 CrossRef CAS PubMed.
- G. M. Forte, M. R. Pool and C. J. Stirling, N-terminal acetylation inhibits protein targeting to the endoplasmic reticulum, PLoS Biol., 2011, 9(5), e1001073 CrossRef CAS PubMed.
- A. Sandomenico, et al., Small peptide inhibitors of acetyl-peptide hydrolase having an uncommon mechanism of inhibition and a stable bent conformation, J. Med. Chem., 2012, 55(5), 2102–2111 CrossRef CAS PubMed.
- K. Shimizu, Y. Kiuchi, K. Ando, M. Hayakawa and K. Kikugawa, Coordination of oxidized protein hydrolase and the proteasome in the clearance of cytotoxic denatured proteins, Biochem. Biophys. Res. Commun., 2004, 324(1), 140–146 CrossRef CAS PubMed.
- G. Palmieri, et al., Acylpeptide hydrolase inhibition as targeted strategy to induce proteasomal down-regulation, PLoS One, 2011, 6(10), e25888 CrossRef CAS PubMed.
- F. Martínez-Jiménez, et al., A compendium of mutational cancer driver genes, Nat. Rev. Cancer, 2020, 20(10), 555–572 CrossRef PubMed.
- A. Pal, M. A. Young and N. J. Donato, Emerging Potential of Therapeutic Targeting of Ubiquitin-Specific Proteases in the Treatment of Cancer, Cancer Res., 2014, 74, 4955–4966 CrossRef CAS PubMed.
- J. Park, J. Cho and E. J. Song, Ubiquitin–proteasome system (UPS) as a target for anticancer treatment, Arch. Pharmacal Res., 2020, 43, 1144–1161 CrossRef CAS PubMed.
- K. Yokogawa, et al., Effect of meropenem on disposition kinetics of valproate and its metabolites in rabbits, Pharm. Res., 2001, 18(9), 1320–1326 CrossRef CAS PubMed.
- E. Suzuki, et al., Identification of valproic acid glucuronide hydrolase as a key enzyme for the interaction of valproic acid with carbapenem antibiotics, Drug Metab. Dispos., 2010, 38(9), 1538–1544 CrossRef CAS PubMed.
- E. Suzuki, et al., Inhibition mechanism of carbapenem antibiotics on acylpeptide hydrolase, a key enzyme in the interaction with valproic acid, Xenobiotica, 2011, 41(11), 958–963 CrossRef CAS PubMed.
- H. Feng, et al., The mechanism of NDM-1-catalyzed carbapenem hydrolysis is distinct from that of penicillin or cephalosporin hydrolysis, Nat. Commun., 2017, 8(1), 2242 CrossRef PubMed.
- R. Yamin, C. Zhao, P. B. O'Connor, A. C. McKee and C. R. Abraham, Acyl peptide hydrolase degrades monomeric and oligomeric amyloid-beta peptide, Mol. Neurodegener., 2009, 4, 33 CrossRef PubMed.
- Z. Zeng, et al., Acylpeptide hydrolase is a component of the cellular response to DNA Damage, DNA Repair, 2017, 58, 52–61 CrossRef CAS PubMed.
- T. Fujino, K. Watanabe, M. Beppu, K. Kikugawa and H. Yasuda, Identification of oxidized protein hydrolase of human erythrocytes as acylpeptide hydrolase, Biochim. Biophys. Acta, 2000, 1478(1), 102–112 CrossRef CAS.
- K. Shimizu, et al., Overexpression of oxidized protein hydrolase protects COS-7 cells from oxidative stress-induced inhibition of cell growth and survival, Biochem. Biophys. Res. Commun., 2003, 304(4), 766–771 CrossRef CAS PubMed.
- R. Erlandsson, et al., The gene from the short arm of chromosome 3, at D3F15S2, frequently deleted in renal cell carcinoma, encodes acylpeptide hydrolase, Oncogene, 1991, 6, 1293–1295 CAS.
- C. A. McGoldrick, et al., Identification of oxidized protein hydrolase as a potential prodrug target in prostate cancer, BMC Cancer, 2014, 14(1), 77 CrossRef PubMed.
- A. Adibekian, et al., Click-generated triazole ureas as ultrapotent, in vivo-active serine hydrolase inhibitors, Nat. Chem. Biol., 2011, 7(7), 469–478 CrossRef CAS PubMed.
- L. Tan, et al., Acylpeptide hydrolase is a novel regulator of KRAS plasma membrane localization and function, J. Cell Sci., 2019, 132(15), jcs232132 CrossRef CAS PubMed.
- L. Polgár, The prolyl oligopeptidase family, Cell. Mol. Life Sci., 2002, 59(2), 349–362 CrossRef PubMed.
- W. Gade and J. L. Brown, Purification and partial characterization of alpha-N-acylpeptide hydrolase from bovine liver, J. Biol. Chem., 1978, 253(14), 5012–5018 CrossRef CAS.
- A. L. Kiss, Z. Szeltner, V. Fülöp and L. Polgár, His507 of acylaminoacyl peptidase stabilizes the active site conformation, not the catalytic intermediate, FEBS Lett., 2004, 571, 17–20 CrossRef CAS PubMed.
- H. Wright, A. L. Kiss, Z. Szeltner, L. Polgár and V. Fülöp, Crystallization and preliminary crystallographic analysis of porcine acylaminoacyl peptidase, Acta Crystallogr., Sect. F: Struct. Biol. Cryst. Commun., 2005, 61(10), 942–944 CrossRef CAS PubMed.
- A. J. Kiss-Szemán, V. Harmat and D. K. Menyhárd, Achieving Functionality through Modular Build-up: Structure and Size Selection of Serine Oligopeptidases, Curr. Protein Pept. Sci., 2019, 20(11), 1089–1101 CrossRef.
- E. F. Pettersen, et al., UCSF Chimera – a visualization system for exploratory research and analysis, J. Comput. Chem., 2004, 25(13), 1605–1612 CrossRef CAS PubMed.
- V. Harmat, et al., Structure and catalysis of acylaminoacyl peptidase: closed and open subunits of a dimer oligopeptidase, J. Biol. Chem., 2011, 286(3), 1987–1998 CrossRef CAS.
- M. Li, C. Chen, D. R. Davies and T. K. Chiu, Induced-fit mechanism for prolyl endopeptidase, J. Biol. Chem., 2010, 285(28), 21487–21495 CrossRef CAS PubMed.
- P. Yadav, et al., Carboxypeptidase in prolyl oligopeptidase family: unique enzyme activation and substrate-screening mechanisms, J. Biol. Chem., 2019, 294(1), 89–100 CrossRef CAS PubMed.
- A. L. Kiss, et al., The Acylaminoacyl Peptidase from Aeropyrum pernix K1 Thought to Be an Exopeptidase Displays Endopeptidase Activity, J. Mol. Biol., 2007, 368(2), 509–520 CrossRef CAS PubMed.
- Z. Szeltner, et al., Characterization of a novel acylaminoacyl peptidase with hexameric structure and endopeptidase activity, Biochim. Biophys. Acta, Proteins Proteomics, 2009, 1794(8), 1204–1210 CrossRef CAS PubMed.
- D. K. Menyhárd, Z. Orgován, Z. Szeltner, I. Szamosi and V. Harmat, Catalytically distinct states captured in a crystal lattice: the substrate-bound and scavenger states of acylaminoacyl peptidase and their implications for functionality, Acta Crystallogr., Sect. D: Biol. Crystallogr., 2015, 71(3), 461–472 CrossRef PubMed.
- D. K. Menyhárd, et al., A self-compartmentalizing hexamer serine protease from Pyrococcus horikoshii: substrate selection achieved through multimerization, J. Biol. Chem., 2013, 288(24), 17884–17894 CrossRef PubMed.
- A. L. Kiss, A. Palló, G. Náray-Szabó, V. Harmat and L. Polgár, Structural and kinetic contributions of the oxyanion binding site to the catalytic activity of acylaminoacyl peptidase, J. Struct. Biol., 2008, 162, 312–323 CrossRef CAS PubMed.
- D. Tsortouktzidis, et al., Acylpeptide hydrolase (APEH) sequence variants with potential impact on the metabolism of the antiepileptic drug valproic acid, Metab. Brain Dis., 2019, 34(6), 1629–1634 CrossRef CAS PubMed.
- D. Pal and P. Chakrabarti, Non-hydrogen Bond Interactions Involving the Methionine Sulfur Atom, J. Biomol. Struct. Dyn., 2001, 19(1), 115–128 CrossRef CAS PubMed.
- J. Jumper, et al., Highly accurate protein structure prediction with AlphaFold, Nature, 2021, 596, 583–589 CrossRef CAS PubMed.
- E. Krissinel and K. Henrick, Inference of macromolecular assemblies from crystalline state, J. Mol. Biol., 2007, 372, 774–797 CrossRef CAS PubMed , Protein interfaces, surfaces and assemblies' service PISA at the European Bioinformatics Institute, http://www.ebi.ac.uk/pdbe/prot_int/pistart.html.
- L. Pravda, et al., MOLEonline: a web-based tool for analyzing channels, tunnels and pores (2018 update), Nucleic Acids Res., 2018, 46(W1), 368–373 CrossRef PubMed.
- D. Kozakov, et al., The FTMap family of web servers for determining and characterizing ligand-binding hot spots of proteins, Nat. Protoc., 2015, 10(5), 733–755 CrossRef CAS PubMed.
- H. Zhou, et al., Toward a comprehensive characterization of a human cancer cell phosphoproteome, Proteome Res., 2013, 12, 260–271 CrossRef CAS PubMed.
- G. K. A. Hochberg, et al., A hydrophobic ratchet entrenches molecular complexes, Nature, 2020, 588, 503–508 CrossRef CAS PubMed.
- V. Fülöp, Z. Böcskei and L. Polgár, Prolyl oligopeptidase: an unusual beta-propeller domain regulates proteolysis, Cell, 1998, 94(2), 161–170 CrossRef.
- J. Macošek, G. Mas and S. Hiller, Redefining Molecular Chaperones as Chaotropes, Front. Mol. Biosci., 2021, 8, 683132 CrossRef PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc02276a |
| This journal is © The Royal Society of Chemistry 2022 |