Open Access Article
Open Access ArticleA pH-independent electrochemical aptamer-based biosensor supports quantitative, real-time measurement in vivo†
Shaoguang
Li
 a,
Andrés
Ferrer-Ruiz
a,
Andrés
Ferrer-Ruiz
 b,
Jun
Dai
b,
Jun
Dai
 c,
Javier
Ramos-Soriano
c,
Javier
Ramos-Soriano
 b,
Xuewei
Du
b,
Xuewei
Du
 a,
Man
Zhu
a,
Man
Zhu
 a,
Wanxue
Zhang
a,
Wanxue
Zhang
 a,
Yuanyuan
Wang
a,
Yuanyuan
Wang
 a,
M. Ángeles
Herranz
a,
M. Ángeles
Herranz
 b,
Le
Jing
b,
Le
Jing
 a,
Zishuo
Zhang
a,
Zishuo
Zhang
 a,
Hui
Li
a,
Hui
Li
 *a,
Fan
Xia
*a,
Fan
Xia
 *a and
Nazario
Martín
*a and
Nazario
Martín
 *bd
*bd
aState Key Laboratory of Biogeology Environmental Geology, Engineering Research Center of Nano-Geomaterials of Ministry of Education, Faculty of Materials Science and Chemistry, China University of Geosciences, Wuhan 430074, China. E-mail: lihui-chem@cug.edu.cn; xiafan@cug.edu.cn
bDepartamento de Química Orgánica, Facultad de Ciencias Químicas, Universidad Complutense de Madrid, 28040 Madrid, Spain
cDepartment of Obstetrics and Gynecology, Tongji Hospital, Tongji Medical College, Huazhong University of Science and Technology, Wuhan 430074, China
dIMDEA-Nanoscience, C/Faraday, 9, Campus de Cantoblanco, 28049 Madrid, Spain. E-mail: nazmar@ucm.es
First published on 27th June 2022
Abstract
The development of biosensors capable of achieving accurate and precise molecular measurements in the living body in pH-variable biological environments (e.g. subcellular organelles, biological fluids and organs) plays a significant role in personalized medicine. Because they recapitulate the conformation-linked signaling mechanisms, electrochemical aptamer-based (E-AB) sensors are good candidates to fill this role. However, this class of sensors suffers from a lack of a stable and pH-independent redox reporter to support their utility under pH-variable conditions. Here, in response, we demonstrate the efficiency of an electron donor π-extended tetrathiafulvalene (exTTF) as an excellent candidate (due to its good electrochemical stability and no proton participation in its redox reaction) of pH-independent redox reporters. Its use has allowed improvement of E-AB sensing performance in biological fluids under different pH conditions, achieving high-frequency, real-time molecular measurements in biological samples both in vitro and in the bladders of living rats.
1. Introduction
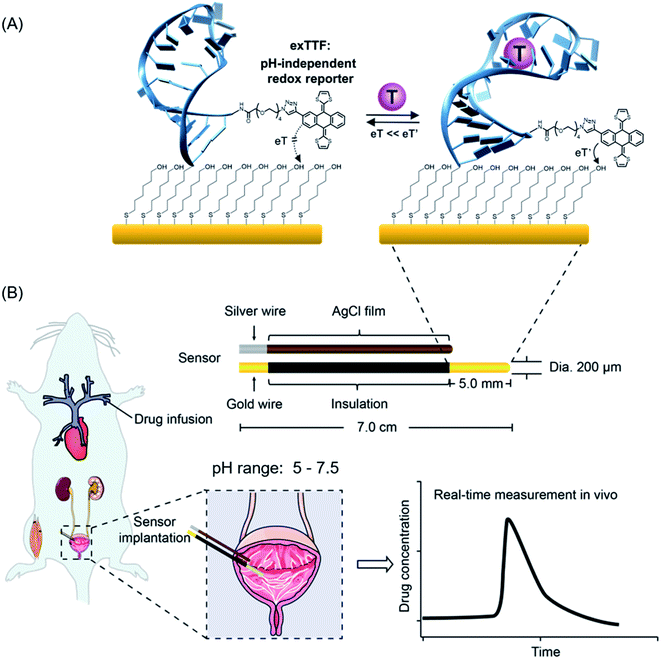
Real-time, continuous monitoring of clinically relevant molecules directly in vivo in the living body would vastly improve our understanding of body health status and our ability to detect, monitor and treat disease.1–6 This ability, for example, could provide the patient-precision pharmacokinetic information required to deliver “the right drug” at the right time and at the right dose.7–12 The development of sensor technology that supports such measurement remains as a challenge.13 First, sensors must achieve a clinically relevant specificity, sensitivity and detection dynamic range. Second, they must function continuously without the need of the addition of exogenous reagents, sample preparation or washing steps. Finally, they must remain stable in complex biological fluids, even so in a rapidly fluctuating environment in the living body, e.g., pH fluctuations.14–16 It is nearly true for most of the biological fluids in the living body and organs and even in subcellular organelles that their pH values are prone to change.17,18 For example, urine and sweat samples exhibited a medically relevant pH range from 5.0 to 7.5.19,20 Even the blood pH, often considered as unaltered under normal conditions, could vary from 6.95 to 7.65 for patients with acidosis or alkalosis disorders.21,22Towards the goal of real-time monitoring of analytes in complex biological fluids, we and others have developed a class of electrochemical aptamer-based (E-AB) biosensors,23–27 a versatile platform supporting the measurement of a variety of analytes. This class of sensors typically employs DNA, RNA or a short peptide aptamer “probe” as its recognition elements (Fig. 1). One terminus of this probe is attached to an interrogating electrode (here, Au electrode), and the other terminus is covalently modified with a redox reporter. Once the probe binds its target, it undergoes a conformational change, which alters the electron transfer kinetics of this redox reporter, thus producing a measurable current change. Due to such a mechanism, sensors of this class are rapid, specific and highly modular, which enables to carry out the analysis of a broad range of target analytes, including small molecules, nucleic acids, proteins, and cells.24,25
Despite their success over the last decade, E-AB sensors suffer from a lack of pH-independent redox reporter, which promises its application in pH-variable body fluids, for example, directly in situ in the bladder of the living body. Several redox reporters (e.g., methylene blue, anthraquinone, Nile blue, and ferrocene) have been explored for E-AB sensors, and very limited success has been achieved,28,29 with only methylene blue (MB) exhibiting good stability. Nevertheless, MB involves two protons for its redox reaction, and thus, the redox potential changes dramatically with pH (see Fig. S1, S2 and eqn (S1)†), dwarfing its function in a pH-variable environment. With respect to the pH-independency, ferrocene is a potential candidate as its redox reaction involves no protons and thus promises its non-pH dependency feature; however, it exhibits an oxidation potential of 0.26 V (vs. Ag/AgCl), which may accelerate the degradation of sensors (see details discussed in the following paragraph). Herein, we propose the use of π-extended tetrathiafulvalene (exTTF) as the redox reporter for E-AB sensors to enable molecular analysis in pH-variable complex media (Fig. 1).
Three main reasons support the choice of exTTF as an efficient reporter: (i) it is chemically and electrochemically stable;30–35 (ii) as a pro-aromatic electron donor, it exhibits a single two-electron oxidation process, and no protons are involved, and thus the redox properties are not altered by pH changes (Fig. S3, S4 and eqn (S2)†); (iii) last but not least, its redox potential is close to “0”, which is a "sweep spot" redox potential with promise for good stability.20 The sweet spot of “0” potential is mainly attributed to the gold substrates employed for E-AB sensors. Despite being most commonly used as an immobilizing electrode in E-AB sensors, gold substrates, nevertheless, limit the electrochemical window in aqueous solutions to the range of −500 mV to 400 mV (vs. Ag/AgCl).36 In an undesirable negative window, oxygen reduction occurs with a pH-dependent onset potential (−100 mV vs. Ag/AgCl, at physiological pH), while in a positive window (+300 mV vs. Ag/AgCl), gold oxidation and etching occur, which may cause the desorption of the DNA probes and SAMs. Given that these two processes cause irreversible damage to sensors, the closer to “0” the redox potential is, the less the E-AB sensors suffer from the above-mentioned two electrocatalytic reactions.
2. Results
As a proof-of-principle, we employed two DNA aptamer sequences, including cocaine- and kanamycin-detecting aptamers, into our E-AB platform. To do so, we first synthesized the precursor exTTF-COOH via a click chemistry reaction (Fig. 2A and Scheme S1†), which was characterized by NMR (1H and 13C) and mass spectrometry (Fig. 2 and S5–S7†). We then prepared DNA-exTTF conjugates via a condensation reaction between DNA sequences with a primary amine and the exTTF derivative endowed with a carboxylic reacting group. Both conjugates were purified using the HPLC technique and characterized by mass spectrometry, observing the respective molecular weights for both exTTF-DNA conjugates (Fig. 2B, C and S8–S14†). Finally, E-AB sensors were fabricated by co-depositing the exTTF-DNA conjugates onto a gold electrode with the 6-mercapto-1-hexanol dilutant.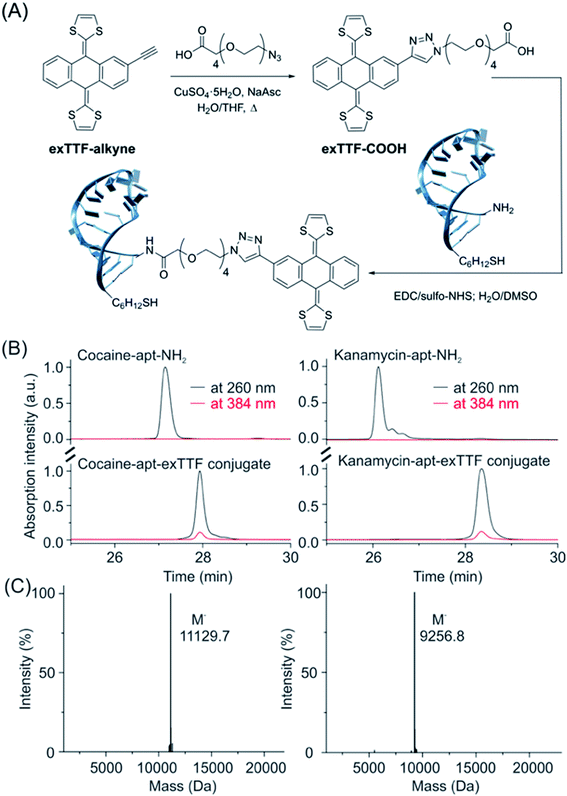 | ||
| Fig. 2 The synthetic route and characterization of the exTTF-aptamer conjugate. (A) Synthesis scheme of exTTF-COOH and the conjugates (Scheme S1†). (B) RP-HPLC trace of the amine-modified cocaine aptamer (cocaine-apt-NH2) and kanamycin aptamer (kanamycin-apt-NH2), and their conjugates. (C) Mass spectrometry analysis of both conjugates confirms the success of the modification reaction between the DNA aptamer and exTTF derivative. | ||
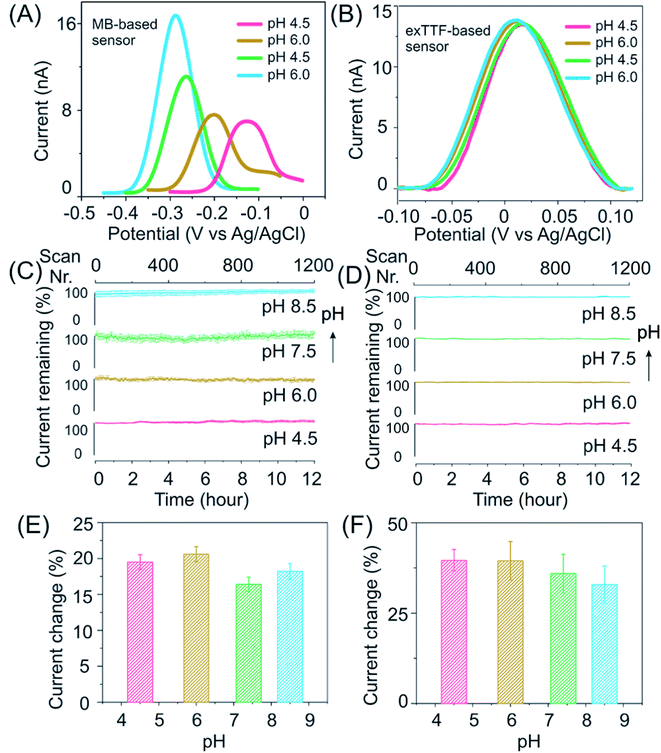
Our E-AB sensors employing exTTF as a redox reporter exhibit good stability and response in buffers over a wide pH range. Upon performing square wave voltammetry to evaluate these freshly prepared kanamycin-detecting sensors in McIlvaine buffer it was observed that while those of MB-based ones differ in both peak positions and height (Fig. 3A), the voltammograms of exTTF-based sensors exhibit a minimal difference peaking at 0.02 V (close to the “sweet spot” at 0 V) when sensors were deployed under varied pH conditions (Fig. 3B). Both kanamycin- and cocaine-detecting sensors exhibited excellent stability, with a less than 10% signal fluctuation under high-frequency measurement conditions (>1200 scans) over a duration of 12 hours (Fig. 3C and D). The current increases upon spiking the system with its target molecules, and it remains similar under different pH conditions (Fig. 3E, F and S15†). For example, upon the addition of 200 nM cocaine, cocaine-detecting sensors exhibit a signal of ∼20% across the pH range from 4.5 to 8.5 (Fig. 3E). Likewise, kanamycin-detecting sensors exhibited a signal change of ∼35% upon the addition of 1 mM kanamycin, with a minimal variation under various pH conditions (Fig. 3F).
After the evaluation of our E-AB sensors in buffers, we then challenge our exTTF-modified sensors in a biological sample, once again observing excellent sensor stability and target response. For example, cocaine-detecting, exTTF-based sensors exhibited excellent stability with a less than 5% signal fluctuation when evaluated in these samples under different pH conditions (Fig. S16†), while the MB-based sensors lost 80% of their original signals. E-AB sensors fabricated from exTTF- and MB-modified DNA sequences exhibited significant variations when they were deployed in real urine samples. The former exhibited a less than 10% signal loss over 12 h, while the latter lost 80% of the original signal under these same conditions (Fig. S17†).
The binding affinities maintain stable for exTTF-based sensors when they are being deployed in biological fluids, while this is not true for MB-based sensors. Taking kanamycin sensors as an example, MB-based sensors exhibited a significant pH-dependence of KD values, ranging from 10 to 200 μM (Fig. 4A and B). In sharp contrast, exTTF-based sensors exhibit negligible differences in their binding affinities and signal gains over a physiological pH range in artificial biological samples (Fig. 4C and D), with similar results achieved for cocaine-detecting sensors (Fig. S18†). Likewise, the kanamycin-detecting, exTTF-based sensors exhibited a steady detection dynamic range and KD values (Fig. 4C and D). Remarkably, the target dose–response curve for the real urine sample matches well with those of artificial biological fluids (Fig. 4E), indicating good reproducibility of sensor performance across biological fluid samples. For further evaluation of their pH-dependency of sensor performance, we then evaluated the kanamycin-detecting sensors at 37 °C (Fig. S19†). Once again, stable binding affinities and signal gains under pH-varying conditions were achieved, despite its higher signal gain of up to ∼80% in comparison to their relative measurements at room temperature. To demonstrate this, we spiked a series of kanamycin solutions varying in concentrations into three different real urine samples (pH of 6.05, 5.50, and 6.50, respectively), obtaining a good recovery rate of 85–90% over a 50-fold concentration span (Fig. 4F).
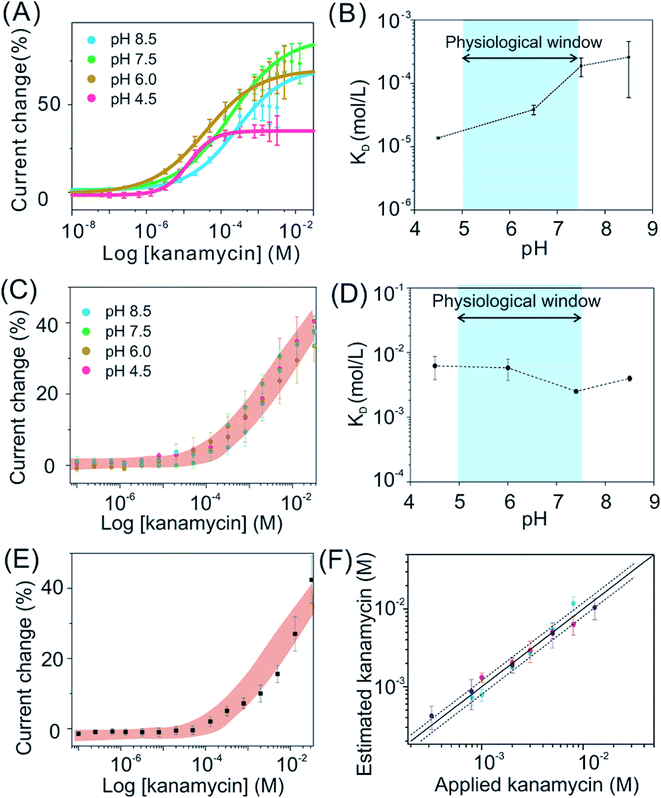 | ||
| Fig. 4 The pH-independent performance likewise holds for exTTF-based sensors deployed in biological systems, greatly improved in comparison to that observed from MB-based sensors. Here we take kanamycin-detecting sensors as an example. (A) The titration plots of MB-based sensors under various pH conditions. (B) These sensors exhibited a variation in KD ranging from 10 μM to 200 μM, over the pH range from 4.5 to 8.5. (C) and (D) In contrast, exTTF-based sensors exhibited pH-independent target-dose responses, with similar results achieved for cocaine-detecting sensors (Fig. S18†). (E) We then deployed exTTF-based sensors in a real urine sample with a series of target concentrations, achieving a target-dose response curve in good accordance with those ranges determined in artificial biological samples. (F) By applying this titration calibration plot to estimate the recovery rate of kanamycin in real urine samples from three healthy individuals, we achieved a good recovery rate of 85–90% over a 50-fold concentration span (the solid line represents 100% recovery, and the dashed lines represent ±20% accuracy bands). | ||
To further confirm the observation of pH-independence of binding affinity and the stability of the exTTF-aptamer conjugate, circular dichroism studies were carried out for the characterization of the target-recognition aptamers. The results suggest that, for both aptamers, the target recognition process produces a conformation change, thus enabling the construction of E-AB sensors. As expected, the binding states remain literally unchanged across a variety of pH changes (Fig. S20 and S21†) (the experimental conditions for CD measurements include 5 μM concentration of the aptamer for both sensors; 0.5 mM kanamycin and 0.1 mM cocaine; 1 cm path length).
Given the promising in vitro performance of these sensors, we were then motivated to test our sensors in vivo in the bladder of a living animal, a pH-varying environment. To do so, we emplaced the sensors for the detection of kanamycin in the bladder of an anesthetized Sprague-Dawley rat and injected the drug via the external jugular vein. Our sensors exhibited excellent baseline stability over the measurement course in vivo (Fig. 5A, see the raw current in Fig. S22†), achieving micromolar-precision measurements with the maximum kanamycin concentration (Cmax) of ∼300 mM and an effective clearance of 90% within 15–20 min, values that closely match with those of previous studies.37,38 Remarkably, the sensors deployed in a rat via an adjusted pH with intravenous sodium bicarbonate therapy exhibited a negligible difference in its performance compared with that deployed in a rat without pH adjustment (Fig. 5B), indicating that our sensors are stable under pH-variable conditions even in the living body.
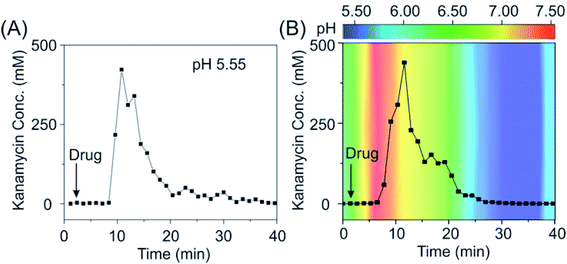 | ||
| Fig. 5 The pH-independent E-AB sensor supports real-time, continuous measurements in vivo. (A) When deploying our sensor in the bladder of a living rat (pH: 5.55), we achieved a micromolar precision and observed an excretion of kanamycin within the duration of half an hour. (B) When deploying our sensors in the bladder of a living rat with pH adjustment, we observed negligible difference in its performance, indicating that our sensors are pH independent. Kanamycin-detecting sensors exhibited a consistent pH-independent performance, with results in good accordance when deploying them in several individual rats (see the raw current and converted concentration information in Fig. S25†). The pH values indicated in B panel were immediately obtained from urine samples collected from the bladder during the in vivo measurements using a pH meter. | ||
To demonstrate that the observed response for in vivo sensor originates from kanamycin, urine samples were simultaneously collected while performing E-AB measurements, and HPLC-MS measurements were conducted for these samples. We observed, as expected, signals from molecular fragments of kanamycin (Fig. S23 and S24†), demonstrating its presence in the bladder. Interestingly, no response was observed when no drug was injected, coupled with these HPLC-MS results, confirming that our sensors probe the concentrations of this drug molecule in the bladder of the living body, rather than something irrelevant.
3. Conclusion
In this study, we have demonstrated the use of exTTF as a pH-independent redox reporter to greatly improve sensing performance under pH-variable conditions both in vitro and in vivo. This exTTF derivative exhibits its redox potential close to ∼0 V (vs. Ag/AgCl), which promises the stability of E-AB sensors when being deployed in buffer, biological fluids and even directly in the living rats. Our sensors exploit the DNA aptamer as a recognition element, which can be readily replaced by aptamers against other targets, promising its modularity of plug-and-play architectures. Thus, we believe that this new redox reporter exTTF proposed here would benefit a wide variety of electrochemical sensor architectures and, even more, this new concept of using a pH-independent biosensor would provide a good route to achieve the precise measurement of clinical molecular analytes in biological systems, which are typically prone to pH changes.39–42 Nevertheless, when designing for such sensors, one should be cautious that the pH-independent sensor performance might be also affected by the specific aptamer structure, target protonation and even the protonation of phosphate groups in DNA strands, etc.Our work could also provide a guideline for other applications relying on the information knowledge of local and precise molecular concentrations, such as drug delivery and targeted cancer therapy. Doxorubicin (Dox), for example, as a potent, clinical commonly-used chemotherapeutic drug for cancer treatment, provokes severe side effects and requires precise control in dose administration.43 Due to the pH gradients of the tumor environment (pH 6.5–7.2),44 it remains challenging to deliver the appropriate dose without knowing the information of the local Dox concentration. Our pH-independent sensors are good candidates to fill this role, as we can readily translate this sensor technology for such a purpose simply by replacing the as-demonstrated aptamers with doxorubicin-recognizing ones. This, in retrospect, would offer an opportunity to integrate biosensors and drug delivery, achieving patient-specific, self-monitoring systems.
Conflicts of interest
There are no conflicts to declare.Author contributions
S. Li, H. Li, M. Á. Herranz, F. Xia and N. Martin conceived the study. S. Li, H. Li, F. Xia and N. Martin designed the experiments. S. Li, A. Ferrer-Ruiz, J. Ramos-Soriano, J. Dai, X. Du and M. Zhu conducted the experiments. S. Li, W. Zhang, Y. Wang, L. Jing and Z. Zhang analyzed and interpreted the data. S. Li and H. Li wrote the manuscript, with input from the rest of the authors.Acknowledgements
This work was supported by the National Natural Science Foundation of China (22090050, 21874121, and 22122410), the National Key Research and Development Program of China (2021YFA1200403 and 2018YFE0206900), the Joint NSFC-ISF Research Grant Program (Grant No. 22161142020), Hubei Provincial Natural Science Foundation of China (2020CFA037), Zhejiang Provincial Natural Science Foundation of China under Grant No. LD21B050001. The authors also thank the Spanish Ministry of Science, Innovation and Universities MCIU (projects PID2020-114653RB-I00, PID2020-115120GB-I00, RED2018-102815-T and Centro de Excelencia Severo Ochoa SEV-2016-0686). We would like to acknowledge Drs Philippe Dauphin-Ducharme and Gabriel Ortega for the development of the MATLAB script used to analyze the square wave voltammetric (SWV) data.References
- M. Bariya, H. Y. Y. Nyein and A. Javey, Nat. Electron., 2018, 1, 160 CrossRef.
- Y. Yang and W. Gao, Chem. Soc. Rev., 2019, 48, 1465 RSC.
- https://labtestsonline.org/tests/urinalysis .
- M. Parviz, P. Toshniwal, H. M. Viola, L. C. Hool, P. M. W. Fear, F. M. Wood, K. Gaus, K. S. Iyer and J. J. Gooding, ACS Sens., 2017, 2, 1482 CrossRef CAS PubMed.
- Y. Yang, M. Cuartero, V. R. Goncales, J. J. Gooding and E. Bakker, Angew. Chem., Int. Ed., 2018, 57, 16801 CrossRef CAS PubMed.
- T. Feng, W. Ji, Y. Zhang, F. Wu, Q. Tang, H. Wei, L. Mao and M. Zhang, Angew. Chem., Int. Ed., 2020, 59, 23445 CrossRef CAS PubMed.
- T. Matsuzaki, D. Scotcher, A. S. Darwich, A. Galetin and A. Rostami-Hodjegan, J. Pharmacol. Exp. Ther., 2019, 368, 157 CrossRef CAS PubMed.
- Z. Sonner, E. Wilder, J. Heikenfeld, G. Kasting, F. Beyette, D. Swaile, F. Sherman, J. Joyce, J. Hagen, N. Kelley-Loughnane and R. Naik, Biomicrofluidics, 2015, 9, 031301 CrossRef CAS PubMed.
- T. Kamei, T. Tsuda, Y. Mibu, S. Kitagawa, H. Wada, K. Naitoh and K. Nakashima, Anal. Chim. Acta, 1998, 365, 259 CrossRef CAS.
- N. De Giovanni and N. Fucci, Curr. Med. Chem., 2013, 20, 545 CAS.
- S. Lin, B. Wang, Y. Zhao, R. Shih, X. Cheng, W. Yu, H. Hojaiji, H. Lin, C. Hoffman, D. Ly, J. Tan, Y. Chen, D. Di Carlo, C. Milla and S. Emaminejad, ACS Sens., 2020, 5, 93 CrossRef CAS PubMed.
- R. Leggett, E. Leesmith, S. M. Jickells and D. A. Russell, Angew. Chem., Int. Ed., 2007, 46, 4100 CrossRef CAS PubMed.
- H. Li, P. Dauphin-Ducharme, N. Arroyo-Currás, C. H. Tran, P. A. Vieira, S. Li, C. Shin, J. Somerson, T. E. Kippin and K. W. Plaxco, Angew. Chem., Int. Ed., 2017, 56, 7492 CrossRef CAS PubMed.
- R. Tavallaie, J. McCarroll, M. Le Grand, N. Ariotti, W. Schuhmann, E. Bakker, R. D. Tilley, D. B. Hibbert, M. Kavallaris and J. J. Gooding, Nat. Nanotechnol., 2018, 13, 1066 CrossRef CAS PubMed.
- M. Labib, B. Green, R. M. Mohamadi, A. Mepham, S. U. Ahmed, L. Mahmoudian, I. H. Chang, E. H. Sargent and S. O. Kelley, J. Am. Chem. Soc., 2016, 138, 2476 CrossRef CAS PubMed.
- M. Labib, Z. Wang, S. U. Ahmed, R. M. Mohamadi, B. Duong, B. Green, E. H. Sargent and S. O. Kelley, Nat. Biomed. Eng., 2021, 5, 41 CrossRef CAS PubMed.
- S. Saha, V. Prakash, S. Halder, K. Chakraborty and Y. Krishnan, Nat. Nanotechnol., 2015, 10, 645 CrossRef CAS PubMed.
- S. Surana, J. M. Bhat, S. P. Koushika and Y. Krishnan, Nat. Commun., 2011, 2, 340 CrossRef PubMed.
- A. Koh, D. Kang, Y. Xue, S. Lee, R. M. Pielak, J. Kim, T. Hwang, S. Min, A. Banks and P. Bastien, Sci. Transl. Med., 2016, 8, 2593 Search PubMed.
- U. M. Jalal, G. J. Jin and J. S. Shim, Anal. Chem., 2017, 89, 13160 CrossRef CAS PubMed.
- R. I. Misbin, Ann. Intern. Med., 1977, 87, 591 CrossRef CAS PubMed.
- V. Gillion, M. Jadoul, O. Devuyst and J. M. Pochet, Acta Clin. Belg., 2019, 74, 34 CrossRef PubMed.
- H. Li, N. Arroyocurras, D. Kang, F. Ricci and K. W. Plaxco, J. Am. Chem. Soc., 2016, 138, 15809 CrossRef CAS PubMed.
- A. A. Lubin and K. W. Plaxco, Acc. Chem. Res., 2010, 43, 496 CrossRef CAS PubMed.
- N. Arroyocurras, J. Somerson, P. A. Vieira, K. L. Ploense, T. E. Kippin and K. W. Plaxco, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 645 CrossRef CAS PubMed.
- E. E. Ferapontova, E. M. Olsen and K. V. Gothelf, J. Am. Chem. Soc., 2008, 130, 4256 CrossRef CAS PubMed.
- H. Hou, Y. Jin, H. Wei, W. Ji, Y. Xue, J. Hu, M. Zhang, Y. Jiang and L. Mao, Angew. Chem., Int. Ed., 2020, 59, 18996 CrossRef CAS PubMed.
- D. Kang, F. Ricci, R. J. White and K. W. Plaxco, Anal. Chem., 2016, 88, 10452 CrossRef CAS PubMed.
- L. Kekedy-Nagy and E. E. Ferapontova, Angew. Chem., Int. Ed., 2019, 58, 3048 CrossRef CAS PubMed.
- A. Ferrer-Ruiz, T. Scharl, P. Haines, L. Rodríguez-Pérez, A. Cadranel, M. A. Herranz, D. M. Guldi and N. Martin, Angew. Chem., Int. Ed., 2018, 57, 1001 CrossRef CAS PubMed.
- S. Liu, I. Perez, N. Martin and L. Echegoyen, J. Org. Chem., 2000, 65, 9092 CrossRef CAS PubMed.
- C. Romero-Nieto, R. Garcia, M. A. Herranz, C. Ehli, M. Ruppert, A. Hirsch, D. M. Guldi and N. Martin, J. Am. Chem. Soc., 2012, 134, 9183 CrossRef CAS PubMed.
- I. A. Wright, P. J. Skabara, J. C. Forgie, A. L. Kanibolotsky, B. Gonzalez, S. J. Coles, S. Gambino and I. D. W. Samuel, J. Mater. Chem., 2011, 21, 1462 RSC.
- J.-Y. Balandier, A. Belyasmine and M. Sallé, Eur. J. Org. Chem., 2008, 269 CrossRef CAS.
- F. G. Brunetti, J. L. López, C. Atienza and N. Martín, J. Mater. Chem., 2012, 22, 4188 RSC.
- N. Arroyo-Currás, P. Dauphin-Ducharme, K. Scida and J. L. Chávez, Anal. Methods, 2020, 12, 1288 RSC.
- G. V. Ling, G. M. Conzelman, C. E. Franti and A. L. Ruby, Am. J. Vet. Res., 1981, 42, 1792 CAS.
- H. Li, S. Li, J. Dai, C. Li, M. Zhu, H. Li, X. Lou, F. Xia and K. W. Plaxco, Chem. Sci., 2019, 10, 10843 RSC.
- S. Emaminejad, W. Gao, E. Wu, Z. A. Davies, H. Yin Yin Nyein, S. Challa, S. P. Ryan, H. M. Fahad, K. Chen, Z. Shahpar, S. Talebi, C. Milla, A. Javey and R. W. Davis, Proc. Natl. Acad. Sci. U. S. A., 2017, 114, 4625 CrossRef CAS PubMed.
- J. Kim, I. Jeerapan, S. Imani, T. N. Cho, A. Bandodkar, S. Cinti, P. P. Mercier and J. Wang, ACS Sens., 2016, 1, 1011 CrossRef CAS.
- W. Gao, H. Y. Y. Nyein, Z. Shahpar, H. M. Fahad, K. Chen, S. Emaminejad, Y. Gao, L.-C. Tai, H. Ota, E. Wu, J. Bullock, Y. Zeng, D.-H. Lien and A. Javey, ACS Sens., 2016, 1, 866 CrossRef CAS.
- J. R. Sempionatto, P. A. Raymundo-Pereira, N. F. B. Azeredo, A. N. De Loyola e Silva, L. Angnes and J. Wang, Chem. Commun., 2020, 56, 2004 RSC.
- F. Meng, Y. Zhong, R. Cheng, C. Deng and Z. Zhong, Nanomedicine, 2014, 9, 487 CrossRef CAS PubMed.
- E. K. Rofstad, B. Mathiesen, K. Kindem and K. Galappathi, Cancer Res., 2006, 66, 6699 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2sc02021a |
| This journal is © The Royal Society of Chemistry 2022 |