Open Access Article
Open Access ArticlePhotosubstitution in a trisheteroleptic ruthenium complex inhibits conjunctival melanoma growth in a zebrafish orthotopic xenograft model†
Quanchi
Chen‡
 ab,
Jordi-Amat
Cuello-Garibo‡
c,
Ludovic
Bretin
c,
Liyan
Zhang
c,
Vadde
Ramu
c,
Yasmin
Aydar
b,
Yevhen
Batsiun
c,
Sharon
Bronkhorst
c,
Yurii
Husiev
ab,
Jordi-Amat
Cuello-Garibo‡
c,
Ludovic
Bretin
c,
Liyan
Zhang
c,
Vadde
Ramu
c,
Yasmin
Aydar
b,
Yevhen
Batsiun
c,
Sharon
Bronkhorst
c,
Yurii
Husiev
 c,
Nataliia
Beztsinna
c,
Lanpeng
Chen
b,
Xue-Quan
Zhou
c,
Claudia
Schmidt
c,
Nataliia
Beztsinna
c,
Lanpeng
Chen
b,
Xue-Quan
Zhou
c,
Claudia
Schmidt
 d,
Ingo
Ott
d,
Martine J.
Jager
d,
Ingo
Ott
d,
Martine J.
Jager
 e,
Albert M.
Brouwer
e,
Albert M.
Brouwer
 f,
B. Ewa
Snaar-Jagalska
*b and
Sylvestre
Bonnet
f,
B. Ewa
Snaar-Jagalska
*b and
Sylvestre
Bonnet
 *c
*c
aDivision of Spine Surgery, Department of Orthopedic Surgery, Nanjing Drum Tower Hospital, The Affiliated Hospital of Nanjing University Medical School, Nanjing, China
bInstitute of Biology, Leiden University, Leiden, The Netherlands. E-mail: b.e.snaar-jagalska@biology.leidenuniv.nl; Tel: +31-71-527-4980
cLeiden Institute of Chemistry, Leiden University, P. O. Box 9502, 2300 RA Leiden, The Netherlands. E-mail: bonnet@chem.leidenuniv.nl; Tel: +31-71-527-4260
dInstitute of Medicinal and Pharmaceutical Chemistry, Technische Universität Braunschweig, Beethovenstrasse 55, D-38106 Braunschweig, Germany
eDepartment of Ophthalmology, Leiden University Medical Center, Leiden, The Netherlands. E-mail: m.j.jager@lumc.nl
fVan't Hoff Institute for Molecular Sciences, University of Amsterdam, Science Park 904, 1098 XH Amsterdam, The Netherlands
First published on 16th May 2022
Abstract
In vivo data are rare but essential for establishing the clinical potential of ruthenium-based photoactivated chemotherapy (PACT) compounds, a new family of phototherapeutic drugs that are activated via ligand photosubstitution. Here a novel trisheteroleptic ruthenium complex [Ru(dpp)(bpy)(mtmp)](PF6)2 ([2](PF6)2, dpp = 4,7-diphenyl-1,10-phenanthroline, bpy = 2,2′-bipyridine, mtmp = 2-methylthiomethylpyridine) was synthesized and its light-activated anticancer properties were validated in cancer cell monolayers, 3D tumor spheroids, and in embryonic zebrafish cancer models. Upon green light irradiation, the non-toxic mtmp ligand is selectively cleaved off, thereby releasing a phototoxic ruthenium-based photoproduct capable notably of binding to nuclear DNA and triggering DNA damage and apoptosis within 24–48 h. In vitro, fifteen minutes of green light irradiation (21 mW cm−2, 19 J cm−2, 520 nm) were sufficient to generate high phototherapeutic indexes (PI) for this compound in a range of cancer cell lines including lung (A549), prostate (PC3Pro4), conjunctival melanoma (CRMM1, CRMM2, CM2005.1) and uveal melanoma (OMM1, OMM2.5, Mel270) cancer cell lines. The therapeutic potential of [2](PF6)2 was further evaluated in zebrafish embryo ectopic (PC3Pro4) or orthotopic (CRMM1, CRMM2) tumour models. The ectopic model consisted of red fluorescent PC3Pro4-mCherry cells injected intravenously (IV) into zebrafish, that formed perivascular metastatic lesions at the posterior ventral end of caudal hematopoietic tissue (CHT). By contrast, in the orthotopic model, CRMM1- and CRMM2-mCherry cells were injected behind the eye where they developed primary lesions. The maximally-tolerated dose (MTD) of [2](PF6)2 was first determined for three different modes of compound administration: (i) incubating the fish in prodrug-containing water (WA); (ii) injecting the prodrug intravenously (IV) into the fish; or (iii) injecting the prodrug retro-orbitally (RO) into the fish. To test the anticancer efficiency of [2](PF6)2, the embryos were treated 24 h after engraftment at the MTD. Optimally, four consecutive PACT treatments were performed on engrafted embryos using 60 min drug-to-light intervals and 90 min green light irradiation (21 mW cm−2, 114 J cm−2, 520 nm). Most importantly, this PACT protocol was not toxic to the zebrafish. In the ectopic prostate tumour models, where [2](PF6)2 showed the highest photoindex in vitro (PI > 31), the PACT treatment did not significantly diminish the growth of primary lesions, while in both conjunctival melanoma orthotopic tumour models, where [2](PF6)2 showed more modest photoindexes (PI ∼ 9), retro-orbitally administered PACT treatment significantly inhibited growth of the engrafted tumors. Overall, this study represents the first demonstration in zebrafish cancer models of the clinical potential of ruthenium-based PACT, here against conjunctival melanoma.
1. Introduction
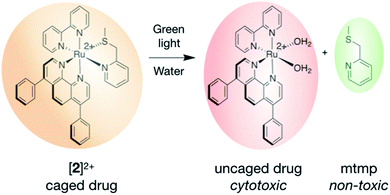
Cisplatin was the first metal-based chemotherapy drug approved by the Food & Drug Administration for the treatment of testicular tumours and ovarian adenocarcinoma, and with the development of carboplatin and oxaliplatin (two derivatives of cisplatin) the use of platinum-based drugs has expanded to the treatment of many different malignancies.1–3 Although the exact mechanism of action of platinum(II) (Pt) complexes is still debated, it is generally accepted that the ultimate event that induces apoptosis in cancer cells is the binding of the heavy metal centre to DNA after thermal hydrolysis of one or two labile ligand(s) of the metal complex.4 DNA binding to Pt inhibits DNA replication and transcription, ultimately leading to cell death.5–7 Spontaneous activation of the drug before it reaches the tumour leads to severe side effects in patients treated with platinum drugs, for example hepato- and nephrotoxicity, which limits the clinical efficacy of these compounds and the patients' quality of life.8–10 Therefore, other metal-based compounds have been considered as anticancer chemotherapy candidates, including those based on ruthenium(II) (Ru).11 Although several of these compounds have reached the stage of clinical trials, the general toxicity of metal-based compounds, due to spontaneous activation of a metal–ligand bond before the drug reaches the tumour, remains an issue.Ruthenium-based PhotoActivated ChemoTherapy (PACT) is a new anticancer phototherapy modality that uses visible light irradiation as an external trigger.12–14 PACT primarily aims at limiting the biological action of the anticancer drug to the location of the tumour by localized, light-induced activation at the tumour site.15 Unlike photodynamic therapy (PDT), a clinically-approved anticancer phototherapy method based on the photochemical activation of dioxygen by an excited photosensitizer, PACT relies on an oxygen-independent photochemical bond cleavage reaction. This process generates a molecular species that is more cytotoxic than the (non-activated) prodrug kept in the dark.16–20 Many examples of PACT agents have been reported in the literature, among which are molecules based on ruthenium.21–27 Ru-Based PACT compounds make use of the versatile and well-understood photochemistry of polypyridyl ruthenium compounds, which, next to energy transfer and electron transfer, comprises light-induced photosubstitution reactions.28–37 When photosubstitution occurs, one of the organic ligands bound to the metal is replaced by loosely-bound solvent molecules. In the dark, the ligand to be photosubstituted serves as a protecting group towards the coordination of biomolecules present in cells. After light irradiation, photosubstitution produces an “uncaged” metal compound that, by analogy with cisplatin, acts as an activated drug, as it can bind to biomolecules and induce cell death (Fig. 1).38–40 For example, blue light-induced photosubstitution of the non-toxic ligand 2-methylthiomethylpyridine (mtmp) in compounds [Ru(dpp)2(mtmp)]2+ ([1]2+, dpp = 4,7-diphenyl-1,10-phenanthroline) and [Ru(bpy)2(mtmp)]2+ ([3]2+, bpy = 2,2′-bipyridine), has recently been demonstrated.41 These two compounds belong to a wide family of complexes [Ru(N–N)2(L–L)]2+, where N–N are non-photocleavable “spectator” polypyridyl ligands, and L–L is a photocleavable chelate.39,41–44 Although the photochemistry of this type of complexes is relatively well-understood, two major challenges remain en-route to their pre-clinical development. On the one hand, the difference between their dark and light toxicity should be maximized; and on the other hand, we need to understand how their molecular structure relates to their toxicity before activation takes place, in particular in vivo. In [1]2+ and [3]2+ for example, the first challenge was not met: [1]2+ bears two very hydrophobic dpp chelates that made the complex taken up by cells in large amounts, which generated high cytotoxicity before light activation. Meanwhile, [3]2+ bears two much less hydrophobic bpy spectator ligands, as a result of which it is too hydrophilic to penetrate significantly into cancer cells, which prevented this compound to show any cytotoxicity even after light activation.40 The second challenge also remains unmet, as in vivo studies on PACT remain scarce,45,46 and no maximum tolerated dose (MTD) has ever been reported yet.
In this work we set out to resolve both challenges. First, we designed a new heteroleptic ruthenium complex, [Ru(dpp)(bpy)(mtmp)]2+ ([2]2+, Fig. 1), characterized by the presence of three different ligands: one dpp and one bpy spectator chelates, to balance the hydrophobicity of the prodrug and optimize cellular uptake; and an mtmp ligand, which serves as a light-cleavable protecting group. Second, we aimed at comparing the photobiological properties of this compound in vitro and in vivo, by testing its maximum tolerated dose (MTD) and light-activated antitumor activity in zebrafish embryo tumour models. Zebrafish tumour models are advantageous for anticancer compound development as they allow for fast compound screening in vivo with low amounts of compound, compared to rodents, and with better statistics.47–49 As zebrafish are transparent, it is especially easy to activate a phototherapeutic compound by light in the whole body of the animal by simply shining light onto the aqueous solution containing the embryos.50–56 The transparency of the embryo makes it easy to quantify the relative tumour burden, using engraftment of human cancer cells which stably express red fluorescent protein (RFP). This property has been used for studying PDT,57 including new clinically tested ruthenium-based photosensitizers such as TLD-1433,58 as well as photoswitchable inhibitors, allowing analysis before and after light activation.53 Zebrafish embryos provide a particularly useful animal model for assessing drug toxicity: acute and chronic toxic effects of metal nanoparticles have been well characterized, with special focus on immunotoxicity, developmental toxicity, neurotoxicity, reproductive toxicity, cardiovascular toxicity, or hepatoxicity.59,60 Systemic drug toxicity to zebrafish embryos has been well described as well.61–63 Here the zebrafish model allowed us to investigate for the first time the toxicity of ruthenium-based PACT compounds in different in vivo models of cancer while respecting the 3Rs principles (reduction, refinement, replacement). As zebrafish embryos had not previously been used for PACT, we tested different protocols of compound administration to find a mode of administration to test anti-cancer efficacy and toxicity. Critically, this work highlights that a high efficacy of a PACT compound in vitro do not necessarily translate into a similarly high efficacy in vivo, while moderate activities in vitro may lead to excellent efficacy in vivo. In animal models, the mode of compound administration really matters, which cannot be assessed in 2D cellular models.
2. Results and discussion
2.1 Synthesis and photoreactivity
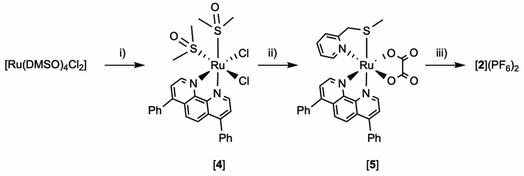
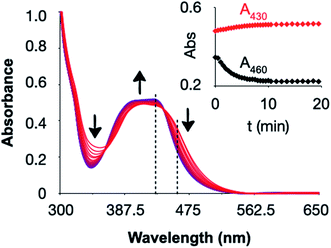
The synthesis of [2]2+ is challenging: as it is a tris-heteroleptic compound, the three different bidentate ligands need to be coordinated to the metal in a controlled fashion (Fig. 2). With most generic synthetic routes, ligand scrambling occurred, i.e. [2]2+ was obtained with traces of [Ru(dpp)(mtmp)2]2+, [Ru(bpy)(mtmp)2]2+, [1]2+, or [3]2+, that were very difficult to remove. The synthesis of [2](PF6)2 was hence adapted from a novel method developed by Keyes et al.64 that involved the sequential coordination, in this order, of dpp, mtmp, and bpy. The novelty of this method relies on the use of an intermediate oxalate ligand (ox2−) during the coordination of the second (mtmp) chelate. This negatively charged chelate prevents the formation of species where two identical ligands coordinate to the metal even when one equivalent of mtmp is used. After purification of the [Ru(dpp)(mtmp)(ox)] ([5]) intermediate complex, oxalate was removed selectively by HClO4 treatment in acetonitrile, after which the last chelate (bpy) was reacted to afford, after counter anion metathesis, [2](PF6)2. Due to the dissymmetry of mtmp and the tri-heteroleptic nature of the final complex, two configurational isomers A and B are expected: one having the sulfur donor atom trans to bpy and another having the sulfur donor atom trans to dpp. These isomers were detected by 1H NMR and initially separated by column chromatography (Fig. S1 and S2†). Isomer [2A](PF6)2 was slightly contaminated with [Ru(dpp)2(bpy)](PF6)2, while isomer [2B](PF6)2 was pure according to 1H NMR but obtained in a low yield (<2%). Later on, as no difference in reactivity could be observed between both isomers, mixtures of [2A](PF6)2 and [2B](PF6)2 were further used in biological studies (Fig. S1–S5†); they are designated below as [2](PF6)2.The photoreactivity of [2](PF6)2 was studied by mass spectrometry and UV-vis spectroscopy in CH3CN. The spectrum of a solution of [2](PF6)2 irradiated for 20 minutes with green light (521 nm, 14 mW cm−2) showed an increase of the intensity of the metal-to-ligand charge-transfer (MLCT) band between 400–430 nm, and a decrease in the valley at 344 nm with clear isosbestic points at 363 and 440 nm (Fig. 3). After 2 min irradiation, the mass spectrum (Fig. S6a†) showed four peaks: at m/z = 336.3 significant amount of the starting compound [2]2+; two strong peaks at m/z = 140.3 and 336.3, corresponding to the free caging ligand {mtmp + H}+ (calc. m/z = 140.1) and to the uncaged photoproduct [Ru(bpy)(dpp)(CH3CN)2]2+ (calc. m/z = 336.1); and a small peak at m/z = 385.4 characteristic for [Ru(bpy)(dpp)(η1-mtmp)(CH3CN)]2+ (calc. m/z = 385.09), i.e. a species where the mtmp chelate is bound to ruthenium by a single heteroatom. After 15 minutes (Fig. S6b†), the reaction reached the steady state and the mass spectrum showed only the peaks of mtmp and [Ru(bpy)(dpp)(CH3CN)2]2+. No traces of free bpy, dpp, or of any ruthenium complex resulting from the photosubstitution of one of the two bis-imine ligands, was observed by mass spectrometry after 15 min. There was also no trace of the starting complex [2]2+, confirming the selective and complete photosubstitution of mtmp upon light irradiation in deaerated CH3CN, to produce [Ru(dpp)(bpy)(MeCN)2]2+ as sole photosubstitution product. The photosubstitution quantum yield, measured by UV-vis spectroscopy was found to be 0.111 in these conditions (Fig. S7†).
For ruthenium polypyridyl compounds photosubstitution reactions compete with phosphorescence and 1O2 generation, which are typically poorly efficient for PACT compounds, while they can be very efficient for ruthenium-based PDT type II compounds.65–67 On the other hand, emission and 1O2 generation might also arise from minute impurities in samples of [2](PF6)2. Such emissive impurities may be generated because of ligand scrambling during the last synthetic step. Indeed, [Ru(dpp)2](bpy)]2+ was detected by HPLC as minute impurity; this species is a decent phosphor68 and probably excellent 1O2 generator.69 To study the intrinsic emission and 1O2 generation properties of [2](PF6)2, an HPLC-purified sample of the compound (Fig. S8a†) was prepared and its 1O2 generation quantum yield (hereafter noted ΦΔ) measured by direct spectroscopic detection of the 1270 nm emission of 1O2 in aerated CD3OD under blue light irradiation (450 nm), using [Ru(bpy)3]2+ as reference (ΦrefΔ = 0.73).69 A low ΦΔ value of 0.03 was found (Fig. S8c†), consistent with [2]2+ being a photosubstitutionally active compound, and comparable to that of [1]2+ (ΦΔ = 0.020 in the same conditions). Both values are much lower than that found for clinically used or tested PDT sensitizers such as Photofrin (0.90),70 Foscan (0.31),71 or TLD-1433 (1.0).72 Consistently, a weak red emission was also observed that was enhanced by a factor 3.2 upon degassing with Argon (Fig. SX2a†), demonstrating its phosphorescent nature. The difference between the absorption and excitation spectra of [2]2+, even after HPLC purification (Fig. S9b†), demonstrated that this emission was due not to [2]2+ itself, but to the presence of an emissive impurity. Time-resolved emission spectroscopy data were measured (in argon-saturated MeOH) to confirm this hypothesis (Fig. S10a and b†), which could be fitted by a bis-exponential decay. The major, slow component (∼660 ns, 76%) of the emission remained independent from prolonged irradiation of the sample, which was consistent with the presence of <2% of a photoinert, phosphorescent impurity, probably minutes traces of [Ru(dpp)2](bpy)]2+, the lifetime of which was reported to be 4.6 μs in O2-free (4 freeze-pump-thawed cycles) acetonitrile solution and 170 ns in aerated acetonitrile solution.68 The minor, faster component (3.1 ns, 24%) of the emission of [2](PF6)2 in MeOH is compatible with the hypothesis of a weak emission of the major species [Ru(dpp)(bpy)(mtmp)]2+ (>98% of the sample), as expected for substitutionally active ruthenium-based compounds. While in photostable compounds such as [Ru(dpp)2(bpy)]2+ the 3MLCT excited states generated photochemically give rise to phosphorescence emission and singlet oxygen generation, for photolabile compounds such as [2]2+ these 3MLCT states are quenched by low-lying metal-centred (3MC) triplet excited states that lead to non-radiative decay and photosubstitution.73–75 Overall, the low ΦΔ value for [2]2+ and a comparatively high photosubstitution quantum yield make of this compound a promising PACT agent, though small amounts (<5%) of a good photosensitizer for 1O2 generation in the samples, also suggested that a photodynamic effect may contribute to the photoactivity of the compound, too (see below).
2.2 Cytotoxicity in vitro and cellular uptake
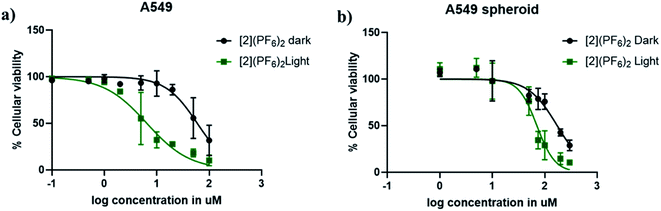
Considering the good photosubstitution properties of [2]2+, its cytotoxicity was first tested in the dark and upon green light activation in a human lung cancer cell line (A549) where the two known analogues [1]2+ and [3]2+ had already been evaluated.15 The protocol is detailed in Hopkins et al.76 The effective concentrations (EC50), defined as the compound concentration (in μM) that reduces cell viability by 50%, compared to untreated cells, are shown in Table 1. In the dark, the EC50 value was 59 μM for [2]2+, which is intermediate between that found, in the same conditions, for [1]2+ (3.4 μM) and for [3]2+ (>150 μM).15 After 15 minutes green light irradiation (520 nm, 21 mW cm−2, 19 J cm−2), the EC50 value decreased to 6.5 μM for [2]2+, respectively, which is also intermediate between the 0.62 μM and >150 found for [1]2+ and [3]2+, respectively (Table 1 and Fig. 4a). The corresponding photoindex (PI) value, defined as PI = EC50,dark/EC50,light, was twice higher for [2]2+ (9.1) than for [3]2+ (5.5), which suggested that the compound design was successful. Qualitatively, cytocytotoxicity is closely related to cellular uptake and subcellular localization, which are in turn closely related to the lipophilicity of the prodrug.77 Typically, the presence of more phenyl groups results in an increase of lipophilicity.78 The intermediate lipophilicity of [2]2+ obtained by balancing the number of dpp and bpy ligands, significantly decreased its dark cytotoxicity, compared to [1]2+, while keeping its cytotoxicity after light activation much higher than for [3]2+. To verify quantitatively that the lower dark cytotoxicity of [2]2+ was related to drug uptake, A549 cells were treated for 24 h with [1]2+ and [2]2+ at their EC50,dark concentrations (3.4 and 59 μM, respectively), after which the ruthenium content was measured using high-resolution continuum source atomic absorption spectrometry (HRCS-AAS, Table 2). In such conditions, the absolute cellular uptake of [2]2+ was found almost equal (2.11 nmol Ru per mg protein) to that of [1]2+ (2.12 nmol Ru per mg protein), although the concentration used for treatment was 20 times higher for [2]2+ than for [1]2+. Also, the concentration found inside the cells was 2 orders of magnitude higher (×99) for [1]2+, compared to the incubation concentration, while for [2]2+ the intracellular concentration was less than 1 order of magnitude higher (×5), compared to incubation concentration. Overall, the intracellular accumulation was ∼20 times lower for [2]2+ than for the more hydrophobic compound [1]2+, suggesting a lower contribution of passive uptake for the less hydrophobic molecule [2]2+. Similar experiments with [3]2+ had demonstrated that this compound was not taken up by A549 cells because of its too high hydrophilicity.15 Thus, the intermediate lipophilicity of [2]2+, i.e., between that of [1]2+ and [3]2+, allowed for moderating cellular uptake, which kept the dark cytotoxicity low while not jeopardizing the cytotoxicity after light activation. Such balanced lipophilicity also allowed [2]2+ to penetrate 3D multicellular tumour spheroids of the same cell line (A549). In such conditions, the activity of [2]2+ remained significantly improved upon light irradiation, with EC50 values in the dark and after light irradiation (520 nm, 21 mW cm−2, 19 J cm−2) of 173 and 70.9 μM, respectively (Fig. 4b). The higher EC50,light value compared to 2D cell monolayer models (70.9 vs. 6.5 μM), is often observed; it can be interpreted as a consequence on the one hand of the more difficult penetration of the compound in a 3D spheroid environment, compared to 2D, and on the other hand, to lower O2 concentrations at the core of large spheroids, which may trigger a hypoxic cellular response that is known to increase resistance to chemotherapy. Most importantly, the viability of the tumour spheroid was almost eradicated at 300 μM upon light irradiation, which highlights the excellent phototoxicity of this compound also in a 3D environment.| Cell line | Light dose (J cm−2) | [1]Cl2 | [2](PF6)2 | [3]Cl2 | Cisplatin | ||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC50 (μM) | 95% CI (μM) | PI | EC50 (μM) | 95% CI (μM) | PI | EC50 (μM) | 95% CI (μM) | PI | EC50 (μM) | 95% CI (μM) | PI | ||
| A549 | 0 | 3.4 | −0.76 | 59 | −13 | >150 | — | — | 3.0 | −0.35 | |||
| 0.97 | 17 | 0.38 | |||||||||||
| 19 | 0.62 | −0.11 | 5.5 | 6.5 | −1.8 | 9.1 | >150 | — | — | 4.3 | −0.54 | 1.4 | |
| 0.14 | 2.4 | 0.61 | |||||||||||
| [1]Cl2 | [2](PF6)2 | |
|---|---|---|
| Treatment concentration [μM] | 3.4 | 65 |
| Cellular uptake (nmol Ru per mg of cell protein) | 2.11 ± 0.12 | 2.12 ± 0.33 |
| Intracellular molar concentration [μM] (accumulation in cells) | 336 ± 19 (x 99) | 338 ± 53 (x 5) |
Encouraged by these results, the cytotoxicity of [2](PF6)2 was further assayed in a wider range of human cancer cell lines: PC3Pro4, a cancer cell line derived from a bone metastasis obtained after injection of PC3 human prostate cancer cells into nude mice;79 the conjunctival melanoma cell lines CRMM1, CRMM2, CM2005.1; and the uveal melanoma cell lines OMM1, OMM2.5 and MEL270 (Fig. S12†). The EC50 values obtained in the dark and after light activation are listed in Table 3. [2](PF6)2 exhibited lower cytotoxicity compared to the PDT compound TLD-1433, both in the dark and after light irradiation (Table 3). The resulting photoindex values were good to excellent, i.e., between 6.3 for OMM1 and >31 for PC3Pro4. Its lowest light EC50 value was observed in PC3Pro4 (3.2 μM) and CRMM1 cells (3.9 μM), while its highest dark EC50 value was obtained in CM2005.1 (184 μM). Overall, the photoreaction shown in Fig. 1 translates, in most cancer cell lines, into a strong light activation of the anticancer activity of [2](PF6)2.
| Cell line | Light dose (J cm−2) | [2](PF6)2a | TLD-1433b | ||||
|---|---|---|---|---|---|---|---|
| EC50 (μM) | 95% CI (μM) | PI | EC50 (μM) | 95% CI (μM) | PI | ||
| a This work. b Values taken from ref. 58. | |||||||
| PC3Pro4 | 0 | >100 | — | — | — | ||
| — | — | ||||||
| 19 | 3.2 | −0.54 | >31 | — | — | — | |
| +0.65 | — | ||||||
| CRMM1 | 0 | 33 | −4 | 0.84 | −0.23 | ||
| +4 | +0.27 | ||||||
| 19 | 3.9 | −0.6 | 8.5 | 0.0059 | −0.00099 | 140 | |
| +0.6 | +0.0012 | ||||||
| CRMM2 | 0 | 97 | −17 | 1.0 | −0.17 | ||
| +23 | +0.19 | ||||||
| 19 | 11 | −1.8 | 8.8 | 0.0048 | −0.00050 | 210 | |
| +1 | +0.00055 | ||||||
| CM2005.1 | 0 | 184 | −34 | 1.1 | −0.22 | ||
| +76 | +0.25 | ||||||
| 19 | 10 | −1.1 | 18 | 0.0058 | −0.00061 | 190 | |
| +2 | +0.00066 | ||||||
| OMM1 | 0 | 150 | −20 | 1.4 | −0.48 | ||
| 30 | +0.95 | ||||||
| 19 | 24 | −4.5 | 6.3 | 0.014 | −0.0016 | 100 | |
| 7.6 | +0.0019 | ||||||
| OMM2.5 | 0 | 100 | −8.4 | 0.64 | −0.16 | ||
| +9.2 | +0.19 | ||||||
| 19 | 14 | −1 | 7.1 | 0.013 | −0.0011 | 49 | |
| 1.2 | +0.0013 | ||||||
| MEL270 | 0 | 140 | −20 | 1.1 | −0.092 | ||
| 27 | +0.097 | ||||||
| 19 | 13 | −1.3 | 11 | 0.010 | −0.0012 | 110 | |
| 1.5 | +0.0013 | ||||||
2.3 Mechanistic investigations
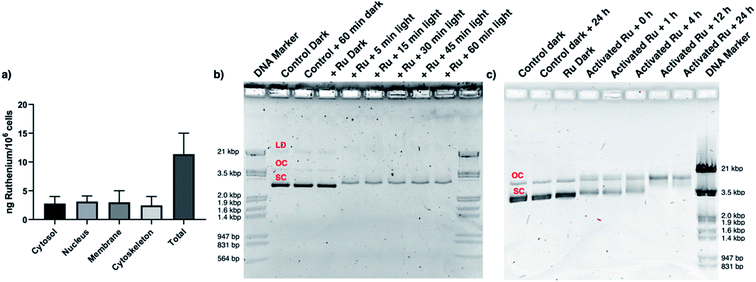
CRMM1 cells were selected to investigate the mechanism of the light-induced cytotoxicity. First, a reactive oxygen species (ROS) generation assay was performed using fluorescence-activated cell sorting (FACS) analysis to check whether either the non-emissive title compound [Ru(dpp)(bpy)(mtmp)]2+, or the emissive, 1O2-generating impurity [Ru(dpp)2(bpy)]2+, identified in low amounts (<5%) by HPLC (Fig. S8a†), would damage the cells by a significant photodynamic effect. The data (Fig. S11 and Table S1†) clearly show that with or without HPLC purification [2](PF6)2 did not generate significant ROS upon light irradiation, while the known green-light activated PDT agent Rose Bengal, used as positive control, did. Hence, the photoinduced cell-killing effect of [2](PF6)2 is not a PDT effect, but it must be related to the photosubstitution reaction triggered by light irradiation. Second, a cell fractionation experiment was performed to gather information on the intracellular localisation of [2]2+ in cells. CRMM1 cells were hence incubated with [2]2+ for 24 h in the dark at the EC50,dark concentration (33 μM). The cells were then harvested, the nuclei, membrane, cytosol and cytoskeleton fractions were separated using a commercial kit, and their ruthenium content was analysed by ICP-MS (Fig. 5a). The results confirmed that [2]2+ was well taken up by these cells (11 ng Ru/106 cells). In addition, Ru distributed evenly between the cytosol (2.8 ng Ru/106 cells), nucleus (3.1 ng Ru/106 cells), membrane (3.0 ng Ru/106 cells), and cytoskeleton (2.5 ng Ru/106 cells). Obviously, from this broad localization the number of possible cellular targets for [2]2+ is very high. However, by analogy with cisplatin, which at 3.3 μM shows 0.6 ng Pt/106 cells in the nucleus,80 and considering the two cis coordination position freed on the metal centre upon light irradiation of [2]2+, we hypothesized that the presence of sufficient amounts of ruthenium in the nucleus may justify a DNA binding and DNA damage study. We hence investigated the ability of this compound to bind to DNA and to do DNA damage. In a cell-free assay using a chloride-free phosphate buffer to model a pseudo intracellular environment, the pUC19 plasmid was incubated with [2](PF6)2 at a DNA base pair (BP) to metal complex (MC) ratio of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and irradiated with different doses of green light (0, 6.3, 19, 38, 57, or 76 J cm−2, Fig. 5b). The pUC19 plasmid exists in three forms: supercoiled (SC, most condensed form), single-nicked open circular (OC, relaxed form of the SC), and linear dimer (LD, see Fig. 5b).81 In the dark, [2](PF6)2 showed no affinity for the plasmid DNA, and no DNA damage was observed. This absence of interaction was also observed at higher concentrations of the ruthenium complex (from 4 to 80 μM, see Fig. S13†). After from as little as 5 min (6.3 J cm−2) to as much as 60 min (76 J cm−2) green light irradiation, the gel showed association of the plasmid with the metal complex, but no DNA cleavage, and this irrespective of the irradiation time (Fig. 5b). In a second experiment, the light dose was fixed at 19 J cm−2 (15 min), but the dark incubation following light activation was varied from 0 to 1, 4, 12, and 24 h (Fig. 5c). In such conditions, it appeared clearly that while 1 h after activation and binding to DNA, there was no significant cleavage of the plasmid, after 4 h some single-strand DNA cleavage was observed, as shown by the higher intensity of the OC form of the plasmid, while at 12 and 24 h the SC form had completely disappeared. According to these data, while in the dark [2]2+ neither binds to DNA nor generate DNA cleavage, after light activation this compound releases the mtmp protecting ligand, which allows for DNA binding to the metal centre to occur quickly (i.e., within 1 h). Strikingly, while with PDT compounds photoinduced 1O2 generation typically leads to light-induced DNA cleavage, with [2](PF6)2 no such light-induced DNA degradation occurs, even when 60 min light irradiation is used. On the other hand, DNA coordination to the activated ruthenium complex leads, after 4 to 12 h incubation in the dark, to significant amounts of single-strand DNA cleavage via a dark process that may explain, at least partly, the cytotoxicity observed in cancer cells following light activation.
1, and irradiated with different doses of green light (0, 6.3, 19, 38, 57, or 76 J cm−2, Fig. 5b). The pUC19 plasmid exists in three forms: supercoiled (SC, most condensed form), single-nicked open circular (OC, relaxed form of the SC), and linear dimer (LD, see Fig. 5b).81 In the dark, [2](PF6)2 showed no affinity for the plasmid DNA, and no DNA damage was observed. This absence of interaction was also observed at higher concentrations of the ruthenium complex (from 4 to 80 μM, see Fig. S13†). After from as little as 5 min (6.3 J cm−2) to as much as 60 min (76 J cm−2) green light irradiation, the gel showed association of the plasmid with the metal complex, but no DNA cleavage, and this irrespective of the irradiation time (Fig. 5b). In a second experiment, the light dose was fixed at 19 J cm−2 (15 min), but the dark incubation following light activation was varied from 0 to 1, 4, 12, and 24 h (Fig. 5c). In such conditions, it appeared clearly that while 1 h after activation and binding to DNA, there was no significant cleavage of the plasmid, after 4 h some single-strand DNA cleavage was observed, as shown by the higher intensity of the OC form of the plasmid, while at 12 and 24 h the SC form had completely disappeared. According to these data, while in the dark [2]2+ neither binds to DNA nor generate DNA cleavage, after light activation this compound releases the mtmp protecting ligand, which allows for DNA binding to the metal centre to occur quickly (i.e., within 1 h). Strikingly, while with PDT compounds photoinduced 1O2 generation typically leads to light-induced DNA cleavage, with [2](PF6)2 no such light-induced DNA degradation occurs, even when 60 min light irradiation is used. On the other hand, DNA coordination to the activated ruthenium complex leads, after 4 to 12 h incubation in the dark, to significant amounts of single-strand DNA cleavage via a dark process that may explain, at least partly, the cytotoxicity observed in cancer cells following light activation.
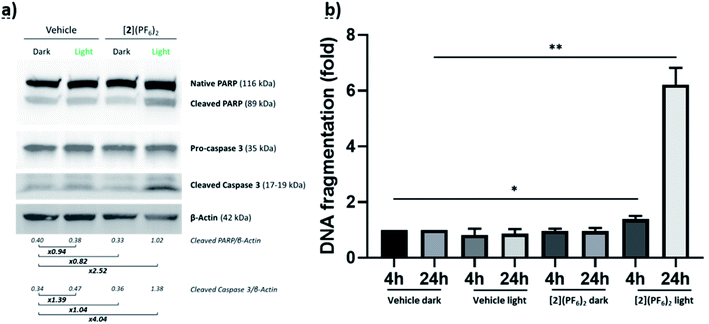
To check whether DNA damage could indeed occur in a cellular context and participate to killing the cell, two cellular assays were conducted. First, the apoptosis-associated proteins native PARP, cleaved PARP, Pro-Caspase-3, and cleaved caspase-3, were quantified by Western blot in CRMM1 cells treated with [2](PF6)2 at EC50,light (4 μM) or vehicle control, and either left in the dark or irradiated during 15 min with green light (520 nm, 19 J cm−2). 48 h after light irradiation, cleaved PARP and cleaved caspase-3 were expressed respectively around 2.5 and 4 times more in the group treated with [2](PF6)2 and irradiated by light compared to all other groups (Fig. 6a). This result suggested that apoptotic cell death occurred following light activation of the ruthenium prodrug. In a second step, an ELISA DNA fragmentation assay was used to confirm this hypothesis. CRMM1 cells were hence treated with [2](PF6)2 at EC50,light (4 μM) or vehicle control, and either left in the dark or exposed to the same dose of green light as in the previous assay. At 4 h or 24 h following light activation, the samples were collected and the Cell Death Detection ELISAPLUS assay was performed, which allowed visualizing histones released from the nucleus upon apoptosis. Both at 4 h and 24 h, the group treated with [2](PF6)2 and activated by light was the only group showing DNA fragmentation, and DNA fragmentation was more than 4 times higher at 24 h, compared to 4 h (Fig. 6b and S14†), which may be due both to DNA damage generated by the DNA-bound Ru complex (Fig. 5), and to the apoptosis process that might have started 24 h after light activation. Overall, these results suggest that nuclear DNA binding and DNA damage may explain at least partly the phototoxicity observed in vitro. The following picture can be drawn: although the full cell killing mechanism is unknown, the compound is notably present in the nucleus, where it cannot bind to DNA or provoke DNA damage in the dark. Following light activation, fast DNA binding occurs that does not immediately generate DNA damage. However, 4 h after activation and DNA binding DNA damage starts and shoots up 24 h after light activation. Such DNA damage most probably contributes to cell death, which at least partly, occurs via apoptosis.
2.4 Maximum tolerated dose of [2](PF6)2 in zebrafish ectopic and orthotopic cancer models
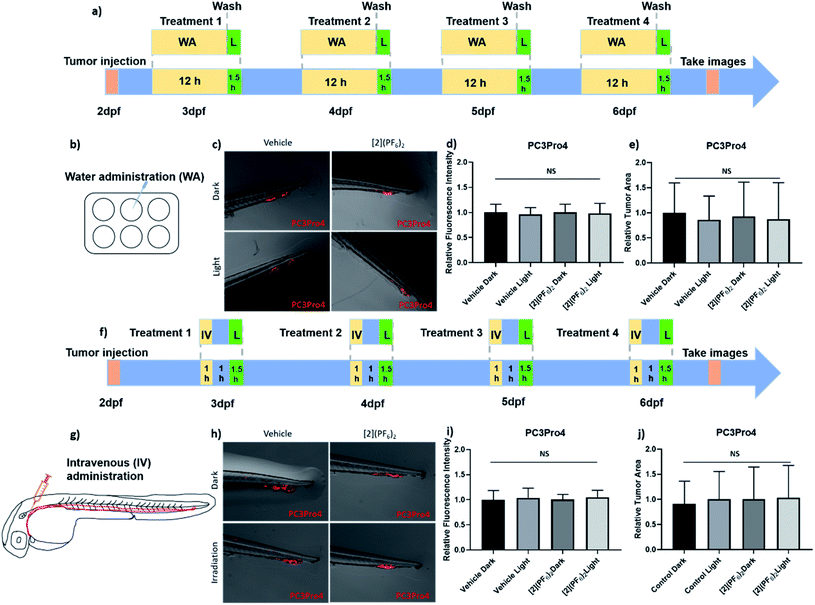
These promising in vitro results led us to test [2](PF6)2 in zebrafish tumour models with cell lines showing high (PC3Pro4) or intermediate (CRMM1, CRMM2) photoindices. Eye (CRMM1, CRMM2) zebrafish embryonic cancer models have recently been established by some of us.82,83 For prostate, androgen-independent osteotropic red-emitting PC3Pro4-mCherry cells were intravenously injected into reporter transgenic zebrafish line with green fluorescent vasculature (GFP) at 2 days post fertilization (dpf) (Fig. 7).84,85 Immediately after injection, cells haematogenously disseminated through the whole circulation. Most of the circulating cells regressed without extravasation or initiating tumour growth. However, within 1 day, some cells were able to extravasate exclusively at the posterior ventral end of caudal hematopoietic tissue (CHT), and invade into the tail fin where they developed perivascular metastatic lesions within 4 dpf (Fig. 7). CHT is an intermediate site of haematopoiesis during zebrafish embryogenesis and is the functional analogue of the foetal liver during mammalian development.86 Metastatic tumours grew around CHT at 6 dpf, as detected by red fluorescence (excitation: 587 nm, emission: 610 nm) that can be quantified, either in terms of emission intensity, or by the relative tumour area in microscopy images; both quantifications are referred below as “relative tumour burden”. This tumour model is called “ectopic” as the CHT site does not represent the organ of origin of these cancer cells. For conjunctive melanoma (CM), we used an orthotopic model recently developed in our group for PDT treatment.58,87 In short, the CM tumours were generated by injection of 200 CRMM1-mCherry or CRMM2-mCherry cells into the retro-orbital site of the embryo at 2 dpf (Fig. 8). From 2 to 6 dpf, the CRMM1 or CRMM2 cells formed local lesions at the injection site behind the eye. This tumour model is called “orthotopic” as the site for tumour growth, i.e. the tissue that surrounds the eye, does represent the area of origin of these cancer cells.In terms of drug treatment modalities, the embryos were subjected to three different protocols (Fig. 7 and 8). For the ectopic prostate cancer model, treatment with [2](PF6)2 was performed either by water administration (WA) or by intravenous injection (IV), while for the orthotopic eye cancer model, treatment was performed either by WA, IV, or retro-orbital (RO) injections. Before testing the anti-tumour efficacy, it was necessary to evaluate the toxicity of the treatment. The toxicity of green light alone (520 nm) was recently reported: at an intensity of 21 mW cm−2, the zebrafish embryos tolerate light irradiation until 6 h without any toxicity or visible developmental defects.58 The toxicity of [2](PF6)2 was then evaluated by measuring its MTD for the different administration modes, both for tumour-free embryos and tumour cell-injected embryos (Table 4 and Fig. S15†). For treatment via water administration, different concentrations (0, 0.1 0.25, 0.5, 1, 2 μM) of [2](PF6)2 were added to the egg water (i.e., the water in which the zebrafish embryo were swimming) at 2.5, 3.5, 4.5 and 5.5 dpf, and incubation was continued overnight for a drug-to-light interval of 12 h. At 3, 4, 5, 6 dpf, excess [2](PF6)2 was washed by drug-free water and the embryos were further irradiated with green light (21 mW cm−2, 90 min, 114 J cm−2, 520 nm). In such conditions, an MTD of 0.5 μM for embryos engrafted with PC-Pro4-mCherry tumours, and of 1 μM for tumour-cell free embryos, was obtained. For treatment via intravenous or retro-orbital administration, 1 nL with different concentrations (0, 50, 100, 200, 300, 500 μM) of [2](PF6)2 was injected into the dorsal vein or retro-orbital site of zebrafish at 3, 4, 5, 6 dpf. After a shorter drug-light interval of 1 h, the zebrafish embryos were irradiated with the same dose of green light (21 mW cm−2, 90 min, 114 J cm−2, 520 nm). The lethality, aberrant morphology and fish length were measured at 6 dpf. Zebrafish embryos tolerated, without any effect on mortality, malformation or fish length, injection of [2](PF6)2, followed by light activation, at a MTD of 200 μM for embryos engrafted with PC-Pro4-mCherry, CRMM1, CRMM2 cells, and of 300 μM for tumour-free embryos (Fig. S15†). These values were lower than that found for TLD-1433 (4.6 mM for intravenous or retro-orbital injection),58 showing the higher toxicity of [2](PF6)2 to zebrafish embryo, when injected, compared to TLD-1433. The MTD values of 0.5 μM (WA) and 1 nL of 200 μM (IV and RO) were further used for assessing the anti-tumour efficacy in the zebrafish tumour models.
| [2](PF6)2 | Maximum tolerated concentration | |
|---|---|---|
| Wild type embryos (μM) | Tumour cells engrafted embryos ectopic and orthotopic model (μM) | |
| a Note that for water administration the Ru compound is added in the large volume of water (200 μL) the ZF embryo swim in, while by IV or RO administration a small volume of solution (1 nL) at the indicated concentration is injected in each embryo. In terms of number of mol of Ru the MTD is hence much higher for WA than for IV or RO administration. b For CRMM1 and CRMM2 xenografts only. | ||
| Water administrationa | 1 | 0.5 |
| Intravenous administration | 300 | 200 |
| Retro-orbital administrationb | 300a | 200a |
2.5 Effect of [2](PF6)2 on PC3Pro4 tumour growth by water and intravenous administration in zebrafish ectopic prostate cancer model
In the PC3Pro4-mCherry zebrafish ectopic model, both WA (0.5 μM) and IV administration (1 nL, 200 μM) of [2](PF6)2 were tested using the previously determined MTD. At 6 dpf, images of the PC3Pro4-mCherry tumours were taken using a stereo microscope. Quantification of the relative tumour burden was performed by measuring either the relative fluorescence intensity or the relative tumour area (Fig. 7). Using a 12 h (WA) or 1 h (IV) drug-to-light interval, green light activation (21 mW cm−2, 90 min, 114 J cm−2, 520 nm) did not change the tumour burden, compared to the dark groups, even when the treatment on each embryo was repeated 4 times (Fig. 7). Usually, WA in zebrafish is acknowledged to mimic the oral route in human patients. Indeed, the compound will first go into the enterohepatic circulation and then disseminate through the blood circulation. The fact that no anti-tumour activity was observed for [2](PF6)2 administered by WA in the prostate cancer zebrafish model, while it showed excellent activity in PC3Pro4 cell monolayers in vitro (Table 3), suggested that in the embryo, the compound may simply not be taken up into the blood circulation. To omit this possible problem we delivered the compound into blood circulation via IV injections, but this treatment had no effect either. The PC3Pro4 are very invasive cells and they extravasated from circulation within 1 day to formed perivascular metastatic lesions in the tail. Presumably the IV delivered drug was not able to reach these metastatic lesions in the sufficiently high concentration to attenuate tumour expansion after irradiation. Alternatively, engrafted prostate cancer cells might have gained chemotherapy resistance in vivo, which they did not have in vitro.88 Overall, these results most probably suggest that more specific targeting strategies would be needed to achieve proper efficacy of this compound in an ectopic prostate cancer model.2.6 Effect of [2](PF6)2 on CRMM1 and CRMM2 tumour growth by retro-orbital administration in the zebrafish orthotopic conjunctival melanoma model
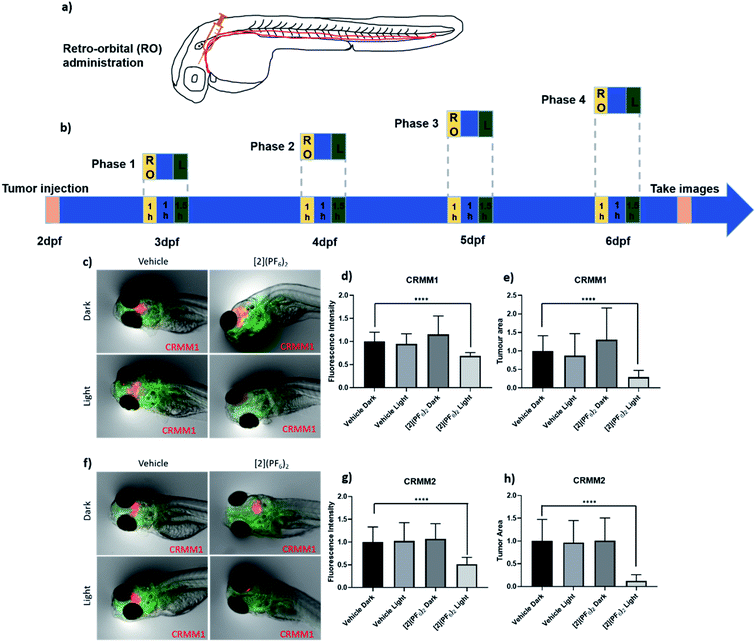
When both the tumour cells and the prodrug are injected into the general blood circulation of the embryo, it should not be taken for granted that the drug properly biodistributes to reach the inside of a tumour at a sufficiently high concentration. One way to address this issue is to use a model where the prodrug is injected near the tumour. The efficacy of [2](PF6)2 was hence examined in the orthotopic model of conjunctival melanoma (CM) described above and in ref. 58. In this model, the tumour develops in tissues surrounding the eye, near the location of the cancer cell injection, and the prodrug is also injected at the same place. A shorter drug-to-light interval (DLI) was hence used (1 h) to avoid prodrug diffusion away from the tumour prior to light activation. In a sense, this model better mimics local PDT treatments performed in human cancer patients. Following our treatment strategy developed for the PDT sensitizer TLD-1433,58 the MTD of [2](PF6)2 (1 nL, 200 μM) was injected retro-orbitally at 3, 4, 5, 6 dpf. After 1 h DLI, the embryos in both light-irradiated groups (vehicle, [2](PF6)2) were irradiated with green light (520 nm, 90 min, 21 mW cm−2, 114 J cm−2), while the two dark groups (vehicle, [2](PF6)2) were kept in the dark. During the experiment, the egg water of engrafted embryos was refreshed before injection and after irradiation. At 6 dpf and 4 consecutive treatments, quantification of the CRMM1 and CRMM2 relative tumour burden was performed by measuring either the relative fluorescence intensity or the relative tumour area using a stereo microscope (Fig. 8 and Tables 5 and 6). In the group treated with [2](PF6)2 and green light (21 mW cm−2, 60 min, 114 J cm−2, 520 nm), the CRMM1 tumour burden was significantly inhibited by 31% (fluorescent intensity) and 71% (tumour area) compared with the dark untreated group, while the CRMM2 tumour burden was inhibited by 49% (fluorescence intensity) and 88% (tumour area), compared with the dark untreated group. These in vivo results were strikingly comparable to that obtained for TLD-1433 at a 1 nL injected volume of 2.3 mM,58 although the EC50 values measured in vitro were significantly higher for [2](PF6)2 than for TLD-1433, both in the dark and under light irradiation, and the photoindexes measured in vitro for TLD-1433 were significantly higher than for [2](PF6)2. This comparison shows that absolute EC50 values in vitro are difficult to translate in antitumor properties in an animal model. Meanwhile, when comparing the excellent results of [2](PF6)2 in an orthotopic CM model with the absence of efficacy of the same compound in the ectopic model for prostate cancer, we envision that local RO administration of [2](PF6)2 generates a higher concentration of the prodrug in the proximity of the tumour, and therefore that green light activation generates sufficient amounts of the activated ruthenium molecules, to attenuate localized CM development in the light-irradiated group (Fig. 8c–h). These results represent the first experimental demonstration that ruthenium-based PACT treatment can inhibit CM growth in an animal tumour model. They also suggest that compound [2](PF6)2 should be further investigated in pre-clinical rodent models.| Route of [2](PF6)2 administration | Relative tumour burden as measured by fluorescence intensity | ||||||
|---|---|---|---|---|---|---|---|
| Ectopic model | Orthotopic model | ||||||
| Light dose (J cm−2) | PI | Light dose (J cm−2) | PI | ||||
| 0 | 19 | 0 | 19 | ||||
| PC3Pro4 | Water | 100% | 98% | 1.0 | |||
| Intravenous | 101% | 105% | 1.0 | ||||
| CRMM1 | Retro-orbital | 116% | 69% | 1.7 | |||
| CRMM2 | Retro-orbital | 107% | 51% | 2.1 | |||
| Route of [2](PF6)2 administration | Relative tumour burden as measured by tumour area | ||||||
|---|---|---|---|---|---|---|---|
| Ectopic model | Orthotopic model | ||||||
| Light dose (J cm−2) | PI | Light dose (J cm−2) | PI | ||||
| 0 | 19 | 0 | 19 | ||||
| PC3Pro4 | Water | 93% | 87% | 1.1 | |||
| Intravenous | 100% | 103% | 1.0 | ||||
| CRMM1 | Retro-orbital | 130% | 29% | 4.5 | |||
| CRMM2 | Retro-orbital | 101% | 12% | 8.4 | |||
2.7 [2](PF6)2 induces CRMM1 cell apoptosis in the zebrafish orthotopic model
To monitor whether the observed inhibition of CM growth in the zebrafish orthotopic model by [2](PF6)2 was occurring via apoptosis, an in situ TUNEL assay was conducted on fixed embryos bearing CRMM1 tumours at 4 dpi (days post injection), which were either kept in the dark or irradiated with green light (520 nm, 90 min, 21 mW cm−2, 114 J cm−2), and treated by RO injection at the MTD (1 nL, 200 μM) either with vehicle control or [2](PF6)2 (Fig. 8a and 9). In the TUNNEL assay, the DNA strand breaks in apoptotic tumour cells were stained with fluorescein and visualized as a green signal in microscopy images. In the dark vehicle group, light vehicle group, and group treated with [2](PF6)2 but not irradiated, no positive green signal was detected (Fig. 9a). Only in the group treated with [2](PF6)2 and irradiated with green light (520 nm, 21 mW cm−2, 90 min, 114 J cm−2), a significant number of cancer cells (Fig. 9b) stained positive for apoptotic signal and turned green, which co-localized with red signal of CRMM1 cells (yellow in overlay, Fig. 9a). The magnitude of this effect is comparable to that reported in neomycin-treated (125 μM) embryos.89 This result indicated that the anti-tumour efficacy of [2](PF6)2 in this PACT regime was at least partially apoptosis-dependent, which fitted well with the western blot analysis in vitro. It should also be noted that there was no apoptotic signal detected in the tissue surrounding the tumours, pointing out that light activated [2](PF6)2 attacked CM tumours but not healthy tissues, which is essential for minimizing side effects.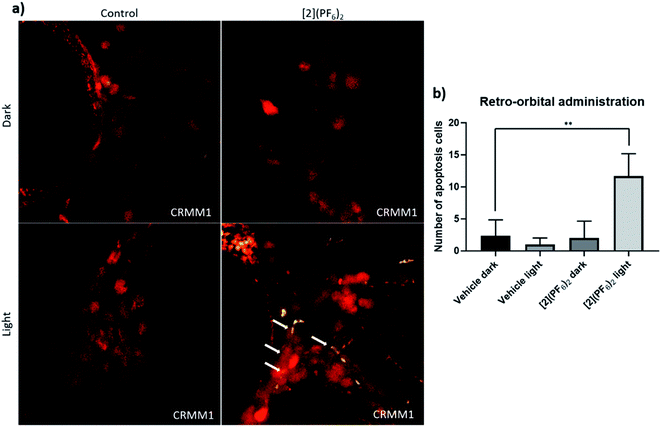 | ||
| Fig. 9 TUNEL assay in the CRMM1 orthotopic tumour model after RO injection of [2](PF6)2. (a) Red fluorescent CRMM1 cells were injected behind the eye of the embryo at 2 dpf, and the embryos were divided into four groups for drug treatment. RO administration of vehicle control and [2](PF6)2 was performed as described in Fig. 8. After dark or light exposure, embryos were fixed and TUNEL staining was performed. (a) Representative overlay images of embryos are shown. In the group treated with [2](PF6)2 and light, nuclear DNA fragmentation in nucleases is detected by co-localization of green (DNA fragments) and red (CM tumour cell) signal, depicted on the overlay as yellow signal marked by white arrows. In the dark control group, light control group, and group treated with [2](PF6)2 and left in the dark, there were no positive green apoptotic tumour cells. The background green signal in the [2](PF6)2 light groups did not co-localize with cytosolic red signal, which is diminished in degraded cells and TUNEL stains only the DNA breaks in these CM apoptotic cells. (b) Quantification of the number of apoptotic tumour cells (yellow dots). Experiment was performed 3 times with a group size of 10 embryos. **P < 0.01. | ||
3. Discussion and conclusions
The new tris-heteroleptic ruthenium-based PACT prodrug [2](PF6)2 is characterized by a well-balanced hydrophobicity in the dark, which allows it to be taken up efficiently in vitro. In 2D and 3D in vitro models the toxicity of this chemical is dramatically enhanced by green light activation, which triggers photosubstitution of the non-toxic mtmp ligand and liberates a ruthenium-based cytotoxic photoproduct. This activated photoproduct can bind to many biomolecules, including nuclear DNA, which leads to apoptotic cell death within 24 to 48 h. As [2](PF6)2 has not been designed for specific targets in tumour cells, it showed a broad range of activity in unrelated cancer cell lines in vitro (i.e., from the lungs, prostate, or eyes). On the one hand, such general phototoxicity may be seen as a potential source of side-effects. On the other hand, it ensures that single mutations in cancer cells would not quench the cytotoxic activity of the light-activated compound, while tumour selectivity will be controlled, in a larger animal, by local light irradiation of the tumour only. In addition, despite no targeting was included in the compound design, differences in EC50 in the dark and after light activation still existed between different cell lines in vitro. In a simplistic approach, one could have predicted that the in vivo activity would qualitatively follow in vitro photoindexes, so that photoindex may bear some predictive value for the in vivo antitumour efficacy of this compound. However, our study clearly demonstrated that tumours from the cell line in which the photoindex in vitro was the highest (PI > 31), i.e., PC3Pro4, were not killed by light-activated [2](PF6)2 in a zebra fish embryo ectopic model, while tumours from cell lines that showed good but more modest photoindexes in vitro (PI ∼ 9), i.e., CRMM1 or CRMM2, were killed efficiently in a zebra fish embryo orthotopic model. These results show the critical role of the tumour model in vivo, and the complicated translation from cell-growth inhibiting EC50 values in vitro to antitumor efficacy in vivo. In an animal model, tumour uptake of a prodrug follows intricate routes compared to in vitro conditions, and a compound that enters cells easily and is very efficient in killing them in 2D cancer cell monolayers, may in an animal never reach the tumour at concentrations that are high enough for generating an antitumor effect. In other words, phototherapy will not work in an animal if the tumour model and/or the way to treat the animal with the light-activated prodrug is inappropriate.In conclusion, we demonstrated for the first time the efficacy of a ruthenium-based PACT prodrug in a conjunctival melanoma zebrafish embryo xenografts.90 Our results also provided the first MTD values of a photosubstitutionally active ruthenium compound, which will serve as an important guideline for future in vivo studies of this family of compounds. Finally, our results highlight the difference between ectopic and orthotopic in vivo models, in particular for photoactivated drugs where one must wait between compound treatment and light activation. While the photoindexes in vitro were higher in prostate cancer cells (PC3Pro4) than in conjunctival melanoma cells (CRMM1, CRMM2), in vivo there was no activity in the ectopic model of prostate cancer, while activity was excellent in the orthotopic model of conjunctival melanoma. Such a difference underscores the interaction between the type of tumour model and the mode of compound administration in tumour xenografts, which cannot be modelled in vitro but dramatically influence both (pro)drug biodistribution, drug uptake by the tumour, and hence the final anti-tumour efficacy of the treatment. Overall, the present validation of the anti-tumour efficacy of retro-orbitally administered ruthenium compound [2](PF6)2 in zebrafish conjunctival melanoma orthotopic models suggests that further pre-clinical development of this new PACT drug should be considered in larger models (rodents) for conjunctival melanoma, where light irradiation can be localized, i.e., limited to the tumour.
4. Method and materials
4.1 Synthesis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 at m/z 400 (mass range m/z = 150–2000) and dioctyphtalate (m/z = 391.28428) as a lock mass. All 1H NMR spectra were recorded on a Bruker DMX-400 spectrometers. Chemical shifts are indicated in ppm relative to the residual solvent peak.
000 at m/z 400 (mass range m/z = 150–2000) and dioctyphtalate (m/z = 391.28428) as a lock mass. All 1H NMR spectra were recorded on a Bruker DMX-400 spectrometers. Chemical shifts are indicated in ppm relative to the residual solvent peak.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5. Three fractions were obtained from a long orange band (Rf ∼ 0.5), from which only the last fraction contained a pure isomer (3.2 mg, 1.5%) (isomer B, [2b](PF6)2). A mixture of isomers A/B in a ratio 0.23
5. Three fractions were obtained from a long orange band (Rf ∼ 0.5), from which only the last fraction contained a pure isomer (3.2 mg, 1.5%) (isomer B, [2b](PF6)2). A mixture of isomers A/B in a ratio 0.23![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 has been used for photochemical analysis and biological testing, further referred to as [2](PF6)2 (60 mg, 28%). 1H NMR (400 MHz, CD3CN) δ 9.63 (d, J = 5.5 Hz, 1HB), 9.39 (d, J = 5.7 Hz, 1HA), 8.61 (d, J = 8.2 Hz, 1HB), 8.58–8.51 (m, 2HA), 8.43 (d, J = 8.1 Hz, 1HB), 8.31 (dd, J = 8.0, 1.5 Hz, 1HB), 8.29–8.23 (m, 1HB + 1HA), 8.22–8.14 (m, 2HB + 2HA), 8.14–8.03 (m, 3HA), 8.02 (d, J = 5.5 Hz, 1HB), 7.99 (d, J = 5.5 Hz, 1HB), 7.93 (ddd, J = 7.8, 6.5, 1.5 Hz, 1HB), 7.86 (td, J = 7.8, 1.6 Hz, 1HA), 7.81–7.51 (m, 15HB + 15HA), 7.48 (dd, J = 5.9, 1.5 Hz, 1HA), 7.32 (ddd, J = 7.1, 5.6, 1.3 Hz, 1HA), 7.24 (d, J = 5.5 Hz, 1HB), 7.17 (td, J = 7.2, 5.6, 1.4 Hz, 1HB + 1HA), 6.98 (ddd, J = 7.7, 5.8, 1.6 Hz, 1HB), 4.82 (d, J = 16.5 Hz, 1HB), 4.74 (d, J = 16.7 Hz, 1HA), 4.28 (dd, J = 16.6, 4.8 Hz, 1HB + 1HA), 1.59 (s, 3HB), 1.32 (s, 3HA). 13C NMR (101 MHz, CD3CN) δ 162.96, 162.63, 158.56, 157.73, 153.51, 153.33, 153.09, 152.90, 151.98, 150.77, 150.61, 150.05, 149.66, 148.72, 139.46, 138.67, 138.55, 136.62, 136.53, 130.79, 130.70, 130.13, 130.06, 129.20, 127.78, 127.26, 127.19, 127.12, 126.92, 125.86, 125.59, 125.55, 124.93, 45.36, 17.04. HR-MS m/z (calcd): 364.5747 (364.5745, [2]2+), 728.1432 (728.1422, [2-H]+), 874.1107 (874.1142, {[2]PF6}+). Anal. calcd for C41H33F12N5P2RuS: C, 48.34; H, 3.26; N, 6.87 found: C, 48.21; H, 3.41; N, 6.82.
1 has been used for photochemical analysis and biological testing, further referred to as [2](PF6)2 (60 mg, 28%). 1H NMR (400 MHz, CD3CN) δ 9.63 (d, J = 5.5 Hz, 1HB), 9.39 (d, J = 5.7 Hz, 1HA), 8.61 (d, J = 8.2 Hz, 1HB), 8.58–8.51 (m, 2HA), 8.43 (d, J = 8.1 Hz, 1HB), 8.31 (dd, J = 8.0, 1.5 Hz, 1HB), 8.29–8.23 (m, 1HB + 1HA), 8.22–8.14 (m, 2HB + 2HA), 8.14–8.03 (m, 3HA), 8.02 (d, J = 5.5 Hz, 1HB), 7.99 (d, J = 5.5 Hz, 1HB), 7.93 (ddd, J = 7.8, 6.5, 1.5 Hz, 1HB), 7.86 (td, J = 7.8, 1.6 Hz, 1HA), 7.81–7.51 (m, 15HB + 15HA), 7.48 (dd, J = 5.9, 1.5 Hz, 1HA), 7.32 (ddd, J = 7.1, 5.6, 1.3 Hz, 1HA), 7.24 (d, J = 5.5 Hz, 1HB), 7.17 (td, J = 7.2, 5.6, 1.4 Hz, 1HB + 1HA), 6.98 (ddd, J = 7.7, 5.8, 1.6 Hz, 1HB), 4.82 (d, J = 16.5 Hz, 1HB), 4.74 (d, J = 16.7 Hz, 1HA), 4.28 (dd, J = 16.6, 4.8 Hz, 1HB + 1HA), 1.59 (s, 3HB), 1.32 (s, 3HA). 13C NMR (101 MHz, CD3CN) δ 162.96, 162.63, 158.56, 157.73, 153.51, 153.33, 153.09, 152.90, 151.98, 150.77, 150.61, 150.05, 149.66, 148.72, 139.46, 138.67, 138.55, 136.62, 136.53, 130.79, 130.70, 130.13, 130.06, 129.20, 127.78, 127.26, 127.19, 127.12, 126.92, 125.86, 125.59, 125.55, 124.93, 45.36, 17.04. HR-MS m/z (calcd): 364.5747 (364.5745, [2]2+), 728.1432 (728.1422, [2-H]+), 874.1107 (874.1142, {[2]PF6}+). Anal. calcd for C41H33F12N5P2RuS: C, 48.34; H, 3.26; N, 6.87 found: C, 48.21; H, 3.41; N, 6.82.
4.2 Photochemistry and photophysics: determination of the photosubstitution and singlet oxygen generation quantum yields, and time-resolved emission
When monitoring photoreactions with UV-vis and mass spectrometry, a Cary 50 Varian spectrometer equipped with temperature control set to 298 K and a LED light source (λex = 521 nm, with a full width at half maximum of 33 nm) with a photon flux of 6.21 × 10−8 mol s−1 was used. The irradiation experiments were performed in a 1 cm optical pathlength quartz cuvette containing 3 mL of solution. A stock solution of the desired complex was prepared in CH3CN, which was then diluted in the cuvette to a working solution concentration (36 μM). The sample was deaerated 15 min by gentle bubbling of dinitrogen and the atmosphere was kept inert during the experiment by a gentle flow of dinitrogen on top of the cuvette. A UV-vis spectrum was measured every 30 s for the first 10 min, every 1 min for the next 10 min, and eventually every 10 min until the end of the experiment. Data were analysed with Microsoft Excel and Glotaran as follows: upon light irradiation, a complex RuL converts into a complex RuY by photosubstitution of a ligand (L) by a solvent molecule (Y). Considering that both metal complexes are thermally stable, the quantum yield of the photosubstition reaction ΦPR can be calculated by monitoring the photoreaction with UV-vis spectroscopy. As explained in detail by Bahreman and Bonnet,91 when the irradiation is performed at a wavelength that is not an isosbestic point, the ΦPR can be obtained from the slope of a plot of the number of mol of RuL (nRuL) vs. the total number of mol of photons absorbed by RuL from t0 till ti (Qi). Qi is calculated according to eqn (1):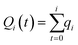 | (1) |
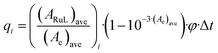 | (2) |
The value of (ARuL)ave, and by extension nRuL, was calculated by modelling the evolution of the UV-vis spectra vs. time using the Glotaran software. We fitted hence the time-dependent evolution of the UV-vis spectroscopy data to a kinetic model based on first-order laws, obtaining two output data sets that can be used for the calculation of ΦPR. The first dataset is a collection of globally fitted absorption spectra of the starting complex and the photoproduct, which makes possible the calculation of the molar extinction coefficient of all the species from that of the starting reagent (Fig. S7a†). The second dataset is the modelled evolution of the relative fractions of the two ruthenium species vs. irradiation time, here as well according to global fitting (Fig. S7b†). From the time evolution of these fractions and the molar absorption coefficient of all species, the time evolution of nRuL can be calculated, as well as Qi. The slope of the plot of nRuLvs. Qi (Fig. S7c†) gives the quantum yield of the reaction.
Singlet oxygen quantum yield measurements were performed by direct spectroscopic detection of the 1275 nm emission, as described by Meijer et al.41
Steady-state absorption spectra (Fig. S9†) were measured with a Shimadzu UV-2700 spectrometer. For photoluminescence measurements (Fig. S9,† right angle configuration) a Spex Fluorolog 3 was used. Time-resolved photoluminescence (Fig. S10†) was recorded using a PI-Max3 time-gated CCD detector. The sample was excited with laser pulses at 440 nm from an Ekspla NT342B laser system. The time delay between laser pulse and detector gate (width 2.9 ns) was incremented in 2 ns steps using a digital delay generator (DG645, Stanford Research Systems, Inc.). The scattered laser light at 440 nm was detected and used to determine the instrument response function (IRF; Gaussian shape, 3.1 ns full width half maximum). The intensity integrated over the emission band was plotted vs. time and fitted to the convolution of the Gaussian IRF and a biexponential decay in which the long-time component due to the impurity was, after separate measurement in a 4 μs time window, fixed to 660 ns.
4.3 Attached cell culture
Human conjunctival malignant melanoma cell lines CRMM1 and CRMM2, isolated by Nareyeck et al.,92 were cultured in F12 Kaighn's modified medium (Hyclone, cat# SH30526.01) supplemented with 10% fetal bovine serum (FBS; Gibco). CM2005.1 established by Keijser et al.93 was cultured in RPMI 1640, Dutch Modified (Life Technologies, cat# 22409-015), supplemented with 10% fetal bovine serum (FBS; Gibco), 3 mM L-glutamine (1%, Life Technologies cat# 35050-038). Human uveal melanoma cell lines OMM1 (provided by Prof. Dr G. P. M. Luyten),94 OMM2.5, and MEL270 (provided by Dr B. R. Ksander)95 were cultured in Ham's F12 medium (Sigma-Aldrich, cat# N3790) supplemented with 10% FCS. Stable fluorescent CRMM1 and CRMM2 cell lines were generated using lentivirus expressing both tandem dimer (td) tomato and blasticidin-S, as previously described.96 PC3Pro4 (provided by Dr Gabriel van der Pluijm) was cultured in DMEM (Sigma-Aldrich, cat# 32160801) supplemented with 10% FCS. Human lung carcinoma A549 was distributed by the European Collection of Cell Cultures (ECACC), and purchased from Sigma Aldrich, cultured in DMEM (Sigma-Aldrich, cat# 32160801) supplemented with 10% FCS. Cells were cultured in either 25 cm2 or 75 cm2 flasks and split at 70–80% confluence. The flasks were incubated in a normoxic incubator at 37 °C at 5.0% CO2 in a PHCbi O2/CO2 incubator, MCO-170 M). The medium was refreshed twice a week. Cells used in all biological experiments were cultured for not more than eight weeks. Trypsin and Opti-MEM® (without phenol red) were purchased from Gibco® Life Technologies. Trypan blue (0.4% in 0.81% sodium chloride and 0.06% potassium phosphate dibasic solution) was purchased from Bio-Rad. Plastic disposable flasks and 96-well plates were obtained from Sarstedt. Cells were counted using a Bio-Rad TC10 automated cell counter with Bio-Rad Cell Counting Slides.4.4 Spheroids cell culture and CellTiter-Glo 3D cell viability assay
1 mM stock solutions of [2](PF6)2 were prepared in OptiMEM; sterilized dimethylsulfoxide (DMSO) was used to dissolve [2](PF6)2 prior to medium addition. DMSO was added in such amounts that the maximum v/v% of DMSO did not exceed 0.5% even at the highest Ru concentration used (150 μM). A549 (500 cells per well) within 100 μL OptiMEM (Gibco® Life Technologies, cat# 11058021) were seeded in the low-attachment 96 well plates (Corning Spheroid microplate 4515) and incubated in normoxia (21% O2). After 24 h, 100 μL per well of diluted [2](PF6)2 with six different concentrations in OptiMEM or OptiMEM for control was added into each well and the cells were further incubated for another 24 h (drug-to-light interval). 100 μL of medium was pipetted out from each well and 100 μL per well of new OptiMEM was added. Then, the plates for [2](PF6)2 treatment with light activation and vehicle with light activation groups were irradiated with green light (21 mW cm−2, 15 min, 19 J cm−2, 520 nm) and the plates for [2](PF6)2 treatment with no light activation and vehicle with no light activation groups were put in the dark box. After treatment, all plates were put back in the incubator for 48 h. Before the CellTiter-Glo 3D cell viability assay, the plates were taken out from the incubator and left out for 20 min to reach the room temperature. 100 μL medium was taken out from each plate and 100 μL of CellTiter Glo 3D was added per well. The plates were put on the shaker for 5 min and left the plates at room temperature without shaking for 25 min. The luminescence of the plates was read by Tecan reader.4.5 Cellular uptake
Cell uptake studies for complexes [1]Cl2 and [2](PF6)2 were conducted on A549 lung cancer cells. 1.6 × 106 cells were seeded at t = 0 h in Opti-MEM complete (10 mL) in a 75 cm2 T-flask. At t = 24 h the media was aspirated and cells were treated with solutions of [1]Cl2, [2](PF6)2 to give a final concentration at the EC50 in the dark (3.4 and 65 μM, respectively) in a total volume of 10 mL. After 24 h of drug incubation at 37 °C and 21% O2, the medium was aspirated and the cells were washed twice with PBS (5 mL). Then, the cells were trypsinized (2 mL), suspended with Opti-MEM (8 mL), and centrifuged (1200 rpm, 4 min). After aspiration of the supernatant, the cells were re suspended in PBS (1 mL) and counted. After a second centrifugation, the supernatant was discarded. For metal and protein quantification, the pellets were resuspended in demineralized water (250 μL) and lysed for 30 min by ultrasonication. The protein content of lysates was determined by the Bradford method, and the ruthenium content was determined by High Resolution Continuum Source – Atomic Absorption Spectroscopy.A contrAA 700 high-resolution continuum-source atomic absorption spectrometer (Analytik Jena AG) was used. Pure samples of the respective complex were used as standard and calibration was done in a matrix-matched manner (meaning all samples and standards were adjusted to the same cellular protein concentration of 1.0 mg mL−1 by dilution with distilled water if necessary). Triton-X 100 (1%, 10 μL) as well as nitric acid (13%, 10 μL), were added to each standard sample (100 μL). Samples were injected (25 μL) into coated standard graphite tubes (Analytik Jena AG) and thermally processed as previously described by Appold et al.97 Drying steps were adjusted and the atomization temperature set to 2400 °C. Ruthenium was quantified at a wavelength of 349.8945 nm. The mean integrated absorbances of double injections were used throughout the measurements. Cell diameters were determined by two different published methods: inverted microscopy and transmission electron microscopy (TEM).98 For these calculations the average of both diameters was used and the intracellular molar concentrations were then calculated according to Ott et al.99 The data from two independent biological replications were used to obtain the uptake values shown in Table 2.
4.6 Cell irradiation setup
The cell irradiation system consisted of a Ditabis thermostat (980923001) fitted with two flat-bottomed microplate thermoblocks (800010600) and a 96-LED array fitted to a standard 96-well plate. The λ = 520 nm LED (OVL3324), fans (40 mm, 24 V DC, 9714839), and power supply (EA PS 2042-06B) were obtained from Farnell as reported in our previous publication.1004.7 Cytotoxicity assay
At day 0, cells were detached using 1 mL of trypsin, resuspended in 4 mL of media and transferred to a 15 mL corning falcon tube. Cells were counted using trypan blue and BioRad® TC20™ automated cell counter. Dilutions of 6000 (CRMM1), 6000 (CRMM2), 8000 (CM2005.1), 6000 (OMM1), 6000 (OMM2.5), 6000 (MEL270) 6000 (A549), and 6000 (PC3Pro4) cells per well were calculated from each cell suspension at a final volume of 6 mL. The cell suspensions were transferred to a 50 mL reservoir and 100 μL of each cell line was seeded at the aforementioned cell densities in triplicate in six 96-well plates. Boarder wells were intentionally filled with PBS media to avoid boarder effects. 1 mM stock solutions of [1]Cl2, [2](PF6)2, or [3]Cl2, were prepared in OptiMEM; sterilized dimethylsulfoxide (DMSO) was used to dissolve [1]Cl2 and [2](PF6)2 prior to medium addition. DMSO was added in such amounts that the maximum v/v% of DMSO did not exceed 0.5% even at the highest Ru concentration used (150 μM). After 24 h, the cells were treated with [2](PF6)2 with six different concentrations. After 24 h of post treatment the cells were exposed to the green light for 15 min (21 mW cm−2, 19 J cm−2, 520 nm). The dark control plate was kept under dark conditions. Then cells were incubated for another 48 h before fixing them with trichloroacetic acid (10% w/w) solution. The fixed cells were kept at 4 °C for 48 h, when TCA was washed out with distilled water before adding the sulphorhodamin B (SRB) (0.6% SRB) dye. The SRB dye was washed out after 30 minutes and plates were air dried overnight. Next day, the dye was dissolved using tri-base (0.25%) and absorbance of SRB at 510 nm was recorded from each well using a Tecan plate reader. The SRB absorbance data was used to calculate the fraction of viable cells in each well (Excel and GraphPad Prism software). The absorbance data were averaged from triplicate wells per concentration. Relative cell viabilities were calculated by dividing the average absorbance of the treated wells by the average absorbance of the untreated wells. Three independent biological replicates were completed for each cell line (three different passage numbers per cell line). The average cell viability of the three biological replicates was plotted versus log(concentration) [μM], with the SD error of each point. By using the dose–response curve for each cell line under dark- and irradiated conditions, the effective concentration (EC50) was calculated by fitting the curves to a non-linear regression function with a fixed maximum (100%) and minimum (0%) (relative cell viability) and a variable Hill slope.584.8 Reactive oxygen species analysis by FACS
Here a 2 mM stock solution of [2](PF6)2 was prepared in 5% DMSO and 95% medium. The generation of ROS (reactive oxygen species) in CRMM1 cell line was measured using the Cellular ROS Assay Kit (Deep Red) ab186029 (Abcam). The ROS Deep Red dye is cell-permeable and generates Deep Red fluorescence when it reacts with ROS. CRMM1 cells (1.75 × 105) were seeded into 6-well plates and were grown for 24 h in the dark under normoxia (37 °C, 5% CO2) or hypoxia (37 °C, 5% CO2, 1% O2) conditions. The cells were then treated or not with 5 μM of [2](PF6)2 compounds or 5 μM of Rose Bengal and labeled as dark or green light groups (at such concentration there was 0.25% of DMSO in each well). After 24 h of incubation the media were refreshed and the green light group was irradiated with 520 nm green light for 15 min (21 mW cm−2, 19 J cm−2). Then the cells were washed with PBS twice, harvested, and centrifuged (5 min × 1300 rpm) to remove supernatant. The cells were resuspended in 250 μL of medium and maintained in the incubator for 2 h. Untreated cells were used as negative controls. 2 h after light irradiation, the cells were stained with the ROS Deep Red dye (1×) for 1 h at 37 °C. The levels of intracellular ROS were then determined using the BD FACSCanto™ II Clinical Flow Cytometry System. Forward versus side scatter (FSC vs. SSC) gating was used to select the population of interest and avoid cell debris. A forward scatter height (FSC-H) vs. forward scatter area (FSC-A) gating was used for doublet exclusion. Fluorescence measurements were acquired with the Cy5 filter given the known excitation/emission wavelengths of the ROS deep red dye (650/675 nm, respectively). All flow cytometry data were processed using FlowLogic software. Fluorescence emission intensity means and medians are expressed with >6000 gated single cells.4.9 Cell-free DNA binding experiment
The experiment was done according to previous work.81 The pUC19 plasmid (2686 bp) used here exists in three forms: supercoiled (SC), single-nicked open circular (OC), and linear dimer (LD). Chloride-free phosphate buffer was used here to mimic a pseudo intracellular environment. All aliquots were prepared with a final volume of 20 μL and prior to loading 4 μL of 6× loading dye was added. The λ DNA-HindIII digest molecular weight (MW) marker was prepared by adding 2 μL (1 μg) of the DNA MW marker, 18 μL PB, and 4 μL 6X loading dye. The MW marker was heated for 3 min at 60 °C prior to loading. In each well, 12 μL (1 μg of pUC19 DNA or 0.5 μg of MW marker) of each sample was loaded. For each gel, the electrophoresis chamber was filled with 50 mL TBA and 210 mL DI H2O. Each gel was run at a constant voltage of 105 V for 90 min. All gels were stained using 10 μL (10 mg mL−1) ethidium bromide in 200 mL DI H2O for 30 min with slight shaking and then de-stained in 200 mL DI H2O for 20 min. Immediately following de-staining, the gel was imaged using a BioRad ChemiDoc imaging system (ethidium bromide setting). Image Lab software was used to process the images.4.10 Cell fractionation ICPMS assay
The intracellular distribution of [2](PF6)2 study was performed on CRMM1 cells. 3 × 106 cells were seeded in Opti-MEM medium in 175 cm2 flasks. After 24 h incubation in the dark, 33 μM [2](PF6)2 was added onto the cells. The flask was further incubated for 24 h in the dark in a total medium volume of 24 mL. Then, the medium was aspirated, the cells were washed with PBS-buffer, trypsinized, counted, and pelleted by centrifugation at 1200 rpm for 5 min. The cells were fractionated according to the suppliers' instructions of FractionPREP cell fractionation kit (BioVision). The cell fractions were digested overnight in super purified nitric acid (>65%) and kept in MilliQ water with nitric acid (5%). The concentration of ruthenium was measured by ICP-MS. Results are presented as means ± SD from three independent experiments.4.11 Western Blot
A 2 mM stock solution of [2](PF6)2 was prepared in 5% DMSO and 95% medium. 175k CRMM1 cells were seeded in 6-well plates, incubated for 24 h, and then treated or not with [2](PF6)2 at 4 μM, i.e., the concentration corresponding to the EC50 value in presence of light (at this concentration there was 0.2% DMSO in each well). After 24 h incubation, cells were irradiated or not during 15 min with green light (520 nm, 21 mW cm−2, 19 J cm−2). 48 h after irradiation, the medium of all wells was collected and remaining cells were detached by 500 μL trypsin for 3 min. Collected medium was added to the wells with detached cells, mixed and centrifuged for 1200 rpm, 5 min. Protein samples were collected by lysing cells with lysis buffer containing phenylmethanesulfonyl fluoride (Cell Signaling Technology) and were separated by SDS-PAGE (Bio-Rad). The concentration of protein from each group was measured by Pierce® BCA protein assay kit (Thermo Scientific, USA). 50 μg proteins were transferred to polyvinylidene difluoride membranes (Millipore). And then the samples on the membranes were incubated with Caspase-3 (Cell Signalling, #9662), cleaved Caspase-3 (Cell Signalling, Asp175), PARP (Cell Signalling, #9542), cleaved PARP (Cell Signalling, Asp214), β-actin (Abcam, ab20272) overnight at 4 °C under shaking. All primary antibodies were diluted 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1000 times according to the manufacturer's protocol. After washing with TBS-T 0.1% 3 times, the samples were incubated with anti-mouse IgG, HRP-lined antibody (Cell Signalling, #7076) or anti-rabbit IgG, HRP-linked antibody (Cell Signalling, #7074) during 2 h at room temperature under shaking. After treating with enhanced chemoluminescence substrate mixture (Cell Signaling Technology), blots were scanned with ChemiDoc XRS+ System (Bio-rad).
1000 times according to the manufacturer's protocol. After washing with TBS-T 0.1% 3 times, the samples were incubated with anti-mouse IgG, HRP-lined antibody (Cell Signalling, #7076) or anti-rabbit IgG, HRP-linked antibody (Cell Signalling, #7074) during 2 h at room temperature under shaking. After treating with enhanced chemoluminescence substrate mixture (Cell Signaling Technology), blots were scanned with ChemiDoc XRS+ System (Bio-rad).
4.12 DNA fragmentation assay
6k CRMM1 cells were seeded in 96-well plates, incubated for 24 h, and then treated or not with [2](PF6)2 at 4 μM, i.e., the concentration corresponding to the EC50 value in presence of light. After 24 h of incubation, cells were irradiated or not during 15 min with green light (520 nm, 21 mW cm−2, 19 J cm−2). Either 4 h or 24 h after irradiation, the plate was centrifuged at 200 g for 10 min and then cells in each well were lysed with 200 μL of lysis buffer during at least 30 minutes. The plate was centrifuged again at 200 g for 10 min and then 20 μL supernatant of each condition was transferred into a new streptavidin coated 96-well plate and analysed by cell death detection ELISAPLUS (Roche, 11774425001). 20 μL supernatants were mixed with 80 μL immunoreagent in each well. The 96-well plate was coved with adhesive foil and incubated on a shaker under gently shaking (300 rpm) for 2 h at 15 to 25 °C. The solution was removed and each well was rinsed 3 times with 250 μL incubation buffer, the solution was removed again. 100 μL ABTS solution was pipetted into each well and incubated on a plate shaker at 250 rpm until the colour development was sufficient for a photometric analysis. 100 μL ABTS stop solution was added into each well and data was measured by spectrophotometry at 405 nm.4.13 Zebrafish maintenance, tumour cells implantation and tumour analysis
Zebrafish lines were kept in compliance with the local animal welfare regulations and European directives. The study was approved by the local animal welfare committee (DEC) of Leiden University (project: “Anticancer compound and target discovery in zebrafish xenograft model”. License number: AVD1060020172410). The Zebrafish Tg(fli1: GFP/Casper)101 were handled in compliance with local animal welfare regulations and maintained according to standard protocols (https://www.ZFIN.org).For cancer cell injection, two days post-fertilization (dpf), dechorionated zebrafish embryos were anaesthetized with 0.003% tricaine (Sigma) and plated on a 10 cm Petri dish covered with 1.5% of solidified agarose. PC3Pro4, CRMM1 and CRMM2 cells were suspended in PBS containing 2% polyvinylpyrrolidone (PVP; Sigma-Aldrich) with a concentration of 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 cells per μL and loaded into borosilicate glass capillary needles (1 mm O.D. × 0.78 mm I.D.; Harvard Apparatus). In the ectopic model, 200 mCherry fluorescent PC3Pro4 or (td) tomato fluorescent CM cells were injected into the Duct of Cuvier at 2 dpf, which led to dissemination through the blood circulation and outgrowth in the head and tail. In orthotopic tumour model, 100 (td) tomato fluorescent CRMM1 or CRMM2 cells were injected retro-orbitally in 2 dpf embryos using a Pneumatic Picopump and a manipulator (WPI). After injection, the embryos were incubated in a 34 °C incubator. Images were acquired at 1-, 2-, 4- and 6-days post injection (dpi) with a Leica M165 FC stereo fluorescence microscope. Tumour growth was quantified by calculating the total fluorescence intensity and area with the ZF4 pixel counting program (Leiden). Each experiment was performed at least 3 times with a group size of >30 embryos.
000 cells per μL and loaded into borosilicate glass capillary needles (1 mm O.D. × 0.78 mm I.D.; Harvard Apparatus). In the ectopic model, 200 mCherry fluorescent PC3Pro4 or (td) tomato fluorescent CM cells were injected into the Duct of Cuvier at 2 dpf, which led to dissemination through the blood circulation and outgrowth in the head and tail. In orthotopic tumour model, 100 (td) tomato fluorescent CRMM1 or CRMM2 cells were injected retro-orbitally in 2 dpf embryos using a Pneumatic Picopump and a manipulator (WPI). After injection, the embryos were incubated in a 34 °C incubator. Images were acquired at 1-, 2-, 4- and 6-days post injection (dpi) with a Leica M165 FC stereo fluorescence microscope. Tumour growth was quantified by calculating the total fluorescence intensity and area with the ZF4 pixel counting program (Leiden). Each experiment was performed at least 3 times with a group size of >30 embryos.
4.14 Maximum tolerated dose (MTD) for wild-type zebrafish and tumour cell-injected zebrafish
For determining the MTD of the water administration (WA) of the [2](PF6)2 solution in wild type zebrafish, solutions of 0.1 μM, 0.25 μM, 0.5 μM, 1 μM, 2 μM were made before the experiment. At 2.5, 3.5, 4.5, 5.5 dpf, [2](PF6)2 was added to the fish water and maintained for 12 h. At 3, 4, 5, 6 dpf, the fish water was refreshed and after 1 h, embryos were exposed to green light for 90 min (21 mW cm−2, 114 J cm−2, 520 nm). For the IV and RO administration, [2](PF6)2 solution (50 μM, 100 μM, 200 μM, 300 μM, 500 μM) was made before the experiment. At 3, 4, 5, 6 dpf, 1 nL of [2](PF6)2 was injected via the dorsal vein or the RO site and maintained for 1 h. Embryos were exposed to green light for 90 min (21 mW cm−2, 114 J cm−2, 520 nm). The images of treated and wild type embryos at 6 dpf were taken using a DFC420C camera coupled to a Leica MZ16FA fluorescence microscope. In order to determine the MTD of tumour cell-bearing zebrafish, 90 min green light activation (21 mW cm−2, 114 J cm−2, 520 nm) was performed according to the same procedure, after [2](PF6)2 was delivered by WA, IV and RA administration as described above for the wild type embryos.4.15 The antitumour efficacy of [2](PF6)2 by WA, IV and RO in zebrafish ectopic and orthotopic tumour models
Fluorescent PC3Pro4 cells were injected at 2 dpf into the Duct of Cuvier (ectopic model) and [2](PF6)2 was delivered by WA and IV administration with or without light treatment as described in 5.9. Fluorescent CRMM1 or CRMM2 cells were injected at 2 dpf into the Duct of Cuvier (ectopic model) and behind the eye (orthotopic model) and [2](PF6)2 was delivered by WA IV and RO administration with or without light treatment as described in 5.9. For the WA administration, the 0.5 μM [2](PF6)2 solution was added to the tumour cells-injected zebrafish at 2.5, 3.5, 4.5, 5.5 dpf and maintained for 12 h. At 3, 4, 5, 6 dpf, the fish water was refreshed, and after 1 h, embryos were exposed to green light for 90 min (21 mW cm−2, 114 J cm−2, 520 nm). For the IV and RO administration, 1 nL of 200 μM [2](PF6)2 solution was injected via the dorsal vein or the RO site at 3, 4, 5, 6 dpf. After 1 h interval, the embryos were exposed to green light for 90 min (21 mW cm−2, 114 J cm−2, 520 nm). After treatment, the embryo images were acquired with a Leica M165 FC stereo fluorescence microscope. Tumour growth was quantified by calculating the total fluorescence intensity and area with the ZF4 pixel counting program (Leiden). Each experiment was performed at least 3 times with a group size of >30 embryos.4.16 TUNEL assay
100 (td) tomato fluorescent CRMM1 cells were injected retro-orbitally in 2 dpf embryos using a Pneumatic Picopump and a manipulator (WPI). After injection, the embryos were incubated in a 34 °C incubator. At 3, 4, 5, 6 dpf, 1 nL of 200 μM [2](PF6)2 solution was injected into the tumour site. After 1 h interval, the embryos were exposed to green light for 90 min (21 mW cm−2, 114 J cm−2, 520 nm). After treatment, the zebrafish larvae were fixed overnight with 4% PFA at 4 °C. Embryos were washed in PBST for five minutes and dehydrated by a graded methanol series until reaching 100% methanol. Embryos were stored at −20 °C for further use. Embryos were gradually rehydrated in PBST (25%, 50%, 75%), washed twice for 10 minutes with PBST and digested by proteinase K (Roche) solution in PBST (10 μg mL−1) at 37 °C for 40 minutes. After two washes in PBST, embryos were post-fixed in 4% PFA for 20 minutes. After washing them again twice in PBST for 10 minutes, 50 μL of TdT reaction mix (Roche) was added to the embryos. Embryos were overnight incubated with the TdT at 37 °C (in the dark). The reaction was stopped by three 15 min washes with PBST at room temperature and embryos were used for high-resolution imaging. Embryos were placed on glass-bottom Petri dishes and covered with 1% low melting agarose containing 0.003% tricaine (Sigma). Imaging was performed using the Leica SP8 confocal microscope. The images were processed with ImageJ software. Each experiment was performed 3 times with a group size of 10 embryos.4.17 Statistical analysis
Determination of the EC50 concentrations in vitro was based on a non-linear regression analysis performed using GraphPad Prism Software. Results are presented as means ± SD from three independent experiments. Significant differences were detected by one-way ANOVA followed by Dunnett's multiple comparisons test implemented by Prism 8 (GraphPad Software, La Jolla, CA, USA). A p-value < 0.05 was considered statistically significant, *: p < 0.05, **: p < 0.01, ***: p < 0.001.Data availability
The data that support the findings of this study are available from the corresponding author, S. B., upon request.Author contributions
Conceptualization: Sylvestre Bonnet, Jordi-Amat Cuello-Garibo, Quanchi Chen, Ewa Snaar-Jagalska, formal analysis: Quanchi Chen, Jordi-Amat Cuello-Garibo, Yurii Husiev, Claudia Schmidt, Albert M. Brouwer, funding acquisition: Ewa Snaar-Jagalska, Sylvestre Bonnet, Albert M. Brouwer, Ingo Ott, investigation: Ludovic Bretin, Liyan Zhang, Vadde Ramu, Yasmin Aydar, Yevhen Batsuin, Sharon Bronkhorst, Yurii Husiev, Nataliia Beztsinna, Lanpeng Chen, Xue-Quan Zhou, Claudia Schmidt, Albert M. Brouwer, methodology: Quanchi Chen, Jordi-Amat Cuello-Garibo, Ludovic Bretin, Ingo Ott, resources: Albert M. Brouwer, Martine J. Jager, Ingo Ott, Ewa Snaar-Jagalska, Sylvestre Bonnet, supervision: Ingo Ott, Ewa Snaar-Jagalska, Sylvestre Bonnet, validation: Albert M. Brouwer, Ewa Snaar-Jagalska, Sylvestre Bonnet, Ingo Ott, visualization: Quanchi Chen, Nataliia Beztsinna, Vadde Ramu, writing – original draft: Quanchi Chen, Jordi-Amat Cuello-Garibo, writing – review & editing: Ewa Snaar-Jagalska, Sylvestre Bonnet.Conflicts of interest
There are no conflicts of interest to declare.Acknowledgements
The European Research Council is kindly acknowledged for a Starting Grant and a Proof-of-Concept grant (Ru4EYE) to S. B. The Dutch Research Council is kindly acknowledged for a VICI grant to S. B. The Chinese Scholarship Council is kindly acknowledged for a PhD grant, National Natural Science Foundation of China (grant no. 82102637) and China Postdoctoral Science Foundation (grant no. 2021M701675) to Q. C. Prof. E. Bouwman is kindly acknowledged for support and scientific discussion.References
- S. Dilruba and G. V. Kalayda, Cancer Chemother. Pharmacol., 2016, 77, 1103–1124 CrossRef CAS PubMed.
- M. Galanski, Recent Pat. Anti-Cancer Drug Discovery, 2006, 1, 285–295 CrossRef CAS PubMed.
- D. Lebwohl and R. Canetta, Eur. J. Cancer, 1998, 34, 1522–1534 CrossRef CAS PubMed.
- J. Pouysségur, F. Dayan and N. M. Mazure, Nature, 2006, 441, 437–443 CrossRef PubMed.
- E. R. Jamieson and S. J. Lippard, Chem. Rev., 1999, 99, 2467–2498 CrossRef CAS PubMed.
- H. N. Fraval, C. J. Rawlings and J. J. Roberts, Mutat. Res., 1978, 51, 121–132 CrossRef CAS PubMed.
- S. Dasari and P. B. Tchounwou, Eur. J. Pharmacol., 2014, 740, 364–378 CrossRef CAS PubMed.
- T. Karasawa and P. S. Steyger, Toxicol. Lett., 2015, 237, 219–227 CrossRef CAS PubMed.
- R. J. Cersosimo, Ann. Pharmacother., 1993, 27, 438–441 CrossRef CAS PubMed.
- N. M. Martins, N. A. Santos, C. Curti, M. L. Bianchi and A. C. Santos, J. Appl. Toxicol., 2008, 28, 337–344 CrossRef CAS PubMed.
- R. G. Kenny and C. J. Marmion, Chem. Rev., 2019, 119, 1058–1137 CrossRef CAS PubMed.
- R. N. Garner, J. C. Gallucci, K. R. Dunbar and C. Turro, Inorg. Chem., 2011, 50, 9213–9215 CrossRef CAS PubMed.
- T. Respondek, R. N. Garner, M. K. Herroon, I. Podgorski, C. Turro and J. J. Kodanko, J. Am. Chem. Soc., 2011, 133, 17164–17167 CrossRef CAS PubMed.
- C. Imberti, P. Zhang, H. Huang and P. J. Sadler, Angew. Chem., Int. Ed., 2020, 59, 61–73 CrossRef CAS PubMed.
- J.-A. Cuello-Garibo, M. S. Meijer and S. Bonnet, Chem. Commun., 2017, 53, 6768–6771 RSC.
- W. R. Wilson and M. P. Hay, Nat. Rev. Cancer, 2011, 11, 393–410 CrossRef CAS PubMed.
- L. N. Lameijer, D. Ernst, S. L. Hopkins, M. S. Meijer, S. H. C. Askes, S. E. Le Dévédec and S. Bonnet, Angew. Chem., Int. Ed., 2017, 56, 11549–11553 CrossRef CAS PubMed.
- S. Bonnet, Dalton Trans., 2018, 47, 10330–10343 RSC.
- D. Havrylyuk, D. K. Heidary, Y. Sun, S. Parkin and E. C. Glazer, ACS Omega, 2020, 5, 18894–18906 CrossRef CAS PubMed.
- J. Roque III, D. Havrylyuk, P. C. Barrett, T. Sainuddin, J. McCain, K. Colón, W. T. Sparks, E. Bradner, S. Monro, D. Heidary, C. G. Cameron, E. C. Glazer and S. A. McFarland, Photochem. Photobiol., 2020, 96, 327–339 CrossRef PubMed.
- C. Mari and G. Gasser, Chimia, 2015, 69, 176–181 CrossRef CAS PubMed.
- N. J. Farrer, L. Salassa and P. J. Sadler, Dalton Trans., 2009, 10690–10701, 10.1039/b917753a.
- K. Arora, M. Herroon, M. H. Al-Afyouni, N. P. Toupin, T. N. Rohrabaugh Jr, L. M. Loftus, I. Podgorski, C. Turro and J. J. Kodanko, J. Am. Chem. Soc., 2018, 140, 14367–14380 CrossRef CAS PubMed.
- L. M. Loftus, K. F. Al-Afyouni and C. Turro, Chem.–Eur. J., 2018, 24, 11550–11553 CrossRef CAS PubMed.
- A. Li, C. Turro and J. J. Kodanko, Acc. Chem. Res., 2018, 51, 1415–1421 CrossRef CAS PubMed.
- T. Joshi, V. Pierroz, C. Mari, L. Gemperle, S. Ferrari and G. Gasser, Angew. Chem., Int. Ed., 2014, 53, 2960–2963 CrossRef CAS PubMed.
- H. Chan, J. B. Ghrayche, J. Wei and A. K. Renfrew, Eur. J. Inorg. Chem., 2017, 2017, 1679–1686 CrossRef CAS.
- Q.-X. Zhou, W.-H. Lei, J.-R. Chen, C. Li, Y.-J. Hou, X.-S. Wang and B.-W. Zhang, Chem.–Eur. J., 2010, 16, 3157–3165 CrossRef CAS PubMed.
- D. Havrylyuk, K. Stevens, S. Parkin and E. C. Glazer, Inorg. Chem., 2020, 59, 1006–1013 CrossRef CAS PubMed.
- L. M. Loftus, J. J. Rack and C. Turro, Chem. Commun., 2020, 56, 4070–4073 RSC.
- M. H. Al-Afyouni, T. N. Rohrabaugh, K. F. Al-Afyouni and C. Turro, Chem. Sci., 2018, 9, 6711–6720 RSC.
- W. Sun, S. Li, B. Häupler, J. Liu, S. Jin, W. Steffen, U. S. Schubert, H.-J. Butt, X.-J. Liang and S. Wu, Adv. Mater., 2017, 29, 1603702 CrossRef PubMed.
- Z. Chen, R. Thiramanas, M. Schwendy, C. Xie, S. H. Parekh, V. Mailänder and S. Wu, Small, 2017, 13, 1700997 CrossRef PubMed.
- F. Battistin, G. Balducci, J. Wei, A. K. Renfrew and E. Alessio, Eur. J. Inorg. Chem., 2018, 2018, 1469–1480 CrossRef CAS.
- N. Karaoun and A. K. Renfrew, Chem. Commun., 2015, 51, 14038–14041 RSC.
- L. Zayat, C. Calero, P. Alborés, L. Baraldo and R. Etchenique, J. Am. Chem. Soc., 2003, 125, 882–883 CrossRef CAS PubMed.
- G. Ragazzon, I. Bratsos, E. Alessio, L. Salassa, A. Habtemariam, R. J. McQuitty, G. J. Clarkson and P. J. Sadler, Inorg. Chim. Acta, 2012, 393, 230–238 CrossRef CAS.
- T. Sainuddin, M. Pinto, H. Yin, M. Hetu, J. Colpitts and S. A. McFarland, J. Inorg. Biochem., 2016, 158, 45–54 CrossRef CAS PubMed.
- K. T. Hufziger, F. S. Thowfeik, D. J. Charboneau, I. Nieto, W. G. Dougherty, W. S. Kassel, T. J. Dudley, E. J. Merino, E. T. Papish and J. J. Paul, J. Inorg. Biochem., 2014, 130, 103–111 CrossRef CAS PubMed.
- B. S. Howerton, D. K. Heidary and E. C. Glazer, J. Am. Chem. Soc., 2012, 134, 8324–8327 CrossRef CAS PubMed.
- M. S. Meijer and S. Bonnet, Inorg. Chem., 2019, 58, 11689–11698 CrossRef CAS PubMed.
- E. Wachter, D. K. Heidary, B. S. Howerton, S. Parkin and E. C. Glazer, Chem. Commun., 2012, 48, 9649–9651 RSC.
- J. Roque 3rd, D. Havrylyuk, P. C. Barrett, T. Sainuddin, J. McCain, K. Colón, W. T. Sparks, E. Bradner, S. Monro, D. Heidary, C. G. Cameron, E. C. Glazer and S. A. McFarland, Photochem. Photobiol., 2020, 96, 327–339 CrossRef PubMed.
- B. A. Albani, B. Peña, K. R. Dunbar and C. Turro, Photochem. Photobiol. Sci., 2014, 13, 272–280 CrossRef CAS PubMed.
- V. H. S. van Rixel, V. Ramu, A. B. Auyeung, N. Beztsinna, D. Y. Leger, L. N. Lameijer, S. T. Hilt, S. E. Le Dévédec, T. Yildiz, T. Betancourt, M. B. Gildner, T. W. Hudnall, V. Sol, B. Liagre, A. Kornienko and S. Bonnet, J. Am. Chem. Soc., 2019, 141, 18444–18454 CrossRef CAS PubMed.
- W. Sun, Y. Wen, R. Thiramanas, M. Chen, J. Han, N. Gong, M. Wagner, S. Jiang, M. S. Meijer, S. Bonnet, H.-J. Butt, V. Mailänder, X.-J. Liang and S. Wu, Adv. Funct. Mater., 2018, 28, 1804227 CrossRef.
- P. Letrado, I. de Miguel, I. Lamberto, R. Díez-Martínez and J. Oyarzabal, Cancer Res., 2018, 78, 6048–6058 CrossRef CAS PubMed.
- M. Kucinska, M. Murias and P. Nowak-Sliwinska, Mutat. Res., 2017, 773, 242–262 CAS.
- J. Xiao, E. Glasgow and S. Agarwal, Trends Cancer, 2020, 6, 569–579 CrossRef CAS PubMed.
- J. F. Amatruda, J. L. Shepard, H. M. Stern and L. I. Zon, Cancer Cell, 2002, 1, 229–231 CrossRef CAS PubMed.
- X. Zhang, L. de Boer, L. Heiliegers, S. Man-Bovenkerk, P. K. Selbo, J. W. Drijfhout, A. Høgset and S. A. J. Zaat, J. Controlled Release, 2018, 283, 214–222 CrossRef CAS PubMed.
- C. Mauriello Jimenez, D. Aggad, J. G. Croissant, K. Tresfield, D. Laurencin, D. Berthomieu, N. Cubedo, M. Rossel, S. Alsaiari, D. H. Anjum, R. Sougrat, M. A. Roldan-Gutierrez, S. Richeter, E. Oliviero, L. Raehm, C. Charnay, X. Cattoën, S. Clément, M. Wong Chi Man, M. Maynadier, V. Chaleix, V. Sol, M. Garcia, M. Gary-Bobo, N. M. Khashab, N. Bettache and J.-O. Durand, Adv. Funct. Mater., 2018, 28, 1800235 CrossRef.
- C. Matera, A. M. J. Gomila, N. Camarero, M. Libergoli, C. Soler and P. Gorostiza, J. Am. Chem. Soc., 2018, 140, 15764–15773 CrossRef CAS PubMed.
- R. Bouchaala, N. Anton, H. Anton, T. Vandamme, J. Vermot, D. Smail, Y. Mély and A. S. Klymchenko, Colloids Surf., B, 2017, 156, 414–421 CrossRef CAS PubMed.
- J. He, Y. Wang, M. A. Missinato, E. Onuoha, L. A. Perkins, S. C. Watkins, C. M. St Croix, M. Tsang and M. P. Bruchez, Nat. Methods, 2016, 13, 263–268 CrossRef CAS PubMed.
- S. P. Li, C. T. Lau, M. W. Louie, Y. W. Lam, S. H. Cheng and K. K. Lo, Biomaterials, 2013, 34, 7519–7532 CrossRef CAS PubMed.
- P. N. Manghnani, W. Wu, S. Xu, F. Hu, C. Teh and B. Liu, Nano-Micro Lett., 2018, 10, 61 CrossRef PubMed.
- Q. Chen, V. Ramu, Y. Aydar, A. Groenewoud, X. Q. Zhou, M. J. Jager, H. Cole, C. G. Cameron, S. A. McFarland, S. Bonnet and B. E. Snaar-Jagalska, Cancers, 2020, 12, 587 CrossRef CAS PubMed.
- C. Bai and M. Tang, J. Appl. Toxicol., 2020, 40, 37–63 CrossRef CAS PubMed.
- L. C. Wehmas, C. Anders, J. Chess, A. Punnoose, C. B. Pereira, J. A. Greenwood and R. L. Tanguay, Toxicol. Rep., 2015, 2, 702–715 CrossRef CAS PubMed.
- C. Gutiérrez-Lovera, J. Martínez-Val, P. Cabezas-Sainz, R. López, J. A. Rubiolo and L. Sánchez, Toxicol. Mech. Methods, 2019, 29, 478–487 CrossRef PubMed.
- B. C. Das, L. McCormick, P. Thapa, R. Karki and T. Evans, Future Med. Chem., 2013, 5, 2103–2116 CrossRef CAS PubMed.
- L. I. Zon and R. T. Peterson, Nat. Rev. Drug Discovery, 2005, 4, 35–44 CrossRef CAS PubMed.
- C. S. Burke and T. E. Keyes, RSC Adv., 2016, 6, 40869–40877 RSC.
- C. Mari, V. Pierroz, R. Rubbiani, M. Patra, J. Hess, B. Spingler, L. Oehninger, J. Schur, I. Ott, L. Salassa, S. Ferrari and G. Gasser, Chem.–Eur. J., 2014, 20, 14421–14436 CrossRef CAS PubMed.
- A. Frei, R. Rubbiani, S. Tubafard, O. Blacque, P. Anstaett, A. Felgentrager, T. Maisch, L. Spiccia and G. Gasser, J. Med. Chem., 2014, 57, 7280–7292 CrossRef CAS PubMed.
- G. Shi, S. Monro, R. Hennigar, J. Colpitts, J. Fong, K. Kasimova, H. Yin, R. DeCoste, C. Spencer, L. Chamberlain, A. Mandel, L. Lilge and S. A. McFarland, Coord. Chem. Rev., 2015, 282–283, 127–138 CrossRef CAS.
- W. R. Browne, P. Passaniti, M. T. Gandolfi, R. Ballardini, W. Henry, A. Guckian, N. O'Boyle, J. J. McGarvey and J. G. Vos, Inorg. Chim. Acta, 2007, 360, 1183–1190 CrossRef CAS.
- D. Garcìa-Fresnadillo, Y. Georgiadou, G. Orellana, A. M. Braun and E. Oliveros, Helv. Chim. Acta, 1996, 79, 1222–1238 CrossRef.
- J. Moan and S. Sommer, Cancer Res., 1985, 45, 1608–1610 CAS.
- C. Hadjur, N. Lange, J. Rebstein, P. Monnier, H. van den Bergh and G. Wagnières, J. Photochem. Photobiol., B, 1998, 45(2–3), 170–178 CrossRef CAS.
- Y. Arenas, S. Monro, G. Shi, A. Mandel, S. McFarland and L. Lilge, Photodiagn. Photodyn. Ther., 2013, 10, 615–625 CrossRef CAS PubMed.
- H. F. Suen, S. W. Wilson, M. Pomerantz and J. L. Walsh, Inorg. Chem., 1989, 28, 786–791 CrossRef CAS.
- J. K. White, R. H. Schmehl and C. Turro, Inorg. Chim. Acta, 2017, 454, 7–20 CrossRef CAS PubMed.
- S. Campagna, F. Puntoriero, F. Nastasi, G. Bergamini and V. Balzani, in Photochemistry and Photophysics of Coordination Compounds I, ed. V. Balzani and S. Campagna, Springer Berlin Heidelberg, Berlin, Heidelberg, 2007, pp. 117–214, DOI: DOI:10.1007/128_2007_133.
- S. L. Hopkins, B. Siewert, S. H. C. Askes, P. Veldhuizen, R. Zwier, M. Heger and S. Bonnet, Photochem. Photobiol. Sci., 2016, 15, 644–653 CrossRef CAS PubMed.
- R. W. Horobin, J. C. Stockert and F. Rashid-Doubell, Histochem. Cell Biol., 2006, 126, 165–175 CrossRef CAS PubMed.
- C. A. Puckett and J. K. Barton, J. Am. Chem. Soc., 2007, 129, 46–47 CrossRef CAS PubMed.
- C. A. Pettaway, S. Pathak, G. Greene, E. Ramirez, M. R. Wilson, J. J. Killion and I. J. Fidler, Clin. Cancer Res., 1996, 2, 1627–1636 CAS.
- V. H. S. van Rixel, A. Busemann, M. F. Wissingh, S. L. Hopkins, B. Siewert, C. van de Griend, M. A. Siegler, T. Marzo, F. Papi, M. Ferraroni, P. Gratteri, C. Bazzicalupi, L. Messori and S. Bonnet, Angew. Chem., Int. Ed., 2019, 58, 9378–9382 CrossRef CAS PubMed.
- V. H. S. van Rixel, B. Siewert, S. L. Hopkins, S. H. C. Askes, A. Busemann, M. A. Siegler and S. Bonnet, Chem. Sci., 2016, 7, 4922–4929 RSC.
- D. Hill, L. Chen, E. Snaar-Jagalska and B. Chaudhry, F1000Research, 2018, 7, 1682 CAS.
- W. van der Ent, C. Burrello, A. F. Teunisse, B. R. Ksander, P. A. van der Velden, M. J. Jager, A. G. Jochemsen and B. E. Snaar-Jagalska, Invest. Ophthalmol. Visual Sci., 2014, 55, 6612–6622 CrossRef CAS PubMed.
- L. Chen, M. De Menna, A. Groenewoud, G. N. Thalmann, M. Kruithof-de Julio and B. E. Snaar-Jagalska, Oncogene, 2020, 39, 1634–1651 CrossRef CAS PubMed.
- L. Chen, M. Boleslaw Olszewski, M. Kruithof-de Julio and B. E. Snaar-Jagalska, Cells, 2020, 9, 797 CrossRef CAS PubMed.
- E. Murayama, K. Kissa, A. Zapata, E. Mordelet, V. Briolat, H.-F. Lin, R. I. Handin and P. Herbomel, Immunity, 2006, 25, 963–975 CrossRef CAS PubMed.
- S. Monro, K. L. Colón, H. Yin, J. Roque 3rd, P. Konda, S. Gujar, R. P. Thummel, L. Lilge, C. G. Cameron and S. A. McFarland, Chem. Rev., 2019, 119, 797–828 CrossRef CAS PubMed.
- G. Wang, D. Zhao, D. J. Spring and R. A. DePinho, Genes Dev., 2018, 32, 1105–1140 CrossRef CAS PubMed.
- J. Chang, J. Choi, Y. C. Rah, M. H. Yoo, K. H. Oh, G. J. Im, S. H. Lee, S. Y. Kwon, H.-C. Park, S. W. Chae and H. H. Jung, PLoS One, 2016, 11, e0151557 CrossRef PubMed.
- E. Reisner, T. C. Abikoff and S. J. Lippard, Inorg. Chem., 2007, 46, 10229–10240 CrossRef CAS PubMed.
- A. Bahreman, J. A. Cuello-Garibo and S. Bonnet, Dalton Trans., 2014, 43, 4494–4505 RSC.
- G. Nareyeck, H. Wuestemeyer, D. von der Haar and G. Anastassiou, Exp. Eye Res., 2005, 81, 361–362 CrossRef CAS PubMed.
- S. Keijser, W. Maat, G. S. Missotten and R. J. de Keizer, Br. J. Ophthalmol., 2007, 91, 1566–1567 CrossRef PubMed.
- G. P. Luyten, N. C. Naus, C. M. Mooy, A. Hagemeijer, J. Kan-Mitchell, E. Van Drunen, V. Vuzevski, P. T. De Jong and T. M. Luider, Int. J. Cancer, 1996, 66, 380–387 CrossRef CAS PubMed.
- P. W. Chen, T. G. Murray, T. Uno, M. L. Salgaller, R. Reddy and B. R. Ksander, Clin. Exp. Metastasis, 1997, 15, 509–518 CrossRef CAS PubMed.
- F. Carlotti, M. Bazuine, T. Kekarainen, J. Seppen, P. Pognonec, J. A. Maassen and R. C. Hoeben, Mol. Ther., 2004, 9, 209–217 CrossRef CAS PubMed.
- M. Appold, C. Mari, C. Lederle, J. Elbert, C. Schmidt, I. Ott, B. Stühn, G. Gasser and M. Gallei, Polym. Chem., 2017, 8, 890–900 RSC.
- R. D. Jiang, H. Shen and Y. J. Piao, Rom. J. Morphol. Embryol., 2010, 51, 663–667 Search PubMed.
- I. Ott, H. Scheffler and R. Gust, ChemMedChem, 2007, 2, 702–707 CrossRef CAS PubMed.
- S. L. Hopkins, B. Siewert, S. H. Askes, P. Veldhuizen, R. Zwier, M. Heger and S. Bonnet, Photochem. Photobiol. Sci., 2016, 15, 644–653 CrossRef CAS PubMed.
- N. D. Lawson and B. M. Weinstein, Dev. Biol., 2002, 248, 307–318 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available: 1H and 13C NMR spectra, mass spectrometry data, HPLC traces, 1O2 generation quantum yield data, FACS analysis for ROS production, steady-state and time-resolved emission spectroscopy data, photosubstitution quantum yield data, dose–response curves in vitro in a range of cancer cell lines, gel electrophoresis data, DNA fragmentation data, and maximum tolerated dose data. See https://doi.org/10.1039/d2sc01646j |
| ‡ These authors contributed equally to the paper. |
| This journal is © The Royal Society of Chemistry 2022 |