Open Access Article
Open Access ArticlePhoto-Brook rearrangement of acyl silanes as a strategy for photoaffinity probe design†
Annika C. S.
Page‡
 ab,
Spencer O.
Scholz‡
ab,
Katherine N.
Keenan
ab,
Spencer O.
Scholz‡
ab,
Katherine N.
Keenan
 ab,
Jessica N.
Spradlin
abc,
Bridget P.
Belcher
ac,
Scott M.
Brittain
bd,
John A.
Tallarico
bd,
Jeffrey M.
McKenna
ab,
Jessica N.
Spradlin
abc,
Bridget P.
Belcher
ac,
Scott M.
Brittain
bd,
John A.
Tallarico
bd,
Jeffrey M.
McKenna
 bd,
Markus
Schirle
bd,
Markus
Schirle
 bd,
Daniel K.
Nomura
bd,
Daniel K.
Nomura
 *abce and
F. Dean
Toste
*abce and
F. Dean
Toste
 *ab
*ab
aDepartment of Chemistry, University of California, Berkeley, California 94720, USA. E-mail: fdtoste@berkeley.edu
bNovartis-Berkeley Center for Proteomics and Chemistry Technologies, University of California, Berkeley, California 94720, USA
cInnovative Genomics Institute, University of California, Berkeley, California 94720, USA
dNovartis Institute for BioMedical Research, Cambridge, Massachusetts 02139, USA
eDepartment of Molecular and Cellular Biology, University of California, Berkeley, California 94720, USA
First published on 11th March 2022
Abstract
Photoaffinity labeling (PAL) is a powerful tool for the identification of non-covalent small molecule–protein interactions that are critical to drug discovery and medicinal chemistry, but this approach is limited to only a small subset of robust photocrosslinkers. The identification of new photoreactive motifs capable of covalent target capture is therefore highly desirable. Herein, we report the design, synthesis, and evaluation of a new class of PAL warheads based on the UV-triggered 1,2-photo-Brook rearrangement of acyl silanes, which hitherto have not been explored for PAL workflows. Irradiation of a series of probes in cell lysate revealed an iPr-substituted acyl silane with superior photolabeling and minimal thermal background labeling compared to other substituted acyl silanes. Further, small molecule (+)-JQ1- and rapamycin-derived iPr acyl silanes were shown to selectively label recombinant BRD4-BD1 and FKBP12, respectively, with minimal background. Together, these data highlight the untapped potential of acyl silanes as a novel, tunable scaffold for photoaffinity labeling.
Introduction
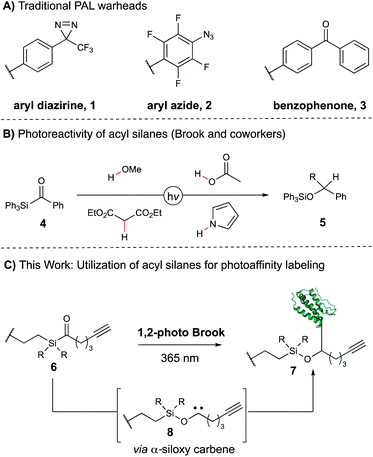
Identifying drivers of on- and off-target phenotypic responses to bioactive small molecules remains a significant challenge in the field of drug discovery, particularly for low-affinity or low-abundance targets.1–4 One of the most powerful approaches to target deconvolution in a native cellular environment is photoaffinity labeling (PAL), which allows for the capture of transient, non-covalent small molecule–protein interactions in a proteome-wide fashion.5 In photoaffinity labeling, a molecule of interest is conjugated to a light-reactive functional group that generates a high-energy intermediate capable of covalently capturing associated proteins when exposed to UV light. Conventional photoaffinity probes employ diazirines,6 aryl azides,7 and diaryl ketones,8 which give rise to carbene, nitrene, and ketyl radical intermediates, respectively (Fig. 1A, 1–3). All three have shown tremendous value as photoaffinity warheads in a variety of PAL applications,5 often in a synthetically modular fashion as exemplified by the suite of “minimalist” photocrosslinking diazirines developed by the Yao group.9 However, not every PAL campaign that utilizes these probes is successful, reflecting the requirements for correct positioning and distance of the photoreactive moiety as well as overall low crosslinking efficiencies typically observed. In addition, the warhead-specific labeling of non-target proteins often results in false positive hits, potentially obscuring true cellular targets of the pharmacophore.10,11 Taken together, the dearth of photocrosslinking functionalities is an inherent limitation for applications of PAL-based workflows—a more varied set of photoreactive motifs would greatly aid target elucidation, both by providing additional tools for capture of protein targets as well as alternative synthetic approaches for a given probe of interest. However, the development of a novel PAL warhead is contingent upon fulfilling a number of rigorous requirements. Ideally, it should possess a long-wavelength UV absorbance profile to minimize cellular and protein damage during irradiation, rapidly generate a short-lived reactive intermediate capable of residue-agnostic crosslinking, and minimally perturb the native substrate–protein interactions. As a result, only a small number of motifs have been recently disclosed meeting these criteria, including aryl tetrazole12 and thienyl ketoamide13 probes. Acyl silanes are an intriguing functionality with ideal properties for use as a photoaffinity warhead. Upon irradiation, acyl silanes undergo a 1,2-photo Brook rearrangement to reveal a stabilized α-siloxy carbene, which has since been shown to insert into a variety of acidic, polar, and non-polarized bonds to yield X–H insertion products in high yields (Figure 1B, 5).14–20 The rearrangement occurs with high quantum yields to the α-siloxy carbene21 at long UV wavelengths (350–420 nm) ideal for photocrosslinking. We became interested in utilizing the 1,2-photo Brook rearrangement as an orthogonal mechanism for carbene generation that could bypass deleterious competing pathways common to other carbene precursors, such as diazo formation from diazirines,22 and provide complementary reactivity to known warheads (Fig. 1C). Moreover, acyl silanes allow for modular substitution on the photoactive moiety itself, offering the potential for direct tuning of desired steric and electronic properties of the probe. Herein, we report the design, synthesis, and evaluation of several acyl silane photoaffinity probes. Upon investigation, an iPr-substituted acyl silane was identified that yielded superior labeling in cellular lysate relative to Me and Ph probe derivatives and avoided unwanted thermal background labeling. Subsequently, we synthesized (+)-JQ1- and rapamycin-derived iPr acyl silanes and confirmed their labeling efficiency and selectivity on pure recombinant proteins. Furthermore, our iPr-substituted acyl silane probe demonstrated similar levels of labeling to a “minimalist” diazirine probe in cellular lysate, suggesting the overall promise of acyl silanes as a novel scaffold for PAL.Results and discussion
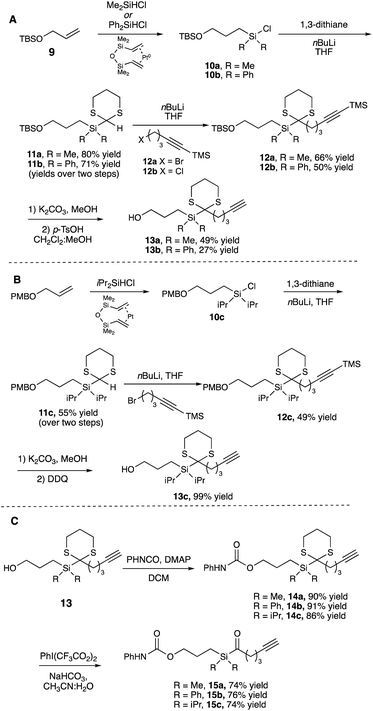
We began by synthesizing a probe that contained an alcohol as a modular site of attachment for conjugation to a small molecule of interest and an alkyne handle, which could be used for either pull-down experiments or visualization after further reaction with a suitable azide dye via Cu-catalyzed azide–alkyne click chemistry.23 Additionally, a series of warheads with varying silane substitution were evaluated to probe the role of sterics on photolabeling. Starting from a TBS-protected allyl alcohol 9, Me- and Ph-substituted silyl dithianes 11a and b were obtained in 80 and 71% yields, respectively, following a Pt-catalyzed hydrosilylation/lithiation sequence. Subsequent alkylation of the lithiated dithianes and deprotection of both the alkyne and TBS moieties yielded alcohols 13a and b (Scheme 1A). A similar sequence was followed in the preparation of iPr-substituted silane 13c from p-methoxybenzyl-protected allyl alcohol (Scheme 1B). Following elaboration of the alcohol, oxidative cleavage of the dithiane with PIFA provided the desired acyl silane probes in high yields (Scheme 1C).24 Following deprotection, we confirmed that the acyl silanes 15a–c each possessed a long-wavelength UV λmax between 364–370 nm, which was consistent with reported values and is ideally suited for utilization as a photoaffinity warhead. The reactivity of acyl silanes 15a–c in MeOH was also evaluated. To avoid possible damage to the protein or proteins of interest, photoaffinity probes should have short irradiation times; consistent with this, all three acyl silanes showed 80% or greater conversion over a 30 minute period upon irradiation with a simple 6 W handheld UV lamp (Fig. 2). Analysis of the irradiation reaction in MeOH with acyl silane 15b confirms the formation of an acetal consistent with α-siloxy carbene insertion into the O–H bond of MeOH, and in accordance with the adducts reported by Brook (Fig. S3†).14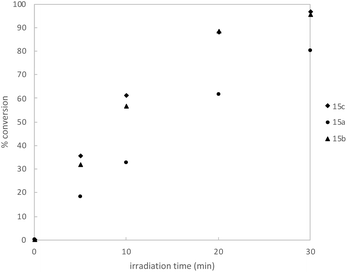 | ||
| Fig. 2 Kinetic profile of irradiation of acyl silane probes in MeOH-d4 (n = 3, average) using a 6 W handheld UV lamp at 365 nm. Conversion was determined by 1H NMR using an internal standard. Full details available in ESI.† | ||
After validating the photoactivity of the acyl silane probes with long-wavelength UV irradiation, their labeling performance in cell lysate was evaluated. Following a 30 minute incubation period, lysates treated with probes 15a–c were irradiated for 30 minutes on ice, followed by a Cu-catalyzed click reaction with rhodamine azide. Labeled proteins were subsequently visualized by in-gel fluorescence. All probes displayed light-dependent labeling of proteins as well as an increase in labeling over DMSO control (Fig. 3A). However, samples treated with iPr-substituted probe 15c showed significantly greater fluorescence over 15a and 15b. While the increased labeling may be the result of increased hydrophobicity of probe 15c, we hypothesized the difference in labeling may be due to the inherent instability of the probes 15a and 15b in cell lysate prior to irradiation. Acyl silanes are known to undergo a slow, thermal 1,2-Brook rearrangement in the presence of primary amines (i.e., lysine), which could not only result in probe decomposition before crosslinking but could also result in uncontrolled background thermal labeling processes.25 A more sterically hindered probe (i.e., 15c) could minimize probe decomposition by slowing the rate of background, thermal 1,2-Brook rearrangement. Thus, to evaluate their stability, probes 15a–c were incubated for either 0, 0.5, 2, or 24 hours in cell lysate prior to 30 minute irradiation, with labeled proteins visualized in a similar fashion as prior (Fig. 3B). Consistent with the previous results, iPr-substituted acyl silane 15c showed no increase in background signal over the course of the experiment and exhibited minimal loss of fluorescence. In contrast 15b, and to a greater extent 15a, showed increased thermal labeling upon extended incubation. Interestingly, all probes showed a decrease of light-dependent labeling over extended incubation, possibly due to instability of the proteome over extended time periods at ambient temperatures.
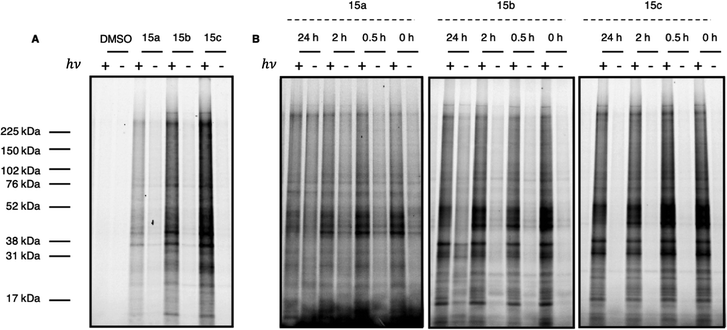 | ||
| Fig. 3 (A) Full gel image of UV-dependent labeling of PAL probes 15a–c in cell lysate. In brief, 231MFP cell lysate in pH 7.4 PBS was treated with either DMSO or 10 μM probe 15a, 15b, or 15c and incubated at ambient temperature for 30 min. Samples were then irradiated for 30 min at 4 °C with a 6 W handheld UV lamp at 365 nm. Labeled protein was visualized following Cu click reaction with Rh–N3. Gel quantitation available in ESI.† (B) Evaluation of probe labeling with variable incubation time. In brief, 231MFP cell lysate in pH 7.4 PBS was treated with 10 μM of probe 15a, 15b, or 15c and incubated at ambient temperature for 0, 0.5, 2, or 24 h. Samples were then irradiated for 30 min at 4 °C with a 6 W handheld UV lamp at 365 nm. Labeled protein was visualized following Cu click reaction with Rh–N3. Gel quantitation available in ESI.† | ||
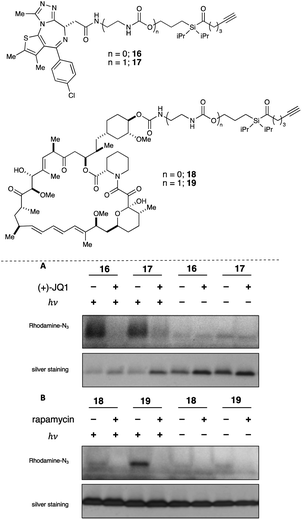
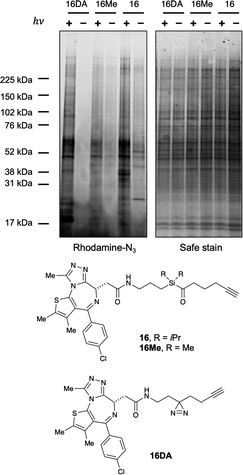
With these data in hand, we sought to further evaluate whether bulkier acyl silane warheads such as iPr derivative 15c could be utilized to capture targeted and specific small molecule–protein interactions; thus, we prepared two small molecule acyl silane probes utilizing the iPr-substitution. The first was based on (+)-JQ1, a known inhibitor of the BET class of bromodomain proteins26 that has seen extensive profiling27 and has come to serve as a key benchmarking tool in the development of novel photoaffinity ligands (Fig. 4, 16 and 17).9 The second probe was based on rapamycin, a potent inhibitor of FKBP12, which, upon binding, forms a ternary complex with mTOR (Fig. 4, 18 and 19).28,29 Recently, Woo and coworkers elegantly demonstrated the ability to probe the formation of this ternary complex utilizing a diazirine-based rapamycin photoaffinity probe.30 Key to both (+)-JQ1- and rapamycin-based probe designs was identification of known points of modification, such that the acyl silane would have minimal impact on protein binding. Thus, pure recombinant BRD4-BD1 and FKBP12 protein were treated with probes 16/17 and 18/19, respectively, followed by irradiation with a 6 W handheld UV lamp at 365 nm (Fig. 4). Covalently labeled protein adducts were subsequently visualized by in-gel fluorescence after Cu-catalyzed click reaction with rhodamine azide. Photochemical labeling was observed for (+)-JQ1 probes 16 and 17 with no thermal background labeling. While more intense fluorescence was observed for the probe with the shorter linker (16), both probes could be inhibited by pretreatment of BRD4-BD1 with excess (+)-JQ1. These data are consistent with labeling being driven by probe-protein binding, not an artifact caused by unspecific association of the acyl silane moiety with the protein target. Interestingly, the linker length for the rapamycin probes 18 and 19 had a more profound effect, as superior labeling of FKBP12 was observed with the longer linker probe 19. Again, minimal background labeling was observed, and for both probes, labeling was inhibited by pretreatment of FKBP12 with excess rapamycin. To evaluate the relative labeling output of acyl silane probes against traditional photoaffinity warheads, we synthesized a “minimalist” diazirine probe modified with (+)-JQ1, 16-DA (Fig. 5). Similar diazirine probes have been used to capture specific interactions in proteins and whole cells.31 Acyl silane (+)-JQ1 probe 16, the analogous dimethyl-substituted probe 16-Me, and diazirine probe 16-DA were incubated in cell lysate and irradiated for 30 min on ice. Following click reaction, visualization by in-gel fluorescence shows intense fluorescence for irradiated samples treated with probes 16 and 16-DA, with minimal thermal labeling observed (Fig. 5). Consistent with previous findings, the less bulky probe 16-Me had both lower labeling after irradiation as well as some thermal background labeling. These results support the promise of acyl silanes as novel probes for photoaffinity labeling, as the bulkier isopropyl derivative labels cell lysate on par with current photoaffinity scaffolds. From these encouraging results, we next sought to confirm that the acyl silane probes were capable of capturing targeted and specific interactions in the proteome. Thus, (+)-JQ1-acyl silane probe 16 was incubated in 231MFP cell lysate, into which exogenous BRD4-BD1 protein was added.32 Following irradiation and click reaction of rhodamine azide, labeled proteins were visualized by in-gel fluorescence. Pretreatment of lysate with excess (+)-JQ1 led to inhibition of BRD4-BD1 labeling, indicating that the (+)-JQ1 acyl silane probe is able to capture the BRD4-BD1 protein in cell lysates (Fig. 6). Initial attempts to identify protein adducts by mass spectrometry approaches were unsuccessful, indicating that further optimization of either the experimental workflow or the acyl silane moiety would be required for their use in target identification and chemoproteomic experiments, a major focus of our ongoing work.
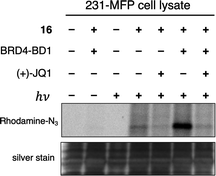 | ||
| Fig. 6 Evaluation of labeling by (+)-JQ1 photoaffinity probe 16 in 231MFP cell lysate by in-gel fluorescence (top image) following Cu click reaction with Rh–N3 after irradiation at 365 nm with a 6 W handheld UV lamp or incubation in the dark. Reactions were performed using 50 μL 231MFP cell lysate (normalized to 1 μg μL−1) spiked with 0.2 μg BRD4-BD1 in PBS with 5 μM 16 and 500 μM (+)-JQ1 for competition. Protein loading was confirmed following silver stain (bottom image) (see ESI† for full gel images). | ||
Conclusions
In summary, we have identified a novel class of photoaffinity warheads with short irradiation times capable of generating α-siloxy carbene intermediates suitable for photoaffinity labeling applications at long UV wavelengths in both cell lysate and with pure protein. Further studies are underway to better understand residue chemoselectivity, as well as an in-depth evaluation of probe structure to allow for the broader use of acyl silanes in proteome profiling and site-of-modification studies.Data availability
The experimental data, including procedures and characterization data, have been uploaded as part of the ESI.†Author contributions
F. D. T. and S. O. S. were responsible for initial acyl silane probe design and experimental validation. A. C. S. P. and S. O. S. synthesized all compounds. K. N. K., J. N. S., and B. P. B. prepared material for experiments in cell lysate. A. C. S. P., S. O. S., K. N. K., J. N. S., and B. P. B. performed experiments in cell lysate and performed gel electrophoresis. Analysis of gels was performed by D. K. N., K. N. K., J. N. S., S. O. S., A. C. S. P., and B. P. B. The initial manuscript was prepared by S. O. S. and A. C. S. P. S. M. B., J. A. T., J. M. M., M. S., D. K. N., and F. D. T. contributed to overall experimental design and manuscript preparation. All authors contributed to manuscript edits.Conflicts of interest
J. A. T., J. M. M., M. S., and S. M. B. are employees of Novartis Institutes for BioMedical Research. This study was funded by the Novartis Institutes for BioMedical Research and the Novartis-Berkeley Center for Proteomics and Chemistry Technologies. D. K. N. is a co-founder, shareholder, and adviser of Frontier Medicines.Acknowledgements
We thank Novartis Institutes for BioMedical Research and the Novartis-Berkeley Center for Proteomics and Chemistry Technologies (NB-CPACT), the Mark Foundation for Cancer Research ASPIRE award, and the National Institutes of Health (R01CA240981, R01GM136945, P42ES004705) for supporting this work, as well as the Catalysis Center (Dr Miao Zhang) and the College of Chemistry's NMR facility supported in part by NIH S10OD024998. A. C. S. P. would like to thank Dr Kacper Skakuj, Dr Thomas O'Connor, and Banruo Huang for fruitful discussions. S. O. S. would like to thank Dr Alec Christian, Dr Patti Zhang, and Dr Carl Ward for useful discussions and advice.References
- S. Ziegler, V. Pries, C. Hedberg and H. Waldmann, Angew. Chem., Int. Ed., 2013, 52, 2744 CrossRef CAS PubMed.
- M. Schenone, V. Dančík, B. K. Wagner and P. A. Clemons, Nat. Chem. Biol., 2013, 9, 232 CrossRef CAS PubMed.
- Y. Su, J. Ge, B. Zhu, Y.-G. Zheng, Q. Zhu and S. Q. Yao, Curr. Opin. Chem. Biol., 2013, 17, 768 CrossRef CAS PubMed.
- M. Schirle and J. L. Jenkins, Drug Discovery Today, 2016, 21, 82 CrossRef CAS PubMed.
- For relevant reviews of use and applications of photoaffinity probes see: (a) F. Kotzyba-Hilbert, I. Kapfer and M. Goeldner, Angew. Chem., Int. Ed., 1995, 34, 1296 CrossRef; (b) J. Sumranjit and S. J. Chung, Molecules, 2013, 18, 10425 CrossRef CAS PubMed; (c) Y. Hatanaka, Chem. Pharm. Bull., 2015, 63, 1 CrossRef CAS PubMed; (d) E. Smith and I. Collins, Future Med. Chem., 2015, 7, 159 CrossRef CAS PubMed; (e) H. A. Flaxman and C. M. Woo, Biochemistry, 2018, 57, 186 CrossRef CAS PubMed.
- R. A. G. Smith and J. R. Knowles, J. Am. Chem. Soc., 1973, 95, 5072 CrossRef CAS PubMed.
- G. W. J. Fleet, R. R. Porter and J. J. Knowles, Nature, 1969, 224, 511 CrossRef CAS.
- R. E. Galardy, L. C. Craig, J. D. Jamieson and M. P. Printz, J. Biol. Chem., 1974, 249, 3510 CrossRef CAS.
- (a) Z. Li, D. Wang, L. Lin, S. Pan, Z. Na, C. Y. J. Tan and S. Q. Yao, J. Am. Chem. Soc., 2014, 136, 9990 CrossRef CAS PubMed; (b) S. Pan, S. J. Jang, D. Wang, S. S. Liew, Z. Li, J. S. Lee and S. Q. Yao, Angew. Chem., Int. Ed., 2017, 56, 11816 CrossRef CAS PubMed.
- J. Park, M. Koh, J. Y. Koo, S. Lee and S. B. Park, ACS Chem. Biol., 2015, 11, 44 CrossRef PubMed.
- P. Kleiner, W. Heydenreuter, M. Stahl, V. S. Korotkov and S. A. Sieber, Angew. Chem., Int. Ed., 2017, 56, 1396 CrossRef CAS PubMed.
- A. H. Herner, J. Marjanovic, T. M. Lewandowski, V. Marin, M. Patterson, L. Miesbauer, D. Ready, J. Williams, A. Vasudevan and Q. Lin, J. Am. Chem. Soc., 2016, 138, 14609 CrossRef CAS PubMed.
- E. Ota, K. Usui, K. Oonuma, H. Koshino, S. Nishiyama, G. Hirai and M. Sodeoka, ACS Chem. Biol., 2018, 13, 876 CrossRef CAS PubMed.
- J. M. Duff and A. G. Brook, Can. J. Chem., 1973, 51, 2869 CrossRef CAS.
- B. G. Ramsey, A. G. Brook, A. R. Bassindale and H. Bock, J. Organomet. Chem., 1974, 3, C41 CrossRef.
- J.-H. Ye, L. Quach, T. Paulisch and F. Glorius, J. Am. Chem. Soc., 2019, 141, 16227 CrossRef CAS PubMed.
- H.-J. Zhang, D. L. Priebbenow and C. Bolm, Chem. Soc. Rev., 2013, 42, 8540 RSC.
- D. L. Priebbenow, Adv. Synth. Catal., 2020, 362, 1927 CrossRef CAS.
- C. Stuckhardt, M. Wissing and A. Studer, Angew. Chem., Int. Ed., 2021, 60, 18605 CrossRef CAS PubMed.
- J. Reimler and A. Studer, Chem.–Eur. J., 2021, 27, 15392 CrossRef CAS PubMed.
- R. A. Bourque, P. D. Davis and J. C. Dalton, J. Am. Chem. Soc., 1981, 103, 697 CrossRef CAS.
- (a) A. V. West, G. Muncipinto, H.-Y. Wu, A. C. Huang, M. T. Labenski, L. H. Jones and C. M. Woo, J. Am. Chem. Soc., 2021, 143, 6691 CrossRef CAS PubMed; (b) J. G. K. O'Brien, A. Jemas, P. N. Asare-Okai, C. W. am Ende and J. M. Fox, Org. Lett., 2020, 22, 9415 CrossRef PubMed.
- A. E. Speers, G. C. Adam and B. F. Cravatt, J. Am. Chem. Soc., 2003, 125, 4686 CrossRef CAS PubMed.
- G. Stork and K. Zhao, Tetrahedron Lett., 1989, 30, 287 CrossRef CAS.
- A. G. Brook and Z. Yu, Organometallics, 2000, 19, 1859 CrossRef CAS.
- P. Filippakopoulos, J. Qi, S. Picaud, Y. Shen, W. B. Smith, O. Fedorov, E. M. Morse, T. Keates, T. T. Hickman, I. Felletar, M. Philpott, S. Munro, M. R. McKeown, Y. Wang, A. L. Christie, N. West, M. J. Cameron, B. Schwartz, T. D. Heightman, N. La Thangue, C. A. French, O. Wiest, A. L. Kung, S. Knapp and J. E. Bradner, Nature, 2010, 468, 1067 CrossRef CAS PubMed.
- L. Anders, M. G. Guenther, J. Qi, Z. P. Fan, J. J. Marineau, P. B. Rahl, J. Lovén, A. A. Sigova, W. B. Smith, T. I. Lee, J. E. Bradner and R. A. Young, Nat. Biotechnol., 2014, 32, 92 CrossRef CAS PubMed.
- E. J. Brown, M. W. Albers, T. B. Shin, K. Ichikawa, C. T. Keith, W. S. Lane and S. L. Schreiber, Nature, 1994, 369, 756 CrossRef CAS PubMed.
- D. M. Sabatini, H. Erdjument-Bromage, M. Lui, P. Tempst and S. H. Snyder, Cell, 1994, 78, 35 CrossRef CAS PubMed.
- H. A. Flaxman, C.-F. Chang, H.-Y. Wu, C. H. Nakamoto and C. M. Woo, J. Am. Chem. Soc., 2019, 141, 11759 CrossRef CAS PubMed.
- L. P. Conway, A. M. Jadhav, R. A. Homan, W. Li, J. Sanchez Rubiano, R. Hawkins, R. M. Lawrence and C. G. Parker, Chem. Sci., 2021, 12, 7839 RSC.
- S.-Y. Wu, C.-F. Lee, H.-T. Lai, C.-T. Yu, J.-E. Lee, H. Zuo, S. Y. Tsai, M.-J. Tsai, K. Ge, Y. Wan and C.-M. Chiang, Mol. Cell, 2020, 78, 1114 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2sc00426g |
| ‡ A. C. S. P and S. O. S contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |