Open Access Article
Open Access ArticleDeterminants for intrinsically disordered protein recruitment into phase-separated protein condensates†
Yongsang
Jo
 ,
Jinyoung
Jang
,
Daesun
Song
,
Hyoin
Park
and
Yongwon
Jung
,
Jinyoung
Jang
,
Daesun
Song
,
Hyoin
Park
and
Yongwon
Jung
 *
*
Department of Chemistry, KAIST, 291 Daehak-ro, Yuseong-gu, Daejeon 34141, Republic of Korea. E-mail: ywjung@kaist.ac.kr; Fax: +82-42-350-2810; Tel: +82-42-350-2817
First published on 16th December 2021
Abstract
Multivalent interactions between amino acid residues of intrinsically disordered proteins (IDPs) drive phase separation of these proteins into liquid condensates, forming various membrane-less organelles in cells. These interactions between often biased residues of IDPs are also likely involved in selective recruitment of many other IDPs into condensates. However, determining factors for this IDP recruitment into protein condensates are not understood yet. Here, we quantitatively examined recruitment tendencies of various IDPs with different sequence compositions into IDP-clustered condensates both in vitro as well as in cells. Condensate-forming IDP scaffolds, recruited IDP clients, and phase separation conditions were carefully varied to find key factors for selective IDP partitioning in protein condensates. Regardless of scaffold sequences, charged residues in client IDPs assured potent IDP recruitment, likely via strong electrostatic interactions, where positive residues could further enhance recruitment, possibly with cation–pi interactions. Notably, poly-ethylene glycol, a widely used crowding reagent for in vitro phase separation, abnormally increased IDP recruitment, indicating the need for careful use of crowding conditions. Tyrosines of IDP clients also strongly participated in recruitment both in vitro and in cells. Lastly, we measured recruitment degrees by more conventional interactions between folded proteins instead of disordered proteins. Surprisingly, recruitment forces by an even moderate protein interaction (Kd ∼ 5 μM) were substantially stronger than those by natural IDP–IDP interactions. The present data offer valuable information on how cells might organize protein partitioning on various protein condensates.
Introduction
Membrane-less organelles (MLOs) are liquid condensates in cells with higher concentrations of specific biomolecules than their surroundings without discrete physical barriers.1 Various MLOs such as nucleoli,2 stress granules3 and p granules4 are observed in diverse locations inside cells. A series of studies suggested that MLOs are deeply related to diverse cellular processes such as DNA damage repair,5 post-transcriptional modification,6 signal transduction,7 development4 or stress response.3 MLOs, which mostly comprise proteins and nucleic acids, show liquid-like properties such as dynamic diffusivity inside, spherical shapes, and abilities to fuse with each other. MLO components can undergo continuous exchange with the outside surroundings, which might contribute to MLO sensitivity against environmental conditions such as temperature, pH, ionic strength,8 and molecular crowding.9 MLO generation and destruction can be spatiotemporally controlled by this environmental dynamicity and specific biomolecules that are interacting with MLO components.Intrinsically disordered proteins (IDPs) are proteins with fully or partially disordered structures. MLOs generally contain various IDPs,10 and IDPs are known to drive protein clustering and subsequent liquid–liquid phase separation (LLPS) with their intrinsically disordered regions (IDRs),11 ultimately to construct MLOs.12,13 IDRs have biased amino acid compositions, particularly those with polar, charged, and aromatic residues. Many studies revealed that weak multivalent interactions between these amino acids such as electrostatic, cation–pi, hydrophobic, or pi–pi interactions are essential driving forces of LLPS.14–17 Also, changes in IDR–IDR interactions by residue mutations,18,19 charge scrambling,16,20 or modifications20–23 could alter LLPS propensities and physicochemical properties of phase-separated liquid droplets. Some of these modifications even transformed protein liquid droplets to disease-related aggregation.24 Various interactions between unstructured IDR residues (e.g. electrostatic, cation–pi, hydrophobic, etc.) are thus believed to be important factors for MLO construction, maintenance, and morphological changes in cells.
Among multiple IDPs in MLOs,10 some of these IDPs dominantly drive LLPS to form MLO condensates (termed scaffolds), while other IDPs are recruited into condensates (termed clients). It is believed that multivalent residual interactions between IDRs of IDPs are one of the key factors for selective IDP recruitment. Multiple studies reported various degrees of IDP recruitment into diverse phase-separated protein condensates. For example, hydrogels25 or liquid droplets26 of full-length or IDR of fused in the sarcoma (FUS) protein could recruit various IDPs with a wide range of recruiting powers. Highly charged IDRs from LAF1 (LAF) and DDX4 proteins were also strongly recruited into various IDP condensates.27,28 Selective recruitment of IDPs and IDP condensates will heavily influence selective compartmentalization (or partitioning) of numerous IDPs and related proteins in cells. However, major determinants for selective protein recruitment into phase-separated condensates have not been fully elucidated.
Here, we designed protein condensate models by clustering phase-separable scaffold proteins such as highly charged LAF or mostly uncharged (but with many tyrosines, Tyr) FUS both in vitro and in cells (Scheme 1). Next, relative recruitment propensities of IDP client variants, the amino acid compositions of which were carefully altered (Table 1), into these condensates were quantitatively measured. We found that charged residues of IDP clients consistently drove strong client recruitment into all tested condensates under diverse in vitro and cellular conditions, indicating a dominant role of electrostatic interactions in IDP recruitment. In addition, positive (particularly arginine, Arg) and Tyr residues, which can offer pi–cation interactions, in IDP clients could further increase recruitment powers. While recruitment patterns are generally similar both in vitro and in cells, recruitment of IDPs with positive (particularly Arg) residues was abnormally enhanced in vitro by poly ethylene-glycol (PEG), which is a widely used crowding reagent to induce biomolecular phase separation. Lastly, we investigated the roles of specific interactions of folded proteins (rather than IDP–IDP interactions) in condensate protein recruitment by introducing specific binding pairs to scaffold and client proteins (Scheme 1). Protein recruitment by a binding pair with ∼5 μM Kd was clearly stronger than recruitment by interactions between IDPs. Moreover, even highly weak binding (Kd > 100 μM) provided protein recruitment comparable to recruitment by natural IDPs. These data indicate that diverse interactions between folded and disordered proteins collectively dictate selective protein partitioning in bimolecular condensates in cells.
| Protein | Dipole–dipole (S, T, N, Q) | + Charge (R, K) | − Charge (D, E) | Aromatic ring (F, Y, W) |
|---|---|---|---|---|
| FUS | 51.9% | 0.5% | 2.3% | 12.6% |
| S51, T10, N7, Q43 | R1, K0 | D5, E0 | F0, Y27, W0 | |
| F4CB | 28.0% | 12.6% | 14.0% | 12.6% |
| S0, T10, N7, Q43 | R27, K0 | D30, E0 | F0, Y27, W0 | |
| F4CB-QR | 28.0% | 12.6% | 14.0% | 12.6% |
| S10, T10, N27, Q13 | R27, K0 | D30, E0 | F0, Y27, W0 | |
| F4CB-YR | 35.5% | 12.6% | 14.0% | 5.1% |
| S10, T10, N13, Q43 | R27, K0 | D30, E0 | F0, Y11, W0 | |
| LAF | 22.0% | 14.3% | 11.9% | 7.1% |
| S12, T0, N23, Q2 | R24, K0 | D17, E3 | F1, Y11, W0 | |
| LCR | 41.7% | 0.0% | 1.8% | 11.9% |
| S45, T0, N23, Q2 | R0, K0 | D0, E3 | F1, Y19, W0 |
Results and discussion
Charged residues of clients potently and universally drive client recruitment
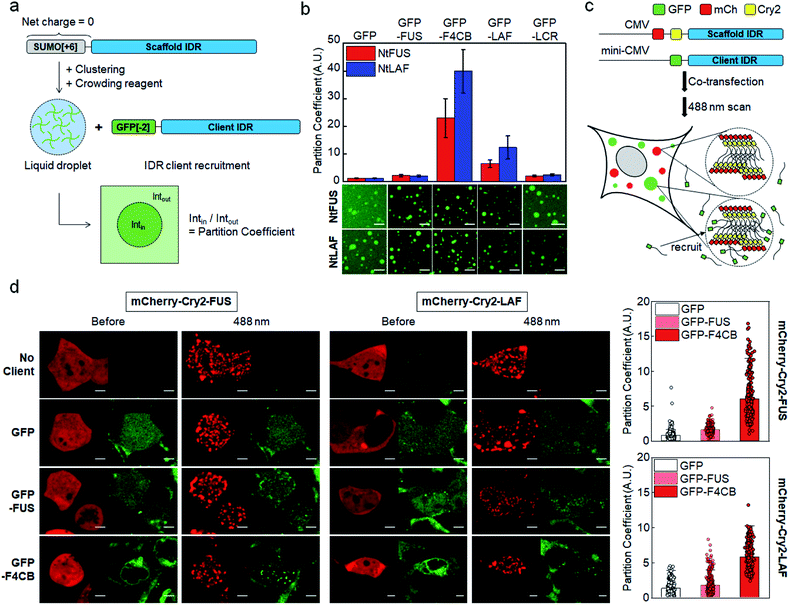
Electrostatic interactions between charged residues are likely one of the strongest among possible non-covalent interactions between disordered IDR residues. Previously,27 we observed strong recruitment of highly charged LAF and DDX IDRs into IDR condensates, but it was weakened by charge deletion. In this previous work, in vitro IDR condensates were constructed by streptavidin-mediated clustering of biotinylated IDRs, which also contained a soluble SUMO protein domain to improve protein stability. However, the biotin-SUMO domain of these IDR scaffolds had a rather biased −14 net charge, which might impede precise evaluation of the roles of charged residues for IDR recruitment into these condensates. Indeed, these SUMO-IDR condensates showed significantly stronger recruitment of positively charged proteins than negatively charged proteins (Fig. S1†). Therefore, we modified the surface charges of the biotin-SUMO domain to have a zero net charge, and fused this neutral domain to two well-known IDRs, FUS and LAF (Fig. 1a). FUS and LAF IDRs were representatively investigated because these IDRs are highly phase separable but contain distinct residue compositions: LAF is highly charged, and FUS is mostly uncharged but contains many serines (Ser), glutamines (Glu), and Tyr (Table 1). The resulting neutral IDR scaffolds (NtFUS and NtLAF) successfully formed liquid condensate droplets with high diffusivity (Fig. S2a†) and, more importantly, biased recruitment for positive charged protein clients was significantly reduced with these neutral scaffolds (Fig. S2b†). We therefore used NtFUS and NtLAF as IDP scaffolds, and GFP with a near neutral charge (−2) was fused to IDR clients for a quantitative recruitment evaluation with minimal perturbation by protein charges other than those from IDRs (Fig. 1a).To comparably test the roles of charged residues in client recruitment, we added four charged blocks to uncharged FUS (F4CB) and removed most charges from LAF (LCR) (Table 1 and Fig. S3†). For a better comparison, the final charge proportion of F4CB was designed to be similar to LAF, and the Ser and Tyr composition of LCR was designed to be similar to FUS. 50 μM of NtFUS and NtLAF scaffolds was clustered and phase separated in the presence of 5% (crowding reagent) PEG. GFP-fused IDR clients (1.5 μM) were included, and client recruitment was quantified by measuring the ratio of the GFP fluorescence signals in and out of condensates (ratio = partition coefficient (PC)) (Fig. 1a). GFP without IDRs barely localized in both NtFUS and NtLAF condensates (PCs ∼ 1.2), but GFP-FUS showed weak but clear recruitment with PCs ∼2 (Fig. 1b). Highly charged GFP-LAF showed significantly higher PCs (∼10). However, when charged residues of LAF were mostly removed (GFP-LCR), recruitment was vastly weakened down to the levels of GFP-FUS, indicating the dominant role of charge residues for client recruitment. Interestingly, recruitment of FUS with charged blocks (F4CB) was even stronger (three- to four-fold) than that of LAF. Compared to LAF, longer charge blocks and more Tyr of F4CB (Table 1 and Fig. S3†) might synergistically contribute to this unusually high recruitment.
Observed strong recruitment of charged clients into IDR condensates was also examined with condensates made by folded proteins without IDRs. Previously reported PRM and SH3 tandem repeat pentamers were mixed to drive phase separation by multiple interactions of PRM and SH3.29 PRM-SH3 condensates were overall neutral while highly charged (PRM pentamer: +31, SH3 pentamer: −32) (Fig. S4†). Strong recruitment for charged LAF and even stronger recruitment for F4CB were also observed with this non-IDR condensate (Fig. S4†), indicating that client IDR charges potently drive recruitment, largely regardless of condensate components.
We also examined the charge-dependent IDR recruitment into IDR condensates in cells. In general, 200–300 mg mL−1 of macromolecules, which are mainly composed of proteins and nucleic acids, are crowded in cells, and they occupy 20–30% of intracellular volume.30 It is clear that designed crowded in vitro conditions, mostly with crowding reagents, are vastly different from crowding environments in cells. For example, 8 kDa PEG is known to occupy ∼40% of volume in vitro with only 40 mg mL−1 (4%) concentration.31 Many recent studies reported that crowding reagents commonly used in vitro can interfere with biomolecular interactions in various ways, affecting diverse biomolecular phase separation related processes.9,12,32,33 In this regard, IDR recruitment must also be carefully investigated in cells to determine meaningful factors for the process.
Light-induced IDR clustering by using Cry2 (with fused red mCherry) was used to generate IDR condensates in cells,28,34 and GFP-fused IDR clients were co-expressed (Fig. 1c). However, unlike in vitro tests, where excess scaffolds (50 μM) were mixed with clients (1.5 μM), simple co-expression of scaffolds and clients might result in a significant portion of condensates being occupied by clients, which would lead to unwanted condensate property variations. More importantly, high client concentrations can reach close to the saturated maximal client concentration inside condensates, and the resulting PCs will not properly indicate recruitment propensity. Therefore, we lowered the expression levels of GFP-IDR clients by using the mini-CMV promoter.35 Compared to the original CMV promoter, protein expression levels were ∼seven-fold lowered with the mini-CMV promoter (Fig. S5 and Table S8†).
Expressed scaffolds (red mCherry) and clients (green GFP) were both uniformly distributed in cells before light illumination. After 488 nm light exposure, FUS and LAF condensates (red) were clearly formed in cells (Fig. 1d). The distribution of GFP clients without IDRs was not altered by condensation, indicating an absence of recruitment. GFP-FUS was only slightly recruited into both FUS and LAF condensates (PCs 1.6–1.8). On the other hand, GFP-F4CB was strongly recruited into cellular condensates with PCs reaching ∼6 (Fig. 1d, right graphs), supporting the influential roles of client charges in condensate recruitment even in cells. Although PC values were highly varied between cells, these values did not show a distinct correlation with the relative scaffold/client expression levels (Fig. S6†). Interestingly, client recruitment PC values in cells were generally much smaller than those obtained in vitro (Fig. 1a, S7, Tables S1 and S2†). The in vitro crowding condition with 5% PEG could magnify client recruitment, compared to cellular environments, which are crowded by various biomolecules.
Positive arginines of clients drive strong recruitment, particularly in vitro
We next examined whether positive (Arg) and negative (aspartate, Asp) residues differently influence client recruitment. Charge blocks of F4CB were used to prepare GFP-fused IDR clients with one or two negative or positive charge blocks (net IDR charges: −34, −19, 10, 24; Fig. 2a). Designed negative and positive blocks have similar lengths, net charges, and overall amino acid compositions (Fig. S3†). Against both NtFUS and NtLAF condensates, positive charged clients showed substantially stronger recruitment than negative clients (Fig. 2b). In fact, condensate recruitment of negative clients was barely observed. This result can be explained by the fact that positive residues, particularly Arg, can have diverse interactions (most notably cation–pi interaction) with other residues in addition to electrostatic interactions,36 whereas negative residues will mostly contribute to electrostatic interactions.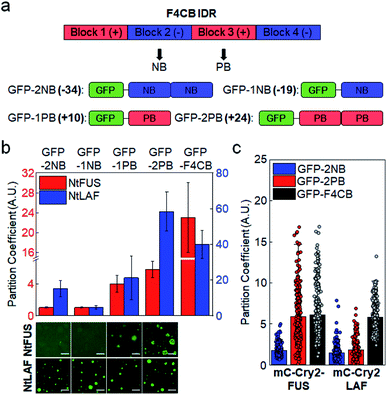 | ||
| Fig. 2 Condensate recruitment for charge biased IDR clients in vitro and in cells. (a) Construction of negative and positive clients from F4CB IDR. Net charges of clients are in bold. (b) Recruitment degrees (PCs) of charge biased IDRs into NtFUS (left y scale, red bars) and NtLAF (right y scale, blue bars) condensates in vitro with representative confocal images. GFP-F4CB values (data from Fig. 1) are included for comparison. Scale bars: 20 μm. Error bars: 1 s.d. (n = 100 from three independent experiments). (c) Recruitment degrees (PCs) of charge biased IDRs and F4CB (data from Fig. 1) into IDR condensates in cells. Error bars: 1 s.d. (n ≥ 30 cells). | ||
Positive-charged GFP-2PB (+24) also showed much stronger recruitment into FUS condensates in cells than negative-charged GFP-2NB (−34) (Fig. 2c). However, both GFP-2PB and GFP-2NB were only moderately recruited to LAF condensates in cells. Compared to LAF (11 Tyr), FUS has much more Tyr (27), which is known to mediate strong cation–pi interactions with positive residues. Cation–pi interactions in FUS proteins are important for FUS phase separation,15,37 and mutations of Tyr to Ser in scaffold FUS increased critical concentrations of phase separation.18 Still, strong recruitment of positive-charged clients into LAF condensates observed in vitro was not observed in cells (Fig. 2c). In-cell data indicate that client recruitment by IDRs in cells is mostly moderate (average PCs ∼ 2) (ESI Table†). It is possible that recruitment by IDR interactions is not overly altered by IDR residue variations in complex cellular environments, unless there are uncommon modifications such as numerous charge blocks (i.e. F4CB and 2 PB: PCs ∼ 6) (Fig. S7 and S8†). On the other hand, recruitment PCs are widely varied in vitro (1–40), suggesting that the relative recruitment power can be more sensitively measured (artificially and more simply crowded) in vitro than in cells.
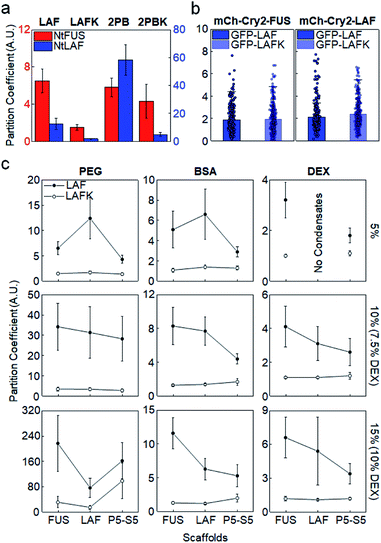
The importance of positive Arg in IDR recruitment was further investigated by mutating Arg to lysine (Lys). Arg and Lys are both positively charged, but multiple studies reported the unique effects of Arg in IDP phase separation via strong cation–pi and pi–pi interactions with other aromatic or even non-aromatic amino acids.16,17,36,38 We mutated all Arg residues to Lys in LAF (LAFK) and 2 PB (2PBK) clients. Under the in vitro (5% PEG) condition, client recruitment was evidently weakened by this Arg–Lys mutation (Fig. 3a), supporting that Arg with its more diverse residual interactions than Lys plays important roles in IDR recruitment into protein condensates. However, again, IDR recruitment was not influenced by this client mutation in cells (Fig. 3b and S9†).
As mentioned earlier, proteins in cells are crowded by a wide range of bio-macromolecules such as proteins, nucleic acids, and polysaccharides with extremely high concentrations. To further validate IDR client recruitment propensities under more diverse phase separation conditions in vitro, we additionally tested two other biomolecule-based crowding reagents, BSA (protein) and dextran (polysaccharide). Crowding reagent concentrations were also increased up to 15% (150 mg mL−1) to reach close to cellular conditions (PEG 5–15%, BSA 5–15% with 2% PEG, dextran 5–10%). 2% PEG was added to BSA since BSA alone was not effective to induce IDR phase separation, and dextran was used up to 10% due to solubility. Again, for all tested in vitro crowding conditions, GFP-LAF clients with Arg showed consistently stronger recruitment into IDR condensates than GFP-LAFK with Arg–Lys mutations (Fig. 3c). Moreover, preferred recruitment of GFP-LAF over GFP-LAFK was also observed with fold protein (PRM-SH3) condensates. Unless high concentrations (10%, 15%) of unnatural PEG were used, GFP-LAFK recruitments were extremely weak, with PCs less than 1.5 for all conditions and condensates (ESI Table†).
Although recruitment PCs with BSA and dextran were closer to those obtained in cells than with PEG, Arg effects were nonetheless observed only in vitro. We suspect that cells are crowded with diverse biomolecules, some of which can non-specifically or even specifically interact with client IDRs, and therefore specific recruitments into condensates are rather moderate, which also makes client compositional variations less influential for recruitment power. Additionally, small PC changes cannot be reliably measured in cells due to high cell-to-cell signal variations. We also note that IDR recruitments were uniquely and vastly increased with PEG increases but not with BSA or dextran (Fig. S10†). In particular, this increase was more evident for charged clients. PEGs occupy high solution volumes, and are also less polar than BSA or dextran. PEG accumulation around amino acid residues (preferably non-polar residues) was theoretically reported.39 We also found that PEG was more concentrated inside IDR condensates (PCs 2–6), while BSA is evenly distributed inside and outside condensates (Fig. S11†). The exact effects of these characteristics of PEG on biomolecular LLPS processes including client recruitment are not yet clear. Nevertheless, in vitro LLPS studies with commonly used PEG must be carefully interpreted by considering various effects of PEG that are different from other crowding reagents and cellular conditions.
Tyrosines synergistically promote client recruitment with positive residues
Among tested IDR variants, F4CB and 2 PB, both of which contain high levels of Arg and Tyr residues, showed consistently strong client recruitment in vitro as well as in cells. In fact, only these IDRs had average recruitment PCs exceeding 5 in cells. Many studies have reported that electrostatic and cation–pi interactions are major driving forces of charge-abundant IDR LLPS, while pi–pi stacking,17 sp2/pi interaction40 and hydrogen bonding can be important driving forces of charge-deficient IDR interactions.41 Tyr plays major roles in various pi interactions, and Tyr residues were essential for LLPS of uncharged IDRs such as FUS.40 To examine the synergistic role of Tyr in observed strong client recruitment of charged IDRs, a large portion of Tyr residues in F4CB were removed (F4CB-YR, Table 1). Strong client recruitment of F4CB into IDR condensates was vastly reduced by this Tyr removal, particularly with crowded BSA conditions (Fig. 4a).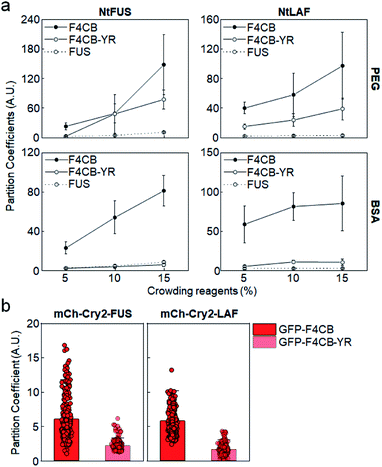 | ||
| Fig. 4 Condensate recruitment for Tyr removed F4CB clients. (a) Recruitment PCs of F4CB, F4CB-YR, and FUS into IDR condensates in vitro with varying concentrations of crowding reagent PEG and BSA. Error bars: 1 s.d. (n = 100 from three independent experiments). (b) Recruitment PCs of GFP-F4CB and GFP-F4CB-YR into IDR condensates in cells. PC values of F4CB client are identical with those in Fig. 1c. Error bars: 1 s.d. (n ≥ 30 cells). | ||
However, F4CB-YR, which contains charge blocks, showed reduced but still high recruitment under PEG conditions, indicating that charged residues alone can drive strong recruitment with crowded PEG. Overall, charge-driven client recruitment was only moderate under BSA-crowded in vitro and in cell conditions. In fact, synergistic strong client recruitment of F4CB in cells also disappeared by Tyr removal in cells (Fig. 4b and S12†). We also removed Gln residues from F4CB, since Gln was also one of the critical residues for FUS LLPS.40 However, recruitment of the resulting F4CB-QR into IDR condensates was mostly similar to that of original F4CB (Fig. S13†). Conclusively, under our experimental in vitro and in cell conditions, consistently high client recruitment was observed only for highly (positive) charged clients with abundant Tyr residues.
Client recruitment via specific interactions between folded proteins
Under our experimental conditions, natural IDR clients were somewhat weakly recruited into protein condensates in cells (PCs ∼ 2). This recruitment was likely driven by slightly stronger residual interactions of unstructured residues of client IDRs with condensate components than with surrounding biomolecules. However, these interactions of disordered residues might not be sufficiently strong or specific to explain highly selective localization (partitioning) of a specific set of proteins inside natural MLOs. Natural IDPs mostly have both unstructured regions (IDRs) and structured regions. One of the key roles of these structured domains in IDPs is to specifically interact with target proteins or nucleic acids. For example, a number of FUS family proteins contain RNA recognition motifs in addition to their disordered regions.37 Specific protein interaction networks are also essential for the composition and miscibility of stress granule and P-body condensates.42 Therefore, it is likely that IDR residue interactions and specific folded protein interactions are both involved in recruitment processes of natural MLOs. However, the relative contribution of these two interactions for client recruitment is still unknown.A direct affinity comparison between IDR interactions and folded protein interactions is not possible yet since the affinities of IDR interactions are not clearly defined or measured. We measured client recruitment by interactions of folded proteins with a specific binding affinity to weigh relative recruitment powers. The well-studied bacterial protein binding pair, SpyCatcher (SC) and SpyTag,43 was used to eliminate unwanted interactions with cellular proteins in tested mammalian cells. D7N SpyTag (D7N), Glu-to-Asn mutated SpyTag, specifically interacts with SC with Kd ∼ 5 μM (Fig. S14†). D7N was fused to the NtFUS scaffold, and the resulting NtFUS-D7N successfully generated phase-separated liquid condensates (Fig. S15†). In MLOs, which contain many different biomolecules, a particular binding protein will be only a fraction of inside components. Therefore, we prepared NtFUS condensates with varying percentages of NtFUS-D7N (from 20% to 0.5%). GFP-fused client SC was strongly recruited into these NtFUS/NtFUS-D7N condensates (PCs ∼ 5–8) (Fig. 5a). Even with only 0.5% NtFUS-D7N, GFP-SC recruitment was comparable to highly charged GFP-LAF recruitment. To examine the affinity–recruitment relation, we reduced the SC-D7N affinity by shortening the D7N tag (D7Ns),43 which showed no clear SC binding signals even at 100 μM (Fig. S14†) (Kd over 100 μM). GFP-SC recruitment was vastly weakened by this D7N shortening, where recruitment PC was ∼2 even with 20% NtFUS-D7Ns (Fig. 5a). These data clearly indicate that specific SC-D7N interactions drive client recruitment.
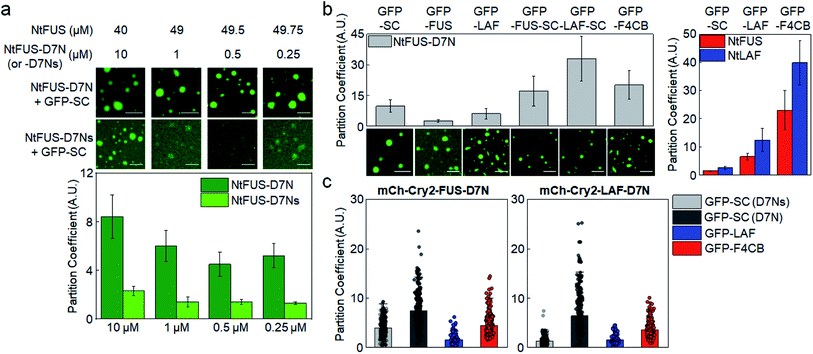 | ||
| Fig. 5 Condensate client recruitment via specific interactions of structured proteins. (a) Recruitment degrees (PCs) of GFP-SC into NtFUS/NtFUS-D7N (or -D7Ns) condensates in vitro with representative confocal images. Concentrations of used scaffold proteins are indicated. Scale bars: 20 μm. Error bars: 1 s.d. (n = 100 from three independent experiments). (b) Recruitment PCs of client proteins with SC or IDR into NtFUS-D7N condensates in vitro with representative confocal images. Recruitment PCs of GFP-SC, GFP-LAF, and GFP-F4CB into NtFUS/NtLAF condensates (data from Fig. 1) are shown in the right graph for comparison. Scale bars: 20 μm. Error bars: 1 s.d. (n = 100 from three independent experiments) (c) Recruitment PCs of client proteins with SC or IDR into mCh-Cry2-IDR-D7N condensates in cells. GFP-SC recruitment into mCh-Cry2-IDR-D7Ns condensates is indicated as GFP-SC (D7Ns) (light gray). Error bars: 1 s.d. (n ≥ 30 cells). | ||
When IDRs (FUS or LAF) were additionally fused to GFP-SC, client recruitment into NtFUS-D7N was further enhanced (Fig. 5b), indicating synergistic effects by both IDR and structured protein interactions. Recruitment PC of GFP-LAF-SC reached ∼30, which is higher than even strong F4CB recruitment (PC ∼ 20). Strong recruitment by SC fusion on various clients was not observed when NtFUS or NtLAF without D7N was used as condensate scaffolds (Fig. S16a†). On the other hand, strong GFP-SC recruitment and even stronger GFP-IDR-SC recruitment were similarly observed under the BSA crowding condition (5% BSA + 2% PEG) (Fig. S16b†).
We also examined client recruitment by SC-D7N binding in cells. FUS-D7N and LAF-D7N condensates were formed in cells again by light-induced Cry2 clustering. Surprisingly, GFP-SC recruitment (via Kd 5 μM binding) was stronger than GFP-F4CB (Fig. 5c). Artificial IDR F4CB showed the strongest recruitment among all tested IDRs in vitro as well as in cells. Moreover, when shortened D7Ns was used, SC recruitment (via Kd > 100 μM binding) was still evident and even comparable to GFP-LAF client recruitment. We also note that recruitment PCs in cells was close to that obtained in vitro for SC-D7N (Fig. S17†). On the other hand, PCs for LAF and F4CB in cells were dramatically lower than in vitro values. These results indicate that interactions between unstructured IDRs can be heavily affected by surrounding molecules, whereas specific interactions between structured motifs were less affected. We envision that interactions of IDRs and folded proteins together orchestrate protein recruitments in MLOs, although specific protein bindings can offer more potent and also consistent recruitment forces.
Conclusions
We examined major environmental or interacting factors for IDR client recruitment into protein condensates by careful design of in vitro and in cell models. A series of mutation tests revealed that charged residues (particularly positive charges) in client IDRs drive strong recruitment into various protein condensates. Electrostatic and cation–pi interactions, which are also important forces for LLPS, played major roles in client recruitment. In fact, client IDRs with high degrees of both Arg and Tyr (two key residues for electrostatic and cation–pi interactions) showed the strongest recruitment under diverse conditions. Overall, residue compositions of client IDRs were less influential for client recruitment in cells than in vitro. We believe that potential interactions of client IDRs with complex mixtures of surrounding biomolecules in cells might weaken selective recruitment into condensate environments. On the other hand, recruitment by specific binding between folded proteins was clearly controlled by binding strength. Even ∼5 μM Kd binding offered stronger recruitment than all tested IDR interactions. Although interactions between disordered proteins are important for LLPS and recruitment, interactions of structured proteins might be dominant factors for selective partitioning of many specific biomolecules in MLOs in cells.IDR recruitment was also strongly affected by crowding reagents used in vitro. PEG induced unnaturally strong client (particularly charged clients) recruitment. In this regard, it must also be noted that observed PC value variations in cells might be altered with other LLPS models that are different from the light-induced clustering system. Further variations such as LLPS-inducing methods, scaffold/client ratios, and condensate components in cellular LLPS models should be investigated. There are also many remaining questions that we wish to address to fully understand selective client recruitment. It is not clear if there are specific interactions between certain IDRs among many IDRs. It will also be interesting to study whether binding strengths are different inside and outside condensates. Lastly, we also plan to investigate if there are threshold driving forces (binding affinities) for effective biomolecule partitioning in cells.
Data availability
All experimental supporting data and procedures are available in the ESI.†Author contributions
Y. Jo conducted or organized all experiments. J. J. assisted Y. Jo for in vitro experiments. D. S. developed the mini-CMV expression system. H. P. performed SPR studies for specific protein interactions. Y. Jo and Y. Jung designed the project and wrote the manuscript with input from all the authors.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by BioNano Health Guard Research Center funded by the Ministry of Science and ICT (MSIT) as Global Frontier Project (H-GUARD_2014M3A6B2060507 (1711073453)) and the National Research Foundation of Korea (NRF) grant funded by MSIT (NRF-2019R1A2C2008558).References
- S. Boeynaems, S. Alberti, N. L. Fawzi, T. Mittag, M. Polymenidou, F. Rousseau, J. Schymkowitz, J. Shorter, B. Wolozin, L. Van Den Bosch, P. Tompa and M. Fuxreiter, Trends Cell Biol., 2018, 28, 420–435 CrossRef CAS PubMed.
- C. P. Brangwynne, T. J. Mitchison and A. A. Hyman, Proc. Natl. Acad. Sci. U. S. A., 2011, 108, 4334–4339 CrossRef CAS PubMed.
- P. Anderson and N. Kedersha, Trends Biochem. Sci., 2008, 33, 141–150 CrossRef CAS PubMed.
- C. P. Brangwynne, C. R. Eckmann, D. S. Courson, A. Rybarska, C. Hoege, J. Gharakhani, F. Julicher and A. A. Hyman, Science, 2009, 324, 1729–1732 CrossRef CAS PubMed.
- M. Altmeyer, K. J. Neelsen, F. Teloni, I. Pozdnyakova, S. Pellegrino, M. Grofte, M. D. Rask, W. Streicher, S. Jungmichel, M. L. Nielsen and J. Lukas, Nat. Commun., 2015, 6, 8088 CrossRef CAS PubMed.
- P. Anderson and N. Kedersha, Nat. Rev. Mol. Cell Biol., 2009, 10, 430–436 CrossRef CAS PubMed.
- X. Su, J. A. Ditlev, E. Hui, W. Xing, S. Banjade, J. Okrut, D. S. King, J. Taunton, M. K. Rosen and R. D. Vale, Science, 2016, 352, 595–599 CrossRef CAS PubMed.
- D. M. Mitrea and R. W. Kriwacki, Cell Commun. Signal., 2016, 14, 1 CrossRef PubMed.
- A. A. M. Andre and E. Spruijt, Int. J. Mol. Sci., 2020, 21, 5908 CrossRef CAS PubMed.
- A. L. Darling, Y. Liu, C. J. Oldfield and V. N. Uversky, Proteomics, 2018, 18, e1700193 CrossRef PubMed.
- L. Malinovska, S. Kroschwald and S. Alberti, Biochim. Biophys. Acta, 2013, 1834, 918–931 CrossRef CAS PubMed.
- P. Yang, C. Mathieu, R. M. Kolaitis, P. Zhang, J. Messing, U. Yurtsever, Z. Yang, J. Wu, Y. Li, Q. Pan, J. Yu, E. W. Martin, T. Mittag, H. J. Kim and J. P. Taylor, Cell, 2020, 181, 325–345.e328 CrossRef CAS PubMed.
- D. S. W. Protter, B. S. Rao, B. Van Treeck, Y. Lin, L. Mizoue, M. K. Rosen and R. Parker, Cell Rep., 2018, 22, 1401–1412 CrossRef CAS PubMed.
- C. P. Brangwynne, P. Tompa and R. V. Pappu, Nat. Phys., 2015, 11, 899–904 Search PubMed.
- S. Qamar, G. Wang, S. J. Randle, F. S. Ruggeri, J. A. Varela, J. Q. Lin, E. C. Phillips, A. Miyashita, D. Williams, F. Strohl, W. Meadows, R. Ferry, V. J. Dardov, G. G. Tartaglia, L. A. Farrer, G. S. Kaminski Schierle, C. F. Kaminski, C. E. Holt, P. E. Fraser, G. Schmitt-Ulms, D. Klenerman, T. Knowles, M. Vendruscolo and P. St George-Hyslop, Cell, 2018, 173, 720–734.e715 CrossRef CAS PubMed.
- S. Das, Y. H. Lin, R. M. Vernon, J. D. Forman-Kay and H. S. Chan, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 28795–28805 CrossRef CAS PubMed.
- R. M. Vernon, P. A. Chong, B. Tsang, T. H. Kim, A. Bah, P. Farber, H. Lin and J. D. Forman-Kay, Elife, 2018, 7, e31486 CrossRef PubMed.
- Y. Lin, S. L. Currie and M. K. Rosen, J. Biol. Chem., 2017, 292, 19110–19120 CrossRef CAS PubMed.
- A. E. Conicella, G. H. Zerze, J. Mittal and N. L. Fawzi, Structure, 2016, 24, 1537–1549 CrossRef CAS PubMed.
- T. J. Nott, E. Petsalaki, P. Farber, D. Jervis, E. Fussner, A. Plochowietz, T. D. Craggs, D. P. Bazett-Jones, T. Pawson, J. D. Forman-Kay and A. J. Baldwin, Mol. Cell, 2015, 57, 936–947 CrossRef CAS PubMed.
- I. Kwon, M. Kato, S. Xiang, L. Wu, P. Theodoropoulos, H. Mirzaei, T. Han, S. Xie, J. L. Corden and S. L. McKnight, Cell, 2013, 155, 1049–1060 CrossRef CAS PubMed.
- W. M. Aumiller, Jr. and C. D. Keating, Nat. Chem., 2016, 8, 129–137 CrossRef PubMed.
- D. T. Murray, M. Kato, Y. Lin, K. R. Thurber, I. Hung, S. L. McKnight and R. Tycko, Cell, 2017, 171, 615–627.e616 CrossRef CAS PubMed.
- A. Patel, H. O. Lee, L. Jawerth, S. Maharana, M. Jahnel, M. Y. Hein, S. Stoynov, J. Mahamid, S. Saha, T. M. Franzmann, A. Pozniakovski, I. Poser, N. Maghelli, L. A. Royer, M. Weigert, E. W. Myers, S. Grill, D. Drechsel, A. A. Hyman and S. Alberti, Cell, 2015, 162, 1066–1077 CrossRef CAS PubMed.
- M. Kato, T. W. Han, S. Xie, K. Shi, X. Du, L. C. Wu, H. Mirzaei, E. J. Goldsmith, J. Longgood, J. Pei, N. V. Grishin, D. E. Frantz, J. W. Schneider, S. Chen, L. Li, M. R. Sawaya, D. Eisenberg, R. Tycko and S. L. McKnight, Cell, 2012, 149, 753–767 CrossRef CAS PubMed.
- Y. Lin, D. S. Protter, M. K. Rosen and R. Parker, Mol. Cell, 2015, 60, 208–219 CrossRef CAS PubMed.
- Y. Jo and Y. Jung, Chem. Sci., 2019, 11, 1269–1275 RSC.
- D. Song, Y. Jo, J. M. Choi and Y. Jung, Nat. Commun., 2020, 11, 5642 CrossRef CAS PubMed.
- P. Li, S. Banjade, H. C. Cheng, S. Kim, B. Chen, L. Guo, M. Llaguno, J. V. Hollingsworth, D. S. King, S. F. Banani, P. S. Russo, Q. X. Jiang, B. T. Nixon and M. K. Rosen, Nature, 2012, 483, 336–340 CrossRef CAS PubMed.
- R. J. Ellis, Curr. Opin. Struct. Biol., 2001, 11, 114–119 CrossRef CAS.
- B. Akabayov, S. R. Akabayov, S. J. Lee, G. Wagner and C. C. Richardson, Nat. Commun., 2013, 4, 1615 CrossRef PubMed.
- T. Kaur, I. Alshareedah, W. Wang, J. Ngo, M. M. Moosa and P. R. Banerjee, Biomolecules, 2019, 9, 71 CrossRef CAS PubMed.
- A. M. Marianelli, B. M. Miller and C. D. Keating, Soft Matter, 2018, 14, 368–378 RSC.
- Y. Shin, J. Berry, N. Pannucci, M. P. Haataja, J. E. Toettcher and C. P. Brangwynne, Cell, 2017, 168, 159–171.e114 CrossRef CAS PubMed.
- J. Wu, S. Zaccara, D. Khuperkar, H. Kim, M. E. Tanenbaum and S. R. Jaffrey, Nat. Methods, 2019, 16, 862–865 CrossRef CAS PubMed.
- K. Kumar, S. M. Woo, T. Siu, W. A. Cortopassi, F. Duarte and R. S. Paton, Chem. Sci., 2018, 9, 2655–2665 RSC.
- J. Wang, J. M. Choi, A. S. Holehouse, H. O. Lee, X. Zhang, M. Jahnel, S. Maharana, R. Lemaitre, A. Pozniakovsky, D. Drechsel, I. Poser, R. V. Pappu, S. Alberti and A. A. Hyman, Cell, 2018, 174, 688–699.e616 CrossRef CAS PubMed.
- B. S. Schuster, G. L. Dignon, W. S. Tang, F. M. Kelley, A. K. Ranganath, C. N. Jahnke, A. G. Simpkins, R. M. Regy, D. A. Hammer, M. C. Good and J. Mittal, Proc. Natl. Acad. Sci. U. S. A., 2020, 117, 11421–11431 CrossRef CAS PubMed.
- G. Settanni, J. Zhou, T. Suo, S. Schottler, K. Landfester, F. Schmid and V. Mailander, Nanoscale, 2017, 9, 2138–2144 RSC.
- A. C. Murthy, G. L. Dignon, Y. Kan, G. H. Zerze, S. H. Parekh, J. Mittal and N. L. Fawzi, Nat. Struct. Mol. Biol., 2019, 26, 637–648 CrossRef CAS PubMed.
- I. Peran and T. Mittag, Curr. Opin. Struct. Biol., 2020, 60, 17–26 CrossRef CAS PubMed.
- D. W. Sanders, N. Kedersha, D. S. W. Lee, A. R. Strom, V. Drake, J. A. Riback, D. Bracha, J. M. Eeftens, A. Iwanicki, A. Wang, M. T. Wei, G. Whitney, S. M. Lyons, P. Anderson, W. M. Jacobs, P. Ivanov and C. P. Brangwynne, Cell, 2020, 181, 306–324 CrossRef CAS PubMed.
- B. Zakeri, J. O. Fierer, E. Celik, E. C. Chittock, U. Schwarz-Linek, V. T. Moy and M. Howarth, Proc. Natl. Acad. Sci. U. S. A., 2012, 109, E690–E697 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d1sc05672g |
| This journal is © The Royal Society of Chemistry 2022 |