Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
The effect of chemical representation on active machine learning towards closed-loop optimization†
A.
Pomberger
a,
A. A.
Pedrina McCarthy
b,
A.
Khan
a,
S.
Sung
c,
C. J.
Taylor
ad,
M. J.
Gaunt
 b,
L.
Colwell
b,
D.
Walz
b,
L.
Colwell
b,
D.
Walz
 e and
A. A.
Lapkin
e and
A. A.
Lapkin
 *ac
*ac
aDepartment of Chemical Engineering and Biotechnology, University of Cambridge, Cambridge CB3 0AS, UK. E-mail: aal35@cam.ac.uk
bYusuf Hamied Department of Chemistry, University of Cambridge, Cambridge CB2 1EW, UK
cCambridge Centre for Advanced Research and Education in Singapore Ltd., CREATE Tower 05-05, 138602 Singapore
dAstex Pharmaceuticals, 436 Cambridge Science Park, Milton, Cambridge CB4 0QA, UK
eBASF SE Data Science for Materials, Carl-Bosch-Strasse 38, 67056 Ludwigshafen am Rhein, Germany
First published on 11th March 2022
Abstract
Multivariate chemical reaction optimization involving catalytic systems is a non-trivial task due to the high number of tuneable parameters and discrete choices. Active machine learning (ML) represents a powerful strategy for automating reaction optimization. However, the translation of chemical reaction conditions into a machine-readable format requires the identification of highly informative features which accurately capture the factors which determine reaction success. Herein, we compare the efficacy of different calculated chemical descriptors for a high throughput experimentation generated dataset to determine the impact on a supervised ML model when predicting reaction yield. Then, the effect of featurization and size of the initial dataset within a closed-loop reaction optimization was examined. Finally, the balance between descriptor complexity and dataset size was considered. Ultimately, tailored descriptors did not outperform simple generic representations, however, a larger initial dataset accelerated reaction optimization.
Introduction
Identifying the optimal reaction conditions to enact a specific transformation is a major challenge for chemists, particularly in the field of small molecule drug synthesis and natural product synthesis.1,2 The field of laboratory automation allows for the rapid and systematic generation of high-quality data, which, when used in combination with ML-directed self-optimization algorithms can become a powerful tool for research.3–8 In order to apply such tools, a chemical reaction must be represented in a machine-readable format. This representation must be composed of descriptors that are simple and relevant enough to avoid the introduction of undesired noise, yet information-rich, enough to account for properties that impact reaction success such as sterics and electronics.Unlike the large datasets used for ML in other disciplines (e.g., image recognition), synthetic chemistry datasets are often extremely small and, to compensate, researchers often develop bespoke descriptors, which are based on expert knowledge such as mechanistic understanding or quantum chemical calculations.9–12 However, it is possible that the descriptors generated contain little relevant information and are simply perceived as distracting noise by the ML model. In this publication, we aim to investigate the relationship between descriptor complexity and ML model performance when predicting the yield of chemical reactions. Furthermore, we aim to explore how the descriptor complexity impacts closed-loop optimization, a strategy that may help to guide synthetic chemists towards optimal reaction conditions.
Whilst it can be challenging for humans to identify complex relationships in large datasets, ML relies on building statistical models that adjust to the given input data and has recently proven to be a powerful tool for the successful identification of nuanced patterns in complex data.13–17 Trained ML models can be used to make predictions when given inputs that lie within the defined parameters of the training dataset, referred to as interpolative tasks. This includes new combinations of already known components, such as catalysts/ligands/additives. In contrast, extrapolative tasks, which are represented by predictions of inputs which are not represented in the training data are challenging, with the predictive power of an ML model decreasing as structural differences between the training and test data increase. These extrapolative tasks require the ML model to learn about the fundamental chemical properties and, as such, the inputs are generally molecular descriptors that are based on fundamental molecular properties such as atomic distances, orbital energies or charge distributions.18
The application of ML as a decision-making tool during reaction optimization represents an effective combination as it accelerates experimental workflows and allows for rapid gains in understanding. Active ML-driven closed-loop optimization uses an initial dataset to make predictions about yet unseen conditions and these predictions can inform decisions about the subsequent experiments. For example, the experiments predicted to deliver the highest yields or the greatest improvement in model performance can be prioritized and conducted. The data gained from running this experiment can be used to re-train the ML model and new predictions are made. This iterative process continues until the desired objective (such as more accurate predictions or increased yield) is fulfilled. ML-based optimizers have the potential to increase the efficiency in which chemical space is navigated, removing operator bias and ultimately reducing the total number of experiments required, thus reducing waste significantly.7,19–21
Previous reports vary in their conclusions on whether the implementation of chemical descriptors, rather than generic one-hot encoding (OHE) representations, truly boosted the predictive performance of their ML models.19,22,23 Pd-catalyzed C(sp3)–H activation is a powerful reaction manifold that enables the facile introduction of functional complexity in small molecules, in addition to the late-stage functionalization of complex molecules like trimipramine (1), a tricyclic antidepressant, as recently demonstrated by one of our groups.24 Hence we chose to explore the parameterization and featurization of the newly developed tertiary amine directed C(sp3)–H bond activation with a HTE-generated dataset, comparing tailored descriptors, based on in silico studies, to understand the influence of descriptor complexity on supervised ML prediction and closed-loop optimization.
Traditional round-bottom flask chemistry is still the major strategy for reaction optimization; however, it is limited by the capacity of a human experimentalist and experimental set-up can vary between chemists, unintentionally introducing sources of error. Hence, we chose to employ high-throughput experimentation (HTE) to conduct experimental arrays in parallel, increasing the rate of data generation and improving reproducibility. To the best of our knowledge, this is the first application of this chemical transformation in HTE.
The reaction contains three discrete reaction parameters which were varied in tandem: mono-N-protected amino acid (MPAA) ligand, the palladium pre-catalyst, and the aryl boronate (Scheme 1). All combinations of 31 ligands (plus one control), three pre-catalysts, and two boronates results in 186 unique conditions that were each run in quadruplicate on a 125 nmol scale using nanoscale HTE, outliers were eliminated, and the repeats averaged to ensure reproducibility. For the active learning studies, this dataset would serve as the navigable chemical space that experiments would be simulated within.
 | ||
| Scheme 1 General conditions for catalytic C(sp3)–H bond activation of tertiary alkylamines. Original conditions: Pd(OAc)2 (10 mol%), (L)-tert-leucine (25 mol%), ArB(OH)2 (1.0 equiv.). | ||
Materials and methods
Molecular parameterization
Developing machine-readable representations that can capture the correlation between structure and reactivity represents a major challenge within computational chemistry.30 A key consideration when choosing a parameterization method is that, depending on the ML model used, the sparsity of input features (i.e., the location and number of ones and zeros in a bit vector) may influence modelling performance by undermining relevant information within the input vector.Within the obtained dataset a varied structure–reactivity relationship was demonstrated by the ligands, with >90% of the variation in yield is attributable to ligand choice, and their role as knock-out criteria for the reaction, we focussed primarily on parameterizing them whilst the boronates and pre-catalysts were encoded by means of OHE and Morgan 2 fingerprints only.
MPAA ligands (Fig. 1), first popularized by Yu and co-workers in 2008,25 are uniquely favoured ligands for Pd-catalyzed C–H activation as both the carboxylate and amide motifs have relatively weak coordination strengths when compared to phosphine or NHC-type ligands, and both are able to bind to Pd as L-type (neutral) and X-type (anionic) ligands enabling them to dynamically adjust between coordination modes throughout the catalytic cycle.26 Diversity within this ligand-set comes primarily from variation of the α-substituent, and with the amide protecting group, enabling a wide range of steric and electronic profiles to be generated. The subtle impact these modifications may have on reactivity, is unintuitive and, as such, it is thought that ML may be able to aid in predicting reaction outcomes for reactions that depend on these ligands.
 | ||
| Fig. 1 An overview of common data representation methods for organic molecules (top) and the diverse ligand space used for screening within this study (bottom). | ||
Although we concede that the conformation of the ligand when alone and when in the catalytic complex differs, we aimed to keep our approach computationally inexpensive and attempted to derive descriptors from the unbound ligands (instead of the ligand–metal complex). We chose three levels of chemical parameterization complexity which contained sequentially more chemical information at the expense of increased computing time. Initially, one-hot encoding (OHE) was used, a computationally simplistic method that merely details the presence or absence of certain reagents whilst encoding no chemical information. For the inclusion of structural information, Morgan fingerprints (radius of 2, see ESI† for discussion) were chosen to encode the molecular fragments that were present in a given reaction mixture.27 These bit vectors contain information on atom types, neighbouring connectivity relationships and bond types. Finally, steric and electronic descriptors were calculated for the set of ligands using density functional theory (DFT) from geometry optimized structures (B3LYP functional and 6-31G(d) basis set, see ESI† for full details). Within this investigation different sub-sets of descriptors were used and combined to develop hybrid molecular representations, containing information of the fingerprints and DFT descriptors.
As discussed previously, there are two main positions of diversity within MPAA ligands: the α-carbon and the acetamide protecting group. Hence, to separate their influence on reaction outcome, both positions were parameterized separately for each ligand rather than using a single descriptor for the entire molecule. The ligand's steric profile was expected to influence reaction yield greatly – preliminary work clearly identified an increase in yield with increasing size of the group on the α-carbon: H → CH3 → C(CH3)3, for more detailed insights see Rodrigalvarez et al.24 Two steric descriptors were introduced, Sterimol and percentage buried volume (%VBur).28,29 Sterimol descriptors quantify steric demands along different principal axes, making them well-suited to describing the steric effects of unsymmetrical substituents. The percentage buried volume is a descriptor that is traditionally used for catalyst-ligand complexes and describes the percentage of the volume of a sphere that is occupied by a given substituent. We used the α-carbon/[N-residue] as centre of our sphere and the calculation was performed considering only the variable residue extending from this position. In addition to this, a number of electronic descriptors were calculated for the ligand molecules as the fine-tuned electronics can impact Lewis basicity of the two binding atoms (N and O) and the aptitude to engage in the key mechanistic step (concerted metalation deprotonation, CMD). To capture the electron density distribution, we calculated the HOMO/LUMO energies and conducted a NBO analysis (natural bond orbital) and a CHELPG analysis (charges from electrostatic potentials using a grid-based method).30,31
Each calculated descriptor was used as a stand-alone input and was also combined with other inputs to develop hybrid features, aiming to deliver a streamlined set of input features which allow for a detailed description of the reaction conditions. A feature importance assessment based on Gini importance method (or mean decrease in impurity, MDI) was conducted and indicated high importance for the ligand fingerprints and the DFT descriptors (see ESI† for more details).
For more detailed information about different parameterization techniques, we refer the reader to these papers on molecular fingerprints,20,32,33 and on DFT-based descriptors.7,9,11,34
Machine learning surrogate models
Following feature engineering, we wanted to compare different ML models and assess their performance given a predictive task, mapping reaction conditions to yield and make predictions for unseen conditions. Different data structures and featurization methods can deliver varying performance with different ML models, something which cannot be predicted a priori, meaning empirical evaluation is required.To evaluate the performance of a ML model, the dataset is partitioned into training and testing data. A model is trained (using the training data), before being given the inputs for the, previously unseen, testing data and asked to make predictions on the outputs. The difference between the predictions and the actual values is given with the root mean squared error (RMSE), a common performance metric. To partition the dataset into training and test sets, we applied two different strategies, a random split, and a designed split. When data for the training and testing partitions are chosen at random it is very likely that the training data contains information that is well distributed amongst the dataset and, as such, is in part representative of the test data. Thus, a random split may be considered as an interpolative prediction task. To simulate out-of-sample prediction the training/test partitions can be chosen with the intention of neglecting a specific part of the dataset, for example by excluding one ligand from the training data via a leave-one group out (LOGO) cross validation (CV). After the model has been trained, it is given the testing data that was poorly represented in the training data and attempts to make predictions. In this way a train/test partition can be designed to simulate an extrapolative prediction of unseen data. To obtain more general results, many different train-test partitions are used and an average RMSE of the predictions based on the test dataset is calculated and used as a performance metric.
Reaction optimization
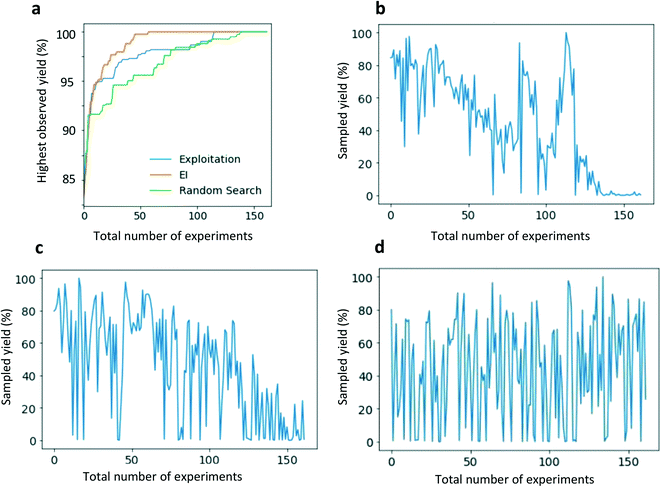
Within this study we applied closed-loop optimization using simulated experiments and assessed the effect of different surrogate models and data representations. In terms of sampling strategy, we conducted both exploitative search (the condition with the highest predicted yield is chosen for experimental evaluation) and Bayesian optimization (BO) based on expected improvement (EI) acquisition function (the condition with the highest expected improvement for yield was chosen for evaluation – incorporating the uncertainty of the prediction). For more detailed information on EI, BO and sampling strategies, refer to the ESI† and published literature.19,35,36Results and discussion
Preliminary studies: supervised ML modelling towards yield prediction
More complex features (that were chosen based on prior knowledge of the reaction) such as DFT and fingerprint-derived descriptors delivered only marginal increases in performance compared to OHE which represents the baseline. RF and GP demonstrated almost equal prediction performance, regardless of which features were used. OHE along with a linear model delivered an RMSE of 9.6% ± 0.7% (standard deviation) yield. The best performance was achieved with feature set 2 and 14 using RF, giving an RMSE of 7.6% ± 1.2% and 7.2% ± 1.3% respectively. As visible in Fig. 2, feature set 14 is a combined input of steric and electronic ligand DFT descriptors (Sterimol, %Bur, NBO, CHELPG), principal components of the Morgan 2 fingerprints, OHE and existence/absence of a proton on the amide nitrogen. RMSE is reported with respect to yield – between 0 and 1. Fig. 2a shows that different features influence the performance of ANN and the linear model significantly and that their overall performance is worse than RF or GP, which is likely due to the small size of the dataset or choice of the features. Fig. 2b illustrates a parity plot of RF regression using feature set 14, illustrating the fitting of the training and test data.
The estimated statistical uncertainty of the datapoints is 2.8%, averaged over all 186 conditions (see ESI†). This serves as a lower limit to the RMSE in the predictions of any model. It may seem surprising that models with more informative hybrid features did not strongly outperform those with OHE. However, since the latter already allows for a qualitatively good performance, there is little room for improvement when using more descriptive features. As discussed, with a random partition for the training/testing data it is likely that every ligand will be represented in the training data and, as such, it is likely that the good performance of OHE results from this ‘data leakage’ since the other two parameters (boronate, pre-catalyst) do not influence the outcome significantly. The magnitude of this effect would likely be smaller if the other two parameters had a greater influence on the reaction outcome.
A graphical representation is shown in Fig. 3a, and a more detailed summary of LOGO CV is presented in the ESI.† After the generation of test RMSE for all data sections, the mean and standard deviation can be calculated and used as indicators for model performance. Fig. 3b illustrates the comparison of different surrogate models and different data representations. Linear regression failed to conduct extrapolative predictions using features containing bit vectors as input and thus was dropped. Expanding the scope of the out-of-sample prediction, we chose to introduce two additional commonly used surrogate models: support vector regression (SVR)40 and adaptive boosting (AdaBoost)41 to experimentally test their ability to fit the chemical reaction data.
 | ||
| Fig. 3 Leave-one-group-out cross validation (CV) (a) a conceptual illustration of LOGO CV. Different shades of blue/brown/green represent datapoints within the same group. For each iteration a different colour is circled which indicates that these datapoints are used as test data for model evaluation and the other remaining datapoints are used as training data (b) LOGO CV results of different ML models with varying input features (for feature description see Fig. 2). Error bars represent the standard deviation. | ||
Comparison of the different models suggests that RF delivered the best overall performance (using Morgan 2 fingerprints as input), achieving the lowest average RMSE of 22.7% ± 18.8% (standard deviation). On the other hand, GP seems to deliver the worst performance for out-of-sample predictions across most of the input features. Feature set 2, consisting of exclusively Morgan 2 fingerprints, delivered the best prediction performance across all models. Additionally, even though the hybrid features (feature sets 11–14) include relevant principal components of the fingerprints and additional steric/electronic information from DFT calculations, they did not outperform the feature set 2. Interestingly, the performance of many of the feature sets was similar to the performance of OHE, which serves as a control (no chemical information), as can be seen in Fig. 3a in which all of the error bars of feature sets 1–14 overlap. Thus, it can be concluded that the additional time and expertise required to generate high-fidelity DFT descriptors, based on mechanistic understanding, is unjustified when the improvement in performance of fingerprints alone is only modest.
Overall, these experiments demonstrate promising predictions with interpolative modelling tasks with the lowest RMSE of 7.2% yield, however extrapolative out-of-sample predictions still represents a significant challenge with the lowest RMSE of 25% yield (or 50% of MAE). The latter results confirm and emphasize the lack of predictive power of ML for reaction condition prediction that are not directly represented within the training data, in low data regimes which are of particular relevance to bench chemists. Within the LOGO CV experiment, even if a diverse chemical space is captured in the training data, the average prediction performance for the held-out test data was limited. The preliminary assessment of these investigation allows for benchmarking of combinations of input representations and surrogate models by showing the most appropriate strategy for similar sized datasets generated by the chemical community. We believe that the performance of extrapolative predictions may be better with larger datasets and, also, may vary depending on the reaction mechanism itself since this affects the learning of structure-reactivity relationships by ML models.
Once the models were trained on the initialization data, yield predictions were generated using the relevant reaction feature-sets. The datapoint (or a batch of datapoints) with the highest yield predictions were selected for “experimental” evaluation and the true yield was transferred from the lookup table (serving as a simulated experiment) to the training dataset. The models are then retrained, and this workflow is repeated until the global optimum reaction yield was identified. We chose to use feature set 14 (unless stated otherwise), for all experiments due to the highest information content. To allow for an easy and fair performance comparison, the initialization was kept the same across all surrogate models. All learning curves shown within this section represent averaged learning trajectories from 10 individual experiments – for insights into standard deviation of those 10 single experiments please refer to ESI.†
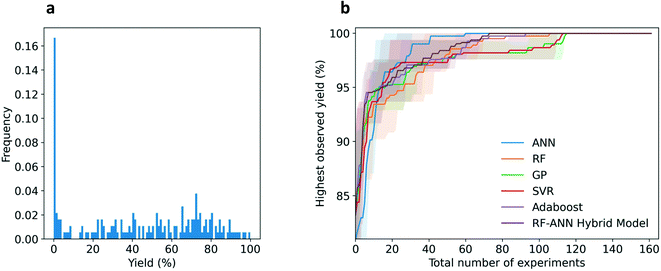
Overall, whilst the rate of improvement in the highest observed yields in the earlier iterations did not significantly vary between the different ML models, the required number of iterations to find the best-performing conditions (=99.9% yield) highlighted the differences between the models. Although the ANN model started initially with the lowest yield, the model achieved the optimal conditions within approximately 60 iterations, the fastest of all models. Tree based models such as AdaBoost and RF required approximately 100 iterations and GP/SVR achieved the ideal conditions within 110 iterations.
We hypothesized that using a combination of active surrogate models within the same optimization strategy may increase performance compared to single models. In detail, we based our investigation on the fact that the current best model (ANN) typically does not perform well at the beginning of the optimization, when very little data is available. Random forest, however, seems to perform better under a low data regime. Within the RF-ANN hybrid model, the rule was set that during the first 10 iterations the decision-making was conducted based on the predictions of the RF and then the ANN continued. Unexpectedly, the performance of the hybrid model did not outperform ANN. Nonetheless, it was the second-best model and achieved the optimal conditions within 70 iterations.
It was observed that OHE clearly outperformed the other representations after 10 iterations were achieved and reached the optimal set of conditions in the fewest number of iterations (Fig. 6). In a similar manner, recent results by Shields et al. observed that OHE delivers approximately equal performance compared to hand-crafted DFT descriptors during Bayesian optimization of organic reaction conditions.19 Initially, we assumed that the superiority of OHE performance could be due to the full factorial chemical space since all possible parameter combinations could be evaluated. Whilst this effect would benefit all representations, we hypothesized that the simplicity of OHE along with a full factorial space could be more beneficial when compared to the effect on other input features (e.g. fingerprints) that are far more complex and might represent a challenge for the model to detect patterns in the data. To test this assumption, we dropped a random selection of the datapoints of the entire dataset (25%), therefore no longer representing a full factorial chemical space. However, we still observed that OHE outperformed the full feature set (see ESI†). Another reason for the good performance of OHE might be that during each optimization experiment the model receive datapoints of the same ligand multiple times (in combination with a different pre-catalyst or boronate). As discussed previously, the impact of the ligand is significantly higher to reaction outcome, compared to the other two parameters. As a result, it is likely that OHE captures the variability between ligands and therefore can efficiently identify high yielding reaction conditions.
All previous experiments were conducted in a sequential design – during each iteration of the active ML algorithm, one decision was made and one single datapoint was sampled from the lookup table. In a real world setting this means that after each single experiment the yield is evaluated and then added to the ML training data. However, typically organic chemists conduct multiple experiments in parallel to accelerate the process of finding optimal conditions. Therefore, batch-sequential sampling within active learning represents a more realistic approach and was also investigated. The ideal batch size is a trade-off: a large batch size typically brings benefits to experimental workflows as HTE equipment can be applied, for example screening 96 conditions at a time. If the experimenter gains more useful data per HTE run, this will often lead to minimizing the total number of HTE runs and be less time intensive. However, large batch sizes at the beginning of the active learning strategy could lead to the acquisition of chemically redundant data as the active learning model may make low informative predictions due to the initially limited number of datapoints used for training. Conversely, a smaller batch size allows the training data of the ML model to be updated more frequently and thus enables better quality predictions. As shown in detail in the ESI† (Fig. S18), the batch size did not significantly vary model performance (most learning trajectories of different batch sizes are overlapping) and thus we propose they should be chosen in accordance with experimental workflows.
To conclude, we believe that prospective research in this area should consider the required complexity of molecular parameterization, due to increased computational time and expense. It could be possible that the low data regime in combination with complex features does not allow the models to efficiently learn from the data and likely over-complicates the task. Moreover, we conclude that instead of over-allocating resources on feature generation, it may be more strategic and resourceful to increase experimental data generation capabilities.
Another important factor for initializing active learning is the size of the initial training data. The choice of the size of the initial dataset represents a trade-off between showing the model enough information so that initial predictions are useful and keeping the dataset sufficiently small to limit the amount of experimental time and resources used. We chose four different sizes of initialization with five, 10, 15 and 20 random datapoints. As shown in Fig. 7b, the results indicate that larger sized initial datasets allow the model to predict conditions that give >99% yield in fewer iterations. Using 20 random datapoints for initializations allowed finding the global optimum on average within 40 iterations while using only five initial datapoints required up to 100 iterations. In total 186 datapoints are available. Whilst the choice of the adequate size of the initialization dataset might vary on the parameter space of the dataset as well as the size and complexity of the prediction space, we assume that a minimum of 15–20 datapoints should be chosen – here this choice allowed for identification of optimal conditions in approximately 45 iterations. Overall, we suspect that the smaller sized initialization datasets are detrimental as biases may get introduced from the beginning, particularly since a greedy search (no exploration) was used. This leads to negative impacts when the active ML is conducting mostly extrapolative predictions (see the low performance of extrapolation described previously). In the case of the small (e.g., five datapoints) initialization datasets, this effect was severe for the learning curves. However, being restricted to a local initialization dataset (data from three ligands) led to very steep initial learning curves and a performance similar to the initialization with a more diverse dataset, thus demonstrating the power of successful navigation through the chemical space by active ML driven closed-loop optimization.
Based on the previous findings on the size of the initial dataset and the different chemical representations, a direct comparison was conducted to assess the performance between complexity of parametrization and size of the initial dataset. Initialization datasets of 10, 15 and 20 datapoints were chosen along with OHE, Morgan 2 fingerprints and hybrid full feature representation (feature set 14, see Fig. 2 for more details). When using different sizes of initialization dataset the remaining data limits the number of possible active learning iterations. To allow for easy comparison of different sized initialization datasets, the data was normalized and, as such, at every location in the x-axis the different models have access to the same number of datapoints and so the effect of initialization dataset is represented.
By comparing extremes such as having no chemical information, but a larger initialization set (Fig. 7c, OHE 20) to using a smaller fully parametrized initialization dataset (full features 10), the effects of parametrization complexity and initialization data size could be more clearly identified. Within the case study, the results clearly indicated that having a dataset parameterized to higher complexity only delivers acceptable learning curves when the size of the initial dataset is sufficiently large. When using 10 datapoints for initialization, the OHE dataset clearly outperformed the fully parameterized dataset, however, when using 20 datapoints for initialization we found less of a difference in performance. When comparing size of the initial dataset against complexity of parameterization, we found that OHE 20 reached the maximum yield within 40 experiments whereas initialization with only 10 datapoints with full features required more than 110 experiments – almost more than three times more experiments where required. Based on these insights, we believe that it is relevant to consider the trade-off between feature complexity and size of the dataset (i.e., number of experiments) when conducting reaction optimization with HTE and active ML. A more detailed case study can be found in the ESI.†
Conclusions
Using an HTE-generated dataset of conditions for the Pd-catalysed C(sp3)–H bond activation of tertiary alkylamines, we investigated the role of parameterization for simulated active ML closed-loop optimization. By using different complexity levels of data representation, we identified the optimum modelling regimes for fitting moderately sized chemical datasets. When using a random split of the data for training/testing partitions, we found that simple OHE delivered already high-quality predictions for reaction yield, however, by adding more complex chemical descriptors we achieved slightly lower prediction error. For out-of-sample predictions we learned that neither fingerprints nor complex DFT-derived descriptors delivered significantly better performance compared to OHE, even though the descriptors were chosen based on mechanistic insights.Then, based on these preliminary findings, we conducted simulated closed-loop optimization experiments wherein the impact of feature complexity on active learning performance was assessed. Unexpectedly, OHE outperformed complex parameterizations that incorporated chemical information even in low data regimes which are used to initialize active learning models. To understand the impact of initialization of the closed-loop optimization, different sized initialization datasets and differently sampled data (random, out-of-sample) were used, showing that initialization with minimal data led to ineffective optimization whilst initialization with out-of-sample data still allows the active ML model to rapidly find ideal conditions. Most importantly, when comparing initialization of the closed-loop optimization with data that included the full feature set (fingerprints, DFT descriptors) to a double-sized dataset that was encoded with OHE (no chemical information), the latter identified the highest yield conditions in fewer experiments. Moreover, we found that increasing complexity of the parameterization requires a larger initialization dataset to deliver comparable performance.
The results of this study clearly indicate that current methods for parameterization are not descriptive enough to capture the factors that govern reaction success even when based on specific and relevant mechanistic insights. It must be noted that the success of different feature sets and models depends on the complexity of chemistry, the dimensionality of the design space and the number of variables. Given a different chemical design space with a larger number of ligands it might be possible that DFT-based descriptors start to outperform OHE because the number of OHE features increase whereas the number of descriptors stays constant. We believe that this work should serve as a challenge for the chemical community, and stimulate discussions about the trade-off between the development of more tailored parameterization methods or more exhaustive screening as two key factors for efficient reaction optimization.
Author contributions
A. Pomberger developed the research question, conducted the molecular parameterization and ML modelling under supervision of A. A. Lapkin. S. Sung, A. Khan, L. Colwell and D. Walz supported the project with their expertise in ML. A. A. Pedrina McCarthy and M. J. Gaunt designed, conducted and analyzed the HTE experiments. C. J. Taylor helped with the structure of the manuscript. Figures were generated by A. Pomberger and A. A. Pedrina McCarthy. All authors discussed the results and prepared the final manuscript. A. A. Lapkin secured funding, conceptualised and supervised the project.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
We are grateful to BASF SE and EPSRC Centre for Doctoral Training, SynTech (EP/S024220) for PhD studentship to A. P., Saudi Aramco for PhD studentship to A. K. and the EPSRC and GSK for a PhD studentship to A. A. P. M. Position of S. Sung was funded by Pharma Innovation Partnership Singapore (PIPS) via “C4” project. We thank Dr. Jesus Rodrigalvarez for useful discussions of the CH activation chemistry. We thank Robert van Putten, Kobi Felton, Daniel Wigh and Ferdinand Kossmann for helpful discussions and providing feedback on the manuscript.References
- A. Y. S. Lam and V. O. K. Li, Memet. Comput., 2012, 4, 3–17 CrossRef.
- A. Cernijenko, R. Risgaard and P. S. Baran, J. Am. Chem. Soc., 2016, 138, 9425–9428 CrossRef CAS PubMed.
- C. W. Coley, N. S. Eyke and K. F. Jensen, Angew. Chem., Int. Ed., 2020, 59, 22858–22893 CrossRef CAS PubMed.
- C. W. Coley, D. A. Thomas, J. A. M. Lummiss, J. N. Jaworski, C. P. Breen, V. Schultz, T. Hart, J. S. Fishman, L. Rogers, H. Gao, R. W. Hicklin, P. P. Plehiers, J. Byington, J. S. Piotti, W. H. Green, A. J. Hart, T. F. Jamison and K. F. Jensen, Science, 2019, 365, 1–9 CrossRef PubMed.
- J. M. Granda, L. Donina, V. Dragone, D.-L. Long and L. Cronin, Nature, 2018, 559, 377–381 CrossRef CAS PubMed.
- S. Steiner, J. Wolf, S. Glatzel, A. Andreou, J. M. Granda, G. Keenan, T. Hinkley, G. Aragon-Camarasa, P. J. Kitson, D. Angelone and L. Cronin, Science, 2019, 363, 1–8 CrossRef PubMed.
- Y. Amar, A. M. Schweidtmann, P. Deutsch, L. Cao and A. Lapkin, Chem. Sci., 2019, 10, 6697–6706 RSC.
- A. Echtermeyer, Y. Amar, J. Zakrzewski and A. Lapkin, Beilstein J. Org. Chem., 2017, 13, 150–163 CrossRef CAS PubMed.
- C. B. Santiago, J.-Y. Guo and M. S. Sigman, Chem. Sci., 2018, 9, 2398–2412 RSC.
- W. Beker, E. P. Gajewska, T. Badowski and B. A. Grzybowski, Angew. Chem., Int. Ed., 2019, 58, 4515–4519 CrossRef CAS PubMed.
- D. T. Ahneman, J. G. Estrada, S. Lin, S. D. Dreher and A. G. Doyle, Science, 2018, 360, 186 CrossRef CAS PubMed.
- G. Skoraczyński, P. Dittwald, B. Miasojedow, S. Szymkuć, E. P. Gajewska, B. A. Grzybowski and A. Gambin, Sci. Rep., 2017, 7, 3582 CrossRef PubMed.
- C. W. Coley, W. H. Green and K. F. Jensen, Acc. Chem. Res., 2018, 51, 1281–1289 CrossRef CAS PubMed.
- E. A. Gerlein, M. McGinnity, A. Belatreche and S. Coleman, Expert Syst. Appl., 2016, 54, 193–207 CrossRef.
- H. Rafiei Mohammad and H. Adeli, J. Constr. Div., Am. Soc. Civ. Eng., 2016, 142, 1–10 Search PubMed.
- A. L. Tarca, V. J. Carey, X. Chen, R. Romero and S. Drăghici, PLoS Comput. Biol., 2007, 3, 953–963 CrossRef CAS PubMed.
- J. VanderPlas, A. J. Connolly, Ž. Ivezi and A. Gray, arXiv preprint, 2014, arXiv:1411.5039v1.
- M. McCartney, M. Haeringer and W. Polifke, J. Eng. Gas Turbines Power, 2020, 142, 1–10 CrossRef.
- B. J. Shields, J. Stevens, J. Li, M. Parasram, F. Damani, J. I. M. Alvarado, J. M. Janey, R. P. Adams and A. G. Doyle, Nature, 2021, 590, 89–96 CrossRef CAS PubMed.
- N. S. Eyke, W. H. Green and K. F. Jensen, React. Chem. Eng., 2020, 5, 1963–1972 RSC.
- P. Jorayev, D. Russo, J. D. Tibbetts, A. M. Schweidtmann, P. Deutsch, S. D. Bull and A. A. Lapkin, Chem. Eng. Sci., 2021, 247, 116938 CrossRef.
- B. Zagidullin, Z. Wang, Y. Guan, E. Pitkänen and J. Tang, Briefings Bioinf., 2021, 22, 1–15 CrossRef PubMed.
- F. Sandfort, F. Strieth-Kalthoff, M. Kühnemund, C. Beecks and F. Glorius, Chem, 2020, 6, 1379–1390 CAS.
- J. Rodrigalvarez, M. Nappi, H. Azuma, N. J. Flodén, M. E. Burns and M. J. Gaunt, Nat. Chem., 2020, 12, 76–81 CrossRef CAS PubMed.
- B.-F. Shi, N. Maugel, Y.-H. Zhang and J.-Q. Yu, Angew. Chem., Int. Ed., 2008, 47, 4882–4886 CrossRef CAS PubMed.
- K. M. Engle, Pure Appl. Chem., 2016, 88, 119–138 CAS.
- H. L. Morgan, J. Chem. Doc., 1965, 5, 107–113 CrossRef CAS.
- A. V. Brethomé, S. P. Fletcher and R. S. Paton, ACS Catal., 2019, 9, 2313–2323 CrossRef.
- L. Falivene, Z. Cao, A. Petta, L. Serra, A. Poater, R. Oliva, V. Scarano and L. Cavallo, Nat. Chem., 2019, 11, 872–879 CrossRef CAS PubMed.
- F. Weinhold, C. R. Landis and E. D. Glendening, Int. Rev. Phys. Chem., 2016, 35, 399–440 Search PubMed.
- C. M. Breneman and K. B. Wiberg, J. Comput. Chem., 1990, 11, 361–373 CrossRef CAS.
- D. Rogers and M. Hahn, J. Chem. Inf. Model., 2010, 50, 742–754 CrossRef CAS PubMed.
- K. Bouhedjar, A. Boukelia, A. Khorief Nacereddine, A. Boucheham, A. Belaidi and A. Djerourou, Chem. Biol. Drug Des., 2020, 96, 961–972 CrossRef CAS PubMed.
- J. De Jesus Silva, M. A. B. Ferreira, A. Fedorov, M. S. Sigman and C. Copéret, Chem. Sci., 2020, 11, 6717–6723 RSC.
- P. I. Frazier, 2018, arXiv:1807.02811.
- K. C. Felton, J. G. Rittig and A. A. Lapkin, Chemistry Methods, 2021, 1, 116–122 CrossRef.
- H. Tin Kam, Proc. 3rd Int. Conf. Doc. Anal. Rec., 1995, vol. 1, pp. 278–282 Search PubMed.
- C. E. Rasmussen and C. K. I. Williams, Gaussian Processes for Machine Learning, MIT Press, 2006 Search PubMed.
- J. Schmidhuber, Neural Netw., 2015, 61, 85–117 CrossRef PubMed.
- C. Cortes and V. Vapnik, Mach. Learn., 1995, 20, 273–297 Search PubMed.
- B. Kégl, arXiv preprint, 2013, arXiv:1312.6086.
- B. Settles, Computer Sciences Technical Report, 2010, 52, 3–8 Search PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: 10.1039/d2re00008c |
| This journal is © The Royal Society of Chemistry 2022 |