Open Access Article
Open Access ArticleDicationic amphiphiles bearing an amino acid head group with a long-chain hydrophobic tail for in vitro gene delivery applications†
Shireesha Manturthia,
Kumar Pranav Narayan*b and
Srilakshmi V. Patri *a
*a
aDepartment of Chemistry, National Institute of Technology Warangal, Hanamkonda, Telangana-506004, India. E-mail: patrisrilaksmi@nitw.ac.in
bDepartment of Biological Science, Bits Pilani-Hyderabad, Hyderabad, Telangana-500078, India. E-mail: pranav@hyderabad.bits-pilani.ac.in
First published on 21st November 2022
Abstract
Amino acid-based cationic lipids, which have proven their efficacy as plasmid DNA nanocarriers, were employed as dicationic forms to transfect genes into cancer and non-cancer cells in this study. Proline, methionine, and serine amino acids are involved as hydrophilic moieties and the hydrocarbon long-chain serves as a hydrophobic tail. In a multicultural investigation, cationic lipids were employed as nano-vectors in conjunction with the helper lipid DOPE. To quantify the lipid efficient size, charge, and pDNA binding, biophysical analyses such as hydrodynamic diameter, zeta potential, agarose gel electrophoresis, and serum stability were done primarily. The liposomal particle composition was examined by scanning electron microscopy (SEM). Synthesized dicationic vector lipoplex formulations with reporter genes were found to be non-toxic to the cells investigated by MTT assay, and in addition, therapeutic gene p53 transfected into oral and brain cancer cells causing cell death was examined. In vitro investigations further validated that the proline-based lipid (C14-P) has high gene knockdown efficacy than methionine-based lipid (C14-M) and serine-based lipid (C14-S) at optimal N/P ratios as measured by β-galactosidase protein and eGFP expression. C14-P lipid shows superior cellular internalization compared to C14-M and C14-S in HEK-293 and CAL-27 cells attested by confocal study. These findings could include the proline-based lipid vector's exceptional gene delivery activity.
1. Introduction
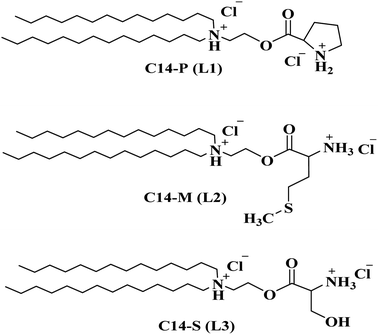
The development of secure and reliable therapeutic gene carriers is still a major problem in gene therapy, which is used to treat a wide range of hereditary illnesses, deadly viral diseases, and malignancies. The two types of gene delivery agents generally in use are viral and non-viral. Despite their remarkable efficiency, recombinant viral vectors have a number of biodefense issues.1–3 Viral vectors can generate erythrogenic and biocompatible reactions to their internal system and produce conceivably clone-competent viruses via related species with the host genetic material.4,5 Moreover, the therapeutic genes that could be placed inside viral vectors get a small insertion size constraint. As a result, a rising number of studies are being published on the creation of non-viral competitors such as cationic amphiphiles, also referred to as cationic transfection lipids,6–8 cationic polymers,9 dendrimers,10 and other non-viral alternatives. Cationic amphiphiles are rising in popularity as innovative non-viral transfection carriers of preference for transporting genes into the human body cells because of their low immunogenicity, durable manufacturing, ability to transfer large chunks of pDNA, and flexibility of dealing and preparation processes.11–15 The biomolecular frameworks of cationic amphiphiles are made up of a polar head group, which is a positively ionised hydrophilic region and a hydrophobic body, which is a nonpolar region (usually consisting of vitamin-E or a cholesterol skeleton or long aliphatic hydrocarbon chains etc.) often fasten conjugation through a linker feature such as amide, amidine, ether, ester, etc. The strategy of intracellular delivery of plasmid DNA to the cytoplasm or nucleus has been investigated using several cationic liposomes to improve gene expression efficacy.16,17 Membrane merging happens between the cationic lipid portion and the membrane of early endosomes (pH 6.0–6.5), whenever lipoplexes are taken hold by the selected cell, resulting in the release of nucleic acids into the cytoplasm. Generally delivered nucleic acids assemble in the nucleus.18 Mostly, hydrophilic domains are generated from amino acids,19–21 heterocyclic moieties,7,22 sugar moieties,23,24 etc., and have been contributing propensity in cancer gene therapy. For decades, long aliphatic hydrocarbon chains have been handled as a hydrophobic tail in gene delivery to several cancer cells by its particular aspect in easily forming liposomal vesicles.25–27Proline (Pro) is the only amino acid containing a ring structure having a secondary α-amino group, and this structural trait affords gene carriers containing the proline group distinct specific chemical and biological features. Hydrolysable proline-rich sequences have been identified as natural sources of cell-permeant vectors that can crack the phospholipid bilayer and transport the associated payloads without inflicting baleful membrane rupture.21,28,29 Serine (Ser) is a natural amino acid that contains a hydroxy functional group, which has been used in in vitro gene delivery as a hydrophilic moiety and shows its optimal transfection results in multiple cancerous cells.25,30 On multiple levels, methionine (Met) metabolism has been linked to cancer. Although they can rapidly manufacture methionine from homocysteine, many cancer cells cannot multiply in a media wherein methionine is substituted by homocysteine.31,32 When methionine is used as a hydrophilic moiety in cationic lipids, its metabolic character may impact gene delivery. Proline, serine, and methionine amino acids having important functional aspects are cyclic amine, hydroxy group, and sulphur group, respectively, which strongly influence gene carrying and delivery by their low toxicity and high biodegradability.
These notable properties of proline, serine, and methionine inspired us to develop lipids with these amino acids in the head group region. Three dicationic lipids, namely C14-P, C14-M, and C14-S, were prepared, carrying the hydrophobic fourteen carbons di long-chain moiety, applied as gene delivery vectors in this work. As part of gene delivery, synthesized vectors could deliver reporter genes as well as therapeutic genes. A tumor-suppressor gene with a well-defined function is the p53 gene. In 1991, the first proof that p53 can trigger apoptosis was reported.33,34 Since a great deal of attention has been paid to the cellular basis that p53 triggers cells to produce apoptosis.35 In this study, synthesized vectors delivering the p53 gene that causes cancer cell apoptosis is reported.
2. Results and discussion
2.1. Chemistry
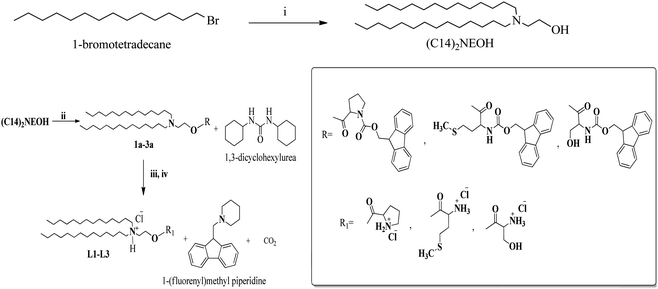
The major hydrophobic elements common to all the three lipids C14-P (L1), C14-M (L2), and C14-S (L3) (Fig. 1) outlined herein are (i) 14 carbon di long-chain as an anchoring group prevalent in all three lipids, (ii) differing hydrophilic groups with three amino acids, and synthetic procedure of dicationic lipids, as shown in Scheme 1. The chemistry required to build these dicationic lipids is as follows. First, the tertiary amine having two hydrophobic long-chain (C14)2NEOH (2-(ditetradecylamino) ethanol) is generated by N-alkylation of ethanolamine using 1-bromo tetradecane reacting with ethanolamine in the presence of potassium carbonate (K2CO3). Then, 2-(ditetradecylamino) ethanol is coupled with Fmoc-amino acid using coupling reagents p-toluene sulfonic acid (PTSA), dicyclohexylcarbodiimide (DCC), and base 4-dimethylaminopyridine (DMAP) derived tertiary amine coupled Fmoc-amino acids (1a, 2a, and 3a). This is followed by deprotection of the Fmoc group deprotected with piperidine in the presence of DMF and then quaternization using HCl to yield C14-P, C14-M, and C14-S dicationic lipids. 1H-NMR, 13C-NMR, and ESI/HRMS mass spectra validated the chemical structures of all the synthetic intermediates and final amphiphiles represented in ESI (Fig. S1–S21†). HPLC analytical chromatogram was used to detect the purity of the final lipids, as described in Fig. S22–S24.† The mobile phase was a mixture of methanol (65%) and water (35%), which are HPLC grade and the stationary phase was a porous graphitic carbon column for the sample run. The lipid (1 mg) was dissolved in 1 mL of methanol solvent and 20 μL of the sample was injected for analysis at 15 min time point. The purity of peaks that appeared at retention times were 8.50, 8.13, and 7.15 for C14-P, C14-M, and C14-S, respectively. All three lipids were analysed as pure, above 90%.2.2. Small uni-lamellar vesicles (SUVs)
Many previous non-viral vector installations demonstrated that merged liposomal formulations (lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) co-lipid) might have improved transfection potentials than simple cationic formulations (lipid alone).36,37 DOPE is a zwitter ionic phospholipid that has been widely employed as a helper lipid in many research studies due to its fusogenic nature and non-bilayer structure development, which aids in the disruption of the endosomal barrier after intracellular uptake of liposome into the cytoplasm.38 Initially, we attempted to create lipid
co-lipid) might have improved transfection potentials than simple cationic formulations (lipid alone).36,37 DOPE is a zwitter ionic phospholipid that has been widely employed as a helper lipid in many research studies due to its fusogenic nature and non-bilayer structure development, which aids in the disruption of the endosomal barrier after intracellular uptake of liposome into the cytoplasm.38 Initially, we attempted to create lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE (liposome) vesicles by mixing DOPE with cationic lipids (C14-P, C14-M, and C14-S) in various molar combinations, but the suspensions were neither transparent nor successful in transfection. We stepped up our attempts to discover a better formulation with DOPE to achieve the plasmid delivery properties of these lipid derivatives. Clear and uniform lipid
DOPE (liposome) vesicles by mixing DOPE with cationic lipids (C14-P, C14-M, and C14-S) in various molar combinations, but the suspensions were neither transparent nor successful in transfection. We stepped up our attempts to discover a better formulation with DOPE to achieve the plasmid delivery properties of these lipid derivatives. Clear and uniform lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) co-lipid formulations were achieved at a 1
co-lipid formulations were achieved at a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 molar ratio without any suspension and were stable at 4 °C for up to 10 weeks.
1 molar ratio without any suspension and were stable at 4 °C for up to 10 weeks.
2.3. Physico-chemical properties of C14-P, C14-M, and C14-S liposomes and their lipoplexes
The physico-chemical properties of cationic lipids play a vital role in achieving effective transfection. The diameter of the lipoplex, the proportion of lipids to plasmid DNA, morphology of the liposome, and the composition of the liposome are all important aspects of cellular nucleic acid transport. The physico-chemical features of liposomes that have been complexed with nucleic acids may differ from those of regular liposomes. The analysis was performed using dynamic light scattering (DLS) with liposomes (lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE-1
DOPE-1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) and complexes (lipid
1) and complexes (lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA) at various N/P ratios of 1
pDNA) at various N/P ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 4
1, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 8
1, and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
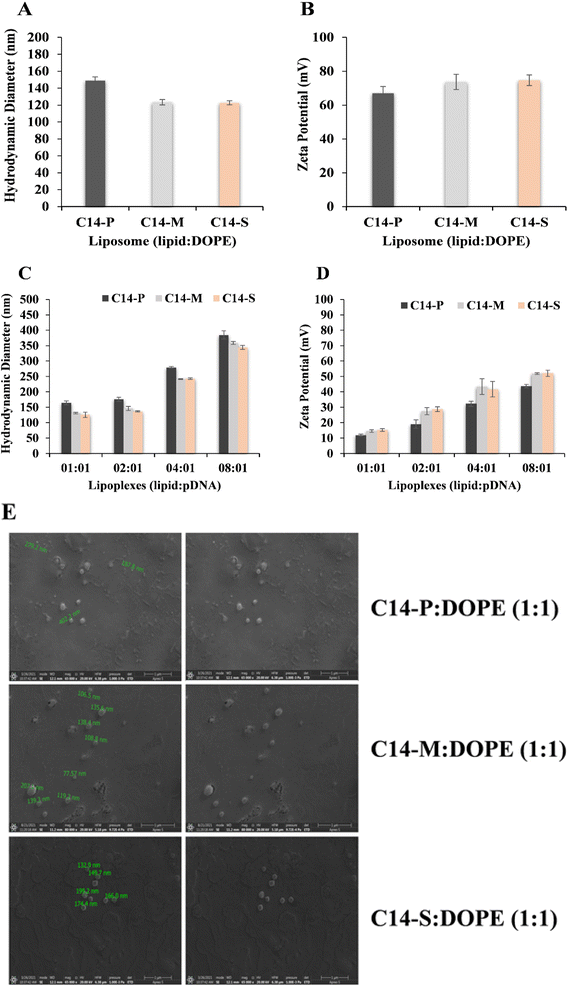
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. The sizes of C14-P, C14-M, and C14-S liposomes were revealed to be 149 nm, 123 nm, and 122 nm, respectively, as shown in Fig. 2A. The sizes of the lipoplexes were observed increase dramatically from 164 nm to 384 nm, 131 nm to 358 nm, and 125 nm to 344 nm as the N/P ratio of lipid
1. The sizes of C14-P, C14-M, and C14-S liposomes were revealed to be 149 nm, 123 nm, and 122 nm, respectively, as shown in Fig. 2A. The sizes of the lipoplexes were observed increase dramatically from 164 nm to 384 nm, 131 nm to 358 nm, and 125 nm to 344 nm as the N/P ratio of lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA complexes of C14-P, C14-M, and C14-S was extended from 1
pDNA complexes of C14-P, C14-M, and C14-S was extended from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 8
1 to 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, respectively (Fig. 2C). The rise in the size with increasing N/P ratio could be explained by better subspaces at higher lipid contents. Furthermore, the maximum range of the size of the lipoplexes is 384 nm, demonstrating that they will be regarded to be optimal for effective gene transfections. The dicationic liposomes of C14-P, C14-M, and C14-S surface charges are +67, +73, and +74 mV, respectively (Fig. 2B). The surface potential of lipoplexes decreased compared to the corresponding liposomes when neutralized with pDNA (Fig. 2D). According to these findings, the proper size and surface potential of lipoplexes are necessary for successful transfection. Scanning electron microscopy was utilized to pictorially assess the DLS findings regarding the liposomal compositions. The visual images in Fig. 2E show that the liposomal particle sizes are substantially identical to the DLS results.
1, respectively (Fig. 2C). The rise in the size with increasing N/P ratio could be explained by better subspaces at higher lipid contents. Furthermore, the maximum range of the size of the lipoplexes is 384 nm, demonstrating that they will be regarded to be optimal for effective gene transfections. The dicationic liposomes of C14-P, C14-M, and C14-S surface charges are +67, +73, and +74 mV, respectively (Fig. 2B). The surface potential of lipoplexes decreased compared to the corresponding liposomes when neutralized with pDNA (Fig. 2D). According to these findings, the proper size and surface potential of lipoplexes are necessary for successful transfection. Scanning electron microscopy was utilized to pictorially assess the DLS findings regarding the liposomal compositions. The visual images in Fig. 2E show that the liposomal particle sizes are substantially identical to the DLS results.
2.4. Interaction of liposomes with pDNA, heparin displacement and serum stability
Gel electrophoretic assay is used to analyse the ionic interactions between the nucleic acid and the current lipids C14-P, C14-M, and C14-S as a function of lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA N/P ratio. For pDNA binding experiments, liposomal formulations of 1
pDNA N/P ratio. For pDNA binding experiments, liposomal formulations of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (lipid
1 (lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) DOPE) molar ratio were utilized. The results of this gel electrophoresis analysis demonstrated that lipids had strong pDNA binding interactions throughout the lipid
DOPE) molar ratio were utilized. The results of this gel electrophoresis analysis demonstrated that lipids had strong pDNA binding interactions throughout the lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA N/P ratios ranging from 2
pDNA N/P ratios ranging from 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 8
1 to 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. It also reveals that the 50% pDNA binding was achieved at 1
1. It also reveals that the 50% pDNA binding was achieved at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
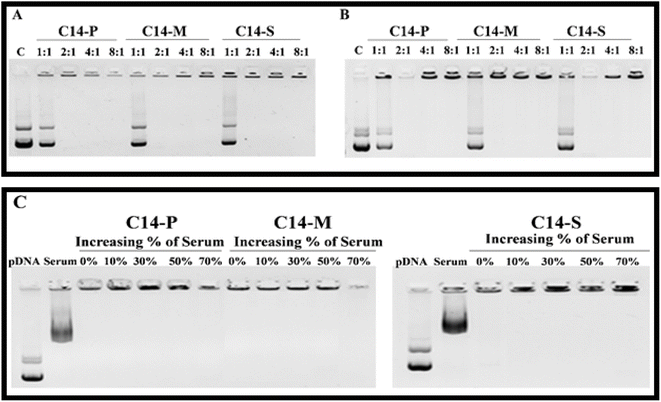
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P ratio (Fig. 3A). The gel retardation experiment revealed that C14-P, C14-M, and C14-S lipids have the capability to bind nucleic acid more reliably. The interaction of the liposome and nucleic acid was found to be aided by the structural amino acid head group.
1 N/P ratio (Fig. 3A). The gel retardation experiment revealed that C14-P, C14-M, and C14-S lipids have the capability to bind nucleic acid more reliably. The interaction of the liposome and nucleic acid was found to be aided by the structural amino acid head group.
Heparin displacement experiments were also performed to substantiate the pDNA binding findings with the liposomal formulations mentioned above. Lipoplexes containing lipids C14-P, C14-M, and C14-S prevented the displacement of nucleic acid with heparin across the lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA interface at N/P ratios 2
pDNA interface at N/P ratios 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–8
1–8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (Fig. 3B). These findings supported the previous report that amino acid head group-based cationic amphiphiles are stable over heparin.21
1 (Fig. 3B). These findings supported the previous report that amino acid head group-based cationic amphiphiles are stable over heparin.21
Lipoplexes were tested for serum resistance by incubating them in various concentrations of serum in DMEM medium. Gel electrophoresis was employed to track the flow of free DNA following incubation at a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P ratio, where the complete binding had happened. Surprisingly, even after incubation with 70% FBS in the medium, lipoplexes of C14-P, C14-M, and C14-S showed strong DNA binding. It has been determined that amino acid-based cationic lipids have acceptable serum resistance.39 Whenever lipoplexes are delivered into the organism, this impact may aid in reducing payload leaking across the circulatory system.40
1 N/P ratio, where the complete binding had happened. Surprisingly, even after incubation with 70% FBS in the medium, lipoplexes of C14-P, C14-M, and C14-S showed strong DNA binding. It has been determined that amino acid-based cationic lipids have acceptable serum resistance.39 Whenever lipoplexes are delivered into the organism, this impact may aid in reducing payload leaking across the circulatory system.40
2.5. Cytotoxicity study
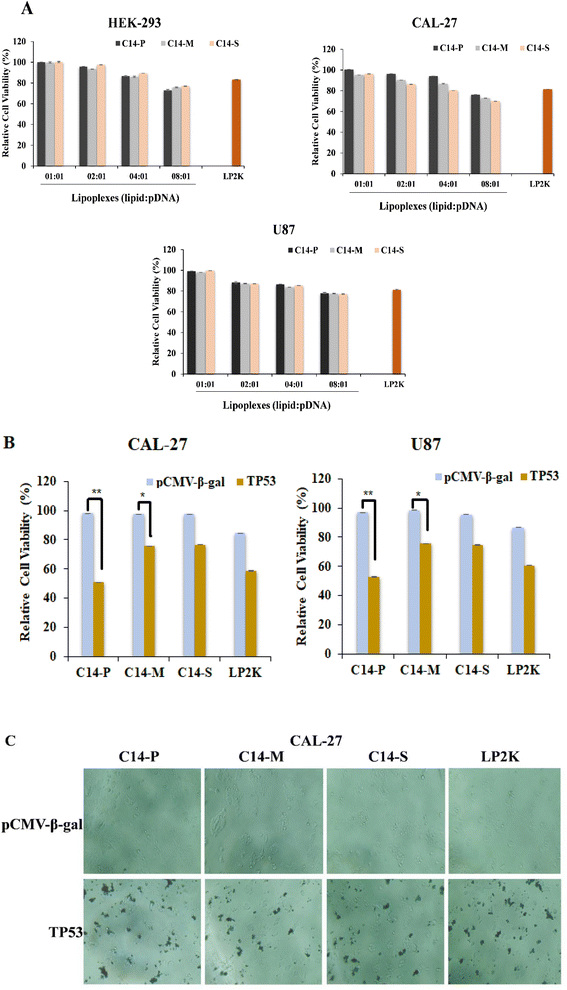
Low cytotoxicity is another crucial trait that increases the likelihood of a synthetic nucleic acid delivery carrier being employed in medical chemistry. The MTT assay was used to determine the safety of the synthetic dicationic lipids under investigation. At lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA N/P ratios of 1
pDNA N/P ratios of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1–8
1–8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, the viability of C14-P/pDNA, C14-M/pDNA, and C14-S/pDNA complexes in HEK-293, CAL-27, and U87 cell lines was investigated. The cytotoxicity of cationic lipids is mostly determined by their positive charge potential.41 Synthesized dicationic lipids have a higher surface potential, not affecting cell viability. The compositions were discovered to have extraordinarily high cell viabilities (more than 85%), especially up to the lipid
1, the viability of C14-P/pDNA, C14-M/pDNA, and C14-S/pDNA complexes in HEK-293, CAL-27, and U87 cell lines was investigated. The cytotoxicity of cationic lipids is mostly determined by their positive charge potential.41 Synthesized dicationic lipids have a higher surface potential, not affecting cell viability. The compositions were discovered to have extraordinarily high cell viabilities (more than 85%), especially up to the lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA N/P ratio 2
pDNA N/P ratio 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 in comparison to the Lipofectamine-2000, which is observed. At higher N/P ratios of 4
1 in comparison to the Lipofectamine-2000, which is observed. At higher N/P ratios of 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 8
1 and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, cell viabilities of lipoplexes were reported to be 85–75% (Fig. 4A). These results revealed that lipid cytotoxicity does not appear to be a factor for differences in gene transfer efficacies. In all three cell lines tested, the C14-P, C14-M, and C14-S formulations were found to be less harmful. As a result, amino acid head group dicationic lipids are finer transfection reagents with reduced cytotoxicity, which is a significant property for in vitro and in vivo applications.
1, cell viabilities of lipoplexes were reported to be 85–75% (Fig. 4A). These results revealed that lipid cytotoxicity does not appear to be a factor for differences in gene transfer efficacies. In all three cell lines tested, the C14-P, C14-M, and C14-S formulations were found to be less harmful. As a result, amino acid head group dicationic lipids are finer transfection reagents with reduced cytotoxicity, which is a significant property for in vitro and in vivo applications.
Natural amino acid-based cationic lipids could carry and transfect the therapeutic nucleic acid to the various cancerous cells.19,42 The current lipids tested for transfer of the anti-cancer gene, p53, into the glioma and oral cancer cells led to cell death. When these cells were treated with lipid/p53 lipoplexes, cell viability was dramatically less compared to lipid/pCMV-β-gal lipoplexes at a 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P ratio (Fig. 4B). Moreover, in the case of C14-P, lipid cell death is more compared to C14-M and C14-S lipids in both U87 and CAL-27 cells. The visual images by optical microscope were recorded after 48 h of the transfection period of CAL-27 cells with lipid/pCMV-β-gal and lipid/p53 lipoplexes. Cell death is clearly recorded in the oral cancer cells for lipid/p53 lipoplexes (Fig. 4C). According to the revealed results, natural amino acid-based lipids typically carry the anticancer gene into cancer cells, leading to cell death.
1 N/P ratio (Fig. 4B). Moreover, in the case of C14-P, lipid cell death is more compared to C14-M and C14-S lipids in both U87 and CAL-27 cells. The visual images by optical microscope were recorded after 48 h of the transfection period of CAL-27 cells with lipid/pCMV-β-gal and lipid/p53 lipoplexes. Cell death is clearly recorded in the oral cancer cells for lipid/p53 lipoplexes (Fig. 4C). According to the revealed results, natural amino acid-based lipids typically carry the anticancer gene into cancer cells, leading to cell death.
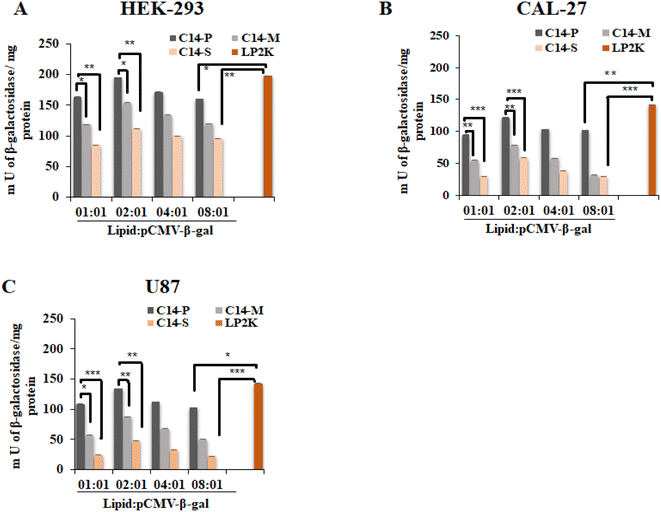
2.6. Transfection biology
The transfection potentials of the lipids C14-P, C14-M, and C14-S were analyzed in three mammalian cultured cells (i.e., HEK-293, CAL-27, and U87). The reporter gene pCMV-β-gal, which encodes the enzyme β-galactosidase, was used to make lipoplexes of C14-P, C14-M, and C14-S lipids with lipid/pDNA N/P ratios ranging from 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 8
1 to 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Despite the fact that the only structural variation between the lipids were the presence of proline, methionine, and serine head groups, C14-P was able to efficiently carry reporter gene into all three cell lines examined, especially when the N/P ratio is 2
1. Despite the fact that the only structural variation between the lipids were the presence of proline, methionine, and serine head groups, C14-P was able to efficiently carry reporter gene into all three cell lines examined, especially when the N/P ratio is 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. Fig. 5 shows that lipoplexes of all three lipids enhanced transfection and displayed the highest percentage of protein production at 2
1. Fig. 5 shows that lipoplexes of all three lipids enhanced transfection and displayed the highest percentage of protein production at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P ratio; this could be attributable to the smallest particle diameter and total binding of the pDNA, as well as the lowest cytotoxicity. More interestingly, the proline head group-based dicationic lipid (C14-P) has demonstrated superior transfection activity in all three cell lines, which is comparable to Lipofectamine-2000. The superior transfection efficacy of C14-P lipid may be due to the presence of a cyclic head group.43,44 Higher transfection efficiency of C14-P/DOPE vesicle indicates that proline-based cationic vectors have a great deal of ability as non-viral gene transfer vectors. The occurrence of cyclic head groups on the proline group of amino acid hybrid-transfecting lipoplexes is, therefore, a crucial characteristic that can considerably alter the gene delivery efficacies of amino acid-based lipids, according to the comparative transfection patterns of C14-P.
1 N/P ratio; this could be attributable to the smallest particle diameter and total binding of the pDNA, as well as the lowest cytotoxicity. More interestingly, the proline head group-based dicationic lipid (C14-P) has demonstrated superior transfection activity in all three cell lines, which is comparable to Lipofectamine-2000. The superior transfection efficacy of C14-P lipid may be due to the presence of a cyclic head group.43,44 Higher transfection efficiency of C14-P/DOPE vesicle indicates that proline-based cationic vectors have a great deal of ability as non-viral gene transfer vectors. The occurrence of cyclic head groups on the proline group of amino acid hybrid-transfecting lipoplexes is, therefore, a crucial characteristic that can considerably alter the gene delivery efficacies of amino acid-based lipids, according to the comparative transfection patterns of C14-P.
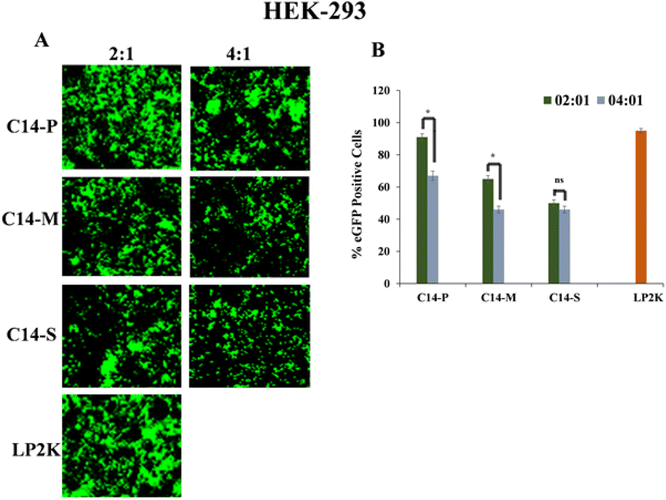
In HEK-293 cells, qualitative and quantitative eGFP (enhanced green fluorescence protein) experiments were performed utilizing lipid/pEGFP mixtures at 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P ratios to support the pCMV-β-gal reporter gene expression results. At this phase, it is crucial to determine whether pDNA expression (eGFP) reaches its maximum value at the chosen N/P ratio (4
1 N/P ratios to support the pCMV-β-gal reporter gene expression results. At this phase, it is crucial to determine whether pDNA expression (eGFP) reaches its maximum value at the chosen N/P ratio (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1). An inverted microscope was used to examine the intracellular expression of eGFP. At the 4
1). An inverted microscope was used to examine the intracellular expression of eGFP. At the 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and 2
1 and 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
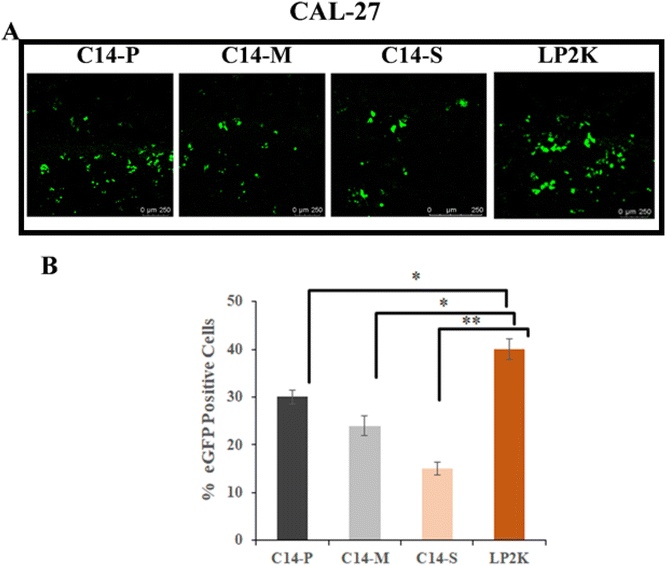
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P ratios, the percentage of eGFP expression in HEK-293 was found to be higher with C14-P and moderate with C14-M and C14-S (Fig. 6A). The percentage of positive eGFP cells of lipids were further confirmed by quantitative measurement of eGFP expression by flow cytometer, which was comparable to Lipofectamine-2000 (Fig. 6B). Further, eGFP expression was studied by qualitative and quantitative analysis in CAL-27 (Fig. 7A and B) and U87 (Fig. S25A and B†) cells at 2
1 N/P ratios, the percentage of eGFP expression in HEK-293 was found to be higher with C14-P and moderate with C14-M and C14-S (Fig. 6A). The percentage of positive eGFP cells of lipids were further confirmed by quantitative measurement of eGFP expression by flow cytometer, which was comparable to Lipofectamine-2000 (Fig. 6B). Further, eGFP expression was studied by qualitative and quantitative analysis in CAL-27 (Fig. 7A and B) and U87 (Fig. S25A and B†) cells at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P ratio. In both cell lines, C14-P has superior transfection compared to C14-M and C14-S. According to the transfection outcomes, it could be inferred that the proline head group on a hydrophobic long chain-based lipid is an effective method for gene delivery.
1 N/P ratio. In both cell lines, C14-P has superior transfection compared to C14-M and C14-S. According to the transfection outcomes, it could be inferred that the proline head group on a hydrophobic long chain-based lipid is an effective method for gene delivery.
2.7. Intracellular uptake
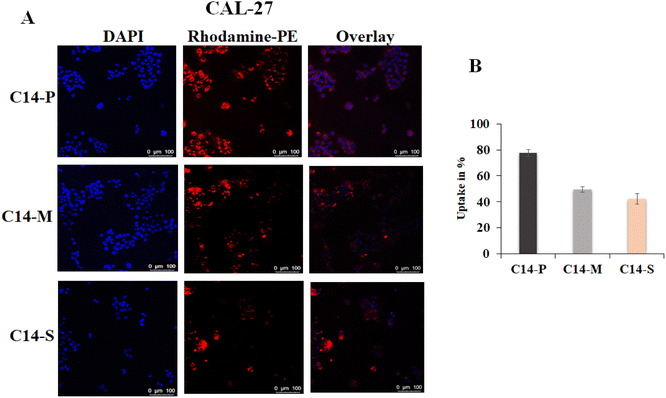
Transfection results demonstrated that C14-P, C14-M, and C14-S lipids exhibited higher transfection efficiency in all three cell lines (HEK-293, CAL-27, and U87) when the lipid/pDNA N/P ratio was 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1; moreover, C14 lipid showed superior expression. HEK-293 and CAL-27 cells were tested with lipoplexes containing pCMV-β-gal and rhodamine-PE labelled liposomes of C14-P, C14-M, and C14-S spanning the lipid/pDNA N/P ratio of 2
1; moreover, C14 lipid showed superior expression. HEK-293 and CAL-27 cells were tested with lipoplexes containing pCMV-β-gal and rhodamine-PE labelled liposomes of C14-P, C14-M, and C14-S spanning the lipid/pDNA N/P ratio of 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to check whether the highest absorption occurs with C14-P lipid. DAPI (4′,6-diamidino-2-phenylindole) was utilized to stain the cell nucleus. Cellular uptake of rhodamine-PE tagged lipoplexes in CAL-27 (Fig. 8A and B) and HEK-293 cells (Fig. S26A and B†) were examined, and the results indicated that C14-P lipid had higher intracellular uptake compared to C14-M and C14-S lipids. These results corroborated the transfection results and revealed that among the three synthesized dicationic lipids, proline-based lipids have the superior capability in in vitro gene delivery.
1 to check whether the highest absorption occurs with C14-P lipid. DAPI (4′,6-diamidino-2-phenylindole) was utilized to stain the cell nucleus. Cellular uptake of rhodamine-PE tagged lipoplexes in CAL-27 (Fig. 8A and B) and HEK-293 cells (Fig. S26A and B†) were examined, and the results indicated that C14-P lipid had higher intracellular uptake compared to C14-M and C14-S lipids. These results corroborated the transfection results and revealed that among the three synthesized dicationic lipids, proline-based lipids have the superior capability in in vitro gene delivery.
2.8. Conclusions
The current study demonstrates the design, synthesis, physicochemical properties, and respective in vitro transfection efficiencies of dicationic lipids (i.e. C14-P, C14-M, and C14-S) tested against three cell lines. Cytotoxicity of the dicationic lipids in three different cell lines was found to be minimal. Furthermore, the green fluorescence and β-galactosidase activity investigations support the findings of the in vitro transfection. Lipoplexes of C14-P, C14-M, and C14-S lipids are compared in terms of serum stability, cytotoxicity, transfection efficiency, and cellular absorption. According to the results, C14-P is a promising nanovesicle as a reliable transfection reagent for a primary cultured cell line with relatively low cytotoxicity.3. Experimental section
3.1. Materials
Fmoc-amino acids, p-toluene sulfonic acid, dicyclohexylcarbodiimide (DCC), 4-dimethylaminopyridine, and 1-bromo tetradecane were delivered from Sigma Aldrich. The organic solvents and chemicals, including ethanolamine, potassium carbonate, piperidine, dimethylformamide (DMF), methylene dichloride (DCM), and ethyl acetate, were purchased from Spectrochem Pvt. Ltd., and Finar Limited. Molecular mass analyses were performed by High-Resolution Mass Spectrometry (HRMS) using Agilent Q-TOF 6230, and the purity of final lipids was characterized by HPLC (Shimadzu LC Solution). 1H-NMR (400 MHz) and 13C-NMR (101 MHz) spectra were recorded on Bruker AVANCE spectrometer, with TMS as an internal standard. DOPE (1,2-dioleoyl-sn-glycerol-3-phosphoethanolamine) was purchased from Sigma Aldrich. Agarose and 6× loading dye were purchased from Hi-media, India. All chemicals for biological applications were procured from Merck, Sigma-Aldrich, Thermo Fischer Scientific, and Invitrogen.3.2. Synthesis
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PE and EtOAc range 0.5–0.7 was purified using 1% ethyl acetate in pet ether eluent column chromatography.
1 PE and EtOAc range 0.5–0.7 was purified using 1% ethyl acetate in pet ether eluent column chromatography.
3.2.2.1 2-(Di-tetradecylamino)-ethyl (Fmoc)-proline (1a). Product: pale white solid, 1.59 mmol (1.23 g), yield: 82%; 1H NMR (CDCl3) [δ/ppm] 7.75 (t, J = 6.0 Hz, 2H), 7.59 (m, 2H), 7.39 (dd, J = 14.4, 7.6 Hz, 2H), 7.31 (dd, J = 7.6, 3.6 Hz, 2H), 4.41 (d, J = 7.2 Hz, 2H), 4.34 (t, J = 5.2 Hz, 1H), 4.31–4.25 (m, 1H), 4.22–4.09 (m, 2H), 3.67 (d, J = 7.6 Hz, 1H), 3.54 (d, J = 7.6 Hz, 1H), 2.64 (s, 2H), 2.39 (t, J = 6.8 Hz, 2H), 2.29 (t, J = 8.0 Hz, 2H), 2.04 (t, J = 7.6 Hz, 2H), 2.00–1.85 (m, 2H), 1.68–1.16 (m, 46H), 0.88 (t, J = 6.8 Hz, 6H); 13C{1H} NMR (CDCl3) [δ/ppm] 169.49, 154.58, 141.63, 139.06, 127.98, 127.60, 127.21, 126.67, 125.53, 124.36, 124.00, 120.59, 119.93, 63.08, 61.31, 60.55, 57.81, 55.94, 54.20, 53.81, 46.35, 46.23, 38.70, 31.93, 29.70, 29.68, 29.66, 29.61, 29.59, 29.45, 29.37, 27.26, 25.92, 24.88, 24.07, 22.70, 14.13; MS m/z: 772.6118, found: 773.6208 [M + H]+.
3.2.2.2 2-(Di-tetradecylamino)-ethyl (Fmoc)-methionine (2a). Product: pale white solid, 1.47 mmol (1.19 g), yield: 79%; 1H NMR (CDCl3) [δ/ppm] 7.68 (d, J = 7.6 Hz, 2H), 7.52 (d, J = 7.2 Hz, 2H), 7.31 (t, J = 7.4 Hz, 2H), 7.23 (t, J = 7.6 Hz, 2H), 4.42 (d, J = 4.4 Hz, 1H), 4.34 (d, J = 7.2 Hz, 2H), 4.14 (t, J = 7.0 Hz, 2H), 4.06–4.01 (m, 1H), 2.62 (t, J = 6.0 Hz, 2H), 2.46 (t, J = 7.2 Hz, 2H), 2.36 (t, J = 7.2 Hz, 2H), 2.01 (s, 3H), 1.95 (s, 2H), 1.50–1.09 (m, 48H), 0.80 (m, 6H); 13C {1H} NMR (CDCl3) [δ/ppm] 168.62, 158.16, 141.33, 140.40, 135.19, 129.36, 129.03, 127.36, 125.32, 124.27, 121.29, 120.91, 120.66, 119.94, 61.64, 58.21, 55.82, 53.92, 46.13, 32.91, 31.82, 29.57, 29.54, 29.24, 27.14, 25.94, 25.89, 22.60, 14.25; MS m/z: 806.5995, found: 807.6127 [M + H]+.
3.2.2.3 2-(Di-tetradecylamino)-ethyl (Fmoc)-serine (3a). Product: pale white solid, 1.75 mmol (1.34 g), yield: 89.33%; 1H NMR (CDCl3) [δ/ppm] 7.65 (d, J = 7.2 Hz, 2H), 7.61 (d, J = 7.6 Hz, 2H), 7.33–7.27 (m, 2H), 7.24–7.20 (m, 2H), 4.40 (d, J = 6.0 Hz, 2H), 4.03 (t, J = 6.2 Hz, 1H), 3.97 (t, J = 6.8 Hz, 1H), 3.55 (t, J = 6.4 Hz, 2H), 2.61 (t, J = 6.2 Hz, 2H), 2.38 (t, J = 7.4 Hz, 4H), 1.96 (d, J = 3.2 Hz, 2H), 1.20 (m, 48H), 0.80 (t, J = 6.8 Hz, 6H); 13C{1H} NMR (CDCl3) [δ/ppm] 166.04, 155.83, 145.87, 141.58, 126.67, 125.53, 124.32, 124.00, 121.01, 120.59, 119.93, 119.82, 119.74, 63.08, 61.31, 57.81, 54.20, 53.81, 46.35, 31.93, 29.70, 29.68, 29.61, 29.59, 29.45, 29.37, 27.26, 25.92, 24.07, 22.70, 14.13; MS m/z: 762.5911, found: 763.6040 [M + H]+.
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 PE/EtOAc was filtered by a column using 60–120 mesh size silica gel and eluted with 3% EtOAc/PE. The obtained Fmoc deprotected amino acid compounds quaternized with 6 N HCl under reflux conditions yielded the final lipid.
1 PE/EtOAc was filtered by a column using 60–120 mesh size silica gel and eluted with 3% EtOAc/PE. The obtained Fmoc deprotected amino acid compounds quaternized with 6 N HCl under reflux conditions yielded the final lipid.
3.2.3.1 2-(Di-tetradecylamino)-ethyl proline (C14-P). Product: light yellow solid oily, yield: 0.82 g (1.48 mmol), 82%; 1H NMR (CDCl3) [δ/ppm] 4.09 (ddd, J = 20.8, 8.4, 3.8 Hz, 1H), 3.80 (t, J = 4.8 Hz, 2H), 3.46 (d, J = 5.0 Hz, 1H), 3.36 (d, J = 10.4 Hz, 1H), 2.98 (t, J = 4.8 Hz, 2H), 2.90 (dd, J = 5.0 Hz, 2H), 2.10 (d, J = 7.8 Hz, 1H), 1.91 (d, J = 12.4 Hz, 1H), 1.74 (t, J = 5.2 Hz, 2H), 1.59 (s, 2H), 1.36–1.12 (m, 48H), 0.81 (t, J = 6.9 Hz, 6H); 13C{1H} NMR (CDCl3) [δ/ppm] 170.94, 62.21, 54.96, 53.20, 52.64, 31.92, 29.70, 29.66, 29.62, 29.53, 29.48, 29.36, 29.18, 27.36, 27.18, 27.01, 22.68, 21.02, 14.11; MS m/z: 552.5583, found: 552.5521 [M]+.
3.2.3.2 2-(Di-tetradecylamino)-ethyl methionine (C14-M). Product: light yellow solid oily, yield: 0.85 g (1.45 mmol), 85%; 1H NMR (CDCl3) [δ/ppm] 4.52 (t, J = 4.0 Hz, 2H), 4.10 (t, J = 6.4 Hz, 1H), 2.62 (t, J = 6.0 Hz, 2H), 2.55 (m, 2H), 2.43 (t, J = 7.6 Hz, 2H), 2.36 (t, J = 7.6 Hz, 2H), 2.03 (s, 3H), 2.02–1.93 (m, 2H), 1.43–1.18 (m, 48H), 0.81 (t, J = 6.4 Hz, 6H); 13C {1H} NMR (CDCl3) [δ/ppm] 174.96, 61.71, 56.58, 56.35, 56.15, 53.67, 31.90, 29.68, 29.66, 29.64, 29.62, 29.58, 29.51, 29.48, 29.44, 29.40, 29.34, 29.07, 26.72, 22.66, 20.79, 14.08; MS m/z: 586.5460, found: 586.5440 [M]+.
3.2.3.3 2-(Di-tetradecylamino)-ethyl serine (C14-S). Product: light yellow solid oily, yield: 0.81 g (1.49 mmol), 81%; 1H NMR (CDCl3) [δ/ppm] 4.31 (t, J = 4.0 Hz, 2H), 4.02 (s, 1H), 3.64 (t, J = 4.8 Hz, 2H), 3.50–3.46 (m, 1H), 3.33–3.28 (m, 1H), 2.71 (t, J = 4.8 Hz, 2H), 2.60 (t, J = 7.6 Hz, 4H), 1.27 (d, J = 10.0 Hz, 48H), 0.87 (t, J = 6.4 Hz, 6H); 13C{1H} NMR (CDCl3) [δ/ppm] 172.63, 62.30, 60.07, 50.58, 49.72, 31.91, 29.69, 29.52, 29.33, 28.86, 26.77, 22.68, 21.37, 14.10; MS m/z: 542.5375, found: 542.5335 [M]+.
3.3. Preparation of liposomes
In an autoclaved dry glass vial, the cationic lipid and the supplementary lipid (DOPE) were both dissolved in dry chloroform (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 mM concentration). The chloroform was evaporated using a thin stream of nitrogen gas, and then the thin film was dried for 5 h under vacuum. The dried lipid film was then swollen for 12 h with 1 mL Milli Q water. After that, the sample was vortexed at ambient temperature for 2–3 min for a clear solution and then kept for sonication in a bath sonicator to form multilamellar vesicles (MLVs). Subsequently, MLVs were sonicated in a cool bath until they were clear, using a Branson Sonifier with a 25 W output voltage. The obtained clear aqueous liposomes were used for lipoplex formation.
1 mM concentration). The chloroform was evaporated using a thin stream of nitrogen gas, and then the thin film was dried for 5 h under vacuum. The dried lipid film was then swollen for 12 h with 1 mL Milli Q water. After that, the sample was vortexed at ambient temperature for 2–3 min for a clear solution and then kept for sonication in a bath sonicator to form multilamellar vesicles (MLVs). Subsequently, MLVs were sonicated in a cool bath until they were clear, using a Branson Sonifier with a 25 W output voltage. The obtained clear aqueous liposomes were used for lipoplex formation.
3.4. Zeta potential (mV) and size (nm) measurements
Zeta sizer 3000HSA equipped using a diode-pumped solid-state laser at 532 nm set at 25 °C was used to quantify the sizes of lipid aggregates and the resultant complexes from the plasmid. From freshly produced liposomes (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 charge ratio), 100 μL were diluted to 900 μL of Milli-Q in quartz cuvette prior to the measurements. Each co-liposomal formulation was used to make lipoplexes with 5 μL (100 ng μL−1) of pDNA (pCMV-β-gal), 30 min before measurement at 1
1 charge ratio), 100 μL were diluted to 900 μL of Milli-Q in quartz cuvette prior to the measurements. Each co-liposomal formulation was used to make lipoplexes with 5 μL (100 ng μL−1) of pDNA (pCMV-β-gal), 30 min before measurement at 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2
1, 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 4
1, 4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, and 8
1, and 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P ratios. Milli-Q water was used to dilute the samples to 1000 μL of the final volume before analysis. The average value of three different measures taken in general mode with the software of a Zeta sizer 3000HSA (Malvern, UK) was taken into consideration. The surface charges of lipoplexes with pCMV-β-gal were measured at a fixed angle of 93° using the same instrument with a different zeta cuvette. Each sample value was taken as the proportion of three different readings.
1 N/P ratios. Milli-Q water was used to dilute the samples to 1000 μL of the final volume before analysis. The average value of three different measures taken in general mode with the software of a Zeta sizer 3000HSA (Malvern, UK) was taken into consideration. The surface charges of lipoplexes with pCMV-β-gal were measured at a fixed angle of 93° using the same instrument with a different zeta cuvette. Each sample value was taken as the proportion of three different readings.
3.5 Scanning electron microscopy (SEM)
A glass slide was applied with 10 μL of liposome, which was placed on a Cu grid with a carbon layer. Using a fresh filter paper, the excess stain was removed and dried at air pressure at room temperature before being examined with an FEI (Apreo LoVac) scanning electron microscope on Mn-kα at a resolution of 127 eV.3.6. Agarose gel electrophoresis
One of the most useful assays for visually predicting the exact pDNA binding capabilities of lipid solutions is gel electrophoresis. Lipoplexes were made by combining 5 μL of supercoiled pDNA (100 ng μL−1) with various amounts of lipid concentrations in a total volume of 24 μL, with the final lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA N/P ratios, 1
pDNA N/P ratios, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 8
1 to 8![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, in a typical experiment. After 30 min of incubation, each lipoplex solution (12 μL) was placed into 1% agarose gel (pre-stained with ethidium bromide) and electrophoresis was performed (100 V, 35 min). The Bio-Rad Gel Doc imaging equipment was used to visualize the bands.
1, in a typical experiment. After 30 min of incubation, each lipoplex solution (12 μL) was placed into 1% agarose gel (pre-stained with ethidium bromide) and electrophoresis was performed (100 V, 35 min). The Bio-Rad Gel Doc imaging equipment was used to visualize the bands.
3.7. Heparin displacement assay
The lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA complexes were produced and incubated for 30 min as described above (agarose gel electrophoresis). These lipid
pDNA complexes were produced and incubated for 30 min as described above (agarose gel electrophoresis). These lipid![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) pDNA complexes were then incubated with heparin sodium salt (0.1 μg) for another 30 min. For heparin displacement analysis, the samples were subjected to electrophoresis in a 1% agarose gel.
pDNA complexes were then incubated with heparin sodium salt (0.1 μg) for another 30 min. For heparin displacement analysis, the samples were subjected to electrophoresis in a 1% agarose gel.
3.8. Serum stability
For serum stability assay, each lipid lipoplexes prepared at 2![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 N/P ratio for five concentrations of serum and incubated for 30 minutes. These lipoplex solutions were added to FBS in DMEM medium at 10–70% of serum concentrations. Then, these complexes were again incubated for 2 h, before subjecting to gel electrophoresis. The gel electrophoresis was conducted at 100 V for 35 min.
1 N/P ratio for five concentrations of serum and incubated for 30 minutes. These lipoplex solutions were added to FBS in DMEM medium at 10–70% of serum concentrations. Then, these complexes were again incubated for 2 h, before subjecting to gel electrophoresis. The gel electrophoresis was conducted at 100 V for 35 min.
3.9. Cells & cell culture
The National Centre for Cell Sciences provided CAL-27 (oral adenosquamous cell carcinoma), HEK-293 (human embryonic kidney) and U87 (human primary glioblastoma) cells (Pune, India). Mycoplasma was not found in any of the cells. The cells were grown in DMEM (Dulbecco's Modified Eagle's Medium) containing 10% fetal bovine serum (FBS). All studies were conducted using cultures that were 85–90% confluent. For cell viability and transfection studies, cell lines were trypsinized, counted, and sub-cultured into 96-well, 24-well, and 12-well plates. Before being utilized in tests, the cells were allowed to adhere for 20–24 h.Conflicts of interest
There are no conflicts to declare.Acknowledgements
SM express gratitude to NIT Warangal, India, for a research fellowship. Dr Srilakshmi V. Patri is grateful to SERB, Department of Science & Technology (DST), Government of India, New Delhi, for financial support (CRG/2018/001049).References
- I. M. Verma and N. Somia, Nature, 1997, 387, 239–242 CrossRef PubMed.
- B. A. Bunnell and R. A. Morgan, Clin. Microbiol. Rev., 1998, 11, 42–56 CrossRef.
- S. Ylä-Herttuala and J. F. Martin, Lancet, 2000, 355, 213–222 CrossRef.
- M. R. Knowles, K. W. Hohneker, Z. Zhou, J. C. Olsen, T. L. Noah, P.-C. Hu, M. W. Leigh, J. F. Engelhardt, L. J. Edwards, K. R. Jones, M. Grossman, J. M. Wilson, L. G. Johnson and R. C. Boucher, N. Engl. J. Med., 1995, 333, 823–831 CrossRef.
- Y. Yang, F. A. Nunes, K. Berencsi, E. Gönczöl, J. F. Engelhardt and J. M. Wilson, Nat. Genet., 1994, 7, 362–369 CrossRef CAS.
- C. McGregor, C. Perrin, M. Monck, P. Camilleri and A. J. Kirby, J. Am. Chem. Soc., 2001, 123, 6215–6220 CrossRef CAS.
- B. Kedika and S. V. Patri, Eur. J. Med. Chem., 2014, 74, 703–716 CrossRef CAS PubMed.
- V. Muripiti, B. Lohchania, V. Ravula, S. Manturthi, S. Marepally, A. Velidandi and S. V. Patri, New J. Chem., 2021, 45, 615–627 RSC.
- D. M. Lynn, D. G. Anderson, D. Putnam and R. Langer, J. Am. Chem. Soc., 2001, 123, 8155–8156 CrossRef CAS PubMed.
- M. X. Tang, C. T. Redemann and F. C. Szoka, Bioconjugate Chem., 1996, 7, 703–714 CrossRef CAS PubMed.
- Y. Lee, H. Koo, Y. B. Lim, Y. Lee, H. Mo and J. S. Park, Bioorg. Med. Chem. Lett., 2004, 14, 2637–2641 CrossRef CAS.
- D. Niculescu-Duvaz, J. Heyes and C. Springer, Curr. Med. Chem., 2005, 10, 1233–1261 CrossRef.
- K. Ewert, A. Ahmad, H. M. Evans, H. W. Schmidt and C. R. Safinya, J. Med. Chem., 2002, 45, 5023–5029 CrossRef CAS.
- M. Gosangi, H. Rapaka, V. Ravula and S. V. Patri, Bioconjugate Chem., 2017, 28, 1965–1977 CrossRef CAS.
- B. Kedika and S. V. Patri, J. Med. Chem., 2011, 54, 548–561 CrossRef CAS PubMed.
- D. Lechardeur and G. L. Lukacs, Curr. Gene Ther., 2002, 2, 183–194 CrossRef.
- I. S. Zuhorn and D. Hoekstra, J. Membr. Biol., 2002, 189, 167–179 CrossRef PubMed.
- Y. Xu and F. C. Szoka, Biochemistry, 1996, 35, 5616–5623 CrossRef PubMed.
- V. Ravula, Y. L. Lo, Y. T. Wu, C. W. Chang, S. V. Patri and L. F. Wang, Mater. Sci. Eng., C, 2021, 126, 112189 CrossRef PubMed.
- B. Kedika and S. V. Patri, Mol. Pharm., 2012, 9, 1146–1162 CrossRef CAS PubMed.
- H. Rapaka, S. Manturthi, P. Arjunan, V. Venkatesan, S. Thangavel, S. Marepally and S. V Patri, ACS Appl. Bio Mater., 2022, 5, 1489–1500 CrossRef CAS PubMed.
- M. Gosangi, T. Y. Mujahid, V. Gopal and S. V. Patri, Org. Biomol. Chem., 2016, 14, 6857–6870 RSC.
- V. Muripiti, B. Lohchania, S. K. Marepally and S. V. Patri, Medchemcomm, 2018, 9, 264–274 RSC.
- V. Ravula, V. Muripiti, S. Manthurthi and S. V. Patri, ChemistrySelect, 2021, 6, 13025–13033 CrossRef CAS.
- J. Sen and A. Chaudhuri, Bioconjugate Chem., 2005, 16, 903–912 CrossRef CAS PubMed.
- P. P. Karmali, V. V. Kumar and A. Chaudhuri, J. Med. Chem., 2004, 47, 2123–2132 CrossRef CAS PubMed.
- J. Wang, X. Guo, Y. Xu, L. Barron and F. C. Szoka, Society, 1998, 2623, 2207–2215 Search PubMed.
- S.-J. Lee and C.-H. Ahn, Macromol. Res., 2006, 14, 129–131 CrossRef.
- S. Pujals and E. Giralt, Adv. Drug Delivery Rev., 2008, 60, 473–484 CrossRef PubMed.
- A. M. Cardoso, C. M. Morais, A. R. Cruz, S. G. Silva, M. L. Do Vale, E. F. Marques, M. C. P. De Lima and A. S. Jurado, Eur. J. Pharm. Biopharm., 2015, 89, 347–356 CrossRef PubMed.
- S. L. Borrego, J. Fahrmann, J. Hou, D. W. Lin, B. J. Tromberg, O. Fiehn and P. Kaiser, J. Lipid Res., 2021, 62, 100056 CrossRef CAS PubMed.
- P. Kaiser, Biomolecules, 2020, 10, 7–9 CrossRef.
- V. Ravula, Y.-L. Lo, Y.-T. Wu, C.-W. Chang, S. V. Patri and L.-F. Wang, Mater. Sci. Eng., C, 2021, 126, 112189 CrossRef CAS.
- E. Yonish-Rouach, D. Resnftzky, J. Lotem, L. Sachs, A. Kimchi and M. Oren, Nature, 1991, 352, 345–347 CrossRef CAS.
- R. W. Johnstone, A. A. Ruefli and S. W. Lowe, Cell, 2002, 108, 153–164 CrossRef CAS PubMed.
- G. Shim, M. G. Kim, J. Y. Park and Y. K. Oh, Asian J. Pharm. Sci., 2013, 8, 72–80 CrossRef CAS.
- H. Elsana, T. O. B. Olusanya, J. Carr-wilkinson, S. Darby, A. Faheem and A. A. Elkordy, Sci. Rep., 2019, 9, 1–17 CrossRef PubMed.
- M. Gosangi, H. Rapaka, T. Y. Mujahid and S. V. Patri, Medchemcomm, 2017, 8, 989–999 RSC.
- L. Li, H. Song, K. Luo, B. He, Y. Nie, Y. Yang, Y. Wu and Z. Gu, Int. J. Pharm., 2011, 408, 183–190 CrossRef.
- M. M. Mady and M. M. Ghannam, Afr. J. Pharm. Pharmacol., 2011, 5, 1898–1905 Search PubMed.
- G. Smistad, J. Jacobsen and S. A. Sande, Int. J. Pharm., 2007, 330, 14–22 CrossRef CAS PubMed.
- V. Ravula, Y. L. Lo, L. F. Wang and S. V. Patri, ACS Omega, 2021, 6, 22955–22968 CrossRef CAS.
- B. K. Majeti, R. S. Singh, S. K. Yadav, S. R. Bathula, S. Ramakrishna, P. V. Diwan, S. S. Madhavendra and A. Chaudhuri, Chem. Biol., 2004, 11, 427–437 CrossRef CAS PubMed.
- H. Gao and K. M. Hui, Gene Ther., 2001, 8, 855–863 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: Experimental procedure, 1H-NMR, 13C-NMR, ESI-HRMS spectra, and HPLC chromatogram purity of compounds are provided. EGFP qualitative and quantitative analysis in U87 cells and cellular uptake qualitative and quantitative analysis in HEK-293 cells are provided. See DOI: https://doi.org/10.1039/d2ra05959b |
| This journal is © The Royal Society of Chemistry 2022 |