Open Access Article
Open Access ArticleThe in vitro fermentation of compound oral liquid by human colonic microbiota altered the abundance of probiotics and short-chain fatty acid production
Ruiming Xiao ab,
Hongzhang Chenab,
Hongbei Hanab,
Guangjuan Luoab and
Ying Lin*ab
ab,
Hongzhang Chenab,
Hongbei Hanab,
Guangjuan Luoab and
Ying Lin*ab
aSouth China University of Technology, School of Bio and Chemical Engineering, Guangdong Key Laboratory of Fermentation and Enzyme Engineering, Guangzhou 510006, People's Republic of China. E-mail: feylin@scut.edu.cn
bSouth China University of Technology, Guangdong Research Center of Industrial Enzyme and Green Manufacturing Technology, School of Bio and Chemical Engineering, Guangzhou 510006, People's Republic of China
First published on 21st October 2022
Abstract
Compound oral liquid (COL), made from functional herbal foods, has gained immense popularity in China for healthcare. However, the interaction between the nutrients in COL and gut microbiota is still unclear. In our study, the content of total flavonoids, polyphenols, and proteins was increased and the total sugar reduced by crushing raw ingredients to 10 mesh (COL-C). After 24 h incubation with supplemented COL by human gut microbiota, the results of 16S rRNA high-throughput sequencing revealed that Faecalibacterium, Collinsella, Bifidobacterium, Megamonas, Lactobacillus, Phascolarctobacterium, and Dialister were enriched by COL. In particular, the latter three genera were observed to be significantly enriched after incubation with COL-C. Meanwhile, the abundance of Dorea, Clostridium XIVa, and Escherichia/Shigella was inhibited by COL. Moreover, the increased levels of acetate, propionate, and butyrate in COL were jointly contributed by supplementary carbohydrates and the enrichment of short-chain fatty acid (SCFA)-producing bacteria. In summary, our results indicated that the optimized extraction facilitated the nutrients to be dissolved out and enhanced the potential prebiotic effects for promoting the abundance of probiotics, suggesting that the nutrients in COL-C might improve the microbial structure by strengthening the metabolism of beneficial bacteria and restricting the conditioned pathogens more efficiently.
Introduction
Functional foods have received wide attention in the past few decades due to their potential in preserving the wellness of human health.1 Beyond their pharmaceutical values, functional foods are also used for healthcare in the form of soups and teas in Southern China.2,3 Currently, traditional herbal medicines, such as ginseng, Siraitia grosvenorii, and Fructus lycii, are common ingredients for making functional foods in China, particularly in the wet and hot districts. Previous studies have shown that the active contents in functional foods, including flavonoids, polyphenols, and non-starch polysaccharides, could improve health conditions by reducing inflammation2,4,5 and protecting the intestinal barrier in colitis mice.6–8 In addition, there is growing evidence that functional foods mediately affected the host health by regulating the gut microbial composition and metabolism.9Since the concept of personal diet and nutrition was introduced to the public,10,11 an increasing number of people started to adopt functional foods to manage their health. Compound oral liquid (COL) is a commercial functional food developed by Infinite Co., Ltd, which mainly consists of the water extracts of Polygoni Multiflori Radix Praeparata (PMRP) and Morinda officinalis (MO). Recent studies have suggested that PMRP could improve the liver mitochondrial function by regulating the mitochondrial metabolic pathways to alleviate glucolipid metabolic disorders, and inulin-type oligosaccharides in root MO was considered to be a potential prebiotic.12,13 Although the health-promoting effects of single herbs are exciting, the impact of their compound nutrition on gut microbiota remained to be investigated.
More recently, the development of sequencing and multi-omics technology has accelerated the comprehension of interaction between the gut microbiota and the nutrients of functional foods. For instance, abundant glucosidases encoded in the genome of Bacteroidetes and Firmicutes helped hydrolyze the non-starch polysaccharides in Lycium barbarum to produce short-chain fatty acids (SCFAs) and contributed to the abundance of probiotics including Faecalibacterium.14,15 Another health tonic oral liquid was confirmed to maintain glucose and lipid metabolism balance, enrich beneficial bacteria and health-related metabolites in rats.16 Hence, gut microbiota are a crucial media when functional foods affect immune responses and gut homeostasis.
In this research, to evaluate the relationship between the major types of bioactive components in COL and probiotics, we assessed the impact of crushing pretreatment on nutrient contents in COL and COL-C, including total sugar, protein, flavonoids, and polyphenols. Then, by incubating COL and COL-C with gut microbiota in vitro, high-throughput sequencing of 16S rRNA gene was applied to evaluate the variations in the probiotic abundance. The gas chromatography assay was conducted to measure SCFA levels in broth. Finally, a correlation analysis of nutrients, bacteria, and SCFAs was carried out to illustrate the relationship between core taxa, nutrients of COL, and metabolites.
Materials and methods
Materials and chemicals
Liquid samples of the COL (ingredients were not crushed) and COL-C (ingredients were crushed) were obtained from Infinite Co., Ltd (Guangdong, China). Bradford Protein Quantification Kit was purchased from Generay biotech Co., Ltd (Shanghai, China). Standards of rutin, gallic acid, SCFAs (acetate, propionate, butyrate, iso-butyrate, valerate, and iso-valerate) and other general chemicals were obtained from Aladdin Industrial Inc. (Shanghai, China).Preparation of the COL and COL-C
The compound oral liquid is a liquid dosage form of health food based on the water extract of Polygoni Multiflori Radix Praeparata (PMRP) and Morinda officinalis (MO). The preparation methods of the COL and COL-C are presented in Fig. 1. In brief, dried PMRP and MO of equal mass were extracted twice with boiling distilled water (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 10, w/v) for 2 h. Then, the extracts were combined and concentrated to obtain compound oral liquid. For the pretreatment of raw ingredients, the COL-C referred to crushing PMRP and MO to 10 mesh for extraction, and COL referred to the extract with raw PMRP and MO used directly.
10, w/v) for 2 h. Then, the extracts were combined and concentrated to obtain compound oral liquid. For the pretreatment of raw ingredients, the COL-C referred to crushing PMRP and MO to 10 mesh for extraction, and COL referred to the extract with raw PMRP and MO used directly.
Measurement of nutrition contents
The in vitro incubation of the oral liquid by gut microbiota was performed according to a previous study with slight modifications.19 In brief, the incubation system was based on a basic nutrient growth medium: 2.0 g L−1 yeast extract, 2.0 g L−1 peptone, 0.1 g L−1 NaCl, 0.04 g L−1 KH2PO4, 0.04 g L−1 K2HPO4, 0.01 g L−1 MgSO4·7H2O, 0.01 g L−1 CaCl2, 2 g L−1 NaHCO3, 0.02 g L−1 haemin, 0.5 g L−1 cysteine-HCl, 0.5 g L−1 bile salts, 2.0 mL L−1 Tween 80, 1.0 mL L−1 1% resazurin solution, and 10 μL L−1 vitamin K. Then, 500 μL of COL and COL-C were added into the BNM medium respectively, and the group with added 500 μL of distilled water was set as the control. Finally, 1.0 mL fecal slurry was inoculated in an airtight serum bottle with 9.0 mL BNM broth under N2 conditions. Incubation was carried out at 37 °C, and samples from 3, 6, 12, and 24 h were collected and saved at −80 °C for further study. Each experiment was performed in triplicate.
The obtained raw data of high-throughput sequencing were subjected to quality control and the clustering results were analyzed using the QIIME 2.0 software (version 2019.7).20 After paired-end read merging and filtering, the remaining clean reads were then clustered into operational taxonomic units (OTUs) at a sequence similarity level of 97%. All OTUs were then annotated based on the Ribosomal Database Project (RDP), and the reads per spike-in OTU were counted. Rarefaction analysis, alpha diversities (Shannon, Simpson, Chao1 and ACE indexes) and beta diversities were analysed using R Project (Vegan package, V 3.3.1).
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 1 min, and 600 μL of the supernatant was acidized with 200 μL 20% H2SO4 (v/v) and vortexed adequately for 1 min. Then, 500 μL n-butanol was added to the acidized fluid for liquid–liquid extraction, and the mixture was vortexed adequately for 2 min and centrifuged at 10
000 rpm for 1 min, and 600 μL of the supernatant was acidized with 200 μL 20% H2SO4 (v/v) and vortexed adequately for 1 min. Then, 500 μL n-butanol was added to the acidized fluid for liquid–liquid extraction, and the mixture was vortexed adequately for 2 min and centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 1 min. Finally, the upper organic phase was filtered through a 0.22 μm filter membrane for further GC detection.
000 rpm for 1 min. Finally, the upper organic phase was filtered through a 0.22 μm filter membrane for further GC detection.An Agilent 7820A GC system (Agilent Technologies, Santa Clara, CA, USA) was equipped with a flame ionization detector (FID) and an automatic sampler, a DB-FFAP capillary column (Agilent, 30 m × 0.25 mm × 0.25 μm) was used for component separation. The temperature operating conditions of the GC system are as follows: initial column temperature, 80 °C, held for 1 min; then 80 °C to 160 °C at a rate of 3 °C min−1; and finally, 160 °C to 230 °C, held for 1 min. Other parameters were set as follows: injection port temperature, 300 °C; FID temperature, 320 °C; and injection volume, 1 μL. The flow rates of dry air, hydrogen and nitrogen were 300, 30, and 20 mL min−1, respectively. Each experiment was performed in triplicate.
Results
Effects of crushing pretreatment on the nutrition constitute of compound oral liquid
The difference of the COL and COL-C was the pretreatment of PMRP and MO. The COL was prepared by raw ingredients, and the COL-C was prepared by 10-meshed crushed ingredients to assess the difference in the dissolving efficiency of contents in the COL. As shown in Table 1, the most abundant nutrient is total sugar. After crushing, the total sugar in COL-C (18.42 ± 0.49 mg mL−1) exhibited a significant decrease compared with COL (20.28 ± 0.61 mg mL−1), but the protein content showed a slight but not significant increase in the COL-C. Meanwhile, the crushing pretreatment obviously contributed to the concentration of total flavonoids and polyphenols, showing increases from 1.56 ± 0.02 to 2.22 ± 0.05 mg mL−1 for flavonoids and 2.22 ± 0.06 to 2.98 ± 0.05 mg mL−1 for polyphenols. Since the crushing pretreatment was the only different craft during extraction, we considered that the variation in nutrients in the COL was mainly caused by crushing the raw ingredients.| Groups | Concentration (mg mL−1) | |||
|---|---|---|---|---|
| Total sugar | Proteins | Flavonoids | Polyphenols | |
| a Different lowercase letters indicate significant differences (P < 0.05) among different groups. | ||||
| COL | 20.28 ± 0.61a | 0.75 ± 0.16a | 1.56 ± 0.02a | 2.22 ± 0.06a |
| COL-C | 18.42 ± 0.49b | 0.98 ± 0.20a | 2.22 ± 0.05b | 2.98 ± 0.05b |
Effect of compound oral liquid fermentation in vitro on gut microbial compositions
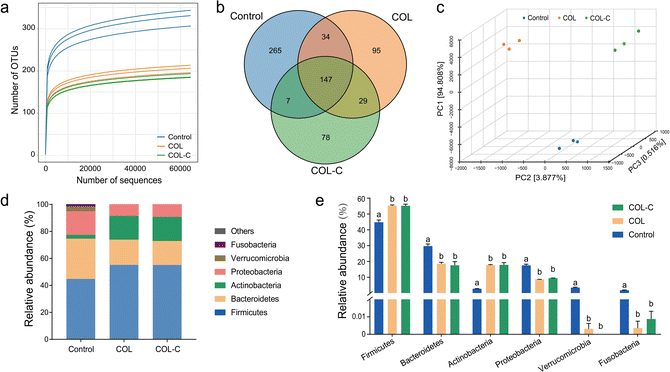
The microbial composition in control, COL, and COL-C was measured by 16S rRNA sequencing, and each measurement was conducted in triplicate. The incubated COL and COL-C groups influenced more OTUs of fecal bacteria and they also showed stronger effects on gut microbial taxonomy. The results of the Shannon rarefaction indicated that the deep sequencing covered most of the operational taxonomic units (OTUs) in each sample (Fig. 2a). As the Venn diagram showed (Fig. 2b), a total of 655 OTUs were discovered after sequencing, in which 265, 95, and 78 OTUs were specifically found in control, COL, and COL-C respectively. Unweighted unifrac distance was calculated to reflect the difference in community composition between groups (Fig. 2c). The clustered points in each group showed a well parallelism, and the distinct distance between groups reflected the variation in microbial communities. Moreover, the alpha diversity results were shown in Table 2. The Chao1, ACE, and Shannon indexes in the COL and COL-C groups were significantly lower than those of the control group, while the Simpson index was higher in the COL and COL-C groups. Particularly, compared with COL, the Chao1 and ACE indexes in COL-C were significantly lower after incubation with gut microbiota.| Groups | Chao1 | ACE | Shannon | Simpson |
|---|---|---|---|---|
| a Different lowercase letters indicate significant differences (P < 0.05) among different groups. | ||||
| Control | 317.93 ± 20.71a | 318.55 ± 20.78a | 4.57 ± 0.03a | (2.04 ± 0.07) × 10−2 a |
| COL | 200.25 ± 9.17b | 200.67 ± 9.30b | 3.86 ± 0.01b | (4.09 ± 0.01) × 10−2 b |
| COL-C | 181.72 ± 4.70c | 181.93 ± 4.54c | 3.83 ± 0.03b | (4.10 ± 0.02) × 10−2 b |
The relative abundance of gut microbiota after incubation is presented in Fig. 2d and e. At the phylum level, gut microbiota consisted mostly of Firmicutes, Bacteroidetes, Actinobacteria, and Proteobacteria representing more than 90% of total bacteria. For these phyla, there were similar variations in COL and COL-C groups in comparison with the control group. The abundance of Bacteroidetes and Proteobacteria were distinctly reduced by the two additives, while the proportions of Actinobacteria and Firmicutes were increased. Besides, other low-abundance phyla such as Fusobacteria and Verrucomicrobia almost disappeared after 24 h incubation.
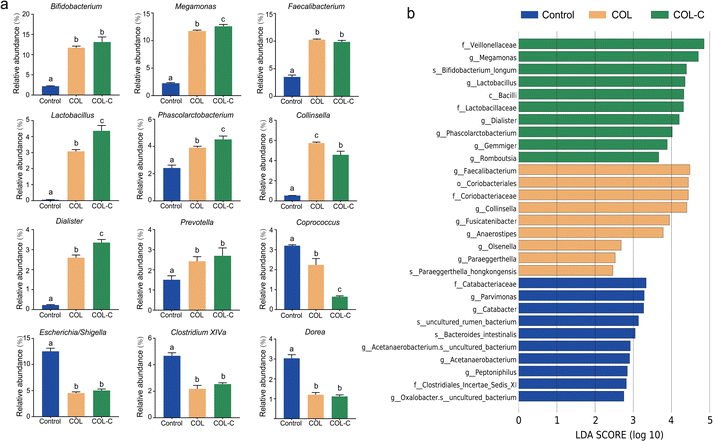
To illustrate the alternations in gut microbiota at a more detailed classification, we further summarized the bacterial abundance changes at the genus level (Fig. 3a). The additive of COL and COL-C both promoted the percentage of several beneficial taxa including Bifidobacterium, Faecalibacterium, and Lactobacillus. In addition, SCFA-producing bacteria such as Megamonas, Phascolarctobacterium, Dialister, and Prevotella were highly enriched, while Coprococcus was decreased after incubation. Particularly, the abundance of Lactobacillus, Phascolarctobacterium, Dialister, and Megamonas had higher enrichment by incubation with the additive COL-C. For the reduced taxa in Proteobacteria, the proportions of Escherichia/Shigella, Clostridium XIVa, and Dorea were decreased in the COL and COL-C groups compared with the control group, however, little significance was observed between COL and COL-C groups.
Accordingly, the advantage taxa in each group were revealed by LEfSe analysis (Fig. 3b). It was confirmed that beneficial bacteria were significantly enriched in the COL-C group. Apart from the above-mentioned genus, Bifidobacterium longum, a well-known probiotic belonging to Bifidobacterium, was also identified to be a core taxon. Overall, the changes in these beneficial bacteria might be related to the alternations of nutrients in the COL after crushing the raw ingredients.
Estimation of the functional pathway in compound oral liquid
The metabolic function of gut microbiota in different groups was further predicted using the PICRUSt 2.0 software. As presented in Fig. 4, the abundance of the KEGG pathway was significantly changed by the COL and COL-C. In particular, the pathways of lysine biosynthesis, alanine, aspartate and glutamate metabolism, aminoacyl-tRNA biosynthesis, pantothenate and CoA biosynthesis, peptidoglycan biosynthesis, and pentose phosphate were highly enriched by the COL and COL-C. Besides, some low-abundant pathways such as protein export, ribosome, and homologous recombination were also improved, whereas the only pathway of biotin metabolism was reduced after incubation. Interestingly, although taxonomy variations were obvious, there were little significant differences in the metabolic functions between the COL and COL-C groups.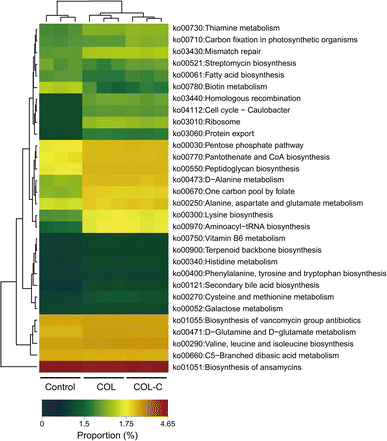 | ||
| Fig. 4 Variation in the gut microbial functional pathway at level 3 by PICRUSt 2 analysis in the control, COL, and COL-C groups. | ||
Effects of compound oral liquid fermentation in vitro on SCFA production
The concentration of the SCFA contents in the broth was detected by the GC assay; the contents of 6 SCFAs, namely, acetate, propionate, butyrate, iso-butyrate, valerate, and iso-valerate were measured for the control, COL, and COL-C groups. As shown in Table 3, acetate, propionate, and butyrate were the predominate SCFAs in the broth. Obviously, the level of acetate in the COL and COL-C groups was significant higher than that in the control group during the whole incubation period. In addition, the COL group had a distinctly higher concentration of acetate than that of COL-C at 6 and 12 h, but the significance disappeared at 24 h. Moreover, additive COL and COL-C significantly improved the levels of propionate and butyrate compared with the control group during the whole incubation period, while no significance was found between them. It was worth noting that the low concentration of iso-butyrate, valerate, and iso-valerate was not found after COL and COL-C incubation. We speculated that the variation in SCFA-producing bacteria changed the fatty acid metabolic flow to the synthesis of acetate, propionate, and butyrate.| SCFAs (mM) | Samples | Anaerobic incubation time (h) | |||
|---|---|---|---|---|---|
| 3 | 6 | 12 | 24 | ||
| a Different lowercase letters indicate significant differences (P < 0.05) among different groups. | |||||
| Acetate | Control | 8.97 ± 0.71a | 11.97 ± 0.16a | 11.55 ± 0.33a | 9.25 ± 0.50a |
| COL | 25.17 ± 0.98b | 25.65 ± 0.43c | 25.74 ± 0.69c | 28.09 ± 0.97b | |
| COL-C | 21.96 ± 1.11b | 23.02 ± 0.48b | 21.98 ± 0.95b | 24.43 ± 1.45b | |
| Propionate | Control | 9.09 ± 0.50a | 13.38 ± 0.19a | 15.81 ± 0.66a | 11.47 ± 0.68a |
| COL | 22.85 ± 1.53b | 26.96 ± 0.78b | 29.45 ± 1.75b | 31.78 ± 0.09b | |
| COL-C | 22.12 ± 0.60b | 25.20 ± 1.16b | 25.28 ± 0.55b | 27.32 ± 1.87b | |
| Butyrate | Control | 2.62 ± 0.04a | 3.50 ± 0.03a | 3.49 ± 0.01a | 2.76 ± 0.07a |
| COL | 5.08 ± 0.31b | 7.88 ± 0.47b | 9.71 ± 0.21b | 9.99 ± 0.11b | |
| COL-C | 4.95 ± 0.12b | 7.73 ± 0.33b | 9.53 ± 0.03b | 9.97 ± 0.16b | |
| Iso-butyrate | Control | Nd | 0.12 ± 0.01 | 1.00 ± 0.02 | 0.72 ± 0.04 |
| COL | Nd | Nd | Nd | Nd | |
| COL-C | Nd | Nd | Nd | Nd | |
| Valerate | Control | Nd | 0.17 ± 0.08a | 0.28 ± 0.07a | 0.12 ± 0.02a |
| COL | Nd | Nd | Nd | Nd | |
| COL-C | Nd | Nd | Nd | Nd | |
| Iso-valerate | Control | 0.07 ± 0.01a | 0.61 ± 0.01a | 2.49 ± 0.10a | 1.68 ± 0.09a |
| COL | Nd | Nd | Nd | Nd | |
| COL-C | Nd | Nd | Nd | Nd | |
Correlations between gut microbial taxa, nutrition contents, and SCFAs
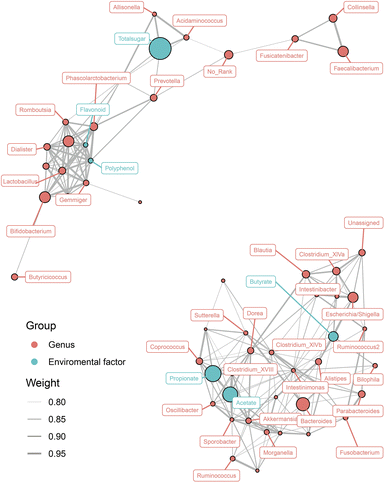
To evaluate whether the SCFA levels and nutrient concentrations were associated with the gut microbiota, Spearman's correlation analysis of a network connection diagram was performed, and is shown in Fig. 5. Total sugar was the most abundant nutrient in both COL and COL-C, it was positively correlated with the enrichment of Prevotella, Allisonella, and Acidaminococcus. Interestingly, though the flavonoids and polyphenols were lower than total sugar, they were highly correlated with the improvement of several increasing taxa such as Phascolarctobacterium, Dialister, Lactobacillus, and Bifidobacterium. The reduced taxa, including Dorea, Sutterella, and Clostridium XIVa, were negatively correlated with acetate, propionate, and butyrate respectively. However, the strengthening interaction between beneficial bacteria was mainly contributed by the enhanced flavonoids and polyphenols; meanwhile, the abundance of potential pathogens was opposite to the concentration of SCFAs, reflecting an interaction relationship between nutrients, metabolites, and gut microbial composition.Discussion
The carbohydrates, flavonoids, and polyphenols were verified to be bioactive molecules in functional foods. For instance, plant polysaccharides, especially non-starch polysaccharides, were identified as potential prebiotics to benefit the gut microbial composition and serve as substrates for SCFA production.8 Flavonoids and polyphenols often exist in herbs in the form of glycosides or aglycone. More recently, diosmetin-7-O-glucoside was mainly converted into diosmetin and acacetin by Escherichia spp. isolated from the human gut.21 Polyphenol compounds had a similar degradation pathway to flavonoids; except hydrolyzing from glycosides, they were metabolized via their aglycones yielding smaller substances such as (−)-epigallocatechin gallate and (−)-gallocatechin gallate.22 Our previous research also indicated that aglycones of glycosides might possess a stronger bioactivity.19In this study, crushing pretreatment promoted the dissolution rate of flavonoids and polyphenols, but total sugar was found to be slightly reduced in COL-C. As known to all, crushing herbal medicines could promote the dissolution rate of contents by increasing the contact area between ingredients and solvents.23 However, as the specific surface area increased, the increase in surface energy might strengthen the adsorbability of granules and affect the dissolution rate of bioactive compounds, which might cause a decrease in total sugar. It was worth noting that herbal flavonoids and polyphenols showed efficient bioactivities in vitro. In particular, the 50% inhibitory concentration (IC50) of flavonoids and polyphenols on free radical and cancer cell line was significantly higher than that of polysaccharides according to previous reports.24,25 As a result, the dissolving-out amount of nutrients in the COL was enhanced by crushing the ingredients.
Gut microbes play a vital role in host health and disease conditions, and they are closely associated with ingested nutrients in daily diet. Compared with the control group, it was found that COL and COL-C both decreased the abundance of Proteobacteria, a phylum containing opportunistic pathogen, especially genera from Dorea, Clostridium XIVa, and Escherichia/Shigella,26,27 suggesting a healthy tendency of the gut microbial structure after incubation. Moreover, although there was a high degree of consistency between COL and COL-C on the variation of phylum level, COL-C contributed to some genera such as Lactobacillus, Phascolarctobacterium, Dialister, and Megamonas with more efficiency. A previous study revealed that Lactobacillus contains abundant probiotics and improved the eradication rate of Helicobacter pylori by using four-probiotics regimen.28,29 Besides, increases of Dialister and Megamonas were commonly found during supplementary of prebiotics.19,30 Recently, another study has put forward that Collinsella may mitigate infections and exacerbation of COVID-19 by generating urso-deoxycholate.31 Prevotella copri, a strictly anaerobic species belonging to Prevotella, was associated with the hypoglycemic effect in diabetic mice.32
Faecalibacterium and Bifidobacterium were the genera enriched both by COL and COL-C. Faecalibacterium prausnitzii, known as a live biotherapeutic product, was confirmed to be negatively related to human diseases including inflammatory bowel disease and type 2 diabetes.33,34 As a common probiotics in the gut tract, Bifidobactrium constituted the main cluster of gut microbiota in early life of infants.35 Meanwhile, it was closely correlated with the metabolism of a variety of prebiotics such as human milk oligosaccharides and galactooligosaccharides.36 It was found that Bifidobactrium longum, Lactobacillus spp., appeared to be core species in the COL-C group, indicating that COL-C had stronger prebiotic effects than those of COL, which might be related to the enhanced contents of nutrition.
Probiotics helped hydrolyzed plant-derived polysaccharides to produce SCFAs, which play an important role in modulating immunity and protecting intestinal barrier integrity.37 Compared with the control group, the additive COL and COL-C conformably improved the concentration of acetate, propionate, and butyrate in the broth. Similar results were also found in incubating inulin with gut microbiota in vitro. SCFAs are major fermentation products of carbohydrates by gut microbiota or they can be transformed from other microbial metabolites such as amino acids.38,39 Hence, we considered that the improved SCFA levels were jointly contributed by supplementary substrates in the COL and metabolism of amino acids, since the synthesis and metabolic function of amino acids were enhanced after incubation. As reported, SCFAs were recognized as a type of health-related metabolite. For instance, acetate and butyrate were confirmed to modulate host immunity through G-protein-coupled receptors and interacted with the immune system via the brain-gut axis.38,40 Propionate significantly stimulated the release of PYY and GLP-1 from human colonic cells to control the appetite.41 Consequently, the results suggested that COL had the potential to improve human health by promoting SCFA levels.
Furthermore, we found that the bacterial genera were also related to changes in SCFAs. It was inferred that the enriched Bifidobacterium and Prevotella increase the acetate production,38 whereas Dialister and Phascolarctobacterium are the major producers of propionate.42 Faecalibacterium was identified as a butyrate producer, leading to the generation of butyrate via the butyryl-CoA![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetate-CoA transferase pathway.43
acetate-CoA transferase pathway.43
Interestingly, the relationship between gut microbiota, nutrients, and SCFAs exhibited polarization. According to Fig. 5, most of beneficial bacteria including Lactobacillus, Bifidobacterium, Dialister, and Phascolarctobacterium were co-enriched with flavonoids and polyphenols in the COL, suggesting that the improved nutrient contents were the major elements for probiotics. For another aspect, a great number of taxa were negatively correlated with SCFA production, reflecting that the gut microbial structure might be altered by metabolites from probiotics. However, the gut microbiota was a complex colony, and the active components in functional foods could directly enrich metabolizable bacteria or indirectly modulated their abundance through microbial metabolites. Our future research will focus on the preservation of gut barrier in disease models such as IBD, since the COL-C showed improvements in gut microbial composition and metabolites in vitro incubation systems.
Conclusions
In summary, our results indicated that the crushing pretreatment contributed to the dissolving out of flavonoids and polyphenols in compound oral liquid. The COL-C showed stronger probiotic effects to enrich Lactobacillus, Bifidobacterium, Dialister, Phascolarctobacterium, Faecalibacterium, and other SCFA-producing bacteria. Meanwhile, the COL-C inhibited conditional pathogens to modulate the microbial structure to a healthy tendency. Furthermore, SCFAs including acetate, propionate, and butyrate in the broth were highly expressed by the COL and COL-C, reflecting the potential for maintaining gut immune homeostasis and providing energy for colon cells. Finally, the flavonoids and polyphenols seem to be crucial factors for promoting the growth of probiotics, and carbohydrates in the COL-C served as available substrates for SCFA fermentation. The results offered a new comprehension of compound oral liquid as functional supplements, and laid the theory foundation for the animal experiments in protecting intestinal health via modulating the gut microbiome.Data availability
The datasets generated during and/or analysed during the current study are available from the corresponding author on reasonable request.Author contributions
Ruiming Xiao and Hongzhang Chen contributed equally to this article, and carried out most of the experiments, designed the project, analyzed microbiome and metabolic data and composed the manuscript. Hongbei Han helped organized data, carried out GC analysis of the short-chain fatty acids of incubation samples. Guangjuan Luo helped collected the faeces samples, organized ethical review document and helped plot and incubation. Ying Lin supervised to carry out most of the experiments, and revised the manuscript for publication.Abbreviations
| COL | Compound oral liquid |
| COL-C | Compound oral liquid crushed |
| SCFA | Short-chain fatty acid |
| PMRP | Polygoni multiflori radix praeparata |
| MO | Morinda officinalis |
| PBS | Phosphate-buffered saline |
| OTU | Operational taxonomic unit |
| LEfSe | Linear discriminant analysis effect size |
| PYY | Peptide tyrosine–tyrosine |
| GLP-1 | Glucagon-like peptide-1 |
| IBD | Inflammatory bowel disease |
Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.Acknowledgements
All the authors are thankful for the financial support of National Key Research and Development Program of China (Grant No. 2017YFD0400300).References
- L. Dominguez Diaz, V. Fernandez-Ruiz and M. Camara, Crit. Rev. Food Sci. Nutr., 2020, 60, 1738–1746 CrossRef CAS PubMed.
- J. Huang, L. Ding, W. Tian, H. Zhi, J. Chen, L. Wu, L. Wang, J. Xie, J. Bai, H. Fan, S. Zhao, K. Zhang and J. Zheng, Microchem. J., 2021, 168, 106399 CrossRef CAS.
- R. Xiao, G. Luo, W. Liao, S. Chen, S. Han, S. Liang and Y. Lin, npj Science of Food, 2022, 6(1), 45 CrossRef PubMed.
- K. Moore, L. Howard, C. Brownmiller, I. Gu, S. O. Lee and A. Mauromoustakos, Food Funct., 2019, 10, 7091–7102 RSC.
- S. L. Johnson, R. D. Kirk, N. A. DaSilva, H. Ma, N. P. Seeram and M. J. Bertin, Metabolites, 2019, 9(4), 78 CrossRef CAS PubMed.
- V.-L. Truong and W.-S. Jeong, Food Sci. Hum. Wellness, 2022, 11, 502–511 CrossRef CAS.
- F. Zhou, Y. L. Li, X. Zhang, K. B. Wang, J. A. Huang, Z. H. Liu and M. Z. Zhu, J. Agric. Food Chem., 2021, 69, 14530–14543 CrossRef CAS PubMed.
- M. Ho Do, Y. S. Seo and H. Y. Park, Crit. Rev. Food Sci. Nutr., 2021, 61, 1212–1224 CrossRef PubMed.
- M. L. Y. Wan, K. H. Ling, H. El-Nezami and M. F. Wang, Crit. Rev. Food Sci. Nutr., 2019, 59, 1927–1936 CrossRef CAS PubMed.
- A. A. Kolodziejczyk, D. Zheng and E. Elinav, Nat. Rev. Microbiol., 2019, 17, 742–753 CrossRef CAS PubMed.
- R. V. Bubnov, M. Y. Spivak, L. M. Lazarenko, A. Bomba and N. V. Boyko, EPMA J., 2015, 6, 14 CrossRef PubMed.
- Y. Q. Yang, F. Y. Meng, X. Liu, M. Zhang, W. Gu, H. L. Yan, J. Yu and X. X. Yang, J. Pharm. Pharmacol., 2021, 73, 796–807 CrossRef PubMed.
- Z. Yang, J. Hu and M. Zhao, Carbohydr. Polym., 2011, 83, 1997–2004 CrossRef CAS.
- A. El Kaoutari, F. Armougom, J. I. Gordon, D. Raoult and B. Henrissat, Nat. Rev. Microbiol., 2013, 11, 497–504 CrossRef CAS PubMed.
- W. Zhu, S. Zhou, J. Liu, R. J. C. McLean and W. Chu, Biomed. Pharmacother., 2020, 121, 109591 CrossRef CAS PubMed.
- L. Du, Y. Sun, Q. Wang, L. Wang, Y. Zhang, S. Li, H. Jin, S. Yan and X. Xiao, Food Res. Int., 2021, 144, 110323 CrossRef CAS PubMed.
- G. Chen, M. Xie, P. Wan, D. Chen, H. Ye, L. Chen, X. Zeng and Z. Liu, Food Chem., 2018, 244, 331–339 CrossRef CAS PubMed.
- J. C. Carmona-Hernandez, M. Le, A. M. Idarraga-Mejia and C. H. Gonzalez-Correa, Molecules, 2021, 26(21), 6431 CrossRef CAS PubMed.
- R. Xiao, W. Liao, G. Luo, Z. Qin, S. Han and Y. Lin, ACS Omega, 2021, 6, 25486–25496 CrossRef CAS PubMed.
- E. Bolyen, J. R. Rideout, M. R. Dillon, N. A. Bokulich, C. C. Abnet, G. A. Al-Ghalith, H. Alexander, E. J. Alm, M. Arumugam, F. Asnicar, Y. Bai, J. E. Bisanz, K. Bittinger, A. Brejnrod, C. J. Brislawn, C. T. Brown, B. J. Callahan, A. M. Caraballo-Rodriguez, J. Chase, E. K. Cope, R. Da Silva, C. Diener, P. C. Dorrestein, G. M. Douglas, D. M. Durall, C. Duvallet, C. F. Edwardson, M. Ernst, M. Estaki, J. Fouquier, J. M. Gauglitz, S. M. Gibbons, D. L. Gibson, A. Gonzalez, K. Gorlick, J. Guo, B. Hillmann, S. Holmes, H. Holste, C. Huttenhower, G. A. Huttley, S. Janssen, A. K. Jarmusch, L. Jiang, B. D. Kaehler, K. B. Kang, C. R. Keefe, P. Keim, S. T. Kelley, D. Knights, I. Koester, T. Kosciolek, J. Kreps, M. G. I. Langille, J. Lee, R. Ley, Y. X. Liu, E. Loftfield, C. Lozupone, M. Maher, C. Marotz, B. D. Martin, D. McDonald, L. J. McIver, A. V. Melnik, J. L. Metcalf, S. C. Morgan, J. T. Morton, A. T. Naimey, J. A. Navas-Molina, L. F. Nothias, S. B. Orchanian, T. Pearson, S. L. Peoples, D. Petras, M. L. Preuss, E. Pruesse, L. B. Rasmussen, A. Rivers, M. S. Robeson II, P. Rosenthal, N. Segata, M. Shaffer, A. Shiffer, R. Sinha, S. J. Song, J. R. Spear, A. D. Swafford, L. R. Thompson, P. J. Torres, P. Trinh, A. Tripathi, P. J. Turnbaugh, S. Ul-Hasan, J. J. J. van der Hooft, F. Vargas, Y. Vazquez-Baeza, E. Vogtmann, M. von Hippel, W. Walters, Y. Wan, M. Wang, J. Warren, K. C. Weber, C. H. D. Williamson, A. D. Willis, Z. Z. Xu, J. R. Zaneveld, Y. Zhang, Q. Zhu, R. Knight and J. G. Caporaso, Nat. Biotechnol., 2019, 37, 852–857 CrossRef CAS PubMed.
- M. Zhao, L. Du, J. Tao, D. Qian, E. X. Shang, S. Jiang, J. Guo, P. Liu, S. L. Su and J. A. Duan, J. Agric. Food Chem., 2014, 62, 11441–11448 CrossRef CAS PubMed.
- B. Chen, J. Zhou, Q. Meng, Y. Zhang, S. Zhang and L. Zhang, Food Funct., 2018, 9, 4858–4864 RSC.
- Y. Fan, C. P. Yan, C. Chen, K. F. So, P. Li and L. W. Qi, J. Pharm. Biomed. Anal., 2014, 95, 213–219 CrossRef CAS PubMed.
- Y. Athukorala, K. N. Kim and Y. J. Jeon, Food Chem. Toxicol., 2006, 44, 1065–1074 CrossRef CAS PubMed.
- Q. Ma, J. G. Jiang, X. Yuan, K. Qiu and W. Zhu, Food Chem. Toxicol., 2019, 125, 422–429 CrossRef CAS PubMed.
- H. C. The, D. P. Thanh, K. E. Holt, N. R. Thomson and S. Baker, Nat. Rev. Microbiol., 2016, 14, 235–250 CrossRef CAS PubMed.
- W. Zhou, Y. Yan, J. Mi, H. Zhang, L. Lu, Q. Luo, X. Li, X. Zeng and Y. Cao, J. Agric. Food Chem., 2018, 66, 898–907 CrossRef CAS PubMed.
- N. Viazis, K. Argyriou, K. Kotzampassi, D. K. Christodoulou, P. Apostolopoulos, S. D. Georgopoulos, C. Liatsos, O. Giouleme, K. Koustenis, C. Veretanos, D. Stogiannou, M. Moutzoukis, C. Poutakidis, Mylonas II, I. Tseti and G. J. Mantzaris, Nutrients, 2022, 14(3), 632 CrossRef CAS PubMed.
- R. Ashraf and N. P. Shah, Crit. Rev. Food Sci. Nutr., 2014, 54, 938–956 CrossRef CAS PubMed.
- J. Mou, Q. Li, W. Shi, X. Qi, W. Song and J. Yang, Carbohydr. Polym., 2020, 228, 115359 CrossRef CAS PubMed.
- M. Hirayama, H. Nishiwaki, T. Hamaguchi, M. Ito, J. Ueyama, T. Maeda, K. Kashihara, Y. Tsuboi and K. Ohno, PLoS One, 2021, 16, e0260451 CrossRef CAS PubMed.
- P. Kovatcheva-Datchary, A. Nilsson, R. Akrami, Y. S. Lee, F. De Vadder, T. Arora, A. Hallen, E. Martens, I. Bjorck and F. Backhed, Cell Metab., 2015, 22, 971–982 CrossRef CAS PubMed.
- M. C. Mentella, F. Scaldaferri, M. Pizzoferrato, A. Gasbarrini and G. A. D. Miggiano, Nutrients, 2020, 12(4), 944 CrossRef CAS PubMed.
- N. Tai, F. S. Wong and L. Wen, Rev. Endocr. Metab. Disord., 2015, 16, 55–65 CrossRef CAS PubMed.
- M. Derrien, A. S. Alvarez and W. M. de Vos, Trends Microbiol., 2019, 27, 997–1010 CrossRef CAS PubMed.
- H. Li, J. A. Lane, J. Chen, Z. Lu, H. Wang, S. Dhital, X. Fu, Q. Huang, F. Liu and B. Zhang, Carbohydr. Polym., 2022, 287, 119322 CrossRef CAS PubMed.
- J. Huo, Z. Wu, W. Sun, Z. Wang, J. Wu, M. Huang, B. Wang and B. Sun, J. Agric. Food Chem., 2022, 70, 711–735 CrossRef CAS PubMed.
- A. Koh, F. De Vadder, P. Kovatcheva-Datchary and F. Backhed, Cell, 2016, 165, 1332–1345 CrossRef CAS PubMed.
- E. P. Neis, C. H. Dejong and S. S. Rensen, Nutrients, 2015, 7, 2930–2946 CrossRef CAS PubMed.
- K. J. O'Riordan, M. K. Collins, G. M. Moloney, E. G. Knox, M. R. Aburto, C. Fulling, S. J. Morley, G. Clarke, H. Schellekens and J. F. Cryan, Mol. Cell. Endocrinol., 2022, 546, 111572 CrossRef PubMed.
- E. S. Chambers, A. Viardot, A. Psichas, D. J. Morrison, K. G. Murphy, S. E. Zac-Varghese, K. MacDougall, T. Preston, C. Tedford, G. S. Finlayson, J. E. Blundell, J. D. Bell, E. L. Thomas, S. Mt-Isa, D. Ashby, G. R. Gibson, S. Kolida, W. S. Dhillo, S. R. Bloom, W. Morley, S. Clegg and G. Frost, Gut, 2015, 64, 1744–1754 CrossRef CAS PubMed.
- P. Louis, G. L. Hold and H. J. Flint, Nat. Rev. Microbiol., 2014, 12, 661–672 CrossRef CAS PubMed.
- E. J. Laserna-Mendieta, A. G. Clooney, J. F. Carretero-Gomez, C. Moran, D. Sheehan, J. A. Nolan, C. Hill, C. G. M. Gahan, S. A. Joyce, F. Shanahan and M. J. Claesson, J. Crohns Colitis, 2018, 12, 204–216 CrossRef PubMed.
| This journal is © The Royal Society of Chemistry 2022 |