Open Access Article
Open Access ArticleCreative Commons Attribution 3.0 Unported Licence
Evaluation of antiviral activity of Carica papaya leaves against SARS-CoV-2 assisted by metabolomic profiling
Amr Adel a,
Mohamed S. Elnaggar
a,
Mohamed S. Elnaggar *b,
Amgad Albohy
*b,
Amgad Albohy c,
Ahmed A. Elrashedyd,
Ahmed Mostafa
c,
Ahmed A. Elrashedyd,
Ahmed Mostafa e,
Omnia Kutkate,
Usama Ramadan Abdelmohsen
e,
Omnia Kutkate,
Usama Ramadan Abdelmohsen fg,
Eman Al-Sayed
fg,
Eman Al-Sayed b and
Mohamed A. Rabehah
b and
Mohamed A. Rabehah
aDepartment of Pharmacognosy, Faculty of Pharmacy, Modern University for Technology and Information, 11865, Cairo, Egypt
bDepartment of Pharmacognosy, Faculty of Pharmacy, Ain Shams University, 11566, Cairo, Egypt
cDepartment of Pharmaceutical Chemistry, Faculty of Pharmacy, The British University in Egypt (BUE), Suez Desert Road, ElSherouk City, 11837, Cairo, Egypt
dNatural and Microbial Chemistry Department, Pharmaceutical and Drug Industries Research Division, National Research Centre (NRC), AlBohoos Street, Dokki, 12311, Cairo, Egypt
eCenter of Scientific Excellence for Influenza Viruses, National Research Centre, 12622, Giza, Egypt
fDepartment of Pharmacognosy, Faculty of Pharmacy, Minia University, Minia, 61519, Egypt
gDepartment of Pharmacognosy, Faculty of Pharmacy, Deraya University, Minia, 61111, Egypt
hDepartment of Pharmacognosy, Faculty of Pharmacy, Cairo University, 11562, Giza, Egypt
First published on 16th November 2022
Abstract
The COVID-19 pandemic caused a huge health crisis all over the globe. SARS-CoV-2 is the virus responsible for the disease and it is highly contagious leaving millions of confirmed infected cases and a dangerous death toll. Carica papaya is a tropical plant known for its antiviral activity since it possesses different classes of compounds that are believed to combat various viral classes. In this study, the extracts prepared from C. papaya leaves cultivated in Egypt were evaluated for their anti-SARS-CoV-2 activity using crystal violet assay and for their cytotoxicity through MTT assay. The total methanolic extract, n-hexane, ethyl acetate, and n-butanol fractions of papaya leaves were used in the study and the results revealed that the n-hexane fraction has a high anti-SARS-CoV-2 activity with an IC50 value = 1.98 μg mL−1. Moreover, it also showed a high selectivity index value = 104.7. Dereplication of the secondary metabolites in the crude methanolic extract of C. papaya leaves revealed the presence of different classes of compounds including sterols, terpenes, fatty acid, alkaloids and flavonoids that are known to possess antiviral activities against various classes of viruses. The current study was assisted by molecular docking, molecular dynamics simulation and MM-PBSA calculations for the annotated compounds against 6 SARS-CoV-2 target proteins. The results of these in silico-based investigations showed high to moderate binding on the targeted proteins. This postulation may instigate further research studies concerning the compounds responsible for this high anti-SARS-CoV-2 activity of the n-hexane fraction of C. papaya leaves.
1. Introduction
Severe acute respiratory syndrome coronavirus (SARS-CoV) and Middle East respiratory syndrome coronavirus (MERS-CoV) caused severe acute respiratory diseases in the last two decades through transmission from animals to humans.1 But in December 2019, a novel severe acute respiratory syndrome coronavirus (SARS-CoV-2) inflicted a disturbance to the global health care and economy systems and affected the lives and health of millions all over the world.Herbal medicines are considered a rich source of natural antiviral compounds that proved their activity against different classes of viruses. Therefore, natural compounds may help in the combat against SARS-CoV-2. C. papaya is an edible tropical fruit native to Central America and Mexico.2 Papaya leaves, seeds, roots, and fruits have been widely used in folk medicine in Africa and Central America in the treatment of various diseases such as dengue fever, jaundice, asthma, gonorrhea, urinary complaints, dressing wounds. Also, in the Ayurvedic literature, it was used as a laxative, diuretic, expectorant, antibacterial, antifungal, counter-irritant and as a treatment for dysentery and chronic diarrhea, also as a paste for treatment of ringworm and psoriasis.3,4 Many pharmacological activities were reported for C. papaya including anti-inflammatory,5 anti-cancer,6 anti-protozoal,7 anti-microbial,8 anti-diabetic,9 anti-fungal,10 anti-hyperlipidemic,11 anti-thrombocytopenic,12 anti-viral,13 anti-gout,14 antihypertensive,15 analgesic16 and hepato-protective activities.17 The wide range of activity is due to the presence of different secondary metabolites including flavonoids, alkaloids, sterols, triterpenoids, isothiocyanates, tannins, and other phenolic compounds.
The different extracts of C. papaya possessed previously reported antiviral activities. For example, it is well known for its activity in the treatment of the dengue virus.18–21 In addition, papaya fruit pulp was recently reported to exert antiviral activity against Zika virus.22 Papaya comprises a wide range of phyto-compounds that may induce antiviral activities against different classes of viruses.23 Fatty acids, sterols, and triterpenoid compounds represent the major compounds present in C. papaya.24–26 Many studies reported that fatty acids mediate antiviral activities via several mechanisms.27–29 Moreover, sterols and triterpenoids had significant antiviral activity against different viral types.30,31 Betulinic acid, oleanolic acid, and stigmasterols were from the major triterpenoid and steroidal compounds present in papaya with reported antiviral activities. Betulinic acid, a pentacyclic triterpene, displayed a broad range of antiviral activities against human immunodeficiency virus (HIV) and herpes simplex virus (HSV).32 In addition, it showed activity against Influenza A and ECHO 6 viruses.33 Another pentacyclic triterpene oleanolic acid demonstrated antiviral activities against HIV-1 and Hepatitis C virus (HCV).34–36 Furthermore, stigmasterol is one of the famous steroidal compounds reported in papaya.37 This sterol is known to have inhibitory activity against the HIV-1 virus's reverse transcriptase.38 These significant antiviral activities initiated the idea of testing the possible antiviral activity of different extracts of the leaves of C. papaya cultivated in Egypt that could contain promising antiviral agents against SARS-CoV-2.
2. Experimental
2.1. Plant material
The plant material of C. papaya leaves was collected in August 2018 from the home garden, Nasr city, Cairo, and was authenticated by Eng. Therease Labib, the consultant at Orman Botanical Garden, Giza, and National Gene Bank at the Ministry of Agriculture, Egypt. A voucher specimen of C. papaya (PHG-P-CP-382) was deposited at the herbarium of the Department of Pharmacognosy, Faculty of Pharmacy, Ain Shams University, Cairo, Egypt. The collected leaves were washed with tap water to remove any dust and contaminants and shade dried for 5 days till complete dryness. The dried leaves were then collected and milled to a fine powder to be ready for extraction.2.2. Chemicals
All the reagents used were of an analytical grade. The chemicals are purchased from Piochem Company and Al-Nasr Company for chemical industries.2.3. Extraction procedure
The powdered leaves of C. papaya (1.7 kg) were extracted by three times maceration with a total of 17 L of methanol at room temperature for 24 h. The extract was then filtered, stored, and protected from sunlight to be evaporated. The crude extract was then evaporated at 50 °C under reduced pressure using a high-capacity rotary evaporator at the research lab unit of the faculty of pharmacy, Ain Shams University. A total dark green crude extract with a weight of (245 g) was produced, collected in a sterile flask for further fractionation.2.4. Fractionation procedure
The crude methanol extract of papaya leaves (245 g) was dissolved in 1 L of methanol/distilled water (10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 90 v/v) solution then transferred to a 2 liter-capacity separating funnel to begin the fractionation process via partitioning method. The crude dissolved extract was then partitioned with 500 mL of n-hexane to separate chlorophyll and non-polar compounds. A total of 7 L of n-hexane fraction were then collected and evaporated at 45 °C under reduced pressure using a rotary evaporator. The n-hexane fraction was evaporated and then freeze-dried via lyophilization giving a total weight of 77 g. The crude extract was further partitioned with ethyl acetate for the isolation of compounds of medium polarity. The crude extract was partitioned with 500 mL of ethyl acetate for twelve runs giving 6 L of ethyl acetate fraction which was subsequently evaporated at 50 °C using a rotary evaporator and then lyophilized to give a solid yellowish-brown ethyl acetate fraction with a net weight of 25 g. As a final fractionation step, the crude extract was partitioned with 500 mL of n-butanol for twelve runs to separate the compounds with higher polarity. A total volume of 6 L of the n-butanol fraction was collected for further evaporation at 59 °C under reduced pressure using a rotary evaporator then lyophilized producing a total weight of 24 g of a dark brown solid n-butanol fraction. The n-hexane, ethyl acetate, n-butanol fraction and the remaining residue of the crude extract were all freeze dried and stored in the refrigerator for further use.
90 v/v) solution then transferred to a 2 liter-capacity separating funnel to begin the fractionation process via partitioning method. The crude dissolved extract was then partitioned with 500 mL of n-hexane to separate chlorophyll and non-polar compounds. A total of 7 L of n-hexane fraction were then collected and evaporated at 45 °C under reduced pressure using a rotary evaporator. The n-hexane fraction was evaporated and then freeze-dried via lyophilization giving a total weight of 77 g. The crude extract was further partitioned with ethyl acetate for the isolation of compounds of medium polarity. The crude extract was partitioned with 500 mL of ethyl acetate for twelve runs giving 6 L of ethyl acetate fraction which was subsequently evaporated at 50 °C using a rotary evaporator and then lyophilized to give a solid yellowish-brown ethyl acetate fraction with a net weight of 25 g. As a final fractionation step, the crude extract was partitioned with 500 mL of n-butanol for twelve runs to separate the compounds with higher polarity. A total volume of 6 L of the n-butanol fraction was collected for further evaporation at 59 °C under reduced pressure using a rotary evaporator then lyophilized producing a total weight of 24 g of a dark brown solid n-butanol fraction. The n-hexane, ethyl acetate, n-butanol fraction and the remaining residue of the crude extract were all freeze dried and stored in the refrigerator for further use.
2.5. Metabolic profiling of papaya leaf methanolic extract
LC-MS profiling was performed on methanol extract C. papaya according to the method described by Haggag et al.39 on an Acquity Liquid Chromatography system coupled to a Synapt G2 HDMS quadrupole time-of-flight hybrid mass spectrometer (Waters, Milford, CT, USA). Chromatographic separation was carried out on a BEH C18 column (2.1 × 100 mm, 1.7 μm particle size, Waters, Milford, USA) with a guard column (2.1 × 50 mm, 1.7 μm particle size). A linear binary solvent gradient of 0–100% eluent B over 6 min at a flow rate of 0.3 mL min−1, using 0.1% formic acid in water (v/v) as solvent A and acetonitrile as solvent B. The injection volume was 2 μL and the column temperature was 40 °C. LC-MS spectra were viewed using Thermo Xcalibur 2.1 (Thermo Scientific, Germany). MSConvert software was used to convert the raw data into separate positive and negative ionization files. MZmine 2.10 software was used for peak picking, deconvolution, deisotoping, alignment and formula predication. The compounds were identified via the Dictionary of Natural Products database.2.6. Anti-SARS-CoV-2 activity
2.7. Molecular docking
Docking was done as reported earlier.47 In brief, ligands 3D structures were downloaded from Pubchem (https://pubchem.ncbi.nlm.nih.gov/) when available or 2D structure is drawn and converted to 3D. Ligands were minimized with 1000 steps of steepest descent algorithm using Avogadro.48 Protein targets were retrieved from protein data bank (http://www.rcsb.org/) under the codes 6LU7, 7BV2, 6W4H, 6M0J, 6VW1 and 6WX4. Proteins were prepared by deleting water molecules, adding hydrogens, merging hydrogens and then computing Gasteiger charges using autodock tools. Docking was done using Autodock vina49 with a grid box of 253 Å3 centred on co-crystalized ligand if available. For 6M0J and 6VW1 where no ligand is available, the grid box was centered on Q493 and E35, respectively. Exhaustiveness of 16 was used for all docking procedures. Co-crystalized ligands were docked when present to validate docking procedure as reported earlier and accepted if RMSD between crystalized and docked ligand is less than 2 Å calculated using DockRMSD server.50,512.8. Molecular dynamics simulation
The chosen complex was then subjected to a 50 ns MD simulation using the AMBER 18 platform.52 The receptor structure topology was made using the Leap of Amber19 tools. The addition of Na+ ions neutralized the simulation system. The neutralized system was then immersed in the water-molecule-containing TIP3P box. Prior to performing 1000 steps of conjugate gradient, the solvated system underwent 1500 steepest descent method iterations of minimization. With gradual heating from 0 K to 300 K at 1 atm pressure, system heating for 100 ps was accomplished. The system was then allowed to reach equilibrium for 100 ps, allowing the exchange of kinetic and potential energies. A 50 ns simulation production run was completed to assess the dynamics of the complex and confirm the stability of the docked ligand conformation. The SHAKE algorithm was used to impose constraints on hydrogen atoms that were covalently bound. The use of canonical ensemble ensured the periodic boundary conditions in the simulation box. Berendsen was used to maintain a constant temperature in the simulation box. The Ewald summation was used to complete MD simulations, and coordinate files were saved every 0.5 ps. The CPPTRAJ module included in Amber18 was used to analyze the trajectory data.2.9. Binding free energy calculation
Molecular Mechanics-Generalized Born Surface Area (MMGBSA) and Molecular Mechanics Poisson-Boltzmann Surface Area (MMPBSA) embedded in the MMPBSA.py module of AMBER18 were utilized to calculate the binding free energy of the docked complex.53 100 frames were processed from the trajectories in total, and the system's net energy was estimated using the following equation:| ΔGbinding = ΔGcomplex − ΔGreceptor − ΔGinhibitor |
Each of the forementioned terms requires the calculation of multiple energy components, including van der Waals energy, electrostatic energy, internal energy from molecular mechanics, and polar contribution to solvation energy.
3. Results and discussion
3.1. Metabolomics profiling of the crude extract of C. papaya leaves
The dereplication of secondary metabolites of the crude methanol extract revealed the presence of different classes of phyto-compounds. The positive and negative modes revealed the presence of phenolic, fatty acids, flavonoids, alkaloid, sterol, and triterpenes (Table 1).| m/z | Retention time (min) | M. wt | Name | Molecular formula | References |
|---|---|---|---|---|---|
| 739.20 | 1.79 | 740.2148625 | Clitorin | C33H40O19 | 54 |
| 611.15 | 8.69 | 610.1526631 | Rutin | C27H30O16 | 54 |
| 595.16 | 9.05 | 594.1576881 | Nictoflorin | C27H30O15 | 54 |
| 755.20 | 12.84 | 756.2090453 | Manghaslin | C33H40O20 | 54 |
| 287.05 | 15.91 | 286.0475676 | Kaempferol | C15H10O6 | 24 |
| 473.36 | 19.25 | 472.3547817 | 2β,3β-Dihydroxy-ursolic acid | C30H48O4 | 55 |
| 487.34 | 19.74 | 488.3500919 | Acacic acid | C30H48O5 | 56 |
| 457.36 | 20.64 | 456.3600077 | Oleanolic acid | C30H48O3 | 57 |
| 457.36 | 23.14 | 456.3600665 | Betulinic acid | C30H48O3 | 55 and 58 |
| 415.38 | 23.31 | 414.3819298 | β-Sitosterol | C29H50O | 26 |
| 479.34 | 26.48 | 478.3418738 | Carpaine | C28H50N2O4 | 54 |
| 479.34 | 26.48 | 478.3418738 | Pseudocarpaine | C28H50N2O4 | 54 |
| 413.36 | 28.45 | 412.3550657 | Stigmasterol | C29H48O | 37 |
3.2. Anti-SARS-CoV-2 activity
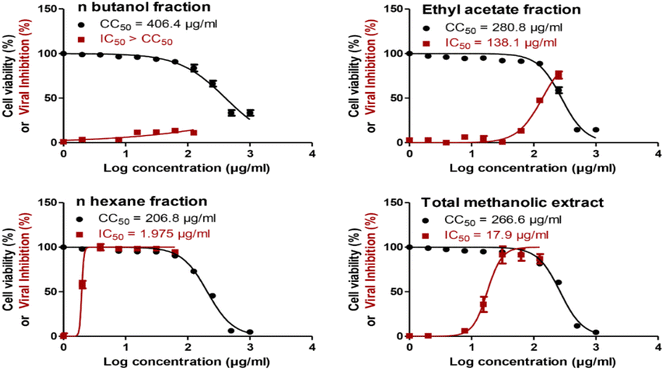
The total methanol extract, n-hexane, ethyl acetate, and n-butanol fractions of papaya leaves were all assessed for their safety against the cells by testing their cytotoxicity on VERO-E6 cells via MTT assay. The CC50 of the total n-butanol extract was the highest with a concentration equal to 406.4 μg mL−1. The anti-SARS-CoV-2 activity was assessed by using the crystal violet assay protocol.44 The n-hexane fraction shows potent antiviral activity against SARS-CoV-2 with an IC50 value = 1.98 μg mL−1. Although the n-butanol extract has the highest CC50 against the cells indicating its relative safety compared to the other fractions, the selectivity indices give a more significant explanation to the safety of the tested extracts relative to their antiviral activity. According to the selectivity indices results, the n-hexane fraction has the highest selectivity index value = 104.70 with a wide difference than any other extract. This indicated that the n-hexane fraction was the most selective to the viral cells without a toxic effect on the other normal cells. The selectivity index was calculated as a ratio that measures the window between cytotoxicity and antiviral activity by dividing the given CC50 value into the IC50 value (Selectivity Index (SI) = CC50/IC50). The results of the cytotoxic activity, antiviral activity, and selectivity indices data are presented in Fig. 1 and Table 2.| Extract | CC50 (μg ml−1) | IC50 (μg ml−1) | Selectivity index (SI) |
|---|---|---|---|
| n-Butanol fraction | 406.4 | >406.4 | <1 |
| Ethyl acetate fraction | 280 | 138.1 | 2.03 |
| n-Hexane fraction | 206.8 | 1.98 | 104.7 |
| Total methanolic extract | 266.6 | 17.9 | 14.89 |
3.3. Molecular docking
The genome of the SARS-CoV-2 virus is a large single-stranded RNA which encodes for four main structural proteins that include the nucleocapsid protein (N), membrane protein (M), spike protein (S), and envelope protein (E). In addition, sixteen non-structural proteins (nsp) are also encoded and are responsible for the replication, survival, transcription and pathogenicity.59 Our current molecular docking study investigate potential interaction between 13 annotated compounds from the total papaya extract against six SARS-CoV-2 target proteins. These targets include four non-structural proteins which are main protease (Mpro), Papain-like protease (PLpro), RNA dependent RNA polymerase (RdRp) as well as O-methyl transferase. Main protease and papain-like proteases are required by the virus to cut the polyprotein chain at specific location to release structural and nonstructural proteins. RdRp is the viral polymerase that is required for viral replication. Furthermore, 2′-O-methyltransferase is essential to protect viral RNA from host immune system including the interferon-induced response.60 In addition to these non-structural proteins, we also investigated potential recognition of our annotated compounds by receptor binding domain (RBD) of structural S-protein and its human target, angiotensin converting enzyme-2 (ACE-2). Binding of inhibitors to any of the last two targets could interrupt the recognition of S-protein by ACE-2 and hence slow down the infection rate. These targets were investigated by several groups including us and we have validated our docking procedure for all of them earlier.50,61,62A variety of compounds annotated exhibited strong docking scores on the targeted proteins (Table 3). For example, flavonoids showed good docking results with Mpro (6LU7). This target protein is responsible for the maturation of replicase and helicase functional enzymes. Therefore, inhibiting Mpro is a crucial step for the inhibition of viral replication.63 Those flavonoidal compounds include nicotiflorin, which showed the score (−9.3 kca/mol), as well as manghaslin (−8.8 kcal mol−1) and rutin (−8.3 kcal mol−1) which are all better than the co-crystalized ligand (−7.7 kcal mol−1). Nicotiflorin was previously reported to have an affinity on SARS-CoV-2 protease in silico.64 In fact, those 3 compounds are flavonoid glycosides which might indicate the importance of attached sugars for the recognition via hydrogen bonding leading to strong binding. These three compounds also showed best results against 2′-O-methyltransferase (6W4H) target protein which is important for viral survival by enhancing viral RNA stability and preventing its degradation by the cellular immunity.65 Furthermore, RdRp is considered one of the most important and successful targets against SARS-CoV-2 due to its responsibility for regulating the viral replication and transcription of single-stranded RNA viruses.65 For this target, 2β,3β-dihydroxy-ursolic acid showed the best docking score (−8.6 kcal mol−1) compared to the remaining tested compounds. In addition, the carpaine alkaloid and rutin flavonoid came next (−8.4 kcal mol−1) as inhibitors of RdRp. It worth to mention here that all these compounds are part of the methanolic extract which has shown more than 100-fold selectivity towards SARS-CoV-2 infected cells compared to non-infected Vero-E6 cells (Table 2). Further mechanistic studies are required to confirm the affected targets that are involved in the anti-SARS-CoV-2 activities.
| Compound | Docking scores (kcal mol−1) | |||||
|---|---|---|---|---|---|---|
| Mpro | RdRp | O-ME transferase | ACE-2 | RBD-S-protein | PLpro | |
| Clitorin | −7.8 | −8.1 | −8.3 | −5.4 | −6.8 | −6.8 |
| Rutin | −8.3 | −8.4 | −8.7 | −5.6 | −6.6 | −7 |
| Nicotiflorin | −9.3 | −8.2 | −9.3 | −5.9 | −7.4 | −6.9 |
| Manghaslin | −8.8 | −8.2 | −8.7 | −5.7 | −7 | −6.8 |
| Kaempferol | −7.9 | −7.5 | −8 | −7 | −6 | −6.8 |
| 2β,3β-Dihydroxy-ursolic acid | −7.2 | −8.6 | −8.1 | −6 | −6.8 | −7 |
| Acacic acid | −7.2 | −7.2 | −8 | −5.8 | −6.6 | −6.9 |
| Oleanolic acid | −7.3 | −7.5 | −8 | −5.8 | −6.5 | −6.8 |
| Betulinic acid | −7.1 | −7.4 | −7.8 | −6 | −6.6 | −7.2 |
| β-Sitosterol | −7.2 | −6.3 | −7.6 | −5.2 | −6.1 | −6.8 |
| Carpaine | −7.8 | −8.4 | −8.5 | −6.3 | −7 | −7.4 |
| Pseudocarpaine | −7.7 | −8 | −8.2 | −6.1 | −7.4 | −7.8 |
| Stigmasterol | −7.2 | −6.3 | −7.7 | −5.8 | −7 | −6.7 |
| Co-crystallization ligand | −7.7 | −6.8 | −8.2 | — | — | −6.8 |
Alkaloids like pseudocarpaine and carpaine showed significant binding scores on all targeted proteins except ACE-2 especially pseudocarpaine which exhibited the best binding score (−7.4 kcal mol−1) on RBD-spike protein (6M0J) which is responsible for the viral attachment and entering the host cell. Therefore, binding to the spike protein could alter the process of viral attachment and prevent the infection process.65 Those alkaloids also significantly showed the best binding scores among all annotated compounds on PLpro (6WX4) enzyme which could help in the prevention of the viral polyproteins processing responsible for the viral spread.66 In addition, terpenes such as betulinic acid, acacic acid and oleanolic acid showed better fitting scores on RNA-dependent RNA polymerase (7BV2) and papain-like protease (PLpro) (6WX4) than the co-crystallized ligands. Sterols like β-sitosterol and stigmatserol exhibited a better fitting score than the Co-Crystallized ligand on PLpro (6WX4) and RBD-Spike protein (6M0J) respectively. Moreover, some annotated flavonoids such as manghaslin, clitorin, rutin and kaempferol showed moderate to high fitting score on the targeted proteins. The docking score results of the annotated compounds are presented in Table 3. Furthermore, a binding pose of rutin on the active site of SARS-CoV-2 RdRp (7BV2) is demonstrated in Fig. 2.
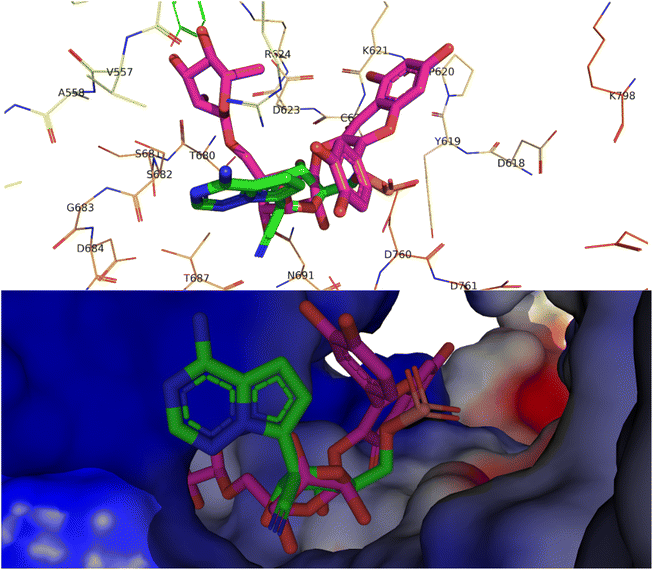 | ||
| Fig. 2 Top, binding pose of rutin (pink) in the active site of SARS-CoV-2 RdRp (7BV2) overlapped with Remdesivir (green). Bottom, surface representation of rutin (pink) in the active site of SARS-CoV-2 RdRp (7BV2) overlapped with Remdesivir (green). | ||
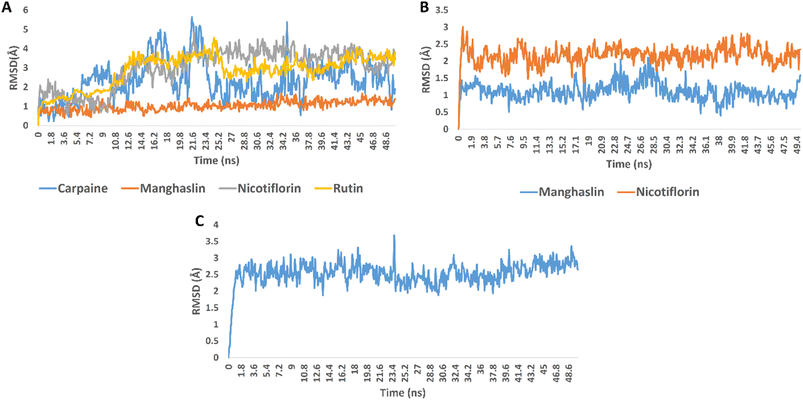
In order to validate the docking results, we subjected the structures of the best scoring compounds to 50 ns-long MDS in addition to MM-PBSA calculation of their binding energy. As shown in Fig. 3, all compounds' structures showed comparable binding stability inside the three proteins' active sites. Regarding OMT, carpaine, nicotiflorin, and rutin were significantly unstable inside the enzyme's active site in comparison with manghalsin (Fig. 3A), where they were highly fluctuating over the course of simulations with high average RMSDs (4.43 Å, 3.89 Å, and 3.45 Å, respectively). In case of manghalsin and, nicotiflorin with Mpro, both structures showed acceptable binding stability inside the enzyme's active site with average RMSDs of 1.42 Å and 2.21 Å, respectively (Fig. 3B). Similarly, dihydroxyursolic acid achieved good binding stability inside RdRp's active site with an average RMSD of 2.53 Å (Fig. 3C).
Moreover, we estimated the binding free energy of each compound inside the corresponding protein targets using the MM-PBSA method.67 The obtained results were in good accordance with that stability profile of each compound. Carpaine and nicotiflorin with OMT were the lowest scoring compounds in terms of binding free energy (Table 4).
| Energy component | Carpaine-OMT | Manghaslin-OMT | Nicotiflorin-OMT | Rutin-OMT | Manghaslin-Mpro | Nicotiflorin-Mpro | Dihydroxyursolic acid-RdRp |
|---|---|---|---|---|---|---|---|
| DELTA G gas | −27.5602 | −54.9876 | −21.8203 | −38.1047 | −48.4837 | −39.3409 | −36.8341 |
| DELTA G solv | 9.5982 | 16.7483 | 11.2943 | 13.6452 | 16.3827 | 14.8213 | 15.3823 |
| DELTA total | −17.962 | −38.2393 | −10.526 | −24.4595 | −32.101 | −24.5196 | −21.4518 |
The in vitro results of this study (Table 2) revealed that the n-hexane fraction had the highest anti-SARS-CoV-2 activity with a significant margin compared to the total methanol extract and the other fractions with an IC50 value = 1.98 μg mL−1 and significantly better selectivity index = 104.7. Therefore, the n-hexane fraction is considered the most effective fraction against SARS-CoV-2 compared to the other fractions and the total extract. According to molecular docking results, it can be deduced that the presence of terpenoids such as 2β,3β-dihydroxy-ursolic acid which showed the best binding score on RdRp and betulinic acid, acacic acid and oleanolic acid which demonstrated good fitting scores on the same protein could help in inhibiting viral replication process. The annotated Sterols had high to moderate binding scores on the targeted PLpro and RBD-spike protein which may contribute to the prevention of the viral attachment to the host cell and the viral spread. This could relate to the high anti-SARS-CoV-2 activity of the n-hexane fraction. In addition, alkaloids such as pseudocarpaine and carpaine showed a good binding on most of the targeted proteins. Studies displayed the presence of such alkaloids in many fractions of papaya leaves including the hexane fraction which explains their role in altering the viral replication and spreading process.7,56,68 Other studies suggest the presence of traces of flavonoids in the hexane fraction.68–70 All these findings indicate that all the compounds that possess high scores on the targeted proteins may contribute to the high in vitro activity of the hexane fraction through synergism. However, further phytochemical and biological investigations are needed to identify more compounds that could have a role in the explanation of the prominent activity of the hexane fraction and the other fractions of papaya leaves against SARS-CoV-2.
4. Conclusion
The previously reported antiviral activities of C. papaya led us to investigate its activity against SARS-CoV-2. The total methanol extract, n-hexane, ethyl acetate, and n-butanol fractions of papaya leaves were subjected to anti-SARS-CoV-2 activity and cytotoxicity assays. The results showed that the n-hexane fraction had the highest anti-SARS-CoV-2 activity besides having the highest selectivity index. This result assumes that the high activity of the n-hexane fraction against SARS-CoV-2 may be due to the presence of some classes of compounds that existed in papaya such as fatty acids, sterols, and triterpenes which were previously reported to have a wide range against different classes of viruses including some compounds that have a reported anti-SARS-CoV-2 activity. In addition, the molecular docking study together with the molecular dynamics simulations and MM-PBSA calculation demonstrated the moderate to high fitting potential of the annotated compounds with the targeted SARS-CoV-2 proteins. The annotated compounds belong to different classes that include sterols and triterpenes, flavonoids, and alkaloids. These findings open the door to further investigations that concern the compounds responsible for the activity of C. papaya leaves against SARS-CoV-2 and the n-hexane extract in particular.Author contributions
Amr Adel: conceptualization, methodology, writing and editing. Mohamed S. Elnaggar: supervision, reviewing and validation. Amgad Albohy: methodology, investigation and editing. Ahmed A. Elrashedy: methodology and investigation. Ahmed Mostafa: Methodology, editing and funding acquisition. Omnia Kutkat: methodology and investigation. Usama Ramadan Abdelmohsen: methodology and investigation. Eman Alsayed: supervision, reviewing and validation. Mohamed A. Rabeh: supervision and reviewing. All authors have read and approved the final manuscript.Conflicts of interest
The authors declare that they have no conflicts of interests.Acknowledgements
The authors would like to thank the Center of Drug Discovery Research and Development at Faculty of Pharmacy, Ain Shams University for their assistance in the experimental part of the project. This research was partially funded by the Egyptian Academy of Scientific Research and Technology (ASRT) within the “Ideation Fund” program under contract numbers 7303.References
- T. Hirano and M. Murakami, Immunity, 2020, 52, 731–733 CrossRef CAS PubMed.
- P. Ding, Tropical Fruits, CAB International, 2016, vol. 3 Search PubMed.
- K. Gulati, A. Ray, P. K. Debnath and S. K. Bhattacharya, J. Nat. Rem., 2002, 2, 121–131 Search PubMed.
- J. B. Harborne, Indian Medicinal Plants. A Compendium of 500 Species, ed. P. K. Warrier, V. P. K. Nambiar and C. Ramankutty, Orient Longman Pvt Ltd, Hyderabad, 2005, vol. 1 Search PubMed.
- B. V. Owoyele, O. M. Adebukola, A. A. Funmilayo and A. O. Soladoye, Inflammopharmacology, 2008, 16, 168–173 CrossRef CAS PubMed.
- Q. V. Vuong, S. Hirun, T. L. K. Chuen, C. D. Goldsmith, S. Murchie, M. C. Bowyer, P. A. Phillips and C. J. Scarlett, Int. J. Food Sci. Technol., 2015, 50, 169–177 CrossRef CAS.
- T. Julianti, M. De Mieri, S. Zimmermann, S. N. Ebrahimi, M. Kaiser, M. Neuburger, M. Raith, R. Brun and M. Hamburger, J. Ethnopharmacol., 2014, 155, 426–434 CrossRef CAS PubMed.
- G. Subramanian, B. B. Tewari and R. Gomathinayagm, Am. J. Pharmacol. Pharmacother., 2014, 1, 25–39 Search PubMed.
- K. Sobia, M. A. Javaid, M. S. Ahmad, Q. Rehmatullah, G. Hina, B. Iram, A. Pervaiz, B. Farhana, J. Nyla and M. Gulfraz, Int. J. Pharm. Sci. Res., 2016, 7, 3658 CAS.
- P. Prabakaran, Int. J. Res. Pharm. Nano Sci., 2014, 3, 552–557 Search PubMed.
- M. Syed and D. S. Rao, Indo Am. J. Pharm. Sci., 2014, 1, 328–332 Search PubMed.
- V. Zunjar, R. P. Dash, M. Jivrajani, B. Trivedi and M. Nivsarkar, J. Ethnopharmacol., 2016, 181, 20–25 CrossRef CAS PubMed.
- N. Sharma, K. P. Mishra, S. Chanda, V. Bhardwaj, H. Tanwar, L. Ganju, B. Kumar and S. B. Singh, Arch. Virol., 2019, 164, 1095–1110 CrossRef CAS PubMed.
- S. M. N. Azmi, P. Jamal and A. Amid, Aust. J. Basic Appl. Sci., 2012, 6, 117–122 CAS.
- G. A. Brasil, S. N. Ronchi, A. M. do Nascimento, E. M. de Lima, W. Romão, H. B. da Costa, R. Scherer, J. A. Ventura, D. Lenz, N. S. Bissoli, D. C. Endringer and T. U. de Andrade, Planta Med., 2014, 80, 1580–1587 CrossRef CAS PubMed.
- P. Hasimun and G. I. Ernasari, Procedia Chem., 2014, 13, 147–149 CrossRef CAS.
- A. Mohammed, S. A. Abubakar and M. S. Sule, Int. J. Pharm. Sci. Rev. Res., 2011, 11, 13–16 Search PubMed.
- B. Joseph, P. Sankarganesh, K. Ichiyama and N. Yamamoto, Front. Life Sci., 2015, 8, 18–22 CrossRef CAS.
- M. R. M. A. Razak, N. A. Norahmad, N. H. M. Jelas, A. Afzan, N. M. Misnan, A. M. Ripen, R. Thayan, M. Zainol and A. F. S. Mohamed, Pathogens, 2021, 10, 501 CrossRef PubMed.
- N. Ahmad, H. Fazal, M. Ayaz, B. H. Abbasi, I. Mohammad and L. Fazal, Asian Pac. J. Trop. Biomed., 2011, 1, 330–333 CrossRef PubMed.
- F. Salim, N. A. Abu, H. Yaakob, L. Kadir, N. Zainol, Z. Taher, S. Shahar, F. Husin, A. S. S. Amri and E. Moghaddam, Sci. Int., 2018, 30, 437–441 CAS.
- J. G. Haddad, V. Carcauzon, O. El Kalamouni, P. Desprès, C. Garcia, F. Remize and C. El Kalamouni, Microorganisms, 2020, 8, 1–11 CrossRef PubMed.
- T. Vij and Y. Prashar, Asian Pac. J. Trop. Dis., 2015, 5, 1–6 CrossRef.
- V. Anjum, P. Arora, S. H. Ansari, A. K. Najmi and S. Ahmad, Pharm. Biol., 2017, 55, 2020–2025 CrossRef PubMed.
- G. Iyappan, D. Daniel and T. Poovanalingam, World J. Pharm. Pharm. Sci., 2014, 3, 942–949 Search PubMed.
- S. U. Ahmed, M. A. Al-Mansur, Y. Ahmed, M. H. Sohrab, A. M. S. Chowdhury and C. M. Hassan, Dhaka Univ. J. Sci., 2010, 58, 265–268 CAS.
- A. Kohn, J. Gitelman and M. Inbar, Arch. Virol., 1980, 66, 301–307 CrossRef CAS PubMed.
- A. Kohn, J. Gitelman and M. Inbar, Antimicrob. Agents Chemother., 1980, 18, 962–968 CrossRef CAS PubMed.
- U. N. Das, Arch. Med. Res., 2020, 51, 282–286 CrossRef CAS PubMed.
- T. Řezanka, L. Siristova and K. Sigler, Anti-Infect. Agents Med. Chem., 2009, 8, 193–210 CrossRef.
- N. H. Shady, K. A. Youssif, A. M. Sayed, L. Belbahri, T. Oszako, H. M. Hassan and U. R. Abdelmohsen, Plants, 2021, 10, 1–34 Search PubMed.
- M. Ghaffari Moghaddam, F. B. H. Ahmad and A. Samzadeh-Kermani, Pharmacol. Pharm., 2012, 03, 119–123 CrossRef.
- N. I. Pavlova, O. V Savinova, S. N. Nikolaeva, E. I. Boreko and O. B. Flekhter, Fitoterapia, 2003, 74, 489–492 CrossRef CAS PubMed.
- V. Khwaza, O. O. Oyedeji and B. A. Aderibigbe, Molecules, 2018, 23, 2300 CrossRef PubMed.
- Y.-M. Zhu, J.-K. Shen, H.-K. Wang, L. M. Cosentino and K.-H. Lee, Bioorg. Med. Chem. Lett., 2001, 11, 3115–3118 CrossRef CAS PubMed.
- L. Kong, S. Li, Q. Liao, Y. Zhang, R. Sun, X. Zhu, Q. Zhang, J. Wang, X. Wu, X. Fang and Y. Zhu, Antiviral Res., 2013, 98, 44–53 CrossRef CAS PubMed.
- N. R. Khaled and F. Gerda, Greener J. Pharm. Pharmacol., 2018, 1, 001–005 Search PubMed.
- T. Akihisa, J. Ogihara, J. Kato, K. Yasukawa, M. Ukiya, S. Yamanouchi and K. Oishi, Lipids, 2001, 36, 507–512 CrossRef CAS PubMed.
- E. G. Haggag, A. M. Elshamy, M. A. Rabeh, N. M. Gabr, M. Salem, K. A. Youssif, A. Samir, A. Bin Muhsinah, A. Alsayari and U. R. Abdelmohsen, Int. J. Nanomed., 2019, 14, 6217–6229 CrossRef CAS PubMed.
- A. Kandeil, A. Mostafa, R. El-Shesheny, M. Shehata, W. H. Roshdy, S. S. Ahmed, M. Gomaa, A. El Taweel, A. E. Kayed, S. H. Mahmoud, Y. Moatasim, O. Kutkat, M. N. Kamel, N. Mahrous, M. El Sayes, N. M. El Guindy, A. Naguib and M. A. Ali, Microbiol. Resour. Announc., 2020, 9(22), e00489-20 CrossRef PubMed.
- L. J. Reed and H. Muench, Am. J. Epidemiol., 1938, 27, 493–497 CrossRef.
- M. Feoktistova, P. Geserick and M. Leverkus, Cold Spring Harb. Protoc, 2016, 2016, 343–346 Search PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
- A. Mostafa, A. Kandeil, Y. A. M. M. Elshaier, O. Kutkat, Y. Moatasim, A. A. Rashad, M. Shehata, M. R. Gomaa, N. Mahrous, S. H. Mahmoud, M. Gaballah, H. Abbas, A. El Taweel, A. E. Kayed, M. N. Kamel, M. El Sayes, D. B. Mahmoud, R. El-Shesheny, G. Kayali and M. A. Ali, Pharmaceuticals, 2020, 13, 1–24 CrossRef PubMed.
- A. Mahmoud, A. Mostafa, A. A. Al-Karmalawy, A. Zidan, H. S. Abulkhair, S. H. Mahmoud, M. Shehata, M. M. Elhefnawi and M. A. Ali, Heliyon, 2021, 7, e07962 CrossRef CAS PubMed.
- G. Indrayanto, G. S. Putra and F. Suhud, Profiles Drug Subst., Excipients, Relat. Methodol., 2021, 46, 273–307 Search PubMed.
- S. S. Ebada, N. A. Al-Jawabri, F. S. Youssef, A. Albohy, S. M. Aldalaien, A. M. Disi and P. Proksch, Inflammopharmacology, 2020, 28, 321–331 CrossRef CAS PubMed.
- M. D. Hanwell, D. E. Curtis, D. C. Lonie, T. Vandermeerschd, E. Zurek and G. R. Hutchison, J. Cheminf., 2012, 4, 1–17 Search PubMed.
- O. Trott and A. J. Olson, J. Comput. Chem., 2010, 31, 455–461 CAS.
- E. M. Zahran, A. Albohy, A. Khalil, A. H. Ibrahim, H. A. Ahmed, E. M. El-Hossary, G. Bringmann and U. R. Abdelmohsen, Mar. Drugs, 2020, 18, 645 CrossRef CAS PubMed.
- E. W. Bell and Y. Zhang, J. Cheminf., 2019, 11, 1–9 CAS.
- D. A. Case, T. A. Darden, T. E. Cheatham, C. L. Simmerling, J. Wang, R. E. Duke, R. Luo, M. Crowley, R. C. Walker and W. Zhang, Amber 10, University of California, 2008 Search PubMed.
- B. R. Miller, T. D. McGee, J. M. Swails, N. Homeyer, H. Gohlke and A. E. Roitberg, J. Chem. Theory Comput., 2012, 8, 3314–3321 CrossRef CAS PubMed.
- M. Raith, M. De Mieri, T. Julianti, M. Neuburger, R. Brun, M. Kaiser, S. Zimmermann, M. Hamburger, S. N. Ebrahimi, M. De Mieri, S. Zimmermann, S. N. Ebrahimi, M. Kaiser, M. Neuburger, M. Raith, R. Brun and M. Hamburger, J. Ethnopharmacol., 2014, 155, 426–434 CrossRef PubMed.
- H. U. C. Hui-fang, Food Sci., 2010, 31, 114–116 Search PubMed.
- M. B. Falana and Q. O. Nurudeen, Not. Sci. Biol., 2020, 12, 57–73 CrossRef.
- Y. Xiu-ling, L. Xiao-ling and L. Zhao-jiang, Guangzhou Food Sci. Technol., 2005, 3, 127–129 Search PubMed.
- S. Ery Rahayu and U. Lestari, IOP Conf. Ser. Earth Environ. Sci., 2019, 276, 1–7 Search PubMed.
- M. Y. Wang, R. Zhao, L. J. Gao, X. F. Gao, D. P. Wang and J. M. Cao, Front. Cell. Infect. Microbiol., 2020, 10, 724 Search PubMed.
- M. Wilamowski, D. A. Sherrell, G. Minasov, Y. Kim, L. Shuvalova, A. Lavens, R. Chard, N. Maltseva, R. Jedrzejczak, M. Rosas-Lemus, N. Saint, I. T. Foster, K. Michalska, K. J. F. Satchell and A. Joachimiak, Proc. Natl. Acad. Sci. U. S. A., 2021, 118, e2100170118 CrossRef CAS PubMed.
- E. M. Zahran, A. M. Sayed, M. F. Abdelwahab, A. Albohy, B. S. Abdulrazik, A. M. Ibrahim, G. Bringmann and U. R. Abdelmohsen, RSC Adv., 2021, 11, 36042–36059 RSC.
- U. R. Abdelmohsen, A. Albohy, B. S. Abdulrazik, S. A. L. Bayoumi, L. G. Malak, I. S. A. Khallaf, G. Bringmann and S. F. Farag, RSC Adv., 2021, 11, 16970–16979 RSC.
- P. K. Agrawal, C. Agrawal and G. Blunden, Nat. Prod. Commun., 2021, 16, 1–12 Search PubMed.
- A. K. Srivastava, A. Kumar and N. Misra, arXiv, 2020, preprint, arXiv:2004.03411 Search PubMed.
- M. A. El Hassab, T. M. Ibrahim, S. T. Al-Rashood, A. Alharbi, R. O. Eskandrani and W. M. Eldehna, J. Enzyme Inhib. Med. Chem., 2021, 36, 727–736 CrossRef CAS PubMed.
- D. Shin, R. Mukherjee, D. Grewe, D. Bojkova, K. Baek, A. Bhattacharya, L. Schulz, M. Widera, A. R. Mehdipour, G. Tascher, P. P. Geurink, A. Wilhelm, G. J. van der Heden van Noort, H. Ovaa, S. Müller, K. P. Knobeloch, K. Rajalingam, B. A. Schulman, J. Cinatl, G. Hummer, S. Ciesek and I. Dikic, Nature, 2020, 587(7835), 657–662 CrossRef CAS PubMed.
- S. Genheden and U. Ryde, Expert Opin. Drug Discovery, 2015, 10, 449 CrossRef CAS PubMed.
- Y. M. Musa, Int. J. Life Sci. Biotechnol. Pharma Res., 2015, 4, 282–284 Search PubMed.
- A. Sharma, A. Bachheti, P. Sharma, R. K. Bachheti and A. Husen, Curr. Res. Biotechnol., 2020, 2, 145–160 CrossRef.
- A. B. Wadekar, M. G. Nimbalwar, W. A. Panchale, B. R. Gudalwar, J. V. Manwar and R. L. Bakal, GSC Biol. Pharm. Sci., 2021, 14, 234–248 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2022 |