Open Access Article
Open Access ArticleOne-pot synthesis and enzyme-responsiveness of amphiphilic doxorubicin prodrug nanomicelles for cancer therapeutics
Ling-Na Han†
 ab,
Kai-Qiang Wang†a,
Zi-Ning Ren†a,
Xue Yanga,
Xiao Duan
ab,
Kai-Qiang Wang†a,
Zi-Ning Ren†a,
Xue Yanga,
Xiao Duan a,
Sasirekha Krishnanc,
Abinaya Jaisankarc,
Jeong-Hui Parkdef,
Khandmaa Dashnyam
a,
Sasirekha Krishnanc,
Abinaya Jaisankarc,
Jeong-Hui Parkdef,
Khandmaa Dashnyam d,
Wujie Zhangg,
José Luis Pedrazhi,
Seeram Ramakrishna
d,
Wujie Zhangg,
José Luis Pedrazhi,
Seeram Ramakrishna j,
Hae-Won Kimdef,
Chang-Feng Li*a,
Li-Hua Song*a and
Murugan Ramalingam
j,
Hae-Won Kimdef,
Chang-Feng Li*a,
Li-Hua Song*a and
Murugan Ramalingam *defk
*defk
aDepartment of Pharmacy, Changzhi Medical College, Changzhi, 046000, Shanxi, People's Republic of China. E-mail: licf1968@qq.com; slh10282001@163.com
bDepartment of Physiology, Changzhi Medical College, Changzhi, 046000, Shanxi, People's Republic of China
cCentre for Biomaterials, Cellular and Molecular Theranostics, School of Mechanical Engineering, Vellore Institute of Technology, Vellore 632014, India
dInstitute of Tissue Regeneration Engineering, Dankook University, Cheonan 31116, Republic of Korea. E-mail: rmurug2000@gmail.com
eDepartment of Nanobiomedical Science, BK21 NBM Global Research Center for Regenerative Medicine, Dankook University, Cheonan 31116, Republic of Korea
fMechanobiology Dental Medicine Research Center, Dankook University, Cheonan 31116, Republic of Korea
gBioMolecular Engineering Program, Physics and Chemistry Department, Milwaukee School of Engineering, Milwaukee, WI 53202, USA
hNanoBioCel Research Group, Laboratory of Pharmacy and Pharmaceutical Technology, Faculty of Pharmacy, University of the Basque Country (UPV/EHU), 01006 Vitoria-Gasteiz, Spain
iNetworking Research Centre of Bioengineering, Biomaterials and Nanomedicine, Institute of Health Carlos III, 28029 Madrid, Spain
jCenter for Nanofibers and Nanotechnology, Department of Mechanical Engineering, National University Singapore, Singapore 119260, Singapore
kSchool of Basic Medical Sciences, Chengdu University, Chengdu 610106, People's Republic of China
First published on 30th September 2022
Abstract
In this study, we report a one-pot synthesis and enzyme-responsiveness of polyethylene glycol (PEG) and glutamic acid (Glu)-based amphiphilic doxorubicin (DOX) prodrug nanomicelles for cancer therapeutics. The nanomicelles were accomplished by esterification and amidation reactions. The nuclear magnetic resonance (NMR) and Fourier transform infrared (FTIR) data confirmed the structure of nanomicelles. The DOX-loaded nanomicelles showed a DLS-measured average size of 107 nm and excellent stability in phosphate-buffered saline (PBS) for 7 days. The drug loading and cumulative release rates were measured by ultraviolet-visible (UV-vis) spectrophotometry at 481 nm. The cumulative release rate could reach 100% in an enzyme-rich environment. Further, the therapeutic efficiency of nanomicelles to cancer cells was determined by cell viability and cellular uptake and distribution using HeLa cells. The cell viability study showed that the DOX-loaded nanomicelles could effectively inhibit the HeLa cell proliferation. The cellular uptake study confirmed that the nanomicelles could be effectively ingested by HeLa cells and distributed into cell nuclei. Based on the collective experimental data, this study demonstrated that the synthesized nanomicellar prodrug of DOX is a potential candidate for cancer therapeutics.
Introduction
The recent decades have witnessed a vast expansion in the area of nanomedicine which has been further advanced by the extensive development of new biomaterials.1–4 Although a massive number of biocompatible materials have successfully contributed to the fabrication of novel nanomedicines for diagnostic and therapeutic purposes, the degradability and exogeneity of such materials often hamper their clinical applications. As yet, only a few nanomedicines have acquired approval from the Food and Drug Administration (FDA) of the United States.5,6 The hindrance in clinical adaptation is further caused by complicated preparation processes and the use of non-FDA approved materials. Most of the studies on biomaterial-based nano-drug deliveries reported multifunctional systems7–12 with a complicated preparation process. For instance, Najafipour and coworkers followed a multi-step synthesis of a poly(N-isopropylacrylamide)-based nano-drug platform for cancer therapy with controlled drug release behaviour.13 Tang and coworkers developed a pH/ATP cascade-responsive nano-courier utilising multiple non-FDA-approved materials and employing a complicated synthesis process.14To expedite regulatory approval and clinical use of nanomedicines, the one-pot synthesis method exploiting endogenous materials is gaining recognition as a simple and easily-applicable strategy to construct novel nanomedicines, especially for drug self-delivery systems.15–19 These drug self-delivery systems have been developed for various drugs such as methotrexate,20–22 doxorubicin,23–30 and melphalan.31 However, their clinical application remained limited owing to the use of exogenous materials.
Considering the above limitations, we developed a one-pot synthesis of an enzyme-responsive prodrug of amphiphilic doxorubicin (DOX) prodrug exploiting polyethylene glycol (PEG) and glutamic acid (Glu). The esterification and amidation reaction was afforded (Scheme 1A) to accomplish PEG- and Glu-based nanomicelles of DOX (hereafter referred to as “DOX–Glu–PEG”). We further demonstrated the functional characteristic features of the DOX–Glu–PEG, their improved stability and water solubility along with enzyme-responsive performance. The results indicated the promising potential of synthesized nanomicelles for industrial production and clinical translation.
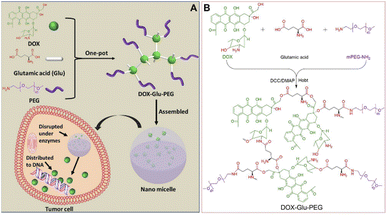 | ||
| Scheme 1 (A) One-pot synthesis method of Glu- and PEG-based DOX-containing nanomicelles. (B) Synthesis route of DOX–Glu–PEG. | ||
Experimental
Materials and instruments
Doxorubicin hydrochloride (DOX, purity: 98%) and porcine pancreatic trypsin (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 250) were obtained from Macklin (Shanghai). L-Glutamic acid (Glu, purity: 99%) and N,N′-dicyclohexylcarbodiimide (DCC, purity: 97%) were purchased from Nine-Dinn Chemistry Co. Ltd. (Shanghai). 1-Hydroxybenzotriazole (HOBt) was purchased from Aladdin Co. China. 4-Dimethylamino pyridine (DMAP) was obtained from TCI Shanghai (China). Amino-PEG (mPEG–NH2, purity: 95%) was purchased from Shanghai Yare Biotech, Inc. Cell Counting Kit-8 (CCK-8) and Hoechst 33258 Staining Kit were purchased from Beyotime Biotechnology Company (China). Neutral protease (200
250) were obtained from Macklin (Shanghai). L-Glutamic acid (Glu, purity: 99%) and N,N′-dicyclohexylcarbodiimide (DCC, purity: 97%) were purchased from Nine-Dinn Chemistry Co. Ltd. (Shanghai). 1-Hydroxybenzotriazole (HOBt) was purchased from Aladdin Co. China. 4-Dimethylamino pyridine (DMAP) was obtained from TCI Shanghai (China). Amino-PEG (mPEG–NH2, purity: 95%) was purchased from Shanghai Yare Biotech, Inc. Cell Counting Kit-8 (CCK-8) and Hoechst 33258 Staining Kit were purchased from Beyotime Biotechnology Company (China). Neutral protease (200![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 U g−1) was obtained from Novocata. Esterase was purchased from Shanghai Xianding Biotech, Inc. Other organic reagents were of analytical grade and were directly used in experiments.
000 U g−1) was obtained from Novocata. Esterase was purchased from Shanghai Xianding Biotech, Inc. Other organic reagents were of analytical grade and were directly used in experiments.
Fourier Transform Infrared (FT-IR) spectra were obtained from Bruker Tensor 27 (Germany). The 1H NMR spectra of DOX and DOX–Glu–PEG were obtained from Bruker Avance 400 spectrometer (Bruker BioSpin, Switzerland) operating at 400 MHz (1H) in DMSO-d6. The DOX–Glu–PEG micelles were measured by dynamic light scattering (DLS) (Malvern Instrument, UK) and transmission electron microscope (TEM) (FEI Tecnai G2 F20). All DLS measurements were taken with an angle detection of 173° at 25 °C. The XRD spectrum was obtained from X-ray powder diffractometer (X'Pert PRO MPD). The drug loading and release properties of DOX–Glu–PEG were measured by a UV-vis spectrophotometer (TU-1901, China). The cell viability was evaluated by CCK-8 and measured by Infinite M200 Pro (TECAN). Leica TCS SP was employed to monitor the distribution of DOX–Glu–PEG and DOX in HeLa cells.
Preparation of amphiphilic poly-DOX prodrug micelle
DOX·HCl (30 mg, 0.0517 mmol), Glu (50.73 mg, 0.3448 mmol), DCC (142.28 mg, 0.6897 mmol), DMAP (21.07 mg, 0.1724 mmol), and HOBt (93.20 mg, 0.6897 mmol) were dissolved in 6 mL of anhydrous DMF in a round-bottom flask and the mixture was allowed to completed the reaction for 6–8 h at room temperature. Then, 68.95 mg (0.0345 mmol) of mPEG–NH2 (MW = 2000) was added to the mixture and successively reacted for three days at room temperature in dark. The mixture solution was then transferred into a dialysis bag (MWCO = 3500) and subsequently immersed in DMF to separate DOX, DOX–Glu and low-molecular-weight DOX–Glu–PEG. The DMF was replaced thrice with a fresh medium over a period of three days. Afterwards, the reaction mixture solution was continuously dialyzed in distilled water for one day with the exchange of water every 2–3 h. Finally, the solution in the dialysis bag was centrifuged at 12![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 min to obtain the supernatant. Thereafter, collected supernatant was passed through a filter membrane (0.45 μm diameter) and the filtered solution was finally freeze-dried providing a yield of 30%.
000 rpm for 2 min to obtain the supernatant. Thereafter, collected supernatant was passed through a filter membrane (0.45 μm diameter) and the filtered solution was finally freeze-dried providing a yield of 30%.
The standard curve of DOX
The calibration curve of absorbance at 481 nm was produced using UV-vis spectroscopy as a function of a series of DOX concentrations in distilled water ranging from 8 μg mL−1 to 36 μg mL−1. The standard curve is A = 0.0186c + 0.0005.Determination of drug loading and cumulative drug release from DOX–Glu–PEG
One milligram of DOX–Glu–PEG was dissolved in 3 mL of distilled water and its absorbance value (Abs) was determined using a UV-vis spectrophotometer. For cumulative drug release, equal portions of DOX–Glu–PEG micelle solution were added into two dialysis bags (MWCO = 3500). A mixed enzyme solution comprising neutral protease, esterase, and trypsin was then introduced into one bag while the PBS was added into the other dialysis bag. These bags were immersed in 8 mL of PBS buffer (pH 7.4) and an aliquot of 3 mL from external solutions of both dialysis bags was withdrawn and replaced by fresh buffer at the experimental time points of 0.5, 1, 2, 4, 6, 8, 12, 24, 48 and 72 h. Finally, the absorbance of aliquots was determined and utilized in the cumulative drug release profile of synthesized nanomicelles.Detection for water solubility of DOX–Glu–PEG
Excessive DOX–Glu–PEG powder was added to 200 μL of distilled water until a small amount of powder was observed as insoluble. The mixture solution was then centrifuged at 15![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 2 min to obtain the supernatant. Then, the 100 μL of supernatant was subjected to freeze-drying and the red powder of DOX–Glu–PEG with 8.4 mg weight was obtained. The maximum water solubility of DOX–Glu–PEG was found to be 84 mg mL−1.
000 rpm for 2 min to obtain the supernatant. Then, the 100 μL of supernatant was subjected to freeze-drying and the red powder of DOX–Glu–PEG with 8.4 mg weight was obtained. The maximum water solubility of DOX–Glu–PEG was found to be 84 mg mL−1.
Cell viability assays
Cell viabilities of free DOX and DOX–Glu–PEG on HeLa cells were measured by CCK-8 assay. HeLa cells were seeded into 96-well plates at a density of 8 × 103 cells per mL and were incubated for 12 h under 5% CO2 at 37 °C. Then the cultivated cells were treated with DOX–Glu–PEG micelles equivalent to DOX concentrations of 5, 15, 30, 50 and 100 μg mL−1 for 24 h and 48 h. Following the removal of the culture medium, 100 μL of PBS solution (pH 7.4) and 10 μL of CCK-8 solution were added to seeded plates which were subsequently incubated for 24 h or 48 h. Cell viability was measured at 450 nm with a microplate reader. Cell survival was determined in terms of percentage using DOX–Glu–PEG as blank control in the following equation;| Cell survival = A_treatment/A_blank × 100 |
Cellular uptake experiment
HeLa cells (5 × 104 cells per mL) were seeded into a confocal dish and cultivated in an incubator at 37 °C; 5% CO2 for 12 h. The cells were then exposed to 50 μg mL−1 of DOX or DOX–Glu–PEG in a concentration equivalent to pristine DOX in 300 μL of culture medium and were incubated for 0.5 h, 2 h and 8 h, subsequently. After that, the culture medium was discarded, nuclear staining solution Hoechst was added, and staining was continued for 10 min. Then the staining solution was removed and the cells were washed three times with sterile PBS solution. The distribution in HeLa cells of free DOX and DOX–Glu–PEG micelles was observed by laser scanning confocal microscope (LSCM).Results and discussion
Synthesis and characterization of DOX–Glu–PEG
The amphiphilic polymer of DOX–Glu–PEG was synthesized by a one-pot method affording esterification and amidation reaction. The Glu with the two carboxyl groups and one amino group was reacted with DOX which contains one amino group and three modifiable hydroxyl groups. Then, the mPEG–NH2 was conjugated on the surface of DOX–Glu to form a hyperbranched amphiphilic polymer (DOX–Glu–PEG) (Scheme 1B). The purpose of employing COOH in excess via reaction with mPEG–NH2 was to accomplish the COOH-containing hyperbranched core of DOX–Glu. The FT-IR spectra showed that the characteristic peaks of DOX, Glu, PEG, and DOX–Glu–PEG could be distinguished from each other (Fig. 1A). The characteristic peak of COOH (1645 cm−1) in Glu was shifted to 1670 cm−1 (ester bonds) and to 1108 cm−1 (ether bonds) in the FT-IR spectrum of DOX–Glu–PEG indicating successful conjugation of Glu and PEG with DOX that led to the formation of DOX–Glu–PEG polymer (Fig. 1A). The structure was further explored through NMR which displayed characteristic peaks of DOX and PEG (Fig. 1B), especially the reactive sites of DOX (position a and b in Fig. 1B, red ellipse) in the DOX–Glu–PEG polymer. The characteristic peak of position a (OH) in DOX disappeared in the DOX–Glu–PEG whereas position b in DOX–Glu–PEG dramatically shifted following the reaction of OH (position a), illustrating the successful synthesis of the polymer of DOX–Glu–PEG. The NMR analysis of DOX–Glu–PEG showed a noticeable P position having one proton and the M position having 193 protons (one molecule PEG2000 has proximately 182 protons) (Fig. 1B). Combining the result of drug loading (13%), we calculated the ratio of DOX, Glu and PEG in the final product which was found to be 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 3
3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1. This final ratio was almost similar to that of the reaction ratio. The UV-vis spectrophotometric peaks of DOX and DOX–Glu–PEG at 481 nm were comparable (Fig. 1C) and thereby indicated that the drug loading and release rates could be determined according to the DOX standard curve at 481 nm. In order to further confirm the structure of DOX–Glu–PEG, we measured the elemental mapping and XRD to analysis the elements of DOX–Glu–PEG. From the Fig. 2A, the elemental mapping results showed that the sample is composed of carbon and oxygen elements and the distribution of C (Fig. 2B) and O (Fig. 2C) is very high on the analysed area. The XRD patterns exhibited strong diffraction peaks at 19.1° and 23.3° which from the PEG phase of the sample (PDF #49-2097) (Fig. 2D).
1. This final ratio was almost similar to that of the reaction ratio. The UV-vis spectrophotometric peaks of DOX and DOX–Glu–PEG at 481 nm were comparable (Fig. 1C) and thereby indicated that the drug loading and release rates could be determined according to the DOX standard curve at 481 nm. In order to further confirm the structure of DOX–Glu–PEG, we measured the elemental mapping and XRD to analysis the elements of DOX–Glu–PEG. From the Fig. 2A, the elemental mapping results showed that the sample is composed of carbon and oxygen elements and the distribution of C (Fig. 2B) and O (Fig. 2C) is very high on the analysed area. The XRD patterns exhibited strong diffraction peaks at 19.1° and 23.3° which from the PEG phase of the sample (PDF #49-2097) (Fig. 2D).
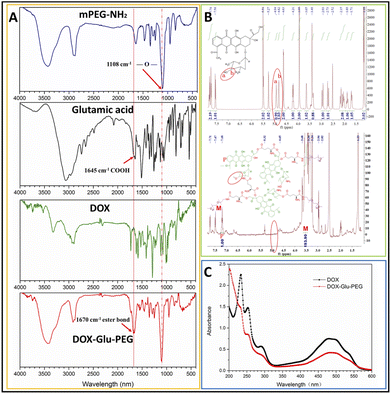 | ||
| Fig. 1 (A) FT-IR spectra of PEG, Glu, DOX, and DOX–Glu–PEG. (B) 1H NMR spectra in DMSO-d6 and (C) UV-vis curves of DOX and DOX–Glu–PEG. | ||
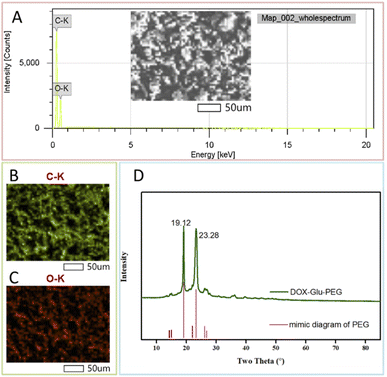 | ||
| Fig. 2 The elemental mapping analysis of DOX–Glu–PEG: (A) wholespectrum, (B) C spectrum and (C) O spectrum; (D) the XRD spectrum of DOX–Glu–PEG (PEG: PDF #49-2097). | ||
Assembly, water-solubility, size stability, drug loading and release properties in vitro
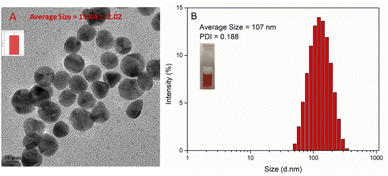
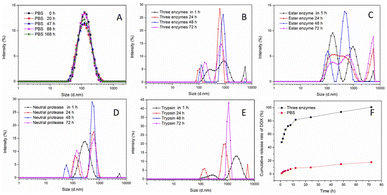
The polymer of DOX–Glu–PEG could be assembled into nano micelle in pure water and PBS. The maximum water solubility of DOX–Glu–PEG could reach 84 mg mL−1. However, the water solubility of DOX could be more than 10 mg mL−1 according to the drug loading (13%) data. The DOX–Glu–PEG micelles exhibited a size of only 13.44 ± 2.02 nm under TEM (Fig. 3A) and a size of 107 nm (PDI = 0.188) and a zeta potential of −19.4 ± 0.231 in pure water through DLS (Fig. 3B). This substantial difference is attributed to the different status of micelles in water and dehydrated condition. The DOX–Glu–PEG micelle of the hydrophilic shell (PEG) would be tightly stuck to the surface of the hyperbranched hydrophobic core (DOX–Glu), resulting in the smaller nanoparticle measured by TEM.24To further evaluate the stability and controllable-instability of DOX–Glu–PEG micelle, the changeable micelle's sizes were measured in pure PBS, PBS with ester enzyme, PBS with trypsin, PBS with neutral protease, and PBS mixed with three enzymes. From Fig. 4A, we can conclude that the DOX–Glu–PEG micelle remained strongly stable for 7 days. Additionally, a significant change occurred in the sizes of the micelles that were exposed to the different enzymatic solutions. Their polydispersity indexes were also increased substantially during 24 h of treatment (Fig. 4B–E). In drug release study of DOX–Glu–PEG, the drug release reached 85.5% during 24 h compared to only 9.8% in PBS (Fig. 4F) indicating that the drug release could be controlled using different enzymes.
Drug loading and release properties were evaluated in vitro and based on the results of UV-vis curves of pure DOX and DOX–Glu–PEG in Fig. 1C, we could plot the standard curve of pure DOX at 481 nm. The standard equation of DOX was A = 0.0186c + 0.0005 and the concentration range was 8 μg mL−1 to 36 μg mL−1. The drug loading was found to be 13% using UV-vis absorption of DOX–Glu–PEG. To evaluate the drug release properties, we determined the drug accumulation rate in the simulated tumour microenvironment enriched with enzymes.32–34 As shown in the Fig. 4F, the DOX accumulation rate reached 47% during 1 h in an enzymatic environment, meaning the hydrophilic PEG can be quickly detached from DOX–Glu–PEG and the partial hyperbranched core of DOX–Glu came out from the dialysis bag. The DOX accumulation rate reached only 17.8% during 72 h in pure PBS whereas the accumulation rate reached approximately 100% in an enzymatic environment composed of trypsin 1 mg mL−1, neutral protease 200 U mL−1, and esterase 15 U mL−1. These results confirmed that the DOX could be released in a controlled manner owing to the abundant availability of ester bonds in DOX–Glu–PEG which could be cleaved by copious esterase and amidase in tumour cells. The results were in line with a previously reported esterase-responsive drug release in an esterase-rich (10 U mL−1) medium.35 Combined instability results of DOX–Glu–PEG micelle in PBS with different enzymes (Fig. 4B–E) further affirmed the controllable release properties of the synthesized nanomicelles.
Cell viability
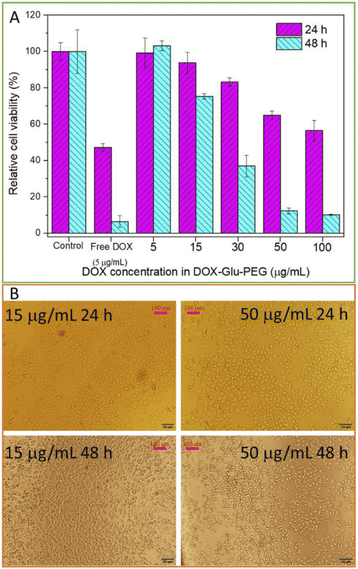
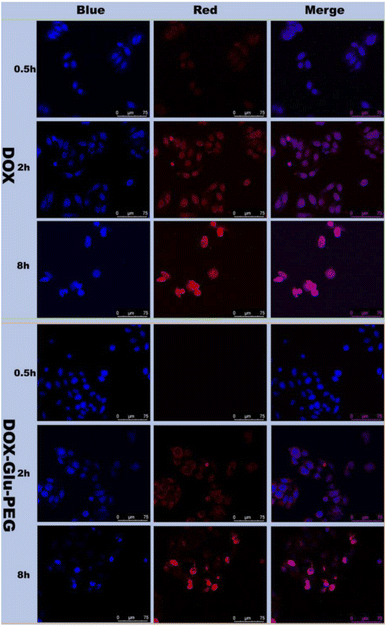
The cytotoxicity of polyprodrug DOX–Glu–PEG and free DOX in HeLa cells was measured by a microplate reader. The free DOX (5 μg mL−1)-treated HeLa cells were almost dead within 48 h (Fig. 5A). Conversely, nanomicelles could effectively inhibit the HeLa cells' proliferation at higher concentrations of the drug. DOX–Glu–PEG having DOX concentrations equivalent to 50 μg mL−1 and 100 μg mL−1 exhibited cell viabilities of 65% and 56%, respectively, at 24 h (Fig. 5A). These results were attributed to the delayed distribution and incomplete disruption of nanomicelles requiring more time to access nuclei and get entirely disrupted. This speculation was maintained by 10% and 12% cell viabilities of HeLa cells at 48 h resulting from DOX equivalent concentrations of 100 μg mL−1 and 50 μg mL−1, respectively. The significant differences of in vitro cell viabilities between free DOX and DOX–Glu–PEG (Fig. 5A) may be attributed to the delayed disruption of DOX–Glu–PEG in esterase overexpressing tumour cells. This esterase-mediated activity of DOX–Glu–PEG offers the advantage of reducing the collateral damage of free DOX on normal tissues.Based on data of cell viabilities, the IC50 value of DOX–Glu–PEG on HeLa cells at 48 h was determined as 28.4 μg mL−1 (standard curve range: 5–50 μg mL−1, standard equation: y = −0.0202x + 1.0744, R2 = 0.9653). Although the drug release rate of DOX–Glu–PEG nanomicelles could reach 85% during 24 h (Fig. 4F), the DOX–Glu–PEG micelles were not completely distributed in the nuclei during 8 h (Fig. 6). The delayed distribution to nuclei of DOX–Glu–PEG micelles compared with free DOX in Fig. 6 may be the main reason associated with the similar cell viabilities that were obtained in cell groups of 100 μg mL−1 and 50 μg mL−1 DOX during 24 h. Following CCK-8 tests, the HeLa cells were observed under microscope and the images were presented for viabilities of two groups of cells (treated with 15 μg mL−1 and 50 μg mL−1 DOX) in Fig. 5B. The dead cells, indicated by the transparent circle, were more apparent in DOX–Glu–PEG-treated cells with a DOX-equivalent concentration of 50 μg mL−1 than that with 15 μg mL−1, at 24 h. As shown in Fig. 5B, the cell morphology further changed significantly after 48 h, displaying more apoptotic cells in the group that was treated with a higher corresponding concentration of DOX (50 μg mL−1).
Cellular uptake experiment
The cellular uptake behaviour and distribution of polyprodrug DOX–Glu–PEG micelles in HeLa cells were observed by LSCM. The red fluorescence represented DOX while the blue fluorescence represented cell nuclei stained with Hoechst. The distribution of DOX–Glu–PEG and free DOX were observed in overlapped fluorescence images of HeLa cells. At 2 h, free DOX was found in the nuclei while DOX–Glu–PEG was distributed in the cytoplasm. However, the most proportion of the nanomicelles reached inside the nuclei at 8 h (Fig. 6). Thus, the cellular uptake pattern, drug release behaviour, and cell viability figures collectively demonstrate that DOX–Glu–PEG were successfully uptaken in cells' nuclei in which the higher concentration of enzymes completely disrupted the structure of nanomicelles resulting in drug release and subsequent cell apoptosis. Moreover, the antiproliferative effect of DOX–Glu–PEG nanomicelles could be improved by using a higher concentration of DOX. These results highlight the possible potential of synthesized nanomicelles to confer in vivo esterase-responsive antitumor activity curtailing the effects of DOX on normal tissues.Conclusions
A DOX prodrug was formulated as nanomicelles by a one-pot synthesis method employing FDA-approved materials i.e. Glu and PEG. The nanomicelles demonstrated their ability to enhance the solubility of the amphiphilic drug, remain stable in PBS, and release the loaded drug in a controllable manner in response to enzymatic activity. The in vitro studies exhibited up to 100% cumulative drug release in 72 h, successful distribution in nuclei of HeLa cells, and effective inhibition of cellular proliferation. Overall, the present study suggested that the fabricated DOX prodrug might serve as a potential candidate for cancer therapeutics.Author contributions
Conceptualization: Lingna Han, Xiao Duan and Murugan Ramalingam; data collection and analysis: Zining Ren, Xue Yang, Changfeng Li, Sasirekha Krishnan, Abinaya Jaisankar, Jeong-Hui Park, Khandmaa Dashnyam; funding acquisition: Changfeng Li; supervision: Lihua Song and Murugan Ramalingam; writing, review & editing: Lingna Han, Kaiqiang Wang, Wujie Zhang, José Luis Pedraz, Seeram Ramakrishna, Hae-Won Kim and Murugan Ramalingam.Conflicts of interest
The authors declare no conflict of interest.Acknowledgements
The present research was supported by the Four “Batches” Innovation Project of Invigorating Medical through Science and Technology of Shanxi Province (No. 2021XM59), Scientific and Technological Innovation Programs of Higher Education Institutions in Shanxi (Grant No. 2021L349 and 2021L355), the Natural Science Foundation for Young Scientists of Shanxi Province, China (Grant No. 201901D211473, 201901D211474 and 201901D211468), the Natural Science Foundation of Shanxi Province, China (Grant No. 201901D111327), Doctoral Scientific Research Foundation of Changzhi Medical College, China (Grant No. BS201909, BS201910 and BS201918), Academic Leader Project of Changzhi Medical College, China (Grant No. XS202001), National College Students' Innovation and Entrepreneurship Training Plan Program, China (Grant No. 20210540), ICTS “NANBIOSIS”, in particular by the Drug Formulation Unit (U10) of the CIBER in Bioengineering, Biomaterials and Nanomedicine (CIBER-BBN) at the University of the Basque Country (UPV/EHU) in Vitoria-Gasteiz, and National Research Foundation of Korea (2018K1A4A3A01064257).Notes and references
- Y. Liu, Y. Q. Liu, J. Zang, A. A. I. Abdullah, Y. Y. Li and H. Q. Dong, ACS Biomater. Sci. Eng., 2020, 6(12), 6510–6527 CrossRef CAS PubMed.
- Y. Zhang, S. Ma, X. M. Liu, Y. D. Xu, J. Y. Zhao, X. H. Si, H. X. Li, Z. C. Huang, Z. X. Wang, Z. H. Tang, W. T. Song and X. S. Chen, Adv. Mater., 2021, 33(7), 2007293 CrossRef CAS PubMed.
- W. Q. Wang, Y. L. Jin, X. Liu, F. M. Chen, X. H. Zheng, T. Q. Liu, Y. M. Yang and H. J. Yu, Adv. Funct. Mater., 2021, 31(26), 2100386 CrossRef CAS.
- M. C. Operti, A. Bernhardt, S. Grimm, A. Engel, C. G. Figdor and O. Tagit, Int. J. Pharm., 2021, 605, 120807 CrossRef CAS.
- X. Liu, N. B. Ye, C. Xiao, X. X. Wang, S. Y. Li, Y. H. Deng, X. Q. Yang, Z. F. Li and X. L. Yang, Nano Today, 2021, 40, 101248 CrossRef CAS.
- X. Luan, H. B. Yuan, Y. D. Song, H. X. Hu, B. Wen, M. He, H. X. Zhang, Y. Li, F. Li, P. Shu, J. P. Burnett, N. Truchan, M. Palmisano, M. P. Pai, S. Zhou, W. Gao and D. X. Sun, Biomaterials, 2021, 275, 120910 CrossRef CAS PubMed.
- M. M. Xu, H. Zhou, Y. N. Liu, J. Sun, W. J. Xie, P. Zhao and J. Liu, ACS Appl. Mater. Interfaces, 2018, 10(39), 32965–32980 CrossRef CAS PubMed.
- Y. Cai, X. Y. Chen, J. X. Si, X. Z. Mou and X. C. Dong, Small, 2021, 2103072 CrossRef CAS PubMed.
- J. C. Li, X. R. Yu, X. Y. Shi and M. W. Shen, Prog. Mater. Sci., 2022, 124, 100871 CrossRef CAS.
- N. Raval, R. Maheshwari, H. Shukla, K. Kalia, V. P. Torchilin and R. K. Tekade, Mater. Sci. Eng., C, 2021, 126, 112186 CrossRef CAS PubMed.
- L. H. Liao, D. Cen, Y. K. Fu, B. Liu, C. Fang, Y. F. Wang, X. J. Cai, X. Li, H. B. Wu and G. R. Han, J. Mater. Chem. B, 2020, 8(17), 3929–3938 RSC.
- Y. Bai, C. P. Liu, X. Song, L. H. Zhuo, H. T. Bu and W. Tian, Chem.–Asian J., 2018, 13(24), 3903–3911 CrossRef CAS.
- A. Najafipour, A. Gharieh, A. Fassihi, H. Sadeghi-Aliabadi and A. R. Mahdavian, Mol. Pharm., 2021, 18(1), 275–284 CrossRef CAS PubMed.
- X. Tang, Q. L. Sheng, C. Q. Xu, M. Li, J. D. Rao, X. H. Wang, Y. Long, Y. Tao, X. He, Z. R. Zhang and Q. He, Nano Today, 2021, 37, 101083 CrossRef CAS.
- P. P. Xu, X. Y. Wang, T. W. Li, L. L. Li, H. H. Wu, J. W. Tu, R. Y. Zhang, L. Zhang, Z. Guo and Q. W. Chen, ACS Appl. Mater. Interfaces, 2021, 13(29), 33926–33936 CrossRef CAS.
- P. Xue, J. Wang, X. F. Han and Y. J. Wang, Colloids Surf., B, 2019, 180, 202–211 CrossRef CAS PubMed.
- H. J. Xiao, Y. P. Guo, H. M. Liu, Y. S. Liu, Y. M. Wang, C. Q. Li, J. Císař, D. Škoda, I. Kuřitka, L. Guo and V. Sedlařík, Biomaterials, 2020, 232, 119701 CrossRef CAS PubMed.
- Q. Wang, M. C. Sun, C. Li, D. Li, Z. M. Yang, Q. K. Jiang, Z. G. He, H. W. Ding and J. Sun, Asian J. Pharm. Sci., 2021, 16(2), 203–212 CrossRef PubMed.
- C. Liu, M. Li, P. Li, W. Chen, H. Li, L. Fan and W. Tian, Biomacromolecules, 2021, 22(6), 2382–2392 CrossRef CAS.
- X. Duan, X. Yang, C. F. Li and L. H. Song, AAPS PharmSciTech, 2019, 20(6), 245 CrossRef CAS PubMed.
- X. Duan, H. Chen, L. Fan and J. Kong, ACS Biomater. Sci. Eng., 2016, 2(12), 2347–2354 CrossRef CAS PubMed.
- C. F. Liu, H. X. Li, P. X. Li, C. P. Liu, Y. Bai, J. Pang, J. X. Wang and W. Tian, Polym. Chem., 2020, 11(36), 5810–5818 RSC.
- X. Duan, J. X. Chen, Y. L. Wu, S. Wu, D. Y. Shao and J. Kong, Chem.–Asian J., 2018, 13(8), 939–943 CrossRef CAS PubMed.
- X. Duan, T. Bai, J. J. Du and J. Kong, J. Mater. Chem. B, 2018, 6(1), 39–43 RSC.
- J. G. Li and P. Liu, Part. Part. Syst. Charact., 2019, 36(7), 1900113 CrossRef.
- J. G. Li, X. M. Li and P. Liu, Colloids Surf., B, 2019, 185, 110608 CrossRef PubMed.
- J. G. Li, X. M. Li and P. Liu, Mol. Pharm., 2020, 17, 710–716 CAS.
- J. G. Li, X. M. Li, M. L. Pei and P. Liu, Colloids Surf., B, 2020, 192, 111064 CrossRef CAS PubMed.
- X. M. Li and P. Liu, Mater. Sci. Eng., C, 2021, 128, 112317 CrossRef CAS PubMed.
- X. M. Li, J. Zhang and P. Liu, Int. J. Pharm., 2021, 606, 120941 CrossRef CAS PubMed.
- X. Y. Zhang, X. Duan, Y. Y. Hu, Z. M. Tang, C. X. Miao, W. Tao and J. Wu, Nano Today, 2021, 37, 101098 CrossRef CAS.
- R. Kumari, M. M. Majumder, J. Lievonen, R. Silvennoinen, P. Anttila, N. N. Nupponen, F. Lehmann and C. A. Heckman, Br. J. Cancer, 2021, 124, 1428–1436 CrossRef CAS PubMed.
- H. Ogasawara, K. Nishio, F. Kanzawa, Y. S. Lee, Y. Funayama, T. Ohira, Y. Kuraishi, Y. Isogai and N. Saijo, Jpn. J. Cancer Res., 1995, 86, 124–129 CrossRef CAS PubMed.
- F. W. Price, Anal. Biochem., 1964, 8, 24–33 CrossRef CAS PubMed.
- B. Surnar and M. Jayakannan, ACS Biomater. Sci. Eng., 2016, 2, 1926–1941 CrossRef CAS PubMed.
Footnote |
| † These authors contributed equally to this work. |
| This journal is © The Royal Society of Chemistry 2022 |