Open Access Article
Open Access ArticleStructural, morphological, and optical properties of carbon nanoparticles unsheathed from date palm fronds†
Shaik Muhammad U. G. Mohiuddin *ab,
Abdulkadir Aydarousa,
Ahmed Alshahrieab,
Abdu Saeed
*ab,
Abdulkadir Aydarousa,
Ahmed Alshahrieab,
Abdu Saeed abc,
Adnan Memićb,
Shittu Abdullahi
abc,
Adnan Memićb,
Shittu Abdullahi abd and
Numan Salah
abd and
Numan Salah *b
*b
aDepartment of Physics, Faculty of Sciences, King Abdulaziz University, 21589, Jeddah, Saudi Arabia. E-mail: smohiuddin@stu.kau.edu.sa
bCenter of Nanotechnology, King Abdulaziz University, 21589, Jeddah, Saudi Arabia. E-mail: nsalah@kau.edu.sa
cDepartment of Physics, Faculty of Science, Thamar University, Thamar, Yemen
dDepartment of Physics, Faculty of Science, Gombe State University, Gombe, Nigeria
First published on 27th September 2022
Abstract
Several studies have reported the synthesis of carbon nanoparticles (CNPs) by various methods. In this study, an easy one-step process to unsheathe CNPs from date palm fronds through a top-down ball milling method has been reported. The CNPs were characterized using various spectroscopic and microscopic methods to determine their structural and morphological features, optical properties, crystallinity, physicochemical properties, and particle stability. Transmission electron microscopy (TEM) revealed that the obtained CNPs' size ranged from 4 to 22 nm in a crystalline form. Scanning electron microscopy (SEM) confirmed their spherical shape, while the maximum photoluminescence (PL) intensity was recorded at 464 nm when excited at 375 nm. The unsheathed CNPs produced a good quantum yield (QY) of 3.24%. Furthermore, the CNPs exhibited high Raman ratios of ID/IG and I2D/IG with values of 0.59 and 0.04, respectively, verifying their multilayer crystalline graphitic nature. These Raman ratios also agree with the X-ray diffractometry (XRD) results. The CNPs' sp2 and sp3 carbon bonds were confirmed by X-ray photoelectron spectroscopy (XPS), with oxygen on the surface forming carboxyl and carbonyl groups with no other observable impurities. Furthermore, the extracted CNPs showed excellent PL properties for up- and down-conversion. These properties are exemplary for low-cost biomass with potential applications in biomedicine. Therefore, the extracted CNPs reported in this study have potential applications in optical imaging.
1. Introduction
Fluorescent nanoparticles (FCNPs) have attracted the attention of the scientific community after the Scrivens group reported a purifying technique for single-walled carbon nanotubes (SWCNTs) in 2004. Their group used an electrophoretic method from an arc discharge process.1 The technique also involved further study of the fluorescent fractions to produce carbon nanomaterials. Since then, several studies have investigated the potential applications of FCNPs in different optoelectronic fields, including biomedical applications,2 while other studies have focused on improving their potential use in theragnostics2 due to their unique PL emission properties.3For the past two decades, numerous experiments and strategies have been developed to determine the efficiency of FCNPs.4–7 The parameters responsible for CNP fluorescence are passivation reagents and size reduction, which are the key factors determining the emission wavelength.4–7 It has been reported that passivating the CNP surface may drastically increase the fluorescence QY%.8 Sun et al. (2006)8 and Liu et al. (2009)9 reported the effects of CNP size on the PL quantum yield percentage (QY%). They stated that large CNP particles present poor QY% and small CNP particles show high QY% in addition to an increasing energy gap approaching that of quantum dots.8,9 However, the preparation technique controlling the CNP fluorescence remains a challenging factor. Previous reports have stated that the important parameters behind the CNP photoluminescence (PL) characteristics are the intensity and wavelengths of the emission and excitation.10 Furthermore, changes in the CNP PL characteristics may be related to the tuning of surface-state properties such as the doping of heteroatoms, functional groups, excitation of carbon, emission traps, structures of the aromatic conjugate and free zigzag regions.10–13
The PL mechanism changes with the different techniques and passivating agents used during CNP extraction.14 The major CNP challenges till date are achieving large-scale production with stable efficiency, their isolation techniques and their purity studies.10,15,16 Retrospective studies indicate that different CNP synthetic techniques may present different QY% values. For example, a technique involving high-energy ion-beam radiation may create point defects in the produced CNPs,17 whereas a thermal decomposition technique may produce CNPs with low QY%. In addition, the gel-electrophoresis technique produces CNPs that emit assorted colors for different carbon materials derived from metal–organic frameworks by the reduction of oxygen.13,18 However, separating these CNP color emissions remains a challenge.10–12,19,20
FCNPs emit distinct colors. For instance, octadecyl amine with carbon nanodiamond functionalization produces a blue color,21 while nitrogen doping produces red fluorescence.22 Furthermore, techniques including thermal decomposition and laser ablation produce ultrafine CNPs with full color emission and low QY%.21–23 FCNPs are important biomedical materials due to their unique characteristics, including sensitivity, water solubility, photovoltaic efficiency, photostability and biocompatibility with the target. It should be noted that the origin and synthetic technique of the FCNPs determine their absorbance and PL efficiency along with their functional groups.10,15,16,20,24,25
Saudi Arabia has more than 25 million date palm trees with 400 different varieties and a date production of approximately 40 kg per tree.26 Therefore, extracting CNPs from these date palms fronds has several benefits including large-scale production in an environmentally friendly, sustainable and renewable manner. Most previous studies have reported CNP preparation using bottom-up methods such as the hydrothermal procedure,11,27–29 with few studies reporting top-down techniques using high-energy ball milling.30 The ball milling technique produces CNPs by crushing large-carbonized particles inside a steel jar. The inner part of the steel jar may accumulate heat and pressure during crushing. CNPs produced using ball milling have features such as excellent graphitic character and low QY%.31–33
In view of the above considerations and to the best of our knowledge, no study has reported on the up-conversion PL properties and chemical structures through Raman spectroscopy for reduced-size CNPs extracted from date palm fronds. Studying the chemical structures of the CNPs through Raman spectroscopy may highlight the graphitic CNPs' purity and presence of defects in the chemical structures. Therefore, this study focuses on extracting CNPs from date palm fronds and evaluating their optical properties, including PL up-conversion, and their structural properties to determine their potential applications in optical imaging.
2. Materials and methods
2.1 Synthesis of the CNPs
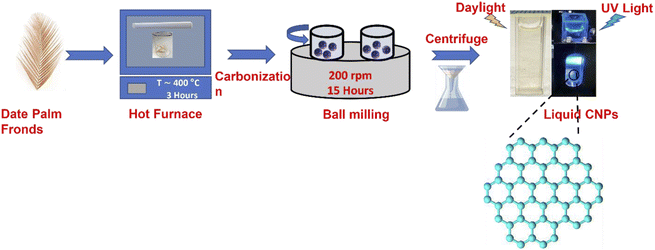
The date palm fronds used in this study were sourced from Jeddah, Saudi Arabia, and prepared following the procedure in Salah et al.34 The fronds were cut into small pieces and carbonized at 400 °C using a muffle furnace. Thereafter, ball milling with a Retsch PM 400 instrument was used to synthesize the novel CNPs by placing the carbonized particles (CPs) in the machine for 15 hours at a speed of 200 rpm.34 The stated CNP preparation procedures are described in Fig. 1.2.2 Unsheathing the PL of the CNPs and their size
The size of the CNPs was further reduced by dissolving 25![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 mg of CNP powder in 1500 ml deionized water (DI) and magnetically stirring for 24 hours at 80 °C. The mixture was filtered using Whatman filter paper to collect particles of homogeneous size. The mixture was further centrifuged at 4000 rpm for 1 hour to collect the smallest homogeneous particles. Hence, the mixture was found to contain a 16.58 ± 0.36 mg ml−1 concentration of ultrafine CNPs at pH 7.23. The mixture was prepared in three replicates to ensure the reproducibility of the results and a yield value of 69.89% ± 0.47% was obtained. Furthermore, the CNP mixture is a light brown aqueous hydrophilic mixture when exposed to sunlight and emitted bright blue light under ultraviolet visible light at an excitation wavelength of 365 nm.
000 mg of CNP powder in 1500 ml deionized water (DI) and magnetically stirring for 24 hours at 80 °C. The mixture was filtered using Whatman filter paper to collect particles of homogeneous size. The mixture was further centrifuged at 4000 rpm for 1 hour to collect the smallest homogeneous particles. Hence, the mixture was found to contain a 16.58 ± 0.36 mg ml−1 concentration of ultrafine CNPs at pH 7.23. The mixture was prepared in three replicates to ensure the reproducibility of the results and a yield value of 69.89% ± 0.47% was obtained. Furthermore, the CNP mixture is a light brown aqueous hydrophilic mixture when exposed to sunlight and emitted bright blue light under ultraviolet visible light at an excitation wavelength of 365 nm.
2.3 Characterizations of the CNPs
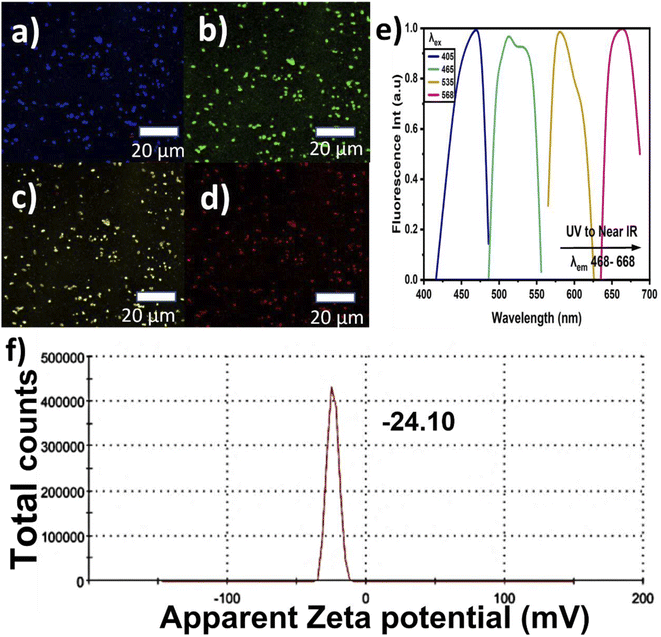
The CNPs' fluorescence images were captured using CM scanning at excitation wavelengths ranging from the blue region to near-infrared (NIR). The samples were prepared by simply drying a few drops of the CNP mixture on glass slides and these glass slides were used for the FTIR, Raman, XRD, and CM studies. Lastly, the stability and colloidal behavior of the CNPs were studied using zeta potential analysis. The samples were prepared in a disposable cuvette that was specific for the instrument and measured at room temperature while monitoring the pH value.
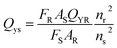 | (1) |
| Sample | Integrated intensity of emission (F) | UV absorbance at the desired excitation nm (A) | Refractive index of the sample (n) | Quantum yield% (QY) | |
|---|---|---|---|---|---|
| Quinine sulfate | 380 | 0.0195 | 1.33 | 54 | 29 |
| CNPs | 375 | 0.399 | 1.33 | 3.24 | This |
3. Results and discussion
3.1 Physiochemical properties of the CNPs
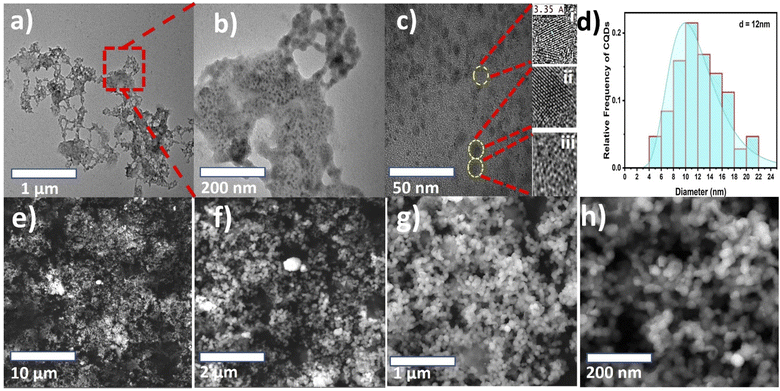
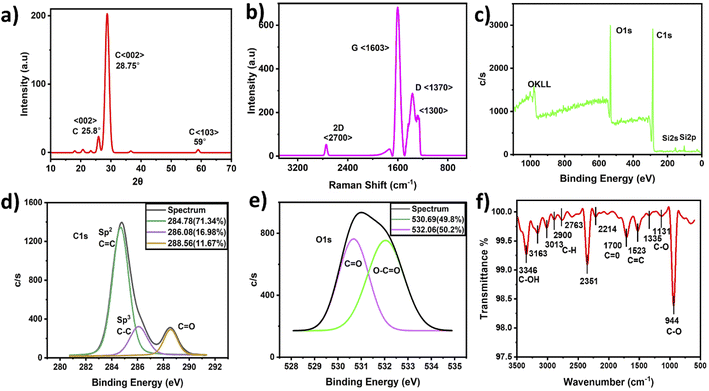
Morphological TEM images were taken at different magnifications and are presented in Fig. 2(a–c). The images show a dendrite-like network at low magnification, as depicted in Fig. 2(a), while at higher magnification, the particles were well separated from each other, as shown in Fig. 2(b). In addition, Fig. 2(c) shows clear lattice fringes, confirming the crystallinity, as depicted in Fig. 2c(i and ii), and the hexagonal rings of carbon in the HR-TEM images, as shown in Fig. 2c(iii). Fig. 2(d) shows the frequency distribution of TEM images with the particle diameter ranging from 4 to 22 nm and an overall average diameter of 12 nm. The CNP particle sizes recorded in this study agree with those of previous studies.27–29 Additionally, the CNP particles in the SEM images depicted in Fig. 2(e–h) appear to be uniformly distributed without agglomeration, as recorded in the low-magnification TEM images. It can be observed that small CNP particles were attached to big particles, which also revealed good porosity. The CNPs were found to be hydrophilic with a transparent color. Moreover, the HR-TEM image revealed an inter-fringe distance of 3.35 Å, corresponding to a carbon material index of (002) and attributed to the crystalline graphitic phase with sp2 graphene (002).37Fig. 3(a) shows the X-ray diffraction patterns of the CNPs. The patterns exhibit a few diffraction peaks with a prominent peak centered at 28.70° and a corresponding interlayer d-spacing of 3.108 Å. The peak also has a shoulder at 25.8° with a d-spacing value of 3.441 Å.38,39 Another peak was recorded at 59° with a d-spacing value of 1.569 Å.40 The sharp peak represents bulk graphite with oxygen functional groups and was recorded due to the small size of the graphitized CNPs' crystalline nature.38 Meanwhile, the small peak recorded at 59° is attributed to graphitic diffraction. Furthermore, the recorded XRD d-spacing value of 3.441 Å with an index value of (002) is identical to the inner lattice spacing also observed in the HR-TEM image.
There are a few studies that have reported the synthesis of CNPs sourced from date palm fronds. For instance, Kavitha et al.27 carbonized date palm fronds through manual grinding and obtained an average size of 32 nm. However, the CNPs' mesoporous form was more amorphous than crystalline in nature. Another study conducted by Athinarayanan et al.28 synthesized CNPs using a hydrothermal technique and achieved CNP sizes ranging from 5 to 15 nm with an inner lattice spacing and XRD d-spacing of 3.36 and 3.34 Å, respectively. In addition, another version of the study reported by Athinarayanan et al.29 synthesized CNPs using the same hydrothermal technique, which yielded CNP sizes of 2 to 15 nm, while inner lattice XRD d-spacings of 0.238 and 0.356 Å were recorded. In contrast, the CNPs reported herein, as shown in Fig. 2c(iii), show the presence of hexagonal rings of graphitic crystalline carbon with excess oxygen functional groups. This was also confirmed through the XRD peak centered at 28.70°, which is attributed to bulk graphitic carbon with 2D hexagonal reflections.41,42 Therefore, the CNPs synthesized from date palm fronds in most previous studies show less graphitic content compared with those reported herein. This indicates that the nature of CNPs depends on the source of the date palm fronds and the synthetic technique.
Raman analysis detects structural disorder and defects in CNPs, including the functional groups, vacancies, doping, and edges.43 Fig. 3(b) shows sharp D and G bands at 1603 cm−1 and 1370 cm−1 with a small 2D band at 2700 cm−1 attributed to graphitic carbon. A peak at 1300 cm−1 near the carbon nanodiamond Raman peak was also recorded36 and a similar FTIR transmittance peak was also recorded at 1335 cm−1, indicating sp3 hybridized carbon atoms and defects. Since the D band indicates impurity and the effects of graphitic carbon, the splitting of the sp3 hybridized D-peak into two peaks is possibly associated with the amount of carbon disorder defects, functionalization effects and CNP edge effects.44 In contrast, the G band indicates the presence of sp2 hybridized carbon atoms. The peak at 1603 cm−1 was assigned to the vibrational stretching bands, describing how the vibrations affect the structural order and purity of graphite. The graphene layers are identified with the 2D band by determining the I2D/IG value, and the value recorded herein is 0.04, corresponding to a multilayer structure. The obtained CNPs ID/IG ratio with a value of 0.59 indicates a graphitic nature. The ID/IG ratio recorded in this study is high compared with that of previous studies.43,45–48 Hence, the CNPs reported in this study are predominantly carbon nanocrystalline particles, which are attributed to crystalline graphitized carbon.43,45–48
Fig. 3(c) shows the elemental structure of the CNPs through their XPS spectra. It can be observed that the CNPs' atomic percent composition was 74% and 24% carbon and oxygen, respectively, which was dissimilar to the original carbon soot before ball milling. The original carbon prior to ball milling contained 96% and 4% carbon and oxygen, respectively. Therefore, the ball milling technique, which involves crushing carbonized carbon particles into CNPs, is an oxide cutting technique, with potential oxidation causing the oxygenated groups to bind with the resultant CNPs, thereby making the CNPs negatively charged carriers.49
The carbon C 1s spectrum presented in Fig. 3(d) shows several peaks, including peaks at 284.78 eV, 286.08 eV and 288.56 eV, which are attributed to sp2(C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), sp3(C–C) and (C
C), sp3(C–C) and (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O) with FWHM (intensity counts) of 1.74 (1360.70), 1.34 (325.24) and 1.30 (298.7),43,50,51 respectively. Fig. 3(e) shows the oxygen O 1s spectrum with peaks at 530.69 eV and 532.06 eV, which are attributed to C
O) with FWHM (intensity counts) of 1.74 (1360.70), 1.34 (325.24) and 1.30 (298.7),43,50,51 respectively. Fig. 3(e) shows the oxygen O 1s spectrum with peaks at 530.69 eV and 532.06 eV, which are attributed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O and C–O, with FWHM (intensity counts) of 1.86 (763.20) and 1.42 (753.86), respectively.35,36 In addition, elemental analysis indicated relatively high oxygen content that may have been caused by oxidation during sample preparation. The FTIR spectra also show similar functionalized carboxylic and carbonyl groups, indicating the dominance of oxygen with carbon.
O and C–O, with FWHM (intensity counts) of 1.86 (763.20) and 1.42 (753.86), respectively.35,36 In addition, elemental analysis indicated relatively high oxygen content that may have been caused by oxidation during sample preparation. The FTIR spectra also show similar functionalized carboxylic and carbonyl groups, indicating the dominance of oxygen with carbon.
It was also observed that the CNPs are mostly composed (71.34%) of the sp2 spectrum, representing the CNPs' graphitic nature, with sp3 hybridized crystalline carbon with a binding energy of 286.08 (eV) covering an area of 16.98%, attributed to π–π* conjugation. These confirm the lower defects in the crystalline graphite.37 However, this could also be formed by carbon nanodiamond (CND) oxygen defects with blue-colored emission. According to previous studies, small CNP particles present high fluorescence.36
Fig. 3(f) presents the CNPs' FTIR spectra. The spectra show a strong band at 3400 cm−1 attributed to the C–OH stretching vibration, while the peaks at 3163, 3013, 2900 and 2763 cm−1 were assigned to C–H stretching vibration of the symmetric and asymmetric hydrocarbons of CH2. Similarly, the sharp peaks recorded at 1523, 1700, 1398, and 1131–1335 cm−1 are assigned to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C
C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–O–C and C–O vibrational stretching.38–42 The strong peak at 2350 cm−1 represents O
O, C–O–C and C–O vibrational stretching.38–42 The strong peak at 2350 cm−1 represents O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C
C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O vibrational stretching. However, some studies have attributed this peak to carbon dioxide,50,51 followed by a weak peak at 2214 cm−1.50,51 The presence of oxygen as a functional group in the CNPs confirms it to be one of the major causes of the increase in the PL property. In previous studies,52,53 CNPs with date palm fronds have shown traces of nitrogen and a low percentage of oxygen and its defects. However, some studies show nearly the same percentage of C and O as the results reported herein, without a trace of nitrogen and with oxygen as the single functional group.43
O vibrational stretching. However, some studies have attributed this peak to carbon dioxide,50,51 followed by a weak peak at 2214 cm−1.50,51 The presence of oxygen as a functional group in the CNPs confirms it to be one of the major causes of the increase in the PL property. In previous studies,52,53 CNPs with date palm fronds have shown traces of nitrogen and a low percentage of oxygen and its defects. However, some studies show nearly the same percentage of C and O as the results reported herein, without a trace of nitrogen and with oxygen as the single functional group.43
The CNPs C 1s spectrum presented in the XPS results with the corresponding peak at 288.5 eV covering an area of 11.67% ascribed to carboxylic groups is analogous to the FTIR results. Therefore, the XPS results indicate that the CNPs reported in this study are composed of crystalline nanodiamonds with structural defects, which agrees with the TEM, Raman and FTIR characterization.36 However, further studies are needed to verify the presence of the carbon nanodiamonds detected in this study.
The results reported in this study could be compared with those of a previous study conducted by Salah et al.34 Salah et al.34 reported a CNP extraction technique from date palm fronds and characterized them using various techniques, including TEM and HR-TEM, SEM, Raman, UV absorption, XPS, XRD and electrical conductivity. They studied the CNPs in the powder form rather than the liquefied form reported in this study. Although they did not mention the exact CNP size they obtained, they mentioned that they were less than 100 nm, which is much larger than the size reported herein. In addition, this study investigated the physiochemical properties using FTIR to assess their chemical composition in addition to the CNPs' Raman intensity ratios of ID/IG and I2D/IG, which are investigated herein to determine the graphitic purity and presence of multilayers, which were not previous reported by Salah et al.34
3.2 Optical properties of the CNPs
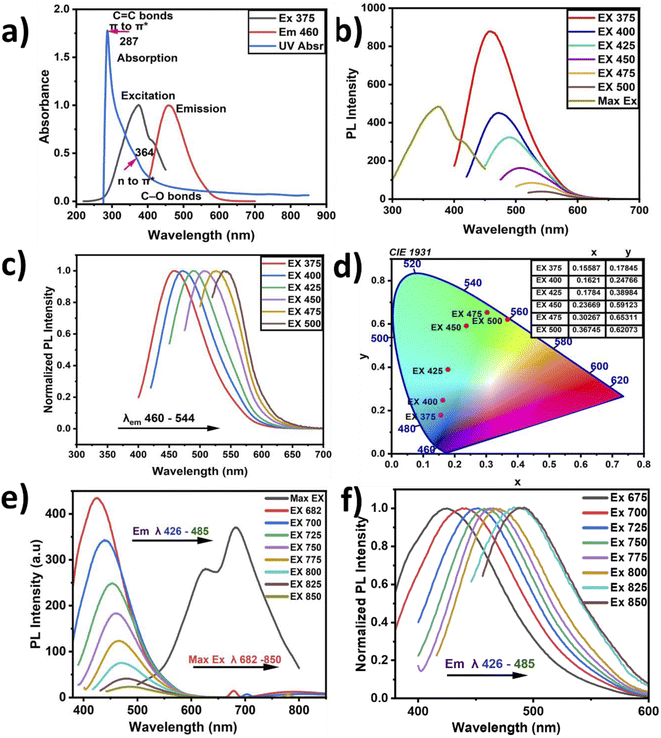
The photon absorption analysis is presented in Fig. 4(a). A sharp absorption peak located at 287 nm with a weak shoulder at around 364 nm was recorded. These peaks described π–π* transitions representing graphitic carbon C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C bonds and n–π* transitions attributed to carboxyl and carbonyl groups (C
C bonds and n–π* transitions attributed to carboxyl and carbonyl groups (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O).37,54 These sharp absorption peaks confirm the delocalization of π-state formation due to the predominance of sp2-like graphitic carbon. In contrast, in the n–π* transitions, the carboxylic and carbonyl functional groups attached to conjugated carbon are responsible for the PL emission at 464 nm, as presented in Fig. 4(b and c). Such CNP behavior has been previously reported in the literature.55 The PL emission spectra show wide broadband emission with a shift from the visible to green-yellow region. The optimum PL excitation was recorded at 375 nm with a corresponding absorption intensity of 0.399. Therefore, the Stokes shift was found to be 89 nm, and it has been previously reported that a high Stokes shift exhibits better fluorescence.55
O).37,54 These sharp absorption peaks confirm the delocalization of π-state formation due to the predominance of sp2-like graphitic carbon. In contrast, in the n–π* transitions, the carboxylic and carbonyl functional groups attached to conjugated carbon are responsible for the PL emission at 464 nm, as presented in Fig. 4(b and c). Such CNP behavior has been previously reported in the literature.55 The PL emission spectra show wide broadband emission with a shift from the visible to green-yellow region. The optimum PL excitation was recorded at 375 nm with a corresponding absorption intensity of 0.399. Therefore, the Stokes shift was found to be 89 nm, and it has been previously reported that a high Stokes shift exhibits better fluorescence.55
There are various sources of CNPs; however, the excitation wavelength of natural CNPs mainly depends on the PL emission wavelength, and this behavior characterizes their structural complexity.56
The CNPs' PL emission is not only dependent on the intrinsic or outer surface emission but it can also explain the CNPs' chemical composition. This is because CNPs have different layers and different sizes, which can lead to selective excitation. The graphitic carbon reported in this study showed the presence of multilayers, describing its imperfection. When the CNPs' PL emission primarily depends on the presence of a conjugated sp2 matrix rather than the CNPs' size, it is referred to as down-conversion photoluminescence.57,58 Furthermore, Fig. 4(c) shows the observed systematic shift when the CNP samples were excited with a wavelength ranging from 375 to 500 nm, and the corresponding the PL emission was recorded at wavelengths ranging from 464 to 560 nm, with the PL intensities normalized for convenient observation of the shifts. The shifts may be attributed to the different particle sizes in the samples, with the abundant oxygen contained as a functional group on the surface causing emission traps. Additionally, Fig. 4(d) depicts the chromaticity diagram displaying the emission color shift of the CNPs along with their x and y axes. Here, the CNPs showed a blue color at the highest emission intensity and near-yellow at the lowest emission. Fig. 4(e) presents the up-conversion PL emission, which is considered one of the vital CNP optical properties. The up-conversion PL emission is recorded using excitation wavelengths ranging from 426 to 485 nm and a considerable red-shift was recorded when the PL emission was recorded with the excitation wavelength ranging from 682 nm to 850 nm. This unique property apparently indicates excitation-independent emission, referred to as up-conversion. This property is required for biomedical applications such as bioimaging, cell labeling and theragnostics.59 The computed CNPs' quantum yield is found to be 3.24%, showing good fluorescence, as presented in Table 1. Incorporation of nitrogen and oxygen can also lead to increased quantum yield according to previous reports. The fluorescence of the defect structure of diamonds is well known from previous studies. Thus, an abundant amount of oxygen and sp3 crystalline diamond-like carbon can possibly be responsible for the blue-colored PL emission.36
There are several ways to produce carbon dots with different optical results and modifications. It has been reported that various CNP emissions originate from the transition of surface electrons to valence holes, emission from the intrinsic band surface to the ground state, and electron–phonon coupling, which occurs due to dipole emission excitations by self-trapping,40 but mostly from the presence of organic molecules. Furthermore, another report on PL emission stated that it depends on excitation, and its characteristics are mostly determined through centers of multiemission.40 Generally, the CNP optical property is dependent on various factors, such as the environment and the interaction of the CNPs with the environment.40
The surface structure of CNPs is responsible for the changes in their optical property, leading to the transfer of electrons from the nanoparticles to other species. It has been reported that the type of solvent also plays a major role in the solubility and interaction of particles. The scientific community has not yet come to a common understanding and agreed explanation of the CNPs' optical properties.40 Fig. 3(b) shows both the sp2 and sp3 bonds of hybridized carbon atoms, which are related to their optoelectronic properties through the delocalized electron sites, number of double bonds and spatial distribution. The PL emission dependent on excitation originates from the CNP cores and the uniformity of their surfaces in addition to the CNP size. Here, the maximum excitation peak is recorded at 375 nm with a corresponding maximum emission at 464 nm. These also present a Stokes shift of 89 nm and a full width half maximum (FWHM) of 94 nm, indicating low energy loss and good peak absorption.49,60
CNPs have a unique electron-transfer property by quenching, whereby the molecules can accept or donate electrons. Although graphene nanoparticles (GNPs) have less defects and better crystallinity than CNPs, CNPs have consistent graphene lattice spacing, graphite, and diamond. CNPs are mainly composed of C and O, which play a significant role in PL enhancement. Having carbonyl and carboxyl surface groups with better solubility also plays a vital role as a functionalizing agent. Overall, it is easy to alter the surface of these functionalized CNPs to improve their PL, solubility, and sensitivity for potential applications in optical imaging and ultimately biomedical applications. It has been reported that the efficiency of the fluorescent quantum yields mostly depends on the incorporation of oxygen elements and the subsequently produced fluorescent centers in the CNPs.61 Oxygen functional group can cause deprotonation in the CNPs due to changes in the environment and pH value. Since the PL also depends on the CNPs' graphitic pH value and the presence of oxygen, a change in pH might lead to a decrease in fluorescence intensity.55 Therefore, a pH value near 7 may be required to achieve excellent luminescence, as reported in this study. GNPs are a subclass of carbon materials produced mostly through top-down methods, having structures with π* conjugation, and containing one or more layers of graphene oxide. In this study, XPS and FTIR analyses confirmed that our CNPs show the presence of excess oxygen components as oxygenated functional groups, and hence it is possible that our CNPs were formed by oxidative routes. The PL chromaticity diagram shows high wavelength excitation, indicating a shift from the blue region to green. A possible reason for this shift might the formation of electron traps due to the oxygenated functional groups, directing the blue emission at 464 nm. At relaxation, this was followed with green emission, which could be due to the large wavelength excitation causing a disruption in the highest unoccupied molecular orbital (HUMO). Therefore, this optical study revealed that the PL emission reported in this study does not depend on excitation. However, other factors such as the functional groups, amount, and preparation method determine several aspects of the CNPs' PL. The CNPs reported herein exhibit light-capturing capability and therefore potential applications in optical imaging.61
As previously reported, the energy attributed to Brownian motion hinders the agglomeration of nanoparticles, thereby making them soluble in water and stable. Optical bioimaging requires nanoparticles that emit at a certain excitation wavelength, are compatible with the environment, and are be water soluble without functionalization after extraction.63 The characterized results obtained for the CNPs are summarized in Table 2.
| λex/λem | FWHM (nm) | QY% | Raman ID/IG | TEM size (nm) | Stokes shift | pH | Zeta potential (mV) | Surface-state functional groups |
|---|---|---|---|---|---|---|---|---|
| 375/464 nm | 94 | 3.24 | 0.59 | 4–22 nm | 89 | 7.23 | −24.10 | C–C/C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C, C C, C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O, C–O–C, C–OH, C–H O O, C–O–C, C–OH, C–H O![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O/O–C O/O–C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O O |
4. Conclusions
We report a simple and easy way to extract CNPs via the ball milling technique. Date palm fronds were employed as an organic source of unsheathed CNPs. The structural and optical properties of the CNPs were studied using various techniques, such as HR-TEM, XRD, XPS and FTIR, Raman, UV, and PL spectroscopy. The extracted CNPs' size was found to range from 4 to 22 nm, having a hydrophilic and crystalline graphitic nature. The crystallinity of the CNPs and the presence of bulk graphite were confirmed by XRD characterization. A Raman ratio (ID/IG) of 0.59 was recorded, confirming the presence of multilayer graphite, indicating an agreement between the Raman and XRD patterns. FTIR spectroscopy confirmed the presence of carboxyl and hydroxyl functional groups. The high percentages of sp2 hybridized graphitic carbon and low sp3 were validated using XPS. Furthermore, the CNPs demonstrated exemplary PL properties, such as down-conversion and up-conversion, justifying the excitation independence of the emission within the blue region to NIR. The QY% was also computed and the value recorded is 3.24% at pH 7.23. The QY% and pH value confirmed the high recorded PL intensity. The extracted CNPs showed excellent optical behavior with particle stability, having a zeta potential value of −24.10 mV, suggesting their potential application in optical imaging. These statements were also verified using confocal microscopy. Therefore, the optical properties of the extracted CNPs reported in this study are favorable for optical bioimaging and biomedical applications. However, their toxicity and interaction with different types of cells need to be further investigated to ensure their potential biomedical applications.Author contributions
Shaik Muhammad U. G. Mohiuddin: methodology, formal analysis, investigation, data curation, writing – original draft, writing – review & editing. Numan Salah: conceptualization, methodology, supervision, resources, investigation, writing – review & editing. Adnan Memic: confocal imaging, validation, data curation, writing – review & editing. Ahmed Alshahrie: validation, writing – review & editing. Abdu Saeed: investigation of the PL of the CNPs, data curation, writing – review & editing. Abdulkadir Aydarous: validation, writing – review & editing. Shittu Abdullahi: validation, writing – review & editing.Conflicts of interest
There are no conflicts to declare.References
- X. Xu, R. Ray, Y. Gu, H. J. Ploehn, L. Gearheart, K. Raker and W. A. Scrivens, J. Am. Chem. Soc., 2004, 126, 12736–12737 CrossRef CAS PubMed.
- L. Zhu, Y. Yin, C. F. Wang and S. Chen, J. Mater. Chem. C, 2013, 1, 4925–4932 RSC.
- J.-Y. Marzin, J.-M. Gérard, A. Izraël, D. Barrier and G. Bastard, Phys. Rev. Lett., 1994, 73, 716–719 CrossRef CAS PubMed.
- L. Zheng, Y. Chi, Y. Dong, J. Lin and B. Wang, J. Am. Chem. Soc., 2009, 131, 4564–4565 CrossRef CAS PubMed.
- M. Bottini, C. Balasubramanian, M. I. Dawson, A. Bergamaschi, S. Bellucci and T. Mustelin, J. Phys. Chem. B, 2006, 110, 831–836 CrossRef CAS PubMed.
- S. T. Yang, J. H. Liu, P. Wang, S. Yang, L. Ge, S. Yan and Y. P. Sun, ChemistrySelect, 2018, 3, 6374–6381 CrossRef CAS.
- Q. Du, J. Zheng, J. Wang, Y. Yang and X. Liu, RSC Adv., 2018, 8, 19585–19595 RSC.
- Y.-P. Sun, B. Zhou, Y. Lin, W. Wang, K. A. S. Fernando, P. Pathak, M. J. Meziani, B. A. Harruff, X. Wang, H. Wang, P. G. Luo, H. Yang, M. E. Kose, B. Chen, L. M. Veca and S.-Y. Xie, J. Am. Chem. Soc., 2006, 128, 7756–7757 CrossRef CAS PubMed.
- R. Liu, D. Wu, S. Liu, K. Koynov, W. Knoll and Q. Li, Angew. Chem., Int. Ed. Engl., 2009, 48, 4598–4601 CrossRef CAS PubMed.
- M. L. Liu, B. B. Chen and C. M. Li, Green Chem., 2019, 21(3), 449–471 RSC.
- M. Ikram, I. Hussain, J. Hassan, A. Haider, M. Imran, M. Aqeel, A. Ul-Hamid and S. Ali, Ceram. Int., 2020, 46, 21073–21083 CrossRef CAS.
- J. Hassan, M. Ikram, A. Ul-Hamid, M. Imran, M. Aqeel and S. Ali, Nanoscale Res. Lett., 2020, 15, 75 CrossRef CAS PubMed.
- X. Wang, A. Dong, Y. Hu, J. Qian and S. Huang, Chem. Commun., 2020, 56, 10809–10823 RSC.
- J. Xia, S. Chen, G.-Y. Zou, Y.-L. Yu and J.-H. Wang, Nanoscale, 2018, 10, 22484–22492 RSC.
- S. Chahal, J. R. Macairan, N. Yousefi, N. Tufenkji and R. Naccache, RSC Adv., 2021, 11, 25354–25363 RSC.
- B. B. Chen, M. L. Liu and L. Zhan, Anal. Chem., 2018, 90(6), 4003–4009 CrossRef CAS.
- L. Bao, Z.-L. Zhang, Z.-Q. Tian, L. Zhang, C. Liu, Y. Lin, B. Qi and D.-W. Pang, Adv. Mater., 2011, 23, 5801–5806 CrossRef CAS.
- L. Chai, Z. Hu, X. Wang, L. Zhang, T.-T. Li, Y. Hu, J. Pan, J. Qian and S. Huang, Carbon, 2021, 174, 531–539 CrossRef CAS.
- M. Ikram, A. Raza, M. Imran, A. Ul-Hamid, A. Shahbaz and S. Ali, Nanoscale Res. Lett., 2020, 15, 0–10 CrossRef PubMed.
- M. Ikram, M. Imran, S. Hayat, A. Shahzadi, A. Haider, S. Naz, A. Ul-Hamid, W. Nabgan, I. Fazal and S. Ali, Nanoscale Adv., 2022, 4, 211–225 RSC.
- S. Sarkar, M. Sudolská, M. Dubecký, C. J. Reckmeier, A. L. Rogach, R. Zbořil and M. Otyepka, J. Phys. Chem. C, 2016, 120, 1303–1308 CrossRef CAS.
- K. Holá, M. Sudolská, S. Kalytchuk, D. Nachtigallová, A. L. Rogach, M. Otyepka and R. Zbořil, ACS Nano, 2017, 11, 12402–12410 CrossRef PubMed.
- Y. Guo, A. Dong, Q. Huang, Q. Li, Y. Hu, J. Qian and S. Huang, J. Colloid Interface Sci., 2022, 606, 1833–1841 CrossRef CAS PubMed.
- M. Ikram, M. I. Khan, A. Raza, M. Imran, A. Ul-Hamid and S. Ali, Phys. E, 2020, 124, 114246 CrossRef CAS.
- U. Qumar, M. Ikram, M. Imran, A. Haider, A. Ul-Hamid, J. Haider, K. N. Riaz and S. Ali, Dalton Trans., 2020, 49, 5362–5377 RSC.
- A. Al-Abbad, M. Al-Jamal, Z. Al-Elaiw, F. Al-Shreed and H. Belaifa, J. Dev. Agric. Econ., 2011, 3, 463–468 Search PubMed.
- T. Kavitha and S. Kumar, Sci. Rep., 2018, 8, 1–10 CAS.
- J. Athinarayanan, V. S. Periasamy and A. A. Alshatwi, ACS Omega, 2022, 7, 19270–19279 CrossRef CAS PubMed.
- J. Athinarayanan, S. A. Almaiman, L. N. Al-Harbi, V. S. Periasamy and A. A. Alshatwi, J. King Saud Univ., Sci., 2021, 33, 101584 CrossRef.
- L. Ge, G. Hu, B. Shi, Q. Guo, L. Li, L. Zhao and J. Li, Appl. Phys. A: Mater. Sci. Process., 2021, 21, 647–650 Search PubMed.
- G. E. LeCroy, K. A. S. Fernando, C. E. Bunker, P. Wang, N. Tomlinson and Y.-P. Sun, Inorg. Chim. Acta, 2017, 468, 300–307 CrossRef CAS.
- F. Zu, F. Yan, Z. Bai, J. Xu, Y. Wang, Y. Huang and X. Zhou, Microchim. Acta, 2017, 184, 1899–1914 CrossRef CAS.
- F. Yan, Y. Jiang, X. Sun, Z. Bai, Y. Zhang and X. Zhou, Mikrochim. Acta, 2018, 185, 424 CrossRef PubMed.
- A. S. A. Salah, N. A. A. Salah and Y. N. A. Alshahrie, U. Pat. U, 2021, 10. 906, 812–813 Search PubMed.
- G. K. Yogesh, E. P. Shuaib, A. Kalai Priya, P. Rohini, S. V. Anandhan, U. M. Krishnan, V. Kalyanavalli, S. Shukla and D. Sastikumar, Opt. Laser Technol., 2021, 135, 106717 CrossRef CAS.
- A. Fujimoto, Y. Yamada, M. Koinuma and S. Sato, Anal. Chem., 2016, 88, 6110–6114 CrossRef CAS PubMed.
- N. Y. Z. P. Russo, A. Hu, G. Compagnini and W. W. Duley, Nanoscale, 2014, 6, 2381–2389 RSC.
- C. Zhu, J. Zhai and S. Dong, Chem. Commun., 2012, 48, 9367–9369 RSC.
- A. M. Alam, B. Y. Park, Z. K. Ghouri, M. Park and H. Y. Kim, Green Chem., 2015, 17, 3791–3797 RSC.
- B. Wang, A. Song, L. Feng, H. Ruan, H. Li, S. Dong and J. Hao, ACS Appl. Mater. Interfaces, 2015, 7, 6919–6925 CrossRef CAS PubMed.
- A. Ambrosi, C. K. Chua, A. Bonanni and M. Pumera, Chem. Rev., 2014, 114, 7150–7188 CrossRef CAS PubMed.
- Z. Qian, J. Ma, X. Shan, H. Feng, L. Shao and J. Chen, Chemistry, 2014, 20, 2254–2263 CrossRef CAS PubMed.
- K. Ganesan, S. Ghosh, N. Gopala Krishna, S. Ilango, M. Kamruddin and A. K. Tyagi, Phys. Chem. Chem. Phys., 2016, 18, 22160–22167 RSC.
- B. Lesiak, L. Kövér, J. Tóth, J. Zemek, P. Jiricek, A. Kromka and N. Rangam, Appl. Surf. Sci., 2018, 452, 223–231 CrossRef CAS.
- S. Li, Z. Guo, R. Feng, Y. Zhang, W. Xue and Z. Liu, RSC Adv., 2017, 7, 4975–4982 RSC.
- Ł. Janus, M. Piatkowski, J. Radwan-Pragłowska, D. Bogdał and D. Matysek, Nanomaterials, 2019, 9, 1–13 CrossRef PubMed.
- Y. Zhou, P. Y. Liyanage, D. L. Geleroff, Z. Peng, K. J. Mintz, S. D. Hettiarachchi, R. R. Pandey, C. C. Chusuei, P. L. Blackwelder and R. M. Leblanc, ChemPhysChem, 2018, 19, 2589–2597 CrossRef CAS PubMed.
- A. Dager, T. Uchida, T. Maekawa and M. Tachibana, Sci. Rep., 2019, 9, 1–10 CrossRef CAS PubMed.
- M. Behi, L. Gholami, S. Naficy, S. Palomba and F. Dehghani, Nanoscale Adv., 2022, 4, 353–376 RSC.
- P. A. Gerakines, W. A. Schutte, J. M. Greenberg and E. F. van Dishoeck, Astron. Astrophys., 1995, 296(43), 810–818 CAS.
- R. Gómez-Hernández, Y. Panecatl-Bernal and M. Á. Méndez-Rojas, Heliyon, 2019, 5, e02139 CrossRef PubMed.
- Y. V. Larichev, P. M. Yeletsky and V. A. Yakovlev, J. Phys. Chem. Solids, 2015, 87, 58–63 CrossRef CAS.
- W. Liu, H. Xu, H. Qin, Y. Lv, G. Zhu, F. Lin, X. Lei, Z. Zhang and L. Wang, Silicon, 2020, 12, 2259–2269 CrossRef CAS.
- K. Bagga, R. McCann, M. Wang, A. Stalcup, M. Vázquez and D. Brabazon, J. Colloid Interface Sci., 2015, 447, 263–268 CrossRef CAS PubMed.
- D. Qu, X. Wang, Y. Bao and Z. Sun, JPhys Mater., 2020, 3, 022003 CrossRef CAS.
- A. B. Siddique, A. K. Pramanick, S. Chatterjee and M. Ray, Sci. Rep., 2018, 8, 1–10 CAS.
- Z. Gan, H. Xu and Y. Hao, Nanoscale, 2016, 8, 7794–7807 RSC.
- J. Xiao, P. Liu, C. X. Wang and G. W. Yang, Prog. Mater. Sci., 2017, 87, 140–220 CrossRef CAS.
- M. Liu, Nanoarchitectonics, 2020, 1, 1–12 CrossRef.
- S. Sarkar, K. Das, M. Ghosh and P. K. Das, RSC Adv., 2015, 5, 65913–65921 RSC.
- Y. Dai, H. Long, X. Wang, Y. Wang, Q. Gu, W. Jiang, Y. Wang, C. Li, T. H. Zeng, Y. Sun and J. Zeng, Part. Part. Syst. Charact., 2014, 31, 509 CrossRef.
- Z. L. Wu, Z. X. Liu and Y. H. Yuan, J. Mater. Chem. B, 2017, 5, 3794–3809 RSC.
- P. Kumar, S. Dua, R. Kaur, M. Kumar and G. Bhatt, RSC Adv., 2022, 12, 4714–4759 RSC.
Footnote |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra04189h |
| This journal is © The Royal Society of Chemistry 2022 |