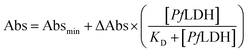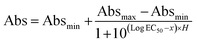Open Access Article
Open Access ArticleEnzyme-linked aptamer-based sandwich assay (ELASA) for detecting Plasmodium falciparum lactate dehydrogenase, a malarial biomarker
Yeon-Jun Kim a and
Jae-Won Choi
a and
Jae-Won Choi *ab
*ab
aDepartment of Biomedical Science, Cheongju University, Cheongju 28160, Republic of Korea. E-mail: jwchoi0211@gmail.com; jwchoi@cju.ac.kr
bDepartment of Bioindustrial Engineering, Cheongju 28503, Republic of Korea
First published on 14th October 2022
Abstract
Herein, we report a sensitive and selective enzyme-linked aptamer-based sandwich assay (ELASA) to detect Plasmodium falciparum lactate dehydrogenase (PfLDH), which is an attractive biomarker for malaria diagnosis and antimalarial medication. We performed the sandwich assay with a single aptamer sequence, called 2008s, owing to the structural properties of the PfLDH tetramer instead of using a conventional sandwich assay with two different aptamers (or antibodies) for capturing and probing a target molecule. First, the biotinylated PfLDH aptamer was linked with immobilized streptavidin on a microwell plate for binding flexibility, and then PfLDH was bound to the aptamer. Next, a horseradish peroxidase-conjugated aptamer of the same sequence was used to analyze PfLDH quantitatively. Using this approach, the limit of detection (LOD) of PfLDH with the naked eye was 100 ng mL−1, and the LOD and limit of quantification from the absorbance measurements were 34.9 ng mL−1 and 95.5 ng mL−1, respectively, based on PfLDH spiked blood samples. Our proposed method selectively binds PfLDH, not human lactate dehydrogenase. Therefore, this method may be a valuable tool for diagnosing, monitoring, and quarantining malaria cases easily and rapidly.
Introduction
Malaria is an infectious disease caused by parasitic protozoans of the Plasmodium species, often occurring in tropical or semitropical regions and commonly transmitted to humans by female mosquitos.1–3 Malaria symptoms appear within a few days of being bitten by a mosquito, including fever, fatigue, sweating, nausea, vomiting, diarrhea, and headache.3 Even though antimalarial medications can cure malaria, The World Malaria Report 2021, published by the World Health Organization, reported over 241 million malaria patients and approximately 627![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 malaria-related fatalities in 85 malaria-endemic countries.1 Therefore, quick diagnosis with an accurate and straightforward platform is essential for obtaining proper medical treatment.
000 malaria-related fatalities in 85 malaria-endemic countries.1 Therefore, quick diagnosis with an accurate and straightforward platform is essential for obtaining proper medical treatment.
Currently, a microscopic examination,4–6 polymerase chain reaction (PCR),7–9 and a rapid diagnostic test (RDT)10–12 are widely utilized for diagnosing malaria. A peripheral blood smear test with a microscopic examination is the most accurate method for confirming the malaria parasites.13 Although the technique is old, it remains the most widely used due to its high accuracy and positioning as a gold standard. However, it requires sophisticated equipment, experienced experts, and several days for high-accuracy analyses. Generally, a PCR assay is employed to detect parasitic DNA with high sensitivity, but its complexity and cost are barriers for general use. PCR also requires equipped laboratory facilities and has difficulties detecting cases of plasmodial genotypic variants.7 RDT diagnoses malaria quickly and easily, but its sensitivity and specificity are manufacturer-dependent, quantitative analysis is impossible, and early diagnosis is difficult.
Enzyme-linked immunosorbent assay (ELISA) is the most actively used format for target molecule detection in liquid or wet samples because of its robust and accurate properties.14–16 ELISAs use antibodies and colorimetric substrates to identify a specific antigen in a sample. Malaria diagnosis is based on various protein biomarkers (or antigens), such as plasmodial lactate dehydrogenase (LDH),10,11,16 histidine-rich protein II,12,17,18 aldolase,19 and glutamate dehydrogenase.20 Among these biomarkers, plasmodial LDH (pLDH) is an attractive target for malaria diagnosis and antimalarial drug design since it produces plasmodial ATP in human red blood cells and has unique amino acids at the active site and a well-understood three-dimensional structure.21–23 Some results have reported successful ELISA-based malaria diagnosis using antibodies against Plasmodium spp. LDH.16,24 However, the ELISA method is an antibody-based detection format; thus, monoclonal or polyclonal antibodies against a specific antigen are necessary. In addition, this format requires sacrificing laboratory animals, such as mice and rabbits, and takes several months to produce the same antibodies. Recently, mass antibody production has trended toward a cell culture system using a bioreactor, eliminating the need for animals. However, this is also not ideal since it requires many additional elements (e.g., medium, antibiotics, and serum), takes time, and can be costly.
An aptamer is an oligonucleic acid that can specifically bind from small molecules such as metal ions and chemicals to large molecules such as proteins. In recent, several aptamers capable of binding to pLDH have been developed by the systematic evolution of ligands by exponential enrichment (i.e., SELEX) for diagnosing malaria.21,25,26 Aptamers can be used to develop aptasensors for diagnosing malaria because they are easily synthesizable and have high specificity and thermal stability. Above all, unlike antibodies, aptamers can be mass-produced by chemical synthesis without animals or bioreactors.
Of the four malaria-causing Plasmodium spp. (P. falciparum, P. vivax, P. malariae, and P. ovale), P. falciparum triggers the version of malaria that is most often fatal and medically severe; this species causes most malarial deaths.1,3 Several aptamers specifically bind to P. falciparum lactate dehydrogenase (PfLDH), but the 2008s aptamer is used the most for detecting PfLDH because it has excellent and stable binding.21,27–29 Herein, we aimed to develop a simple yet effective aptamer-based PfLDH detection method and report an enzyme-linked aptamer-based sandwich assay (ELASA) for rapid and sensitive detection of PfLDH using the 2008s aptamer.
Experimental
Reagents and materials
Streptavidin from Streptomyces avidinii, horseradish peroxidase (HRP), lysozyme, imidazole, Tris powder, 1-fluoro-2,4-dinitrobenzene (FDNB), phenylmethylsulfonyl fluoride (PMSF), sodium bicarbonate, sodium carbonate, sodium phosphate, sodium periodate, and sodium borohydride were purchased from Sigma-Aldrich (St. Louis, MO, USA). Ampicillin and isopropyl β-D-1-thiogalactopyranoside (IPTG) were purchased from LPS Solution (Daejeon, Republic of Korea). All enzymes for plasmid construction were purchased from Enzynomics (Daejeon, Republic of Korea). Pyridoxal phosphate and phosphate buffered saline (pH 7.4) were obtained from AMRESCO (Solon, OH, USA), and ethylene glycol was purchased from Junsei Chemical (Tokyo, Japan). Bovine serum albumin (BSA) was purchased from Roche (Basel, Switzerland). The bicinchoninic acid (BCA) protein assay kit was purchased from Thermo Fisher Scientific (Waltham, MA, USA). The 3,3′,5,5′-tetramethylbenzidine (TMB) substrate was purchased from Kementec (Taastrup, Denmark). The single-stranded DNA aptamer that specifically binds to PfLDH called ‘2008s’ (5′-CTGGGCGGTAGAACCATAGTGACCCAGCCGTCTAC-3′) was synthesized by Integrated DNA Technologies (Singapore, Republic of Singapore), and biotin or HRP was conjugated at 3′-end of aptamer, respectively. Human LDH (hLDH) was purchased from Abcam (Cambridge, UK).PfLDH expression and purification
A recombinant PfLDH plasmid was constructed in pET-21a(+) and transformed into the BL21(DE3) strain of Escherichia coli. BL21(DE3) cells carrying pET-21a(+)-PfLDH plasmids were grown in lysogeny broth containing 50 μg mL−1 of ampicillin by an OD600 of 0.6. Recombinant PfLDH proteins were induced with 1 mM IPTG at 20 °C for 16 h. Cells were harvested by centrifugation and then lysed using lysis buffer (20 mM Tris–Cl [pH 7.9], 5 mM imidazole, 500 mM NaCl, 1 mM PMSF, and 10 mg g−1 [lysozyme/pellet]) by sonication. The lysates were centrifuged at 9000 rpm for 30 min at 4 °C. After centrifugation, the supernatant was loaded on a nickel–nitrilotriacetic acid (Ni–NTA) resin-packed column. The column was washed with washing buffer (20 mM Tris–Cl [pH 7.9], 40 mM imidazole, 500 mM NaCl, and 1 mM PMSF) and then eluted with elution buffer (20 mM Tris–Cl [pH 7.9], 500 mM imidazole, and 500 mM NaCl). The eluted PfLDH proteins were dialyzed into a dialysis buffer containing 20 mM sodium phosphate, 500 mM NaCl, and 0.5 mM pyridoxal phosphate for 16 h at 4 °C. Finally, the PfLDH concentration was confirmed by BCA protein assay kit.Polyacrylamide gel electrophoresis (PAGE) of PfLDH and hLDH
We performed PAGE to confirm the purification procedures of PfLDH using a 12% polyacrylamide gel under reduced conditions containing sodium dodecyl sulfate (SDS) and β-mercaptoethanol (β-ME). PAGE was performed in both reduced and native conditions to identify purified PfLDH and hLDH proteins. In the reduced condition, 4× reducing sample buffer containing 250 mM Tris–Cl (pH 6.8), 8% SDS, 40% glycerol, 8% β-ME, and 0.02% bromophenol blue was used. Then, SDS-PAGE was performed using a 15% polyacrylamide gel and 1× electrophoresis buffer containing 0.01% SDS. In the native condition, 4× native sample buffer containing 250 mM Tris–Cl (pH 6.8), 40% glycerol, and 0.02% bromophenol blue was used. Native PAGE was performed using a 10% polyacrylamide gel without SDS and 1× electrophoresis buffer without SDS.HRP conjugation with PfLDH
HRP (2.5 mg) was dissolved in 300 mM sodium carbonate buffer (pH 8.1). Then, FDNB was added to the HRP solution and incubated for 1 h at room temperature. Next, the solution was incubated with 80 mM of sodium periodate for 1 h at room temperature, and then 160 mM of ethylene glycol was added. After 1 h, the activated HRP solution was dialyzed into a 10 mM sodium bicarbonate buffer (pH 9.5) for 16 h at 4 °C. For PfLDH, buffer exchange was performed by extensively dialyzing against the 10 mM sodium bicarbonate buffer (pH 9.5). Finally, 1 mg of PfLDH was mixed with the HRP solution for 2 h at room temperature, and then this mixture was incubated with 5 mg mL−1 of sodium borohydride for 2 h at 4 °C. The HRP–PfLDH conjugates were purified via Ni–NTA resin and concentrated by a centrifugal filter unit from Merck Millipore (Billerica, MA, USA). The concentration of HRP–PfLDH conjugates was determined by a NanoDrop 2000c spectrophotometer (Thermo Fisher Scientific).Aptamer and PfLDH binding affinity measurements
A 1 μg mL−1 streptavidin solution was added to a 96-well microwell plate and incubated at 37 °C for 1 h. After streptavidin coating, all microwells were blocked with a blocking solution containing 5% BSA, 5% sucrose, and 0.05% Tween-20 in phosphate-buffered saline (PBS) at 37 °C for 1 h. Then, the blocking solution was removed from the microwells, and the plate was dried at 37 °C for 1 h. Next, 10 nM of the biotin-conjugated PfLDH aptamer was added to the streptavidin-coated microwell plate and incubated at 37 °C for 1 h. At this time, the aptamer was allowed to self-form a secondary structure through the cooling process at room temperature after denaturation at 90 °C for 30 seconds for structuration prior to being added into the microwell plate. Three different buffer conditions were used to determine the best structuration (buffer A: PBS, pH 7.4, buffer B: 25 mM Tris–Cl (pH 7.5), 100 mM NaCl, and 20 mM imidazole, and buffer C: 10 mM Tris–Cl (pH 7.4), 10 mM NaCl, and 0.2 mM MgCl2). After incubation, all microwells were washed three times using 0.05% Tween-20 containing PBS (0.05% PBS-T). Finally, different concentrations of HRP-conjugated PfLDH (serially diluted from 2 μM) were added to the microwells and incubated at 37 °C for 1 h. Finally, 100 μL of TMB substrate was added per microwell after washing for 15 min at room temperature to develop the colorimetric reaction. The reaction step was stopped via a 0.5 N H2SO4 solution, and the absorbance was measured at 450 nm using a Bio-Rad microplate reader (Hercules, CA, USA). The binding affinity between PfLDH and the aptamer was calculated from the extracted absorbances per PfLDH concentration. A microwell plate coated only with BSA without streptavidin was used as a negative control.Aptamer and streptavidin binding affinity measurements
A 96-well plate was coated, blocked, and dried as described above. Next, different concentrations of structured HRP-conjugated PfLDH aptamer (serially diluted from 1 μM) were added to the microwells and incubated at 37 °C for 1 h. After incubation, all microwells were washed three times using 0.05% PBS-T. The colorimetric reaction and spectrophotometric measurements were performed as described above.Multiple sequence alignment of PfLDH and hLDH amino acids
The homology of PfLDH and hLDH amino acids was analyzed using the ‘Align’ mode in UniProt (https://www.uniprot.org/align). The UniProt entry ID corresponding to this experiment's PfLDH amino acids sequence was Q76NM3 (entry name: Q76NM3_PLAF7, gene name: PF3D7_1324900), and the UniProt entry ID for hLDH was P00338 (entry name: LDHA_HUMAN, gene name: LDHA_PIG19).PfLDH and hLDH detection by ELASA
A 1 μM streptavidin solution was added to a 96-well microwell plate and incubated at 37 °C for 1 h. The 96-well microwell plate was blocked, and dried as described above. Then, 1 μM of structured biotin-conjugated PfLDH aptamer was added to the streptavidin-coated microwell plate and incubated at 37 °C for 1 h. After incubation, all microwells were washed three times using washing buffer. Next, purified PfLDH in buffer, hLDH in buffer, or spiked PfLDH in mouse blood of various concentrations (0, 10−4, 10−3, 10−2, 10−1, 100, 101, and 102 μg mL−1) were loaded into the wells then incubated at 37 °C for 1 h. Then, after washing, 1 μM of structured HRP-conjugated PfLDH aptamer was added to the microwell plate and incubated at 37 °C for 1 h. After washing, 100 μL of TMB substrate was added per microwell and then incubated at room temperature for 15 min. The reaction step was stopped using a 0.5 N H2SO4 solution, and the absorbance was measured at 450 nm using a microplate reader.Results and discussion
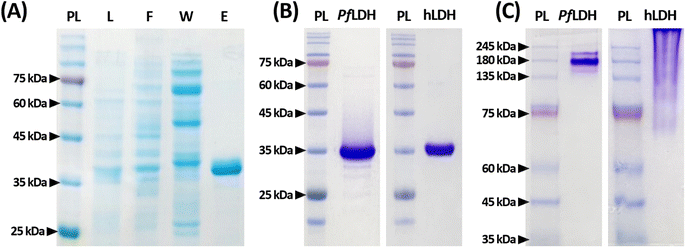
PfLDH and hLDH confirmation by PAGE
Before examining PfLDH and the 2008s aptamer binding, we confirmed that purified recombinant PfLDH was obtained by reduced and native PAGE analyses (Fig. 1); we found that PfLDH was selectively separated (Fig. 1(A)). PfLDH and hLDH were highly purified in the reduced condition, as indicated by a single band on 15% polyacrylamide gels at approximately 36 kDa (Fig. 1(B)). In the native condition on 10% polyacrylamide gels, we observed bands above 140 kDa for both proteins, while no bands were observed around 36 kDa (Fig. 1(C)).The PfLDH and hLDH molecular weights do not match in the native condition due to differences in their isoelectric points (pIs). The PfLDH and hLDH pIs are 7.26 and 8.64, respectively. In native gels, it is natural that hLDH migrates less than PfLDH in a resolving gel with a pH of 8.8. On the other hand, since the protein ladder molecules are stored in a buffer containing 20 mM Tris–phosphate (pH 7.5), 2% SDS, 3.6 M urea, 0.2 mM dithiothreitol, and 15% glycerol, they are located in appropriate molecular weight. These results suggest that the PfLDH and hLDH proteins used in this experiment were well-composed of tetramers.
Optimizing the reaction conditions and binding affinity measurements
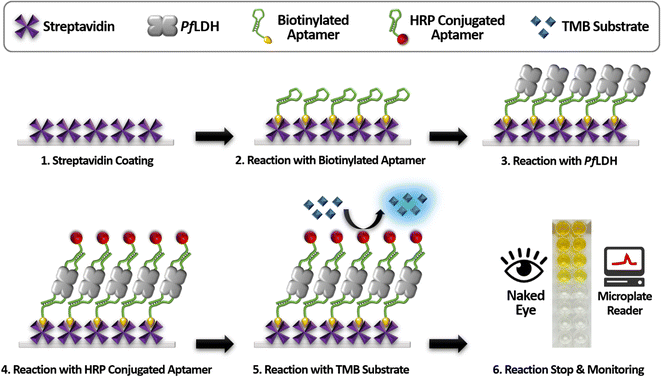
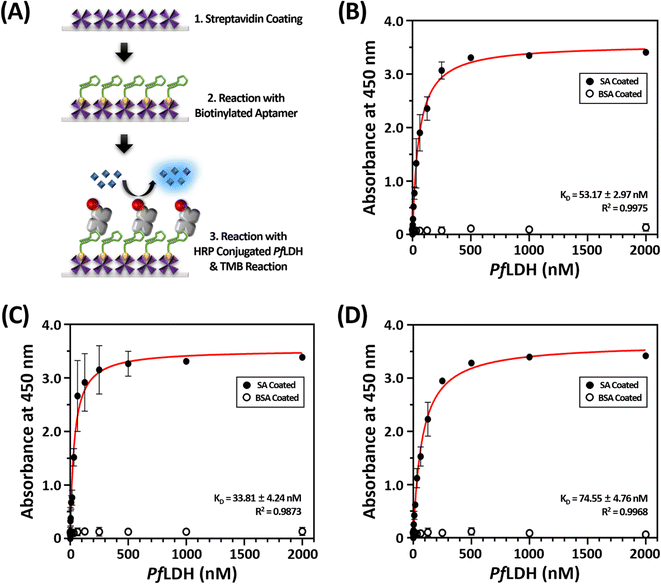
Scheme 1 illustrates the proposed ELASA procedure for detecting PfLDH. Prior to perform the ELASA, we examined the binding affinity between PfLDH and the 2008s aptamer. The 2008s aptamer against PfLDH was adopted by Cheung et al., who performed a structural study that defined the PfLDH binding site.21 Direct immobilization of the 2008s aptamer on a microwell plate may interrupt PfLDH binding. Therefore, biotin was conjugated at the 3′-end of the aptamer sequence for flexible binding with PfLDH. Also, the aptamer's primary binding sequence is at the 5′-end, and thus biotin was conjugated at the 3′-end.First, we coated the microwell plates with 1 μg mL−1 of streptavidin, then bound 10 nM of structured biotinylated PfLDH aptamer through biotin–streptavidin pairing (Fig. 2(A)). Next, the HRP-conjugated PfLDH was serially diluted to different concentrations (0 to 2 μM) and incubated with the coated the 2008s aptamer. Here, we used three different buffer conditions to determine the best reaction: buffer A: PBS, pH 7.4, buffer B: 25 mM Tris–Cl (pH 7.5), 100 mM NaCl, and 20 mM imidazole, and buffer C: 10 mM Tris–Cl (pH 7.4), 10 mM NaCl, and 0.2 mM MgCl2. Each microwell was treated with TMB substrate to trigger a color reaction based on the HRP-conjugated PfLDH remaining in the microwell, which was terminated with a 0.5 N H2SO4 solution. Finally, colorization was monitored by the naked eye, and the absorbance was measured at 450 nm using a microplate reader.
The dissociation constant (KD) was calculated by extracting the absorbance from each PfLDH concentration using the following equation:
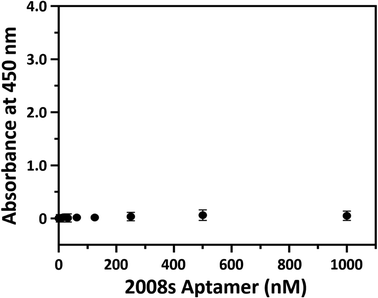
HRP-conjugated PfLDH binding did not occur in the BSA-coated microwells without streptavidin (white circles in Fig. 2(B–D)), indicating that the absorbance increase with increasing PfLDH concentrations resulted from specific binding between PfLDH and the 2008s aptamer linked to streptavidin in the wells. Also, we examined the reaction between the 2008s aptamer and streptavidin as a negative control, which confirmed that the absorbance did not increase with increasing HRP-conjugated 2008s aptamer concentrations in the streptavidin-immobilized microwells (Fig. 3). Therefore, PfLDH and the 2008s aptamer bind specifically in a microwell plate.
PfLDH aptamer sensitivity in the ELASA platform
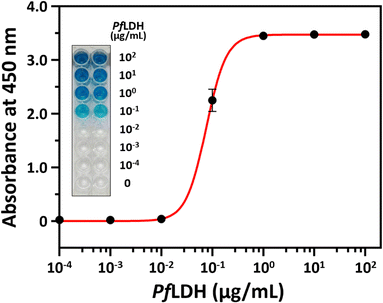
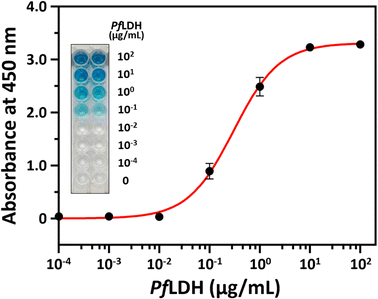
Next, we performed a PfLDH detection experiment using the ELASA method (Scheme 1). First, we saturated the microwells with 1 μM of streptavidin for efficient PfLDH molecule capture (i.e., the coating step). Then, 1 μM of biotinylated aptamer was bound with the coated streptavidin through biotin–streptavidin pairing. Next, 100 μg mL−1 of purified PfLDH was 10× serially diluted and then added to the wells to calculate the quantifiable range. Then, 1 μM of HRP-conjugated 2008s aptamer was bound with the captured PfLDH in the microwells and subsequently treated with TMB substrate to trigger colorization (terminated with 0.5 N H2SO4 solution). Finally, absorbance was measured at 450 nm using a microplate reader.We observed by the naked eye a significant color change at 100 ng mL−1 of PfLDH. Furthermore, we determined the limit of detection (LOD) and limit of quantification (LOQ) based on the absorbances (Fig. 4). In general, the LOD is an absorbance value that is 3× higher than that of the blank (negative control), and the LOQ is the absorbance value 10× higher than that of the blank. We calculated the LOD and LOQ in buffer as 21.3 ng mL−1 and 44.0 ng mL−1, respectively, using the following equation:
PfLDH aptamer selectivity in the ELASA platform
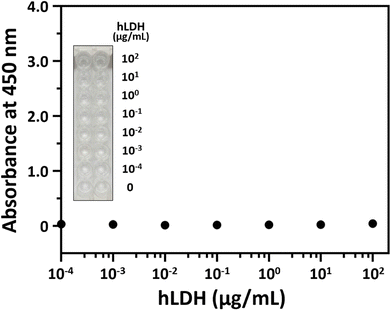
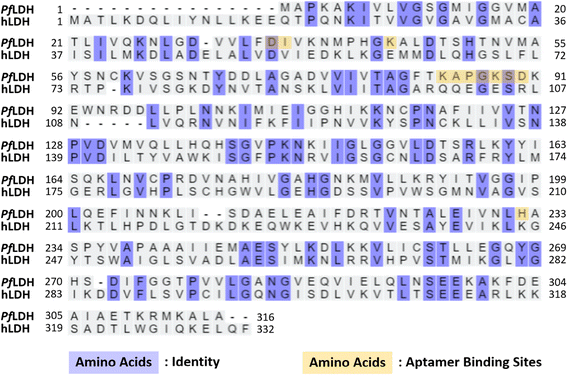
The hLDH detection experiment was performed to examine the selectivity of our ELASA method using the 2008s aptamer. All experimental conditions were adopted from the ELASA for PfLDH detection, except that hLDH was used instead of PfLDH. The hLDH was 10× serially diluted from 100 μg mL−1 and measured by ELASA to confirm whether non-specific binding between hLDH and the 2008s aptamer exists. hLDH did not bind the 2008s aptamer, evidenced by similar absorbances between the blank and the 100 μg mL−1 concentration (Fig. 5).Moreover, multiple sequence alignment was performed on PfLDH and hLDH amino acids using UniProt (Fig. 6). Of the 332 amino acid positions, 89 amino acids existed at the same positions in aligned sequence (purple color in Fig. 6), and the homology between PfLDH and hLDH was 26.8%. Furthermore, we marked the primary binding site of the 2008s aptamer in PfLDH based on the information based by Cheung et al.,21 which confirmed a difference in the amino acids sequence of PfLDH and hLDH (yellow color in Fig. 6). A molecule of the 2008s aptamer binds to two monomers of PfLDH that form a tetramer. Specifically, the 2008s aptamer binds to Asp35, Ile36, and Lys84 to Asp90 of PfLDH monomer, and binds to Lys44 and His232 of another PfLDH monomer. This difference contributes to the structural stability of the 2008s aptamer–PfLDH complex, which has a different charge and polarity than hLDH. Together, these results indicate that our ELASA selectively captures only PfLDH, and thus it can be distinguished from hLDH in P. falciparum-infected blood samples for malaria diagnosis.
A quantitative analysis of PfLDH in a blood sample using the ELASA platform
Finally, we performed an experiment to validate that PfLDH can be selectively detected in the blood with high sensitivity. First, we spiked mouse blood with PfLDH to final concentrations of 0, 10−4, 10−3, 10−2, 10−1, 100, 101, and 102 μg mL−1 of PfLDH. Then, performed the ELASA as previously described using the spiked samples.We detected 100 ng mL−1 of PfLDH by the naked eye, which aligned with the purified PfLDH sensitivity result. Furthermore, the LOD and LOQ in matrix were 34.9 ng mL−1 and 95.5 ng mL−1, respectively (Fig. 7). These results imply that PfLDH detection is possible in blood samples from patients. The mean pLDH concentration in the blood of malaria patients is 681.4 ng mL−1 (range, 219.1–1803.1 ng mL−1).30 Moreover, since expression of this biomarker is drastically increased in all asexual stages of erythrocytic cycle of P. falciparum parasite, it is attractive for the diagnosis of malaria.31,32 Therefore, our ELASA method has sufficient sensitivity and selectivity to diagnose malaria from clinical samples with the naked eye or by a spectrophotometer.
Conclusions
We developed a novel ELASA platform, similar to the sandwich ELISA method, using a single-stranded DNA aptamer (called 2008s) that selectively binds PfLDH (a malarial biomarker). Therefore, it enables robust, simple, and rapid PfLDH detection. Also, our method, using aptamer instead of antibodies, does not require sacrificing animals, saving time and costs since monoclonal and polyclonal antibody preparations are not required. Additionally, chemical synthesis makes mass production of aptamers possible, and key raw materials for production are readily available, unlike antibodies that require laboratory animals and/or bioreactors.Above all, our ELASA is distinct because this sandwich assay uses a single aptamer sequence that can bind two sites owing to the tetrameric characteristics of PfLDH. Therefore, less time is required to screen capture and probe molecule pairs using the ELASA method than a conventional sandwich assay. Moreover, using our method, 100 ng mL−1 of PfLDH can be detected with the naked eye or by a spectrophotometer (LOD: 34.9 ng mL−1; LOQ: 95.5 ng mL−1 based on absorbance measurement), as indicated by PfLDH spiked blood samples. Finally, our ELASA method was selective for PfLDH and thus suitable for malaria diagnosis.
During the coronavirus disease 2019 crisis, eradicating malaria has been especially difficult. However, the ELASA platform features make it an attractive option for clinical application. Using this ELASA platform may improve P. falciparum infection diagnosis and could be extended to other malaria parasites.
Author contributions
Conceptualization, J.-W. C.; data curation, Y.-J. K.; methodology, Y.-J. K. and J.-W. C.; resources, Y.-J. K. and J.-W. C.; formal analysis, J.-W. C.; visualization, Y.-J. K. and J.-W. C.; investigation, J.-W. C.; supervision, J.-W. C.; writing-original draft, Y.-J. K. and J.-W. C.; writing-review and editing, Y.-J. K. and J.-W. C.; funding acquisition, J.-W. C. Both authors have read and agreed to the published version of the manuscript.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by Regional Innovation Strategy (RIS) and Basic Research Program through the National Research Foundation (NRF) of Korea funded by the Ministry of Education (MOE) (No. 2021RIS-001) and Ministry of Science and ICT (MSIT) (No. 2021R1F1A1062994), respectively. This work was also supported by Bio-Convergence Technology Education Program through the Korea Institute for Advancement of Technology (KIAT) funded by the Ministry of Trade, Industry and Energy (MOTIE) (No. P0017805).Notes and references
- World Health Organization (WHO), World Malaria Report 2021, Geneva, 2021 Search PubMed.
- L. Neves Borgheti-Cardoso, M. San Anselmo, E. Lantero, A. Lancelot, J. L. Serrano, S. Hernández-Ainsa, X. Fernàndez-Busquets and T. Sierra, J. Mater. Chem. B, 2020, 8, 9428–9448 RSC.
- M. A. Phillips, J. N. Burrows, C. Manyando, R. H. van Huijsduijnen, W. C. Van Voorhis and T. N. C. Wells, Nat. Rev. Dis. Primers, 2017, 3, 17050 CrossRef PubMed.
- W. M. Fong Amaris, C. Martinez, L. J. Cortés-Cortés and D. R. Suárez, Malar. J., 2022, 21, 74 CrossRef CAS PubMed.
- P. U. Eze and C. O. Asogwa, Bioengineering, 2021, 8, 150 CrossRef CAS PubMed.
- G. S. Alemayehu, K. Blackburn, K. Lopez, C. Cambel Dieng, E. Lo, D. Janies and L. Golassa, Malar. J., 2021, 20, 109 CrossRef CAS PubMed.
- T. Hänscheid and M. P. Grobusch, Trends Parasitol., 2002, 18, 395–398 CrossRef.
- K. Kassegne, S. W. Fei, K. Ananou, K. S. Noussougnon, K. Komi Koukoura, E. M. Abe, X. K. Guo, J. H. Chen and X. N. Zhou, Front. Microbiol., 2021, 12, 732923 CrossRef PubMed.
- K. van Bergen, T. Stuitje, R. Akkers, E. Vermeer, R. Castel and T. Mank, Malar. J., 2021, 20, 314 CrossRef CAS PubMed.
- C. Morang'a, C. Ayieko, G. Awinda, R. Achilla, C. Moseti, B. Ogutu, J. Waitumbi and E. Wanja, Malar. J., 2018, 17, 10 CrossRef PubMed.
- M. L. Gatton, A. Chaudhry, J. Glenn, S. Wilson, Y. Ah, A. Kong, R. L. Ord, R. R. Rees-Channer, P. Chiodini, S. Incardona, Q. Cheng, M. Aidoo and J. Cunningham, Malar. J., 2020, 19, 392 CrossRef CAS PubMed.
- A. Badiane, J. Thwing, J. Williamson, E. Rogier, M. A. Diallo and D. Ndiaye, Int. J. Infect. Dis., 2022, 121, 92–97 CrossRef CAS PubMed.
- A. Calderaro, S. Montecchini, M. Buttrini, G. Piccolo, S. Rossi, M. C. Arcangeletti, B. Farina, F. De Conto and C. Chezzi, Microorganisms, 2021, 9, 2265 CrossRef CAS PubMed.
- P. E. Brasil, L. De Castro, A. M. Hasslocher-Moreno, L. H. Sangenis and J. U. Braga, BMC Infect. Dis., 2010, 10, 337 CrossRef PubMed.
- R. M. Lequin, Clin. Chem., 2005, 51, 2415–2418 CrossRef CAS PubMed.
- P. S. Atchade, C. Doderer-Lang, N. Chabi, S. Perrotey, T. Abdelrahman, C. D. Akpovi, L. Anani, A. Bigot, A. Sanni and E. Candolfi, Malar. J., 2013, 12, 279 CrossRef CAS PubMed.
- I. M. Ali, A. M. Nji, J. C. Bonkum, M. N. Moyeh, G. K. Carole, A. Efon, S. Dabou, V. P. K. Tchuenkam, C. Tah, J. C. Kengne, D. F. Achu, J. D. Bigoga and W. F. Mbacham, Diagnostics, 2021, 11, 1556 CrossRef CAS PubMed.
- Y. Lo, Y. W. Cheung, L. Wang, M. Lee, G. Figueroa-Miranda, S. Liang, D. Mayer and J. A. Tanner, Biosens. Bioelectron., 2021, 192, 113472 CrossRef CAS PubMed.
- N. S. Gopal and R. Raychaudhuri, Malar. Res. Treat., 2017, 2017, 9062514 Search PubMed.
- L. D. Kori, N. Valecha and A. R. Anvikar, Sci. Rep., 2020, 10, 6307 CrossRef CAS.
- Y. W. Cheung, J. Kwok, A. W. Law, R. M. Watt, M. Kotaka and J. A. Tanner, Proc. Natl. Acad. Sci. U. S. A., 2013, 110, 15967–15972 CrossRef CAS.
- J. D. Wirth, J. I. Boucher, J. R. Jacobowitz, S. Classen and D. L. Theobald, Biochemistry, 2018, 57, 6434–6442 CrossRef CAS PubMed.
- S. Khrapunov, A. Waterman, R. Persaud and E. P. Chang, Biochemistry, 2021, 60, 3582–3595 CrossRef CAS PubMed.
- L. P. Sousa, L. A. Mariuba, R. J. Holanda, J. P. Pimentel, M. E. Almeida, Y. O. Chaves, D. Borges, E. Lima, J. L. Crainey, P. P. Orlandi, M. V. Lacerda and P. A. Nogueira, BMC Infect. Dis., 2014, 14, 49 CrossRef PubMed.
- S. Lee, K. M. Song, W. Jeon, H. Jo, Y. B. Shim and C. Ban, Biosens. Bioelectron., 2012, 35, 291–296 CrossRef CAS.
- K. A. Frith, R. Fogel, J. P. D. Goldring, R. G. E. Krause, M. Khati, H. Hoppe, M. E. Cromhout, M. Jiwaji and J. L. Limson, Malar. J., 2018, 17, 191 CrossRef PubMed.
- R. M. Dirkzwager, A. B. Kinghorn, J. S. Richards and J. A. Tanner, Chem. Commun., 2015, 51, 4697–4700 RSC.
- Y. W. Cheung, R. M. Dirkzwager, W. C. Wong, J. Cardoso, J. D'Arc Neves Costa and J. A. Tanner, Biochimie, 2018, 145, 131–136 CrossRef CAS.
- G. Figueroa-Miranda, Y. Liang, M. Suranglikar, M. Stadler, N. Samane, M. Tintelott, Y. Lo, J. A. Tanner, X. T. Vu, J. Knoch, S. Ingebrandt, A. Offenhäusser, V. Pachauri and D. Mayer, Biosens. Bioelectron., 2022, 208, 114219 CrossRef CAS PubMed.
- M. M. Plucinski, P. D. McElroy, P. R. Dimbu, F. Fortes, D. Nace, E. S. Halsey and E. Rogier, Parasites Vectors, 2019, 12, 293 CrossRef.
- D. J. Bzik, B. A. Fox and K. Gonyer, Mol. Biochem. Parasitol., 1993, 59, 155–166 CrossRef CAS.
- L. Vivas, A. Easton, H. Kendrick, A. Cameron, J. L. Lavandera, D. Barros, F. G. de las Heras, R. L. Brady and S. L. Croft, Exp. Parasitol., 2005, 111, 105–114 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |