Open Access Article
Open Access ArticleDesign, synthesis, bio-evaluation, and in silico studies of some N-substituted 6-(chloro/nitro)-1H-benzimidazole derivatives as antimicrobial and anticancer agents†
Em Canh Pham *a,
Tuong Vi Thi Leb and
Tuyen Ngoc Truong
*a,
Tuong Vi Thi Leb and
Tuyen Ngoc Truong *c
*c
aDepartment of Medicinal Chemistry, Faculty of Pharmacy, Hong Bang International University, 700000 Ho Chi Minh City, Vietnam. E-mail: canhem112009@gmail.com; empc@hiu.vn
bDepartment of Pharmacology – Clinical Pharmacy, Faculty of Pharmacy, City Children's Hospital, 700000 Ho Chi Minh City, Vietnam
cDepartment of Organic Chemistry, Faculty of Pharmacy, University of Medicine and Pharmacy at Ho Chi Minh City, 700000 Ho Chi Minh City, Vietnam. E-mail: truongtuyen@ump.edu.vn
First published on 3rd August 2022
Abstract
A new series of 6-substituted 1H-benzimidazole derivatives were synthesized by reacting various substituted aromatic aldehydes with 4-nitro-o-phenylenediamine and 4-chloro-o-phenylenediamine through condensation using sodium metabisulfite as the oxidative reagent. The N-substituted 6-(chloro/nitro)-1H-benzimidazole derivatives were prepared from the 6-substituted 1H-benzimidazole derivatives and substituted halides using potassium carbonate by conventional methods as well as by exposure to microwave irradiation. Seventy-six 1H-benzimidazole derivatives have been synthesized in moderate to excellent yields with the microwave-assisted method (40 to 99%). Compounds 1d, 2d, 3s, 4b, and 4k showed potent antibacterial activity against Escherichia coli, Streptococcus faecalis, MSSA (methicillin-susceptible strains of Staphylococcus aureus), and MRSA (methicillin-resistant strains of Staphylococcus aureus) with MIC (the minimum inhibitory concentration) ranging between 2 and 16 μg mL−1 as compared to ciprofloxacin (MIC = 8–16 μg mL−1), in particular compound 4k exhibits potent fungal activity against Candida albicans and Aspergillus niger with MIC ranging between 8 and 16 μg mL−1 compared with the standard drug fluconazole (MIC = 4–128 μg mL−1). In addition, compounds 1d, 2d, 3s, 4b, and 4k also showed the strongest anticancer activity among the synthesized compounds against five tested cell lines with IC50 (half-maximal inhibitory concentration) ranging between 1.84 and 10.28 μg mL−1, comparable to paclitaxel (IC50 = 1.38–6.13 μM). Furthermore, the five most active compounds showed a good ADMET (absorption, distribution, metabolism, excretion, and toxicity) profile in comparison to ciprofloxacin, fluconazole, and paclitaxel as reference drugs. Molecular docking predicted that dihydrofolate reductase protein from Staphylococcus aureus is the most suitable target for both antimicrobial and anticancer activities, and vascular endothelial growth factor receptor 2 and histone deacetylase 6 are the most suitable targets for anticancer activity of these potent compounds.
1. Introduction
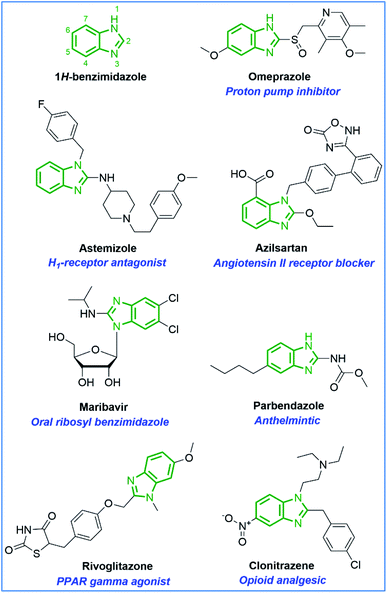
Benzimidazole is a naturally occurring bicyclic compound consisting of fused benzene and imidazole rings and is an integral part of the structure of vitamin B12. It is a cyclic ring that has two nitrogens as heteroatoms and is called a heterocyclic aromatic compound. In addition, benzimidazole is a remarkable scaffold of medicinal importance possessing promising pharmacological activities like anticancer,1–4 antimicrobial,5–7 anti-inflammation,8 antiviral,9 anti-hypertensive,10 antihistamine,11 antitubercular,12 antiulcer,13 analgesic,14 anthelmintic,15 antiprotozoal,16 antiamoebic,17 anticonvulsant,18 and antiparasitic.19Moreover, many important drugs used therapeutically in the research area contain a benzimidazole ring such as antiulcer (omeprazole, lansoprazole, rabeprazole, pantoprazole), antihistamines (astemizole, clemizole, and emedastine), antihypertensives (telmisartan, candesartan, and azilsartan), anthelmintics (thiabendazole, parbendazole, mebendazole, albendazole, cambendazole, and flubendazole), antiviral (maribavir), antidiabetic (rivoglitazone), analgesic (clonitazene), especially antifungal (systemic fungicide, e.g. benomyl) and anticancer (antimitotic agent, e.g. nocodazole, PAR inhibitor, e.g. veliparib) (Fig. 1).20 In addition, the potency of drugs like carbendazim, and dovitinib containing benzimidazole moiety has been recognized against various types of cancer cell lines.21,22
There are many different synthetic pathways to build the 1H-benzimidazole structures with different substituents at positions C-2 and C-5/6. However, the simplest synthesis pathway is the condensation of o-phenylenediamines and carboxylic acids (or their derivatives such as nitriles, chlorides, and orthoesters) in the presence of an acid or aldehydes using sodium metabisulfite (Na2S2O5).3,4 In addition, the N-1 derivatives were synthesized using 1H-benzimidazole derivatives and substituted halides in the presence of a base.23 The highlight of our study is the application of microwaves in the whole synthesis process of 1H-benzimidazole derivatives. This is a green chemical method that contributes to environmental protection.
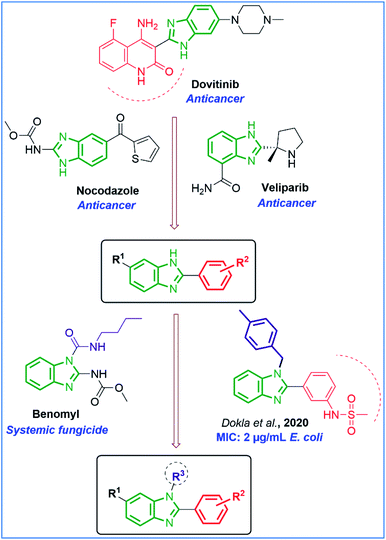
Rationale and structure-based design as antimicrobial and anticancer agents: Structure–activity relationship studies of benzimidazole ring system suggested the N-1, C-2, C-6 positions are very much important for the pharmacological effect.24,25 Especially, the N-1 position can increase chemotherapeutic activity when attached to different substituents, for example, benzyl groups similar to clemizole and candesartan drugs. Since N-substitutions in benzimidazole exhibit biologically active compounds,23,26 we were interested in designing compounds containing them (Fig. 2). Our designed derivatives and anticancer drug dovitinib, antifungal drug benomyl, and antibacterial derivatives of Dokla et al., 2020 (minimal inhibitory concentration (MIC) on E. Coli strain at 2 μg mL−1)27 share three common essential structural features (i) a planar benzimidazole moiety. (ii) Aromatic ring with different substituted groups at the C-2 position. (iii) The different substituted groups at the N-1 position. Moreover, the C-6 position with different substituents such as –Cl and –NO2 were designed in order to examine their effects on antimicrobial and anticancer activities.
The mechanism of action of one pharmacological activity is expressed through one or more different receptors.28,29 Furthermore, a receptor may also exhibit more than one pharmacological activity. A good example is dihydrofolate reductase (DHFR) which is a potential receptor for both antitumor and antimicrobial activities.20,30 Therefore, the in silico studies were the potential approach to confirm the ligand–target interaction in many different receptors. In recent years there has been significant progress to improve the receptor flexibility in docking,31–33 in silico studies are able to rank the compound potency or precisely predict the target after having experimental in vitro results.
Amoxicillin, norfloxacin, and ciprofloxacin are the most commonly used antibacterial drugs, as well as docetaxel, cyclophosphamide, 5-fluorouracil, and epirubicin are the most commonly used anticancer drugs but are related to severe side effects. Besides, the continued increase in the number of infections caused by bacteria resistant to one or multiple antibiotic classes and cancer resistance is a significant threat and can lead to treatment failure and complications. This has resulted in research and development in search of new antibiotics and anticancer drugs to maintain an effective drug supply at all times.34 It is important to find out newer, safer, and more effective antibiotics and anticancer drugs with multiple effects, especially showing both good anticancer and anti-microbial activities. This is very beneficial for cancer patients due to their weakened immunity and susceptibility to microbial attack.
Therefore, the purpose of this study is to synthesize novel N-substituted 6-(chloro/nitro)-1H-benzimidazole derivatives with various substituents at positions N-1, C-2, and C-6, and evaluation of their antibacterial, antifungal, and anticancer activities. The synthesized derivatives will be investigated in silico to understand the potential for drug–receptor interaction.
2. Results and discussion
2.1. Chemistry
The benzene-1,2-diamine derivatives with a 4-Cl or 4-NO2 group are the starting material for the preparation of N-substituted 6-(chloro/nitro)-1H-benzimidazole derivatives. The process of synthetic research consists of two steps (Scheme 1). Firstly, a series of 6-substituted 1H-benzimidazole derivatives (1a–1q and 2a–2q) have been synthesized by condensing benzene-1,2-diamine derivatives with substituted aromatic aldehydes using conventional heating and microwave-assisted methods. Thirty-four derivatives have been synthesized in good to excellent yields with the reflux method (70 to 91%) and excellent yields with the microwave-assisted method (90 to 99%). The reaction time has been dramatically reduced, as using conventional heating the reaction is carried out in 6–12 h compared with 10–15 min heating in the microwave. In addition, the reaction yield has increased ranging between 7 to 22% with microwave assistance (Table 1). Secondly, a series of N-substituted 6-(chloro/nitro)-1H-benzimidazole derivatives (3a–3x and 4a–4r) have been synthesized by reacting 6-substituted 1H-benzimidazole derivatives with substituted halides using conventional heating and microwave-assisted methods. Compounds 3a–3x and 4a–4r showed nearly identical reaction yields. Forty-two derivatives have been synthesized in moderate yields with the reflux method (26 to 43%) and moderate to excellent yields with the microwave-assisted method (40 to 50%). The reaction time also has been dramatically reduced, as using conventional heating the reaction is carried out in 12–24 h compared with 20–60 min heating in the microwave. Furthermore, the reaction yield has increased ranging between 7 to 15% with microwave assistance (Table 2). All compounds have physical–chemical properties of fragments (Mw < 500) or lead-like (Mw < 350) that follow Lipinski's rules which could lead to potent compounds for further development.35,36 Especially, twenty-nine derivatives (3 and 4) are new compounds.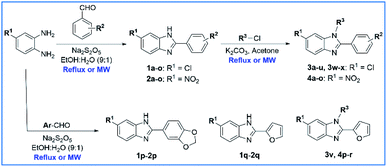 | ||
| Scheme 1 Construction of N-substituted 6-(chloro/nitro)-1H-benzimidazole derivatives (MW: microwave irradiation, EtOH: ethanol). | ||
| Entry | R group | Code | Physicochemical parameters | Yield | |||
|---|---|---|---|---|---|---|---|
| R1 | R2 | Re | MW | ||||
| a Re and MW – yields of conventional heating (or reflux) and microwave-assisted method (%), Re – reflux, MW – microwave, Mw – molecular weight, NHA – number of hydrogen bond acceptor, NHD – number of hydrogen bond donor, NRB – number rotatable bond, PSA – polar surface area (Angstroms squared). | |||||||
| 1 | 6-Cl | 2-Cl | 1a | Mw: 263.12 | NRB: 1 | 87 | 96 |
| NHA: 1 | LogP: 3.96 | ||||||
| NHD: 1 | TPSA: 28.68 | ||||||
| 2 | 6-Cl | 4-Cl | 1b | Mw: 263.12 | NRB: 1 | 91 | 99 |
| NHA: 1 | LogP: 4.01 | ||||||
| NHD: 1 | TPSA: 28.68 | ||||||
| 3 | 6-Cl | 2,4-Cl2 | 1c | Mw: 297.57 | NRB: 1 | 77 | 90 |
| NHA: 1 | LogP: 4.46 | ||||||
| NHD: 1 | TPSA: 28.68 | ||||||
| 4 | 6-Cl | 3,4-Cl2 | 1d | Mw: 297.57 | NRB: 1 | 81 | 91 |
| NHA: 1 | LogP: 4.51 | ||||||
| NHD: 1 | TPSA: 28.68 | ||||||
| 5 | 6-Cl | 2-Cl, 6-F | 1e | Mw: 281.11 | NRB: 1 | 78 | 92 |
| NHA: 2 | LogP: 4.33 | ||||||
| NHD: 1 | TPSA: 28.68 | ||||||
| 6 | 6-Cl | 3,4-(OCH3)2 | 1f | Mw: 288.73 | NRB: 3 | 75 | 90 |
| NHA: 3 | LogP: 3.40 | ||||||
| NHD: 1 | TPSA: 47.14 | ||||||
| 7 | 6-Cl | 4-OC2H5 | 1g | Mw: 272.73 | NRB: 3 | 76 | 90 |
| NHA: 2 | LogP: 3.80 | ||||||
| NHD: 1 | TPSA: 37.91 | ||||||
| 8 | 6-Cl | 3-OC2H5, 4-OH | 1h | Mw: 288.73 | NRB: 3 | 79 | 93 |
| NHA: 3 | LogP: 3.40 | ||||||
| NHD: 2 | TPSA: 58.14 | ||||||
| 9 | 6-Cl | 4-F | 1i | Mw: 246.67 | NRB: 1 | 82 | 94 |
| NHA: 2 | LogP: 3.79 | ||||||
| NHD: 1 | TPSA: 28.68 | ||||||
| 10 | 6-Cl | 3-OH | 1j | Mw: 244.68 | NRB: 1 | 76 | 95 |
| NHA: 2 | LogP: 3.07 | ||||||
| NHD: 2 | TPSA: 48.91 | ||||||
| 11 | 6-Cl | 3-OCH3 | 1k | Mw: 258.70 | NRB: 2 | 75 | 90 |
| NHA: 2 | LogP: 3.47 | ||||||
| NHD: 1 | TPSA: 37.91 | ||||||
| 12 | 6-Cl | 3-OH, 4-OCH3 | 1l | Mw: 274.70 | NRB: 2 | 90 | 98 |
| NHA: 3 | LogP: 3.04 | ||||||
| NHD: 2 | TPSA: 58.14 | ||||||
| 13 | 6-Cl | 3-NO2 | 1m | Mw: 273.67 | NRB: 2 | 81 | 97 |
| NHA: 3 | LogP: 2.86 | ||||||
| NHD: 1 | TPSA: 74.50 | ||||||
| 14 | 6-Cl | 4-NO2 | 1n | Mw: 273.67 | NRB: 2 | 79 | 96 |
| NHA: 3 | LogP: 2.86 | ||||||
| NHD: 1 | TPSA: 74.50 | ||||||
| 15 | 6-Cl | 4-N(CH3)2 | 1o | Mw: 271.74 | NRB: 2 | 81 | 93 |
| NHA: 1 | LogP: 3.47 | ||||||
| NHD: 1 | TPSA: 31.92 | ||||||
| 16 | 6-Cl | 1p | Mw: 272.69 | NRB: 1 | 80 | 90 | |
| NHA: 3 | LogP: 3.29 | ||||||
| NHD: 1 | TPSA: 47.14 | ||||||
| 17 | 6-Cl | 1q | Mw: 218.64 | NRB: 1 | 83 | 91 | |
| NHA: 2 | LogP: 2.82 | ||||||
| NHD: 1 | TPSA: 41.82 | ||||||
| 18 | 6-NO2 | 2-Cl | 2a | Mw: 273.67 | NRB: 2 | 84 | 93 |
| NHA: 3 | LogP: 2.90 | ||||||
| NHD: 1 | TPSA: 74.50 | ||||||
| 19 | 6-NO2 | 4-Cl | 2b | Mw: 273.67 | NRB: 2 | 88 | 95 |
| NHA: 3 | LogP: 2.94 | ||||||
| NHD: 1 | TPSA: 74.50 | ||||||
| 20 | 6-NO2 | 2,4-Cl2 | 2c | Mw: 308.12 | NRB: 2 | 82 | 91 |
| NHA: 3 | LogP: 3.32 | ||||||
| NHD: 1 | TPSA: 74.50 | ||||||
| 21 | 6-NO2 | 3,4-Cl2 | 2d | Mw: 308.12 | NRB: 2 | 85 | 96 |
| NHA: 3 | LogP: 3.46 | ||||||
| NHD: 1 | TPSA: 74.50 | ||||||
| 22 | 6-NO2 | 2-Cl, 6-F | 2e | Mw: 291.66 | NRB: 2 | 81 | 90 |
| NHA: 4 | LogP: 3.23 | ||||||
| NHD: 1 | TPSA: 74.50 | ||||||
| 23 | 6-NO2 | 3,4-(OCH3)2 | 2f | Mw: 299.28 | NRB: 4 | 82 | 97 |
| NHA: 5 | LogP: 2.26 | ||||||
| NHD: 1 | TPSA: 92.96 | ||||||
| 24 | 6-NO2 | 4-OC2H5 | 2g | Mw: 283.28 | NRB: 4 | 74 | 90 |
| NHA: 4 | LogP: 2.71 | ||||||
| NHD: 1 | TPSA: 83.73 | ||||||
| 25 | 6-NO2 | 3-OC2H5, 4-OH | 2h | Mw: 299.28 | NRB: 4 | 70 | 92 |
| NHA: 5 | LogP: 2.09 | ||||||
| NHD: 2 | TPSA: 103.96 | ||||||
| 26 | 6-NO2 | 4-F | 2i | Mw: 257.22 | NRB: 2 | 83 | 93 |
| NHA: 4 | LogP: 2.72 | ||||||
| NHD: 1 | TPSA: 74.50 | ||||||
| 27 | 6-NO2 | 3-OH | 2j | Mw: 255.23 | NRB: 2 | 76 | 94 |
| NHA: 4 | LogP: 1.82 | ||||||
| NHD: 2 | TPSA: 94.73 | ||||||
| 28 | 6-NO2 | 3-OCH3 | 2k | Mw: 269.26 | NRB: 3 | 75 | 92 |
| NHA: 4 | LogP: 2.37 | ||||||
| NHD: 1 | TPSA: 83.73 | ||||||
| 29 | 6-NO2 | 3-OH, 4-OCH3 | 2l | Mw: 285.25 | NRB: 3 | 78 | 90 |
| NHA: 5 | LogP: 1.74 | ||||||
| NHD: 2 | TPSA: 103.96 | ||||||
| 30 | 6-NO2 | 3-NO2 | 2m | Mw: 284.23 | NRB: 3 | 83 | 95 |
| NHA: 5 | LogP: 1.64 | ||||||
| NHD: 1 | TPSA: 120.32 | ||||||
| 31 | 6-NO2 | 4-NO2 | 2n | Mw: 284.23 | NRB: 3 | 86 | 95 |
| NHA: 5 | LogP: 1.65 | ||||||
| NHD: 1 | TPSA: 120.32 | ||||||
| 32 | 6-NO2 | 4-N(CH3)2 | 2o | Mw: 282.30 | NRB: 3 | 74 | 91 |
| NHA: 3 | LogP: 2.41 | ||||||
| NHD: 1 | TPSA: 77.74 | ||||||
| 33 | 6-NO2 | 2p | Mw: 283.24 | NRB: 2 | 77 | 92 | |
| NHA: 5 | LogP: 2.17 | ||||||
| NHD: 1 | TPSA: 92.96 | ||||||
| 34 | 6-NO2 | 2q | Mw: 229.19 | NRB: 2 | 80 | 91 | |
| NHA: 4 | LogP: 1.77 | ||||||
| NHD: 1 | TPSA: 87.64 | ||||||
| Entry | R group | Code | Physicochemical parameters | Yield | ||||
|---|---|---|---|---|---|---|---|---|
| R1 | R2 | R3 | Re | MW | ||||
| a (#) – 1-(2-ethoxy-2-oxoethyl) (–CH2COOC2H5), Re and MW – yields of conventional heating (or reflux) and microwave-assisted method (%), Re – reflux, MW – microwave, Mw – molecular weight, NHA – number of hydrogen bond acceptor, NHD – number of hydrogen bond donor, NRB – number rotatable bond, PSA – polar surface area (Angstroms squared). | ||||||||
| 1 | 6-Cl | 2-Cl | Allyl | 3a | Mw: 303.19 | NRB: 3 | 31 | 43 |
| NHA: 1 | LogP: 4.58 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 2 | 6-Cl | 4-Cl | Allyl | 3b | Mw: 303.19 | NRB: 3 | 35 | 46 |
| NHA: 1 | LogP: 4.58 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 3 | 6-Cl | 2,4-Cl2 | Allyl | 3c | Mw: 337.63 | NRB: 3 | 30 | 41 |
| NHA: 1 | LogP: 5.11 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 4 | 6-Cl | 3,4-Cl2 | Allyl | 3d | Mw: 337.63 | NRB: 3 | 36 | 45 |
| NHA: 1 | LogP: 5.11 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 5 | 6-Cl | 3,4-(OCH3)2 | Allyl | 3e | Mw: 328.79 | NRB: 5 | 27 | 40 |
| NHA: 3 | LogP: 4.01 | |||||||
| NHD: 0 | TPSA: 36.28 | |||||||
| 6 | 6-Cl | 4-OC2H5 | Allyl | 3f | Mw: 312.79 | NRB: 5 | 29 | 42 |
| NHA: 2 | LogP: 4.37 | |||||||
| NHD: 0 | TPSA: 27.05 | |||||||
| 7 | 6-Cl | 4-F | Allyl | 3g | Mw: 286.73 | NRB: 3 | 36 | 48 |
| NHA: 2 | LogP: 4.36 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 8 | 6-Cl | 3-NO2 | Allyl | 3h | Mw: 313.74 | NRB: 4 | 34 | 41 |
| NHA: 3 | LogP: 3.42 | |||||||
| NHD: 0 | TPSA: 63.64 | |||||||
| 9 | 6-Cl | 4-NO2 | Allyl | 3i | Mw: 313.74 | NRB: 4 | 42 | 50 |
| NHA: 3 | LogP: 3.45 | |||||||
| NHD: 0 | TPSA: 63.64 | |||||||
| 10 | 6-Cl | 2-Cl | Benzyl | 3j | Mw: 353.24 | NRB: 3 | 31 | 40 |
| NHA: 1 | LogP: 5.22 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 11 | 6-Cl | 4-Cl | Benzyl | 3k | Mw: 353.24 | NRB: 3 | 41 | 49 |
| NHA: 1 | LogP: 5.30 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 12 | 6-Cl | 2,4-Cl2 | Benzyl | 3l | Mw: 387.69 | NRB: 3 | 35 | 44 |
| NHA: 1 | LogP: 5.78 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 13 | 6-Cl | 3,4-Cl2 | Benzyl | 3m | Mw: 387.69 | NRB: 3 | 38 | 47 |
| NHA: 1 | LogP: 5.79 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 14 | 6-Cl | 3,4-(OCH3)2 | Benzyl | 3n | Mw: 378.85 | NRB: 5 | 26 | 41 |
| NHA: 3 | LogP: 4.68 | |||||||
| NHD: 0 | TPSA: 36.28 | |||||||
| 15 | 6-Cl | 4-OC2H5 | Benzyl | 3o | Mw: 362.85 | NRB: 5 | 34 | 43 |
| NHA: 2 | LogP: 5.07 | |||||||
| NHD: 0 | TPSA: 27.05 | |||||||
| 16 | 6-Cl | 4-F | Benzyl | 3p | Mw: 336.79 | NRB: 3 | 33 | 46 |
| NHA: 2 | LogP: 5.08 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 17 | 6-Cl | 3-NO2 | Benzyl | 3q | Mw: 363.80 | NRB: 4 | 37 | 48 |
| NHA: 3 | LogP: 4.15 | |||||||
| NHD: 0 | TPSA: 63.64 | |||||||
| 18 | 6-Cl | 4-NO2 | Benzyl | 3r | Mw: 363.80 | NRB: 4 | 40 | 50 |
| NHA: 3 | LogP: 4.13 | |||||||
| NHD: 0 | TPSA: 63.64 | |||||||
| 19 | 6-Cl | 4-N(CH3)2 | Benzyl | 3s | Mw: 361.87 | NRB: 4 | 27 | 40 |
| NHA: 1 | LogP: 4.76 | |||||||
| NHD: 0 | TPSA: 21.06 | |||||||
| 20 | 6-Cl | 4-Cl | 2-Chlorobenzyl | 3t | Mw: 387.69 | NRB: 3 | 29 | 42 |
| NHA: 1 | LogP: 5.73 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 21 | 6-Cl | 3,4-Cl2 | 4-Chlorobenzyl | 3u | Mw: 422.13 | NRB: 3 | 40 | 49 |
| NHA: 1 | LogP: 6.29 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 22 | 6-Cl | 4-Chlorobenzyl | 3v | Mw: 343.21 | NRB: 3 | 43 | 50 | |
| NHA: 2 | LogP: 4.58 | |||||||
| NHD: 0 | TPSA: 30.96 | |||||||
| 23 | 5-Cl | 4-Cl | Benzyl | 3w | Mw: 378.85 | NRB: 5 | 35 | 47 |
| NHA: 3 | LogP: 4.69 | |||||||
| NHD: 0 | TPSA: 36.28 | |||||||
| 24 | 5-Cl | 3,4-(OCH3)2 | 4-Chlorobenzyl | 3x | Mw: 387.69 | NRB: 3 | 39 | 46 |
| NHA: 1 | LogP: 5.76 | |||||||
| NHD: 0 | TPSA: 17.82 | |||||||
| 25 | 6-NO2 | 2,4-Cl2 | Allyl | 4a | Mw: 348.18 | NRB: 4 | 35 | 45 |
| NHA: 3 | LogP: 3.94 | |||||||
| NHD: 0 | TPSA: 63.64 | |||||||
| 26 | 6-NO2 | 3,4-Cl2 | Allyl | 4b | Mw: 348.18 | NRB: 4 | 42 | 49 |
| NHA: 3 | LogP: 3.99 | |||||||
| NHD: 0 | TPSA: 63.64 | |||||||
| 27 | 6-NO2 | 3,4-(OCH3)2 | Allyl | 4c | Mw: 339.35 | NRB: 6 | 40 | 48 |
| NHA: 5 | LogP: 2.90 | |||||||
| NHD: 0 | TPSA: 82.10 | |||||||
| 28 | 6-NO2 | 4-OC2H5 | Allyl | 4d | Mw: 323.35 | NRB: 6 | 40 | 47 |
| NHA: 4 | LogP: 3.24 | |||||||
| NHD: 0 | TPSA: 72.87 | |||||||
| 29 | 6-NO2 | 4-F | Allyl | 4e | Mw: 297.28 | NRB: 4 | 32 | 44 |
| NHA: 4 | LogP: 3.22 | |||||||
| NHD: 0 | TPSA: 63.64 | |||||||
| 30 | 6-NO2 | 4-N(CH3)2 | Allyl | 4f | Mw: 322.36 | NRB: 5 | 38 | 46 |
| NHA: 3 | LogP: 2.77 | |||||||
| NHD: 0 | TPSA: 66.88 | |||||||
| 31 | 6-NO2 | 3,4-Cl2 | 4-Chlorobenzyl | 4g | Mw: 432.69 | NRB: 4 | 39 | 49 |
| NHA: 3 | LogP: 5.10 | |||||||
| NHD: 0 | TPSA: 63.64 | |||||||
| 32 | 6-NO2 | 3,4-(OCH3)2 | 4-Chlorobenzyl | 4h | Mw: 423.85 | NRB: 6 | 40 | 49 |
| NHA: 5 | LogP: 4.01 | |||||||
| NHD: 0 | TPSA: 82.10 | |||||||
| 33 | 6-NO2 | 3-OC2H5, 4-OH | 4-Chlorobenzyl | 4i | Mw: 423.85 | NRB: 6 | 35 | 43 |
| NHA: 5 | LogP: 3.92 | |||||||
| NHD: 1 | TPSA: 93.10 | |||||||
| 34 | 6-NO2 | 3-OH | 4-Chlorobenzyl | 4j | Mw: 379.80 | NRB: 4 | 32 | 44 |
| NHA: 4 | LogP: 3.55 | |||||||
| NHD: 1 | TPSA: 83.87 | |||||||
| 35 | 6-NO2 | 4-N(CH3)2 | 4-Chlorobenzyl | 4k | Mw: 406.86 | NRB: 5 | 38 | 49 |
| NHA: 3 | LogP: 4.05 | |||||||
| NHD: 0 | TPSA: 66.88 | |||||||
| 36 | 6-NO2 | 4-Cl | # | 4l | Mw: 359.76 | NRB: 6 | 37 | 46 |
| NHA: 5 | LogP: 2.85 | |||||||
| NHD: 0 | TPSA: 89.94 | |||||||
| 37 | 6-NO2 | 4-N(CH3)2 | # | 4m | Mw: 368.39 | NRB: 7 | 35 | 48 |
| NHA: 5 | LogP: 2.39 | |||||||
| NHD: 0 | TPSA: 93.18 | |||||||
| 38 | 6-NO2 | 4-F | # | 4n | Mw: 343.31 | NRB: 6 | 37 | 47 |
| NHA: 6 | LogP: 2.60 | |||||||
| NHD: 0 | TPSA: 89.94 | |||||||
| 39 | 6-NO2 | 3-O#, 4-OCH3 | # | 4o | Mw: 457.43 | NRB: 12 | 40 | 48 |
| NHA: 9 | LogP: 2.51 | |||||||
| NHD: 0 | TPSA: 134.70 | |||||||
| 40 | 6-NO2 | Allyl | 4p | Mw: 269.26 | NRB: 4 | 36 | 45 | |
| NHA: 4 | LogP: 2.28 | |||||||
| NHD: 0 | TPSA: 76.78 | |||||||
| 41 | 6-NO2 | 4-Chlorobenzyl | 4q | Mw: 353.76 | NRB: 4 | 36 | 48 | |
| NHA: 4 | LogP: 3.41 | |||||||
| NHD: 0 | TPSA: 76.78 | |||||||
| 42 | 6-NO2 | # | 4r | Mw: 315.28 | NRB: 6 | 37 | 48 | |
| NHA: 6 | LogP: 1.72 | |||||||
| NHD: 0 | TPSA: 103.08 | |||||||
IR, 1H NMR, 13C NMR, and mass spectra of the synthesized compounds are in accordance with the assigned structures. The IR spectra of all the synthesized displayed a medium absorbance band in the ν 1535–1374 cm−1 region which is distinctive of the aromatic ring as well as a strong absorbance band in the ν 1646–1505 cm−1 region characteristic of imine (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N) of imidazole nucleus of 1H-benzimidazole derivatives. Compounds 4a–4r displayed a strong absorbance band in the ν 1355–1215 cm−1 region which is distinctive of the NO2 group. In addition, 1H NMR spectra of compounds 1 and 2 indicated the characteristic NH protons of 1H-benzimidazole as a singlet in the δ 13.92–12.71 ppm region, as well as the distinctive aromatic proton in the δ 9.00–6.72 ppm region. On the other hand, 1H NMR spectra of compounds 3 and 4 revealed the appearance of a singlet in the 5.90–4.70 ppm region of methylene (–CH2–) moiety of allyl (–CH2–CH
N) of imidazole nucleus of 1H-benzimidazole derivatives. Compounds 4a–4r displayed a strong absorbance band in the ν 1355–1215 cm−1 region which is distinctive of the NO2 group. In addition, 1H NMR spectra of compounds 1 and 2 indicated the characteristic NH protons of 1H-benzimidazole as a singlet in the δ 13.92–12.71 ppm region, as well as the distinctive aromatic proton in the δ 9.00–6.72 ppm region. On the other hand, 1H NMR spectra of compounds 3 and 4 revealed the appearance of a singlet in the 5.90–4.70 ppm region of methylene (–CH2–) moiety of allyl (–CH2–CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 1-(2-ethoxy-2-oxoethyl) (–CH2COOC2H5), and arylmethyl (–CH2–Ar) groups, as well as the distinctive aromatic proton in the δ 8.80–6.65 ppm region. Furthermore, the C
CH2), 1-(2-ethoxy-2-oxoethyl) (–CH2COOC2H5), and arylmethyl (–CH2–Ar) groups, as well as the distinctive aromatic proton in the δ 8.80–6.65 ppm region. Furthermore, the C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N group (δ 164.0–145.0 ppm), CAr (δ 171.5–101.5 ppm), and the methylene moiety of allyl, 1-(2-ethoxy-2-oxoethyl), and arylmethyl groups (δ 57.1–46.1 ppm) were identified in the 13C NMR spectrum of compounds 3 and 4. The molecular ion peak M (m/z) of compounds 1–4 was observed in the mass spectrum, confirming the hypothesized structure.
N group (δ 164.0–145.0 ppm), CAr (δ 171.5–101.5 ppm), and the methylene moiety of allyl, 1-(2-ethoxy-2-oxoethyl), and arylmethyl groups (δ 57.1–46.1 ppm) were identified in the 13C NMR spectrum of compounds 3 and 4. The molecular ion peak M (m/z) of compounds 1–4 was observed in the mass spectrum, confirming the hypothesized structure.
2.2. In vitro antibacterial and antifungal activities
Antimicrobial activities (exhibited by MIC values) including antibacterial activities at two strains of Gram-negative (EC – Escherichia coli and PA – Pseudomonas aeruginosa) and three strains of Gram-positive (SF – Streptococcus faecalis, MSSA, MRSA) and antifungal activities (CA – Candida albicans and AN – Aspergillus niger) of all synthesized compounds are summarized in Table 3 and 4.| Entry | Code | Antibacterial | Antifungal | Anticancer | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC | PA | SF | MSSA | MRSA | CA | AN | HepG2 | MDA-MB-231 | MCF7 | C26 | RMS | ||
| a -: MIC ≥ 1024 μg mL−1, ND – not determined, EC – Escherichia coli ATCC 25922, PA – Pseudomonas aeruginosa ATCC 27853, SF – Streptococcus faecalis ATCC 29212, MSSA – Methicillin-susceptible strains of Staphylococcus aureus ATCC 29213, MRSA – Methicillin-resistant strains of Staphylococcus aureus ATCC 43300, CA – Candida albicans ATCC 10321, AN – Aspergillus niger ATCC 16404, Cipro – ciprofloxacin, Flu – fluconazole, MIC (μg mL−1) ± 0.5 μg mL−1. PTX – paclitaxel, HepG2 – human hepatocyte carcinoma cell line, MDA-MB-231 – human breast adenocarcinoma cell line, MCF7 – human breast cancer cell line, C26 – colon carcinoma cell line, RMS – human rhabdomyosarcoma cell line. IC50 ± SEM (μM, SEM – standard error of the mean). The values in bold highlight the best compounds with the best MIC and IC50 values compared to positive controls. | |||||||||||||
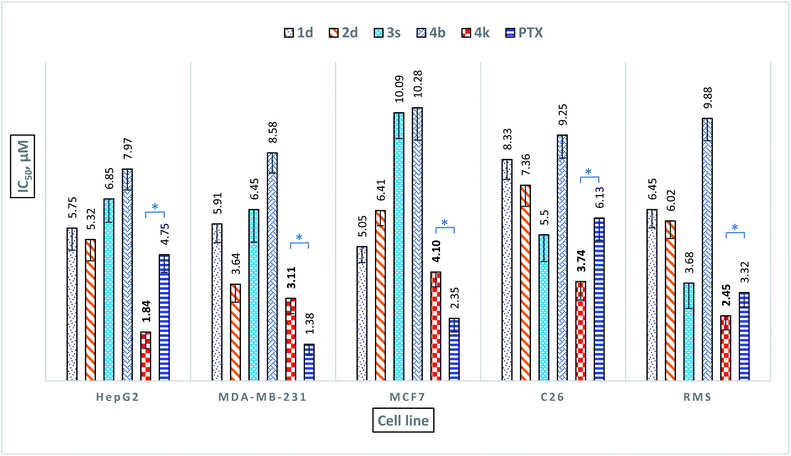
| 1 | 1a | 64 | — | 128 | 64 | 128 | — | — | 73.19 ± 3.71 | 52.28 ± 2.66 | 47.89 ± 2.92 | 37.45 ± 3.03 | 33.41 ± 2.44 |
| 2 | 1b | 16 | — | 8 | 8 | 8 | 64 | 64 | 19.56 ± 1.24 | 10.32 ± 1.60 | 17.04 ± 1.71 | 7.35 ± 0.88 | 8.33 ± 1.07 |
| 3 | 1c | 64 | — | 64 | 64 | 128 | 512 | 512 | 36.74 ± 2.42 | 30.24 ± 2.70 | 18.53 ± 1.62 | 35.29 ± 2.54 | 19.36 ± 1.72 |
| 4 | 1d | 8 | — | 8 | 4 | 8 | 32 | 32 | 5.75 ± 0.83 | 5.91 ± 0.71 | 5.05 ± 0.60 | 8.33 ± 0.75 | 6.45 ± 0.69 |
| 5 | 1e | 128 | — | 128 | 64 | 128 | — | — | 51.34 ± 3.36 | 29.70 ± 1.73 | >100 | 35.82 ± 2.35 | 38.91 ± 2.40 |
| 6 | 1f | 64 | — | 128 | 128 | 256 | 512 | 512 | 15.72 ± 1.23 | 12.09 ± 0.94 | 17.54 ± 1.15 | 10.46 ± 0.85 | 13.60 ± 1.03 |
| 7 | 1g | 256 | — | 256 | 128 | 256 | — | — | 21.68 ± 1.49 | 23.05 ± 1.21 | >100 | 36.44 ± 2.52 | 31.37 ± 2.73 |
| 8 | 1h | 64 | — | 64 | 32 | 32 | 512 | 512 | 46.51 ± 2.35 | 40.24 ± 2.19 | 52.34 ± 2.79 | 41.23 ± 2.38 | 35.48 ± 1.99 |
| 9 | 1i | 64 | — | 64 | 64 | 128 | 512 | 512 | 64.63 ± 3.02 | 81.29 ± 3.50 | 34.73 ± 2.48 | 75.20 ± 3.84 | 28.17 ± 1.45 |
| 10 | 1j | 64 | — | 64 | 32 | 64 | 256 | 256 | 29.04 ± 1.78 | 22.39 ± 1.32 | 34.59 ± 1.93 | 22.18 ± 1.65 | 31.07 ± 1.95 |
| 11 | 1k | 128 | — | 128 | 128 | 256 | 256 | 256 | 17.89 ± 1.92 | 10.44 ± 1.64 | 37.55 ± 2.39 | 14.62 ± 1.75 | 23.84 ± 1.79 |
| 12 | 1l | 64 | — | 64 | 64 | 128 | 512 | 512 | 28.09 ± 2.53 | 37.46 ± 2.97 | 44.81 ± 3.03 | 32.73 ± 1.95 | 41.43 ± 2.21 |
| 13 | 1m | 128 | — | 128 | 32 | 64 | — | — | 55.08 ± 3.12 | >100 | 26.36 ± 1.70 | 64.47 ± 2.67 | 28.82 ± 1.50 |
| 14 | 1n | 64 | — | 64 | 32 | 64 | 256 | 256 | 8.91 ± 1.02 | 7.37 ± 1.41 | 10.22 ± 0.98 | 8.16 ± 1.14 | 11.92 ± 1.05 |
| 15 | 1o | 64 | — | 64 | 64 | 128 | 128 | 128 | 18.50 ± 1.43 | 10.61 ± 1.34 | 12.78 ± 1.01 | 20.38 ± 1.76 | 16.04 ± 1.65 |
| 16 | 1p | 128 | — | 64 | 64 | 128 | 512 | 512 | 56.11 ± 3.02 | 62.35 ± 2.81 | 33.47 ± 3.23 | 63.34 ± 3.01 | 36.60 ± 2.92 |
| 17 | 1q | 8 | - | 8 | 4 | 8 | 32 | 32 | 52.63 ± 2.43 | 74.62 ± 2.53 | 54.65 ± 3.35 | 28.39 ± 2.17 | 47.05 ± 2.28 |
| 18 | 2a | 64 | — | 128 | 64 | 64 | — | — | 63.24 ± 3.19 | 36.88 ± 2.74 | >100 | 55.73 ± 3.41 | 42.03 ± 2.54 |
| 19 | 2b | 16 | - | 16 | 8 | 8 | 32 | 32 | 16.64 ± 1.36 | 11.25 ± 1.52 | 41.68 ± 3.83 | 8.04 ± 0.84 | 9.87 ± 1.19 |
| 20 | 2c | 64 | — | 64 | 64 | 128 | 256 | 256 | 25.05 ± 1.87 | 33.50 ± 1.69 | 30.08 ± 1.78 | 16.78 ± 0.98 | 22.74 ± 1.95 |
| 21 | 2d | 8 | - | 8 | 8 | 8 | 32 | 32 | 5.32 ± 0.80 | 3.64 ± 0.68 | 6.41 ± 0.57 | 7.36 ± 0.79 | 6.02 ± 0.66 |
| 22 | 2e | 64 | — | 128 | 64 | 128 | 512 | 512 | 51.34 ± 3.36 | 29.70 ± 1.73 | >100 | >100 | 38.91 ± 2.40 |
| 23 | 2f | 64 | — | 128 | 128 | 128 | 512 | 512 | 23.86 ± 1.62 | >100 | 36.64 ± 1.56 | 27.94 ± 1.44 | 30.39 ± 2.22 |
| 24 | 2g | 256 | — | 256 | 128 | 256 | — | — | 24.93 ± 1.38 | 29.70 ± 1.51 | >100 | 44.63 ± 2.83 | 34.74 ± 1.96 |
| 25 | 2h | 64 | — | 64 | 64 | 64 | 256 | 256 | 66.28 ± 3.12 | 52.84 ± 2.10 | >100 | 42.78 ± 2.77 | 47.66 ± 2.08 |
| 26 | 2i | 64 | — | 64 | 64 | 128 | — | — | 28.36 ± 1.35 | 31.32 ± 1.33 | 44.16 ± 1.94 | 38.49 ± 1.87 | 40.27 ± 1.74 |
| 27 | 2j | 64 | — | 64 | 32 | 32 | 128 | 128 | 42.56 ± 1.76 | >100 | 49.91 ± 2.03 | 36.27 ± 1.43 | 31.13 ± 1.85 |
| 28 | 2k | 128 | — | 128 | 128 | 256 | 256 | 256 | 64.53 ± 2.24 | >100 | 84.91 ± 4.31 | 26.12 ± 1.38 | 55.49 ± 2.63 |
| 29 | 2l | 64 | — | 64 | 64 | 128 | 256 | 256 | 76.85 ± 3.16 | >100 | >100 | >100 | 47.79 ± 2.44 |
| 30 | 2m | 128 | — | 64 | 32 | 64 | 512 | 512 | 58.91 ± 2.65 | >100 | 83.57 ± 4.08 | 13.44 ± 0.89 | 34.09 ± 1.98 |
| 31 | 2n | 64 | — | 64 | 32 | 64 | 512 | 512 | 8.91 ± 1.02 | 7.37 ± 1.41 | 10.22 ± 0.98 | 8.16 ± 1.14 | 11.92 ± 1.05 |
| 32 | 2o | 64 | — | 64 | 32 | 64 | 128 | 128 | 23.45 ± 1.84 | 47.02 ± 2.60 | 10.36 ± 1.25 | 19.53 ± 1.58 | 21.09 ± 1.36 |
| 33 | 2p | 128 | — | 64 | 64 | 128 | — | — | 64.25 ± 3.50 | 66.22 ± 2.77 | 41.83 ± 3.05 | 68.20 ± 2.71 | 39.53 ± 2.85 |
| 34 | 2q | 16 | - | 8 | 8 | 16 | 64 | 64 | 90.14 ± 4.07 | >100 | >100 | >100 | 87.42 ± 4.21 |
| 35 | Cipro | 16 | 16 | 8 | 8 | 16 | ND | ND | ND | ND | ND | ND | ND |
| 36 | Flu | ND | ND | ND | ND | ND | 4 | 128 | ND | ND | ND | ND | ND |
| 37 | PTX | ND | ND | ND | ND | ND | ND | ND | 4.75 ± 0.67 | 1.38 ± 0.42 | 2.35 ± 0.51 | 6.13 ± 0.83 | 3.32 ± 0.55 |
| Entry | Code | Antibacterial | Antifungal | Anticancer | |||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| EC | PA | SF | MSSA | MRSA | CA | AN | HepG2 | MDA-MB-231 | MCF7 | C26 | RMS | ||
| a -: MIC ≥ 1024 μg mL−1, ND – not determined, EC – Escherichia coli ATCC 25922, PA – Pseudomonas aeruginosa ATCC 27853, SF – Streptococcus faecalis ATCC 29212, MSSA – Methicillin-susceptible strains of Staphylococcus aureus ATCC 29213, MRSA – Methicillin-resistant strains of Staphylococcus aureus ATCC 43300, CA – Candida albicans ATCC 10321, AN – Aspergillus niger ATCC 16404, Cipro – ciprofloxacin, Flu – fluconazole, MIC (μg mL−1) ± 0.5 μg mL−1. PTX – paclitaxel, HepG2 – human hepatocyte carcinoma cell line, MDA-MB-231 – human breast adenocarcinoma cell line, MCF7 – human breast cancer cell line, C26 – colon carcinoma cell line, RMS – human rhabdomyosarcoma cell line. IC50 ± SEM (μM, SEM – standard error of the mean). The values in bold highlight the best compounds with the best MIC and IC50 values compared to positive controls. | |||||||||||||
| 1 | 3a | 128 | — | 128 | 32 | 64 | 512 | 512 | 25.92 ± 2.13 | 25.33 ± 1.91 | 21.27 ± 1.63 | 33.50 ± 2.30 | 65.50 ± 3.04 |
| 2 | 3b | 16 | — | 16 | 16 | 32 | 64 | 64 | 35.59 ± 2.74 | 32.52 ± 1.96 | 54.24 ± 2.35 | 9.59 ± 0.82 | 11.17 ± 2.62 |
| 3 | 3c | 8 | 512 | 16 | 16 | 16 | 64 | 64 | 35.03 ± 1.48 | 30.44 ± 2.21 | 22.68 ± 1.86 | 10.68 ± 0.84 | 66.35 ± 3.18 |
| 4 | 3d | 16 | 256 | 16 | 8 | 8 | 32 | 32 | 32.40 ± 1.71 | 48.52 ± 1.80 | 61.78 ± 3.12 | 35.86 ± 1.93 | 51.11 ± 2.55 |
| 5 | 3e | 128 | — | 64 | 64 | 64 | 256 | 256 | 44.59 ± 2.78 | 28.15 ± 1.44 | 47.03 ± 2.36 | 26.14 ± 1.51 | 32.26 ± 1.62 |
| 6 | 3f | 16 | - | 16 | 8 | 8 | 32 | 32 | 25.58 ± 1.53 | 28.91 ± 1.76 | 40.64 ± 2.38 | 47.60 ± 2.29 | 16.76 ± 0.99 |
| 7 | 3g | 128 | 512 | 128 | 128 | 256 | 256 | 256 | 25.63 ± 1.46 | 22.60 ± 1.37 | 58.11 ± 2.71 | 30.54 ± 1.84 | 51.95 ± 2.20 |
| 8 | 3h | 128 | — | 64 | 64 | 64 | 512 | 512 | 68.37 ± 3.49 | 29.98 ± 1.60 | 25.89 ± 1.65 | 34.67 ± 1.77 | 32.49 ± 2.33 |
| 9 | 3i | 64 | — | 64 | 32 | 64 | 128 | 128 | 12.91 ± 0.62 | 13.26 ± 0.58 | 10.48 ± 0.56 | 8.65 ± 0.70 | 9.73 ± 0.53 |
| 10 | 3j | 16 | 512 | 16 | 16 | 16 | 64 | 64 | 48.86 ± 2.24 | 27.74 ± 1.74 | 31.16 ± 2.03 | 9.86 ± 0.89 | 35.22 ± 1.66 |
| 11 | 3k | 64 | — | 128 | 32 | 32 | — | — | 30.87 ± 2.38 | 29.07 ± 1.63 | >100 | 42.43 ± 1.87 | 43.77 ± 2.78 |
| 12 | 3l | 64 | 512 | 64 | 64 | 64 | 256 | 256 | 32.16 ± 1.38 | 24.33 ± 1.31 | 29.63 ± 1.65 | 21.49 ± 1.82 | 20.65 ± 1.43 |
| 13 | 3m | 64 | — | 64 | 32 | 32 | 128 | 128 | 36.77 ± 2.40 | 48.77 ± 3.34 | 23.22 ± 1.37 | 26.99 ± 1.56 | 57.39 ± 3.29 |
| 14 | 3n | 16 | 256 | 16 | 4 | 8 | 64 | 64 | 26.60 ± 1.36 | 19.19 ± 1.42 | 23.19 ± 2.38 | 14.91 ± 0.88 | 28.16 ± 2.43 |
| 15 | 3o | 64 | — | 64 | 64 | 64 | — | — | 42.76 ± 2.58 | 46.65 ± 3.06 | 29.19 ± 1.30 | 38.16 ± 2.41 | 36.60 ± 1.47 |
| 16 | 3p | 64 | 256 | 64 | 32 | 64 | 32 | 32 | 25.25 ± 1.65 | 24.60 ± 2.09 | 37.84 ± 1.78 | 28.97 ± 1.68 | 52.86 ± 3.23 |
| 17 | 3q | 64 | — | 64 | 32 | 64 | — | — | 86.12 ± 3.67 | 79.77 ± 4.02 | 27.35 ± 1.59 | 23.39 ± 1.61 | >100 |
| 18 | 3r | 64 | — | 64 | 16 | 32 | 256 | 256 | 18.74 ± 1.47 | 22.61 ± 1.13 | 17.36 ± 1.52 | 14.05 ± 0.92 | 16.34 ± 1.07 |
| 19 | 3s | 8 | 128 | 8 | 4 | 4 | 32 | 32 | 6.85 ± 0.88 | 6.45 ± 1.23 | 10.09 ± 0.97 | 5.50 ± 1.01 | 3.68 ± 0.95 |
| 20 | 3t | 64 | — | 64 | 64 | 64 | 64 | 64 | 87.74 ± 3.11 | 51.01 ± 2.45 | 47.45 ± 1.96 | 15.77 ± 1.33 | 68.33 ± 2.36 |
| 21 | 3u | 64 | 128 | 64 | 32 | 64 | 128 | 128 | 61.25 ± 3.36 | 54.06 ± 2.91 | 44.38 ± 2.67 | 34.62 ± 3.55 | 32.79 ± 3.02 |
| 22 | 3v | 128 | — | 128 | 128 | 128 | 256 | 256 | 53.27 ± 2.45 | 48.06 ± 3.79 | 31.23 ± 1.75 | 26.93 ± 1.80 | 34.65 ± 2.03 |
| 23 | 3w | 64 | — | 64 | 32 | 32 | 512 | 512 | 39.32 ± 1.48 | 36.29 ± 1.51 | 40.27 ± 2.11 | 34.70 ± 1.69 | 30.09 ± 1.46 |
| 24 | 3x | 16 | 128 | 16 | 8 | 8 | 32 | 32 | 40.94 ± 1.63 | 31.55 ± 1.24 | 38.42 ± 2.04 | 29.85 ± 1.72 | 32.32 ± 1.52 |
| 25 | 4a | 32 | — | 32 | 32 | 32 | 64 | 64 | 37.48 ± 2.37 | 33.61 ± 1.59 | >100 | 37.19 ± 1.36 | 31.90 ± 1.33 |
| 26 | 4b | 16 | - | 16 | 8 | 8 | 32 | 32 | 7.97 ± 0.78 | 8.58 ± 0.76 | 10.28 ± 1.22 | 9.25 ± 0.87 | 9.88 ± 0.84 |
| 27 | 4c | 128 | — | 64 | 64 | 128 | 512 | 512 | 67.98 ± 3.14 | 59.05 ± 2.87 | >100 | 42.81 ± 2.25 | 46.11 ± 2.35 |
| 28 | 4d | 64 | — | 64 | 32 | 64 | — | — | 36.48 ± 2.47 | 40.32 ± 1.90 | 47.58 ± 2.34 | 89.91 ± 3.79 | 54.02 ± 2.22 |
| 29 | 4e | 128 | — | 128 | 128 | 256 | 256 | 256 | 39.36 ± 2.47 | 30.16 ± 1.54 | 25.96 ± 1.18 | 43.38 ± 2.01 | 26.97 ± 1.60 |
| 30 | 4f | 32 | — | 32 | 16 | 32 | 64 | 64 | 13.32 ± 0.85 | 15.90 ± 1.04 | 18.92 ± 1.37 | 10.98 ± 0.94 | 11.83 ± 0.94 |
| 31 | 4g | 64 | 128 | 64 | 32 | 64 | 128 | 128 | 65.32 ± 2.95 | 47.24 ± 2.68 | 51.23 ± 2.37 | 29.71 ± 1.76 | 23.31 ± 1.80 |
| 32 | 4h | 64 | — | 64 | 64 | 64 | 256 | 256 | 33.84 ± 1.96 | 39.01 ± 1.60 | 40.18 ± 2.03 | 25.40 ± 1.70 | 27.73 ± 2.23 |
| 33 | 4i | 64 | — | 64 | 64 | 64 | 256 | 256 | 48.64 ± 1.83 | 27.43 ± 1.47 | 21.04 ± 1.21 | 41.12 ± 2.33 | 32.73 ± 1.39 |
| 34 | 4j | 64 | — | 64 | 64 | 64 | 512 | 512 | 53.09 ± 2.31 | 36.42 ± 1.77 | 28.13 ± 1.34 | 26.85 ± 2.04 | 37.54 ± 1.55 |
| 35 | 4k | 4 | 64 | 4 | 2 | 4 | 8 | 16 | 1.84 ± 0.62 | 3.11 ± 0.58 | 4.10 ± 0.56 | 3.74 ± 0.70 | 2.45 ± 0.53 |
| 36 | 4l | 128 | — | 128 | 128 | 256 | 512 | 512 | 65.97 ± 3.65 | >100 | 54.88 ± 2.35 | 60.05 ± 3.14 | 56.38 ± 2.47 |
| 37 | 4m | 32 | — | 64 | 32 | 64 | 256 | 256 | 78.83 ± 3.13 | >100 | 46.73 ± 2.33 | 53.49 ± 2.08 | 49.50 ± 2.26 |
| 38 | 4n | 128 | — | 128 | 128 | 256 | — | — | 88.05 ± 3.49 | >100 | 73.25 ± 3.29 | 58.37 ± 1.86 | 45.58 ± 2.36 |
| 39 | 4o | 64 | — | 128 | 64 | 128 | 512 | 512 | >100 | >100 | 78.34 ± 3.51 | 61.78 ± 3.42 | 64.45 ± 3.30 |
| 40 | 4p | 64 | — | 128 | 64 | 128 | 256 | 256 | 42.33 ± 1.77 | 33.64 ± 1.65 | 49.10 ± 2.42 | 74.19 ± 2.47 | 37.62 ± 1.81 |
| 41 | 4q | 64 | — | 64 | 64 | 128 | 256 | 256 | 39.17 ± 1.24 | 42.90 ± 1.98 | >100 | >100 | 31.25 ± 1.84 |
| 42 | 4r | 64 | — | 128 | 64 | 128 | 512 | 512 | >100 | >100 | 80.11 ± 3.64 | 56.88 ± 3.35 | 67.72 ± 3.57 |
| 43 | Cipro | 16 | 16 | 8 | 8 | 16 | ND | ND | ND | ND | ND | ND | ND |
| 44 | Flu | ND | ND | ND | ND | ND | 4 | 128 | ND | ND | ND | ND | ND |
| 45 | PTX | ND | ND | ND | ND | ND | ND | ND | 4.75 ± 0.67 | 1.38 ± 0.42 | 2.35 ± 0.51 | 6.13 ± 0.83 | 3.32 ± 0.55 |
With antimicrobial activities of a series of 6-substituted 1H-benzimidazole derivatives, all compounds are totally inactive at the Gram-negative strain PA (MIC ≥ 1024 μg mL−1). Compounds 1a, 1c, 1e–1p, 2a, 2c and 2e–2p showed weak to moderate activities at 4 strains of bacteria (EC, SF, MSSA, and MRSA) and 2 strains of fungi (MIC ≥ 32 μg mL−1). Compounds 1b (6-chloro, 4-chlorophenyl), 1d (6-chloro, 3,4-dichlorophenyl), 1q (6-chloro, furan-2-yl), 2d (6-nitro, 3,4-dichlorophenyl) showed potent antibacterial activities against the Gram-positive strains SF, MSSA and MRSA with MIC ranging between 4 to 8 μg mL−1 as compared to ciprofloxacin (Cipro, MIC = 8–16 μg mL−1), but showed moderate activities at the fungi strains CA, and AN (MIC 32–64 μg mL−1) as compared to fluconazole (Flu, MIC of 4 μg mL−1 at CA and 128 μg mL−1 at AN). In addition, these compounds also showed good antibacterial activities against the Gram-negative strain EC with MIC ranging between 8 to 16 μg mL−1. Compound 2b (6-nitro, 4-chlorophenyl), 2q (6-nitro, furan-2-yl) showed good antibacterial activities against EC, SF, MSSA, and MRSA with MIC ranging between 8 to 16 μg mL−1 as compared to Cipro and moderate fungal activities against CA and AN (MIC 32–64 μg mL−1) as compared to Flu. The results suggested that the 4-chloro and 3,4-dichloro groups of the aromatic ring and furan nucleus at position 2 of the 1H-benzimidazole scaffold enhanced antibacterial activities against EC, SF, MSSA, and MRSA strains.
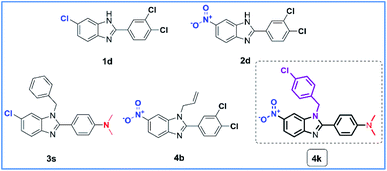
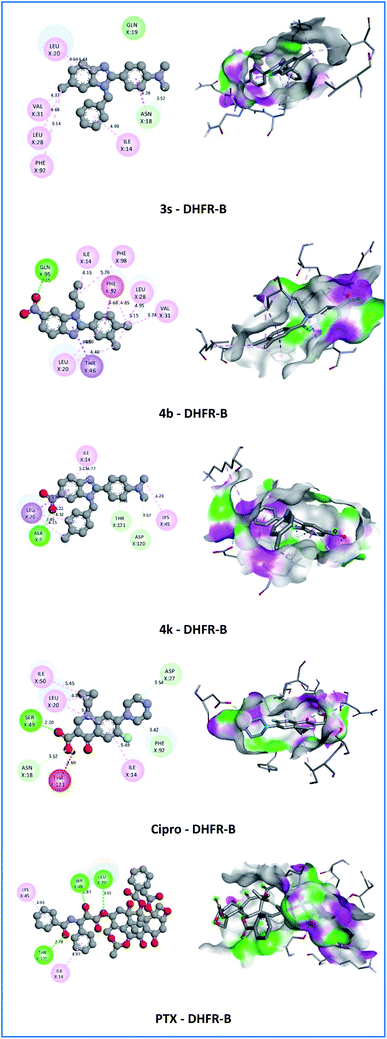
With antimicrobial activities of a series of N-substituted 6-(chloro/nitro)-1H-benzimidazole derivatives, compounds 3a, 3e, 3g–3i, 3k–3m, 3o–3q, 3t–3w, 4a, 4c–4e, 4g–4j and 4l–4r showed weak to moderate activities at 5 strains of bacteria and 2 strains of fungi (MIC ≥ 32 μg mL−1). Compounds 3b (6-chloro, 4-chlorophenyl, N-allyl), 3c (6-chloro, 2,4-dichlorophenyl, N-allyl), 3j (6-chloro, 2-chlorophenyl, N-benzyl) 3r (6-chloro, 4-nitrophenyl, N-benzyl), and 4f (6-nitro, N,N-dimethylaminophenyl, N-allyl) showed good antibacterial activities against Gram-positive strain MSSA with MIC of 16 μg mL−1 but showed weak to moderate activities at PA, CA and AN with MIC ≥ 32 μg mL−1. Compounds 3c and 3j also showed good antibacterial activities against three bacteria strains EC, SF, and MRSA with MIC ranging between 8 to 16 μg mL−1 as compared to Cipro. Compounds 3b showed good antibacterial activities against EC with MIC of 16 μg mL−1 while showed moderate antibacterial activities against MRSA with MIC of 32 μg mL−1. Besides, compounds 3d (6-chloro, 3,4-dichlorophenyl, N-allyl), 3f (6-chloro, 4-ethoxyphenyl, N-allyl), 3n (6-chloro, 3,4-dimethoxyphenyl, N-benzyl), 3x (5-chloro, 3,4-dimethoxyphenyl, N-(4-chlorobenzyl)), and 4b (6-nitro, 3,4-dichlorophenyl, N-allyl) showed good antibacterial activities at Gram-positive strains MSSA and MRSA with MIC ranging between 4 to 8 μg mL−1, strains EC and SF with MIC value at 16 μg mL−1, and strain AN with MIC value at 32 μg mL−1. However, these compounds showed weak to moderate activities at PA and CA (MIC ≥ 32 μg mL−1) as compared to Cipro and Flu. Moreover, compounds 3s (6-chloro, N,N-dimethylaminophenyl, N-benzyl), and 4k (6-chloro, N,N-dimethylaminophenyl, N-(4-chlorobenzyl)) exhibited the strongest activity among the synthesized compounds against the Gram-positive strains MSSA and MRSA with MIC ranging between 2 to 4 μg mL−1 and strains EC and SF with MIC ranging between 4 to 8 μg mL−1 as compared to Cipro (MIC = 8–16 μg mL−1), but showed weak activity at PA. In particular, compound 4k also showed potent fungal activities against CA and AN with MIC of 8 and 16 μg mL−1, respectively as compared to Flu (MIC of 4 μg mL−1 at CA and 128 μg mL−1 at AN). From the structure–activity relationship (SAR), the presence of the N,N-dimethylamino (-N(CH3)2) group in the aromatic ring at position 2 and chloro/nitro group at position 6 of the 1H-benzimidazole scaffold is more desirable for enhanced antibacterial and antifungal activities in 3s and 4k (Fig. 3).
In published studies, the 5,6-dichloro-1H-benzimidazole derivative with 4-fluoro and 4-chloro substituent on the phenyl ring or N-cyclopentyl and 4-benzyloxy on the phenyl ring exhibited the potent antibacterial activity with MIC 3.12 mg mL−1 against S. aureus.5 In addition, the 1H-benzimidazole-5-carbohydrazide derivative with a 4-nitro substituent on the phenyl ring showed good inhibitory activity against lanosterol 14α-demethylase (CYP51) with IC50 value at 0.19 μg mL−1 compared to fluconazole as reference IC50 value at 0.62 μg mL−1.37 The pyridin-3-yl-1H-benzimidazole-5-carboxylate derivative was found to be the potent active with MIC of 0.112 μM against Mycobacterium tuberculosis H37Rv and 6.12 μM against INH-resistant Mycobacterium tuberculosis, respectively.38 Especially, the 6-fluoro-1H-benzimidazole derivative showed potent antibacterial activities against the Gram-positive strains MSSA and MRSA with MIC of 4 and 2–8 μg mL−1, respectively.7 Some of our synthesized compounds (3s and 4k) also exhibited potential antibacterial activity with MICs of 2–4 μg mL−1 against MSSA and MSRA when compared with compounds of Tunçbilek et al. (2009) and Malasala et al. (2021).5,7 This may be due to the structure of compound 3s and 4k with the presence of N,N-dimethylamino group at position 4 on the phenyl ring of the 1H-benzimidazole nucleus has shown a similar role to the 4-chloro/4-nitro group that of Tunçbilek et al. (2009) and Morcoss et al. (2020).5,37
2.3. Anticancer activity
The synthesized compounds 1a–1q, 2a–2q, 3a–3x, and 4a–4r were also tested for their potent anticancer activity using MTT assay against hepatocellular carcinoma cell line (HepG2), human breast cancer cell line (MDA-MB-231 and MCF7), colon carcinoma cell line (C26) and the aggressive and highly malignant rhabdomyosarcoma cell line (RMS) using paclitaxel (PTX) as a non-selective positive control. The results are summarized in Table 3 and 4In both series of 1H-benzimidazole derivatives, fifty-six compounds 1a, 1c, 1e, 1g–1j, 1l–1m, 1p–1q, 2a, 2c, 2e–2l, 2p–2q, 3a, 3d–3h, 3k–3m, 3o–3q, 3t–3x, 4a, 4c–4j, and 4l–4r exhibited moderate activity (IC50 = 15.0–50.0 μM) or weak activity (IC50 > 50 μM) toward HepG2, MDA-MB-231, MCF7, RMS, and C26. Compounds 1b, 1f, and 2b showed good anticancer activity against the MDA-MB-231, C26, and RMS cell lines with IC50 ranging between 7.35 to 13.60 μM as compared to PTX (IC50 = 1.38–6.13 μM) and weak to moderate anticancer activities against HepG2 and MCF7 cell lines (IC50 > 15.0 μM). Compound 1k showed good anticancer activity against the MDA-MB-231 and C26 cell lines with IC50 values at 10.44 and 14.62 μM, respectively but exhibited weak moderate anticancer activities against HepG2, MCF7, and RMS cell lines (IC50 = 17.89–37.55 μM). Compound 1o showed good anticancer activity against the MDA-MB-231 and MCF7 cell lines with IC50 values at 10.61 and 12.78 μM, respectively, and weak moderate anticancer activities against HepG2, C26, and RMS cell lines with IC50 ranging between 16.04 to 20.38 μM. Besides, compound 3b showed good anticancer activity against the C26 and RMS cell lines with IC50 values at 9.59 and 11.17 μM, respectively but exhibited weak moderate anticancer activities against HepG2, MDA-MB-231, and MCF7 cell lines (IC50 = 32.52–54.24 μM). On the other hand, some compounds showed good anticancer activity at only one cell line such as compounds 2m, 3c, 3j, 3n, and 3r against C26 and 2o against MCF7 with IC50 ranging between 9.86 to 14.91 μM as compared to PTX. Particularly, eight compounds 1d (6-chloro, 3,4-dichlorophenyl), 1n (6-chloro, 4-nitrophenyl), 2d (6-nitro, 3,4-dichlorophenyl), 2n (6-nitro, 4-nitrophenyl), 3i (6-chloro, 4-nitrophenyl, N-allyl), 3s (6-chloro, N,N-dimethylaminophenyl, N-benzyl), 4b (6-nitro, 3,4-dichlorophenyl, N-allyl), and 4k (6-chloro, N,N-dimethylaminophenyl, N-(4-chlorobenzyl)) showed the strongest anticancer activity among the synthesized compounds against all tested cell lines with IC50 ranging between 1.84 to 13.26 μg mL−1 comparable to PTX (IC50 = 1.38–6.13 μM). Moreover, compound 4k showed the strongest anticancer activity among all active compounds against HepG2, MDA-MB-231, MCF7, RMS, and C26 with IC50 of 1.84, 3.11, 4.10, 3.74, and 2.45 μM, respectively as compared to PTX. Compound 4k exhibited a weaker anticancer activity than PTX on the MCF7 cell line but exhibited better anticancer activity than PTX on HepG2, MDA-MB-231, C26, and RMS cell lines (Fig. 4), and especially also showed the strongest antimicrobial activities (Table 4). Target engagement with electron-donating substituent 4-N(CH3)2 on the phenyl ring and N-(4-chlorobenzyl) substituent of 1H-benzimidazole scaffold may be responsible for its anticancer activity as compared to other compounds.
In published studies with similar structures, the 6-benzoyl-1H-benzimidazole derivative with a 3-hydroxy substituent on the phenyl ring was found to be a potent multi-cancer inhibitor against human lung adenocarcinoma epithelial (A549), MDA-MB-231, and human prostate cancer (PC3) cell lines with IC50 values 4.47, 4.68, and 5.50 μg mL−1, respectively.4 Besides, the 2,6-disubstituted benzoimidazolyl quinazolinamine derivative with a 4-fluoro substituent on the phenyl ring was found to be potent against tyrosine-protein kinase Met (c-Met) and vascular endothelial growth factor receptor 2 (VEGFR-2) with of IC50 of 0.05 μM and 0.02 μM respectively.39 In addition, the 2,6-disubstituted benzimidazole–oxindole conjugate derivative with 3,5-difluoro substituent on the phenyl ring showed 43.7% and 64.8% apoptosis against MCF-7, respectively, at 1 and 2 μM concentrations.40 The presence of the halogen groups of our active 1H-benzimidazole derivatives (1d, 2d, and 4b) is the similarity to the reported potent compounds.
On the other hand, the N,2,5-trisubstituted 1H-benzimidazole derivative with N-phenyl group and 4-dimethylamino on the phenyl ring showed Sirtuins inhibitory activity for SIRT1 (IC50 = 54.21 μM) and for SIRT2 (IC50 = 26.85 μM). Cell proliferation assay demonstrated that this compound had pronounced antitumor activity against three different types of cancer cells (breast MDA-MB-468, colon HCT-116, and blood-leukemia CCRF-CEM).41 Moreover, the N,2,5-trisubstituted 1H-benzimidazole derivative with N-(3-phenylpropyl) group showed good antitumor activity against MCF7 cell line with IC50 value at 5.73 ± 0.95 μM and induces obvious autophagy in MCF7 cells by fluorescence microscope assays and western blot analysis.42 The 5-chloro-N-benzyl-1H-benzimidazole also showed potent antitumor activity with an IC50 of 7.01 ± 0.20 μM and arresting MCF-7 cell growth at the G2/M phase and S phase.43 Compounds 3s and 4k have similarities to the active compound of Yoon et al. (2014) with the –N(CH3)2 group at position 4 on the 2-phenyl ring and the N-aryl group at position 1 of the 1H-benzimidazole nucleus. However, compound 4k exhibited more potential antitumor activity against five different types of cancer cells when compared with compounds of Yoon et al. (2014) and Zhang et al. (2017).41,42 This may be due to the structure of 4k having the presence of a 4-N(CH3)2 substituent on the phenyl ring, 6-nitro, and N-(4-chlorobenzyl) groups on the 1H-benzimidazole scaffold.
2.4. In silico ADMET profile
In the present study, a computational study of the five most active compounds was conducted to determine the surface area and other physicochemical properties according to the directions of Lipinski's rule.28 Lipinski suggested that the absorption capacity of a compound is much better if the molecule achieves at least three out of four of the following rules: (i) HB donor groups ≤ 5; (ii) HB acceptor groups ≤ 10; (iii) Mw less than 500; (iv) logP less than 5. In this study, compounds 1d, 2d, 3s, 4b, and 4k follow all Lipinski's rules. All the highest active derivatives have a number of hydrogen bonding acceptor groups ranging between 1 to 3, and hydrogen bonding donor groups ranging between 0 to 1. Also, molecular weights ranging from 297.57 to 406.86, and logP values ranging between 3.46 to 4.76, and all these values agree with Lipinski's rules.After assessing ADMET profiles of active compounds (Table 5), we can suggest that these derivatives have the advantage of better intestinal absorption in humans than Cipro, Flu, and PTX, as all compounds showed Caco-2 permeability higher than the control drugs and higher than −5.15 log unit. Besides, compounds 2d, 3s, 4b, and 4k showed high passive MDCK permeability (>20 × 10−6 cm s−1) as compared to the reference drugs. This preference may attribute to the superior lipophilic of the designed ligands, which would facilitate passage along different biological membranes.28 Accordingly, they may have remarkably good bioavailability after oral administration. Compounds 3s, 4b, and 4k are highly likely to be Pgp-inhibitor similar to the PTX reference drug. This is a therapeutic approach for overcoming multidrug resistance in cancer. In addition, all compounds showed good plasma protein binding capacity with PPB > 98.5% as compared to Cipro (PPB = 37%), Flu (PPB = 62%), and PTX (95%). Studying the BBB (Blood–Brain Barrier) permeability, compounds 3s and 4b demonstrated the best ability to penetrate the BBB as compared to Flu, while other compounds, Cipro and PTX are unable to penetrate. Therefore, the treatment of brain tumors and brain infections is a great advantage of compounds 4s and 4b compared with reference drugs.
| Co. | DHFR-B | GyrB | DHFR-F | NMT | VEGFR-2 | FGFR-1 | HDAC6 | |||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| a | b | a | b | a | b | a | b | a | b | a | b | a | b | |
| a Co. – compound. The bacterial targets consist of DHFR-B: Dihydrofolate Reductase-Bacteria, GyrB: Gyrase B. The fungal targets consist of DHFR-F: Dihydrofolate Reductase-Fungi, NMT: N-myristoyl Transferase. The cancer targets include all seven receptors. Hydrogen bonds include conventional (not a symbol), carbon (#), and π–donor (*) categories. Cipro: ciprofloxacin, Flu: fluconazole, PTX: paclitaxel. a: affinity (Kcal mol−1), b: hydrogen bond (number, position). | ||||||||||||||
| 1d | −8.6 | 1, THR121* | −7.4 | 2, GLU58, GLY85 | −7.7 | 0 | −10.1 | 1, HIS227 | −9.7 | 2, ASP1046, GLU885 | −8.2 | 1, LYS514* | −7.4 | 1, HIS232* |
| 2d | −8.9 | 2, ASN18, ILE14 | −7.7 | 0 | −8.4 | 5, ALA11, ILE19, VAL10#, GLY114#, THR147* | −10.1 | 1, HIS227 | −9.1 | 3, ASP1046, GLU885, LYS868 | −8.1 | 3, ASP641, PHE642, LYS514* | −8.1 | 3, HIS192, HIS193, LYS330* |
| 3s | −9.2 | 1, ASN18# | −7.7 | 1, ASP81# | −8.4 | 0 | −9.6 | 0 | −9.4 | 1, VAL914# | −9.6 | 1, GLU531# | −7.6 | 1, SER150 |
| 4b | −9.2 | 1, GLN95 | −7.6 | 1, ASN54# | −7.7 | 3, GLY23, GLY114#, THR147* | −9.9 | 1, HIS227 | −7.5 | 3, ASP1046, ILE1025#, HIS1026# | −8.0 | 1, GLY567# | −7.8 | 5, HIS192, HIS193, SER150, TYR363, GLY361* |
| 4k | −9.4 | 3, ALA7, ASP120#, THR121* | −8.0 | 1, ASN54# | −8.2 | 1, TYR118 | −9.9 | 1, ASP110# | −9.6 | 3, HIS1026#, VAL914# | −8.5 | 1, LYS514* | −8.6 | 5, HIS192, HIS193, SER150, TYR363, GLY361* |
| Cipro | −9.1 | 1, SER49 | −7.3 | 2, ASP81, SER55 | — | — | — | — | — | — | — | — | — | — |
| Flu | — | — | — | — | −7.0 | 4, ALA115, GLU116, LYS57 | −7.9 | 1, TYR225 | — | — | — | — | — | — |
| PTX | −10.0 | 3, LEU20, SER49, THR121 | −7.8 | 5, ASN54, ARG84, GLY85, THR173 | −8.5 | 2, ARG28 | −11.4 | 1, GLY213 | −7.8 | 1, GLY1048 | −10.5 | 3, ASN628, GLU486, THR658 | −8.8 | 4, LYS330, SER150, VAL151 |
The less skin permeant is the molecule, the more negative the log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp (with Kp in cm s−1). Therefore, all active compounds (log
Kp (with Kp in cm s−1). Therefore, all active compounds (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp in the range of −5.03 to −4.54) showed better skin permeation than Cipro (log
Kp in the range of −5.03 to −4.54) showed better skin permeation than Cipro (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp at −9.09) and Flu (log
Kp at −9.09) and Flu (log![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) Kp −7.92). The cytochrome enzymes could be moderate to strong inhibited under the effect of active compounds especially CYP1A2, CYP2C19, CYP2C9, and CYP2D6, while Cipro and Flu couldn't. Compounds 2d, 3s, 4b, and 4k also showed the effect of CYP3A4 inhibition compared with PTX.
Kp −7.92). The cytochrome enzymes could be moderate to strong inhibited under the effect of active compounds especially CYP1A2, CYP2C19, CYP2C9, and CYP2D6, while Cipro and Flu couldn't. Compounds 2d, 3s, 4b, and 4k also showed the effect of CYP3A4 inhibition compared with PTX.
The CL (clearance) is a significant parameter in deciding dose intervals as a tool for the assessment of excretion. Compounds 3s and 4k (6.55–7.91 mL min−1 kg−1) and Flu (CL = 5.69 mL min−1 kg−1) was classified as a moderate clearance level ranging between 5 to 15 mL min−1 kg−1. In contrast, compounds 1d (4.89 mL min−1 kg−1), 2d (3.98 mL min−1 kg−1), and 4b (4.48 mL min−1 kg−1), Cipro (3.21 mL min−1 kg−1) and PTX (3.42 mL min−1 kg−1) showed lower CL values and were classified as low clearance levels (CL < 5 mL min−1 kg−1).
Toxicity is the last parameter examined in the ADMET profile. All the new ligands did not show H-HT (human hepatotoxicity), rat oral acute toxicity, skin sensitization, and eye corrosion. However, all the new ligands showed eye irritation, and the maximum recommended daily dose similar to the reference drug. In particular, the most potent compounds 3s and 4k showed lower carcinogenicity than the reference drug Cipro and lower respiratory toxicity than the reference drug PTX. Moreover, compound 4k exhibited good “Tox21 pathway” and “Toxicophore rules” profiles as compared to the reference drug PTX.
2.5. In silico molecular docking studies
After ADMET profiling, docking studies were carried out to predict the most suitable binding pose and inhibition mechanism of good active compounds. Based on the principle that similar compounds tend to bind to the same proteins, we predicted several reported protein targets against reference compounds (Cipro – ciprofloxacin, Flu – fluconazole, and PTX – paclitaxel) and docked our active compounds against them. Four different target proteins were selected for antimicrobial activity including dihydrofolate reductase (DHFR-F) and N-myristoyl transferase (NMT) from Candida albicans as fungal targets together with dihydrofolate reductase (DHFR-B) and gyrase B (GyrB) from Staphylococcus aureus as bacterial targets.28 Both seven target proteins were selected for anticancer activity including DHFR-B, GyrB, DHFR-F, NMT, vascular endothelial growth factor receptor 2 (VEGFR-2), fibroblast growth factor receptor 1 (FGFR-1), and histone deacetylase 6 (HDAC6). The protein–ligand complex is formed through the electrostatic interactions of the binding interface including hydrogen bonds (both from side chains and backbones), salt bridges, and π–π stacking. Hydrogen bonding provides stability to protein molecules and selected protein–ligand interactions, thus being one of the most important for biological macromolecule interactions. In addition, hydrogen bonds are divided into different types such as conventional, carbon, and π-donor, in which conventional hydrogen bonds are the strongest interactions.Among all these seven proteins, two proteins (DHFR-B and NMT) as both antimicrobial and antitumor targets presented good binding affinity with a higher affinity than −8.6 Kcal mol−1.44 On the other hand, two proteins (VEGFR-2 and HDAC6) as antitumor targets presented good interactions with affinity in the range of −7.4 to −9.7 Kcal mol−1 compared with reference drug PTX (−7.8 Kcal mol−1 at VEGFR-2 and -8.8 Kcal mol−1 at HDAC6), while FGFR-1 showed weaker interactions with affinity in the range of −8.0 to −9.6 Kcal mol−1 compared with PTX (−10.5 Kcal mol−1) (Table 5).
On the DHFR-B receptor, compounds 3s and 4b established one hydrogen bond with the affinity of −9.2 Kcal mol−1. Compound 4b established a conventional hydrogen bond (2.05 Å) with GLN95 amino acid but compound 3s only established a carbon–hydrogen bond (3.52 Å) with ASN18 amino acid. In particular, compound 4k being the most potent antimicrobial and antitumor agent against DHFR-B displayed the highest negative affinity of −9.4 Kcal mol−1 which is comparable to Cipro (−9.1 Kcal mol−1) and PTX (−10.0 Kcal mol−1). Besides, compound 4k established three hydrogen bonds that were similar to PTX with ALA7 (conventional), ASP120 (carbon), and THR121 (π-donor) amino acids with bond lengths of 2.46, 3.57, and 2.98 Å, respectively. On the other hand, compound 4k showed hydrophobic interactions (π–σ, alkyl, π–alkyl) with LEU20, LYS45, ILE14, and ALA7 with the crucial residue of the DHFR-B protein from S. aureus that resembles the co-crystallization ligand, Cipro, and PTX. As illustrated in Fig. 5, the N,N-dimethylamino (–N(CH3)2) group in the 2-phenyl ring of a 1H-benzimidazole nucleus of compound 4k were engaged in the formation of carbon–hydrogen bond with ASP120 and alkyl interactions with LYS45 amino acid with a bond length of 4.26 Å. Moreover, the N-(4-chlorobenzyl) group and 1H-benzimidazole nucleus displayed π–σ, alkyl, and π–alkyl interactions with the crucial residue LEU20, ILE14, and ALA7 of the target protein with a bond length in the range of 3.78–5.23 Å. These results have demonstrated that compound 4k has the most potential in vitro antibacterial and antitumor activities.
On the GyrB receptor, all active compounds showed good interactions with affinity in the range of −7.4 to −8.0 Kcal mol−1 compared with the standard drug Cipro (−7.3 Kcal mol−1) and PTX (−7.8 Kcal mol−1) but showed less hydrogen bonding. Similarly, all active compounds also showed good interactions with affinity in the range of −7.7 to −8.4 Kcal mol−1 compared with the standard drug Flu (−7.0 Kcal mol−1) and PTX (−8.5 Kcal mol−1) on DHFR-F receptor, but compounds 1d, 3s, 4b, and 4k have not formed or have formed hydrogen bonds in lesser quantities at amino acid sites other than the reference drug Flu. However, compound 2d displayed the best negative affinity of −8.4 Kcal mol−1 with five hydrogen bonds (2.13–3.62 Å) with ALA11, ILE19, VAL10, GLY114, and THR147 amino acids.
On the NMT receptor, compounds 1d, 2d, and 4b established one conventional hydrogen bond (2.30–2.44 Å) with a good affinity (−9.9 to −10.1 Kcal mol−1) with HIS227 amino acid compared with Flu (−7.9 Kcal mol−1), PTX (−11.4 Kcal mol−1). On the contrary, compound 3s did not establish hydrogen bonds with affinity at −9.6 Kcal mol−1 and compound 4k only establish one carbon–hydrogen bond with a bond length of 3.79 Å with a good affinity of −9.9 Kcal mol−1.
On the VEGFR-2 receptor, compounds 1d, 2d, 3s, and 4k showed stronger interactions with the affinity between −9.1 and −9.7 Kcal mol−1 compared with reference drug PTX (−7.8 Kcal mol−1). Compounds 2d, 4b, and 4k established three hydrogen bonds with bond lengths in the range of 1.87–3.75 Å. However, compounds 1d and 4k showed the strongest interactions with the affinity of −9.7 and −9.6 Kcal mol−1, respectively. Compound 1d established two conventional hydrogen bonds (1.97–2.29 Å) with ASP1046 and GLU885 amino acids, electrostatic (π–cation) interactions with bond lengths in the range of 4.33 to 4.50 Å with LYS868 amino acid. In particular, compound 4k established three carbon–hydrogen bonds (3.29–3.65 Å) at 6-nitro and N,N-dimethylamino groups with HIS1026 and VAL914 amino acids. Besides, compound 4k showed electrostatic (π–cation) interactions with LYS868 and HIS1026 amino acids with bond lengths in the range of 4.45 to 4.49 Å and electrostatic (π–anion) interactions with GLU885 amino acid with a bond length of 3.67 Å. In addition, compound 4k showed hydrophobic interactions (π–σ, π–π T-shaped, alkyl, π–alkyl) with LEU889, VAL899, HIS1026, LYS868, VAL916, ILE892, LEU1019, and ILE888 amino acids with bond lengths in the range of 3.79 to 5.44 Å (Fig. 6).
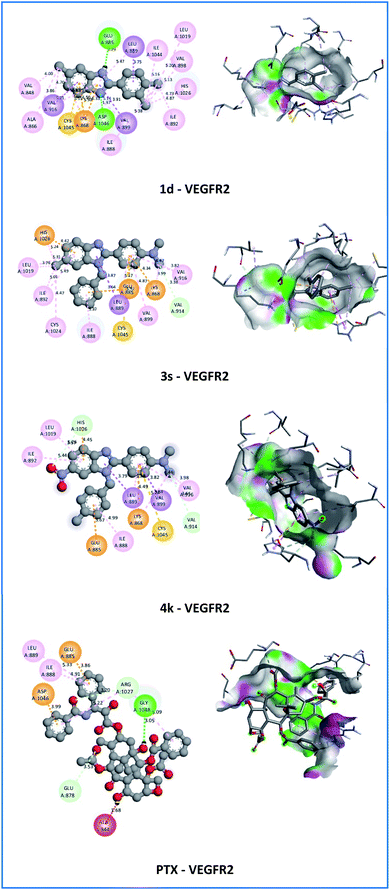 | ||
| Fig. 6 2D and 3D representation of the interaction of the synthesized molecules 1d, 3s, 4k, and paclitaxel (PTX) with vascular endothelial growth factor receptor 2 (unit of interaction distance – Å). | ||
On the FGFR-1 receptor, all active compounds established one π-donor hydrogen or carbon–hydrogen bond (3.00–3.66 Å) except for 2d established three hydrogen bonds (2.17–3.01 Å) with ASP641, PHE642, and LYS514 amino acids. In addition, these compounds showed weaker interactions with the affinity between −8.0 and −9.6 Kcal mol−1 compared with the reference drug PTX (−10.5 Kcal mol−1). Compound 3s displayed the highest negative affinity of −9.6 Kcal mol−1 among active compounds against FGFR-1. Moreover, compound 3s established one carbon–hydrogen bond with a bond length of 3.20 Å with GLU531 amino acid, electrostatic (π–anion) interaction with a bond length of 4.54 Å with ASP641 amino acid. Compound 3s also showed hydrophobic interactions (π–σ, π–π T-shaped, alkyl, π–alkyl) with LEU889, VAL899, HIS1026, LYS868, VAL916, ILE892, LEU1019, and ILE888 amino acids with bond lengths in the range of 3.79 to 5.44 Å.
On the HDAC6 receptor, all active compounds showed weaker interactions with the affinity between −7.4 and −8.6 Kcal mol−1 compared with the reference drug PTX (−8.8 Kcal mol−1). However, compounds 4b and 4k exhibited more hydrogen bonds than PTX (Fig. 7). These compounds established four conventional hydrogen bonds (1.84–2.96 Å) with SER150, HIS192, HIS193, and TYR363 amino acids and one carbon–hydrogen bond (3.59–3.69 Å) with GLY361 amino acid at the nitro group and 1H-imidazole nucleus. In particular, compound 4k demonstrated the strongest affinity (−8.6 Kcal mol−1) among all active derivatives against HDAC6. In addition, compound 4k showed metal–acceptor interaction with ZN2001 with a bond length of 2.44 Å and hydrophobic interactions (π–π stacked, π–π T-shaped, π–alkyl) with PHE202, PHE260, TRP261, and LYS330 amino acids with bond lengths in the range of 4.16 to 5.34 Å at the aromatic rings. Hydrophobic interaction with TRP261 of 4k compound is similar to that of PTX. These results suggested that HDAC6 also are the most likely target for the anticancer activity of the 4k compound.
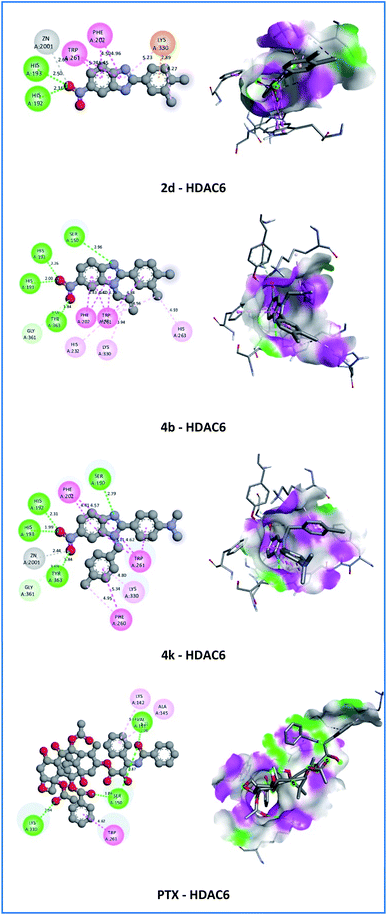 | ||
| Fig. 7 2D and 3D representation of the interaction of the synthesized molecules 2d, 4b, 4k, and paclitaxel (PTX) with histone deacetylase 6 (unit of interaction distance – Å). | ||
In summary, from the in silico molecular docking study results, it can be concluded that compound 4k is considered the best dock conformation in antibacterial and antitumor targets such as DHFR-B, VEGFR-2, and HDAC6.
3. Conclusion
In summary, thirty-four 6-substituted 1H-benzimidazole and forty-two N-substituted 6-(chloro/nitro)-1H-benzimidazole derivatives including twenty-nine new compounds have been designed, synthesized, and evaluated for their antimicrobial and anticancer activities. The microwave-assisted method has contributed to a significant reduction in reaction time and a significant increase in product yield. In addition, compounds 1d, 2d, 3s, 4b, and 4k showed potent antibacterial activity against EC, SF, MSSA, and MRSA with MIC ranging between 2 to 16 μg mL−1 compared with standard drug Cipro, especially compound 4k is potent fungal activity against CA and AN with MIC ranging between 8 to 16 μg mL−1 compared with standard drug Flu. Moreover, these compounds also exhibited potent anticancer activity with IC50 < 10 μM against all tested cell lines (HepG2, MDA-MB-231, MCF7, C26, and RMS) compared with reference drug PTX. From the structure–activity relationship, the presence of the N-benzyl/N-(4-chlorobenzyl) group and the chloro/N,N-dimethylamino moiety in the aromatic ring at position 2 of the 1H-benzimidazole scaffold is more desirable for enhanced antibacterial activity as well as antitumor activity in 1d, 2d, 3s, 4b, and 4k, and antifungal activity in 4k. Molecular docking predicted that dihydrofolate reductase protein from S. aureus is the most suitable target for antibacterial and anticancer activities, as well as vascular endothelial growth factor receptor 2 and histone deacetylase 6 are the most suitable targets for anticancer activity. Compound 4k being the most potent antimicrobial and anticancer displayed good interactions against DHFR-B, VEGFR2, and HDAC6 with the affinity of −9.4, −9.6, and −8.6 Kcal mol−1, respectively. Especially, this compound showed electrostatic and hydrophobic interactions that resemble the co-crystallization ligand and reference drugs. ADMET profile was evaluated for the five most active compounds in comparison to ciprofloxacin and fluconazole, and paclitaxel as reference drugs. The obtained results predicted that our derivatives may show a good ADMET profile. All compounds show physical–chemical properties of fragment and lead-like compounds which are of great interest for further drug development. This work paved the way for the synthesis of more potent compounds based on N-benzyl-1H-benzimidazole scaffolds and explore their various and potential biological activities as well as their mechanism of action.4. Experimental section
4.1. Materials
All chemicals and solvents were purchased from commercial suppliers Merck and Acros. All the reactions were carried out under an inert atmosphere of nitrogen. TLC was performed on pre-coated aluminum sheets of silica (60 F254 nm) and visualized by shortwave UV light at λ 254. Column chromatography uses 0.040–0.063 mm granular silica gel (Merck).The microwave reactor used was the Microwave synthesizer – CEM Discover, USA, fitted with a magnetic stirrer for continuous stirring and an infrared temperature sensor, which enabled and controlled the temperature. Melting points (mp) were determined on a Sanyo-Gallenkamp melting point apparatus. UV-Vis absorption spectra were recorded on a PerkinElmer Lambda 40p spectrometer. IR spectra were recorded on an IRAffinity-1S. NMR spectra were recorded on a Bruker Avance 500 NMR Spectrometer (1H NMR 500 MHz, 13C NMR 125 MHz). Chemical shifts were measured in δ (ppm). Mass spectrometry was measured on 1100 series LC-MS Trap Agilent.
4.2. Experimental procedures
The reflux method. A mixture of 4-chlorobenzene-1,2-diamine or 4-chlorobenzene-1,2-diamine (5 mmol), the substituted aromatic aldehydes (5 mmol), and Na2S2O5 (20 mmol) was dissolved in a mixture of absolute alcohol and water at a ratio of 9
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v, 30 mL) and refluxed for 6–12 h at 80 °C. After cooling, the reaction crude was poured on a mixture of ice/water to give a solid that was filtered off in a Büchner funnel. The resulting solid was purified by column chromatography on silica gel using hexane/ethyl acetate as eluent. Reaction yields ranged within 70–91%.
1 (v/v, 30 mL) and refluxed for 6–12 h at 80 °C. After cooling, the reaction crude was poured on a mixture of ice/water to give a solid that was filtered off in a Büchner funnel. The resulting solid was purified by column chromatography on silica gel using hexane/ethyl acetate as eluent. Reaction yields ranged within 70–91%.
The microwave-assisted method. A mixture of 4-chlorobenzene-1,2-diamine or 4-chlorobenzene-1,2-diamine (5 mmol), the substituted aromatic aldehydes (5 mmol), Na2S2O5 (20 mmol), and a mixture of absolute alcohol and water at a ratio of 9
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 (v/v, 10 mL) was placed in a microwave oven and irradiated at a power of 300 W for 10–15 min at 80 °C. After cooling, the reaction crude was poured on a mixture of ice/water to give a solid that was filtered off in a Büchner funnel. The resulting solid was purified by column chromatography on silica gel using hexane/ethyl acetate as eluent. Reaction yields ranged within 90–99%.
1 (v/v, 10 mL) was placed in a microwave oven and irradiated at a power of 300 W for 10–15 min at 80 °C. After cooling, the reaction crude was poured on a mixture of ice/water to give a solid that was filtered off in a Büchner funnel. The resulting solid was purified by column chromatography on silica gel using hexane/ethyl acetate as eluent. Reaction yields ranged within 90–99%.
6-Chloro-2-(2-chlorophenyl)-1H-benzo[d]imidazole (1a). White solid, mp 122–123 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 12.92 (1H, s, –NH–), 7.91 (1H, dd, J = 7.5, 2.0 Hz, HAr), 7.78–7.70 (1H, m, HAr), 7.65 (1H, dd, J = 7.0, 1.5 Hz, HAr), 7.60–7.52 (3H, m, HAr), 7.27 (1H, d, J = 4.0 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 150.4, 144.0, 133.5, 132.1, 131.6, 130.4, 129.4, 127.5, 122.8, 120.5, 118.5, 113.1, 111.3. LC-MS (m/z) [M − H]− calcd for C13H7Cl2N2 260.9992, found 260.9912; [M + H]+ calcd for C13H9Cl2N2 263.0137, found 263.0160.
6-Chloro-2-(4-chlorophenyl)-1H-benzo[d]imidazole (1b). White solid, mp 227–228 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.14 (1H, s, –NH–), 8.19 (2H, d, J = 8.5 Hz, HAr), 7.64–7.61 (2H, m, HAr), 7.40 (2H, d, J = 8.5 Hz, HAr), 7.23 (2H, d, J = 8.0 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.6, 134.9 (2C), 129.1 (2C), 128.5 (2C), 128.3 (2C), 126.6 (2C), 122.6 (2C). LC-MS (m/z) [M − H]− calcd for C13H7Cl2N2 260.9992, found 260.9881; [M + H]+ calcd for C13H9Cl2N2 263.0137, found 263.0148.
6-Chloro-2-(2,4-dichlorophenyl)-1H-benzo[d]imidazole (1c). White solid, mp 245–246 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 12.97 (1H, s, –NH–), 7.94 (1H, d, J = 8.5 Hz, HAr), 7.84 (1H, d, J = 2.0 Hz, HAr), 7.77–7.68 (1H, m, HAr), 7.67–7.59 (2H, m, HAr), 7.30–7.26 (1H, m, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 149.4, 135.3, 133.2, 132.6 (2C), 129.9 (2C), 128.3 (2C), 127.8 (2C), 122.8 (2C). LC-MS (m/z) [M − H]− calcd for C13H6Cl3N2 294.9602, found 294.9624; [M + H]+ calcd for C13H8Cl3N2 296.9748, found 296.9725.
6-Chloro-2-(3,4-dichlorophenyl)-1H-benzo[d]imidazole (1d). White solid, mp 233–234 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.26 (1H, s, –NH–), 8.37 (1H, d, J = 2.0 Hz, HAr), 8.13 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.83 (1H, d, J = 8.5 Hz, HAr), 7.74–7.55 (2H, m, HAr), 7.25 (1H, d, J = 8.5 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 150.2, 132.7, 131.9, 131.3 (2C), 130.2 (2C), 128.1 (2C), 126.6 (2C), 122.9 (2C). LC-MS (m/z) [M − H]− calcd for C13H6Cl3N2 294.9602, found 294.9566; [M + H]+ calcd for C13H8Cl3N2 296.9748, found 296.9752.
6-Chloro-2-(2-chloro-6-fluorophenyl)-1H-benzo[d]imidazole (1e). White solid, mp 177–178 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 12.87 (1H, s, –NH–), 7.68 (1H, q, J = 8.5 Hz, HAr), 7.65–7.60 (1H, m, HAr), 7.56 (1H, d, J = 8.0 Hz, HAr), 7.51 (1H, t, J = 8.5 Hz, HAr), 7.46 (1H, d, J = 9.0 Hz, HAr), 7.15 (1H, t, J = 8.5 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 161.4, 159.2, 148.5, 136.8, 134.3, 134.2, 133.5, 133.6, 126.0, 125.97, 118.9, 118.7, 118.3, 115.3, 115.2. LC-MS (m/z) [M − H]− calcd for C13H6Cl2FN2 278.9898, found 278.9682; [M + H]+ calcd for C13H8Cl2FN2 281.0043, found 281.0109.
6-Chloro-2-(3,4-dimethoxyphenyl)-1H-benzo[d]imidazole (1f). White solid, mp 215–218 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 12.93 (1H, s, –NH–), 7.75–7.73 (2H, m, HAr), 7.53–7.51 (2H, m, HAr), 7.19 (1H, d, J = 8.0 Hz, HAr), 7.13 (1H, d, J = 8.5 Hz, HAr), 3.88 (3H, s, –OCH3), 3.84 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.1, 150.6, 144.8, 142.7, 135.7, 126.4, 122.2, 121.8, 119.6, 118.1, 112.3, 110.7, 109.8, 55.62, 55.60. LC-MS (m/z) [M − H]− calcd for C15H12ClN2O2 287.0593, found 287.0522; [M + H]+ calcd for C15H14ClN2O2 289.0738, found 289.0718.
6-Chloro-2-(4-ethoxyphenyl)-1H-benzo[d]imidazole (1g). White solid, mp 253–254 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 12.91 (1H, s, –NH–), 8.09 (2H, d, J = 8.5 Hz, HAr), 7.66–7.61 (1H, m, HAr), 7.49 (1H, d, J = 9.0 Hz, HAr), 7.18 (1H, t, J = 8.5 Hz, HAr), 7.09 (2H, d, J = 9.0 Hz, HAr), 4.12 (2H, q, J = 7.0 Hz, –CH2–), 1.36 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 160.2, 128.2, 122.0 (2C), 119.7, 117.8 (2C), 114.8 (2C), 111.3 (2C), 110.7 (2C), 63.3, 14.6. LC-MS (m/z) [M − H]− calcd for C15H12ClN2O 271.0644, found 271.0628; [M + H]+ calcd for C15H14ClN2O 273.0789, found 273.0771.
4-(6-Chloro-1H-benzo[d]imidazole-2-yl)-2-ethoxyphenol (1h). White solid, mp 215–217 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 12.82 (1H, s, –NH–), 9.51 (1H, s, –OH), 7.71 (1H, s, HAr), 7.68–7.40 (3H, m, HAr), 7.17 (1H, dd, J = 8.5, 1.5 Hz, HAr), 6.93 (1H, d, J = 8.0 Hz, HAr), 4.14 (2H, q, J = 7.0 Hz, –CH2–), 1.39 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.2, 149.1, 147.0, 125.9 (2C), 121.8 (2C), 120.8 (2C), 119.9 (2C), 115.8, 111.7, 64.0, 14.7. LC-MS (m/z) [M − H]− calcd for C15H12ClN2O2 287.0593, found 287.0478; [M + H]+ calcd for C15H14ClN2O2 289.0738, found 289.0708.
6-Chloro-2-(4-fluorophenyl)-1H-benzo[d]imidazole (1i). White solid, mp 219–221 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.18 (1H, s, –NH–), 8.18 (2H, d, J = 8.5 Hz, HAr), 7.67–7.73 (1H, m, HAr), 7.64 (2H, d, J = 8.5 Hz, HAr), 7.57 (1H, s, HAr), 7.24 (1H, s, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.1, 148.7, 147.8, 124.2 (2C), 120.8 (2C), 108.7 (2C), 106.4 (2C), 101.5 (2C). LC-MS (m/z) [M − H]− calcd for C13H7ClFN2 245.0287, found 245.0210; [M + H]+ calcd for C13H9ClFN2 247.0433, found 247.0277.
3-(6-Chloro-1H-benzo[d]imidazole-2-yl)phenol (1j). Brown solid, mp 289–290 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.01 (1H, s, –NH–), 9.76 (1H, s, –OH), 7.71–7.52 (4H, m, HAr), 7.35 (1H, t, J = 8.0 Hz, HAr), 7.21 (1H, d, J = 7.0 Hz, HAr), 6.92 (1H, dd, J = 8.0, 1.5 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 157.8, 152.9, 144.7, 142.6, 135.7, 133.8, 130.9, 126.7, 122.5, 120.0, 118.2, 113.4, 110.9. LC-MS (m/z) [M − H]− calcd for C13H8ClN2O 243.0331, found 243.0196; [M + H]+ calcd for C13H10ClN2O 245.0476, found 245.0427.
6-Chloro-2-(3-methoxyphenyl)-1H-benzo[d]imidazole (1k). White solid, mp 179–181 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.07 (1H, s, –NH–), 7.75–7.73 (2H, m, HAr), 7.70–7.52 (2H, m, HAr), 7.47 (1H, t, J = 8.0 Hz, HAr), 7.23 (1H, d, J = 6.5 Hz, HAr), 7.08 (1H, dd, J = 8.5, 1.5 Hz, HAr), 3.86 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 159.6, 152.1, 144.6, 142.5, 135.7, 130.9, 126.9, 122.6, 120.1, 118.9, 116.2, 112.6, 111.5, 55.3. LC-MS (m/z) [M − H]− calcd for C14H10ClN2O 257.0487, found 257.0487; [M + H]+ calcd for C14H12ClN2O 259.0633, found 259.0684.
5-(6-Chloro-1H-benzo[d]imidazole-2-yl)-2-methoxyphenol (1l). White solid, mp 253–254 °C. IR (ν, cm−1): 1605.2 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1439.0 (C
N), 1439.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1340.5 (N
C), 1340.5 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.66–7.60 (3H, m, HAr), 7.53 (1H, d, J = 9.0 Hz, HAr), 7.21 (1H, t, J = 8.5 Hz, HAr), 7.14 (1H, d, J = 8.5 Hz, HAr), 3.96 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 155.6, 149.8, 147.4, 138.8, 126.3 (2C), 122.1 (2C), 121.2 (2C), 120.1, 115.8, 112.7, 56.9. LC-MS (m/z) [M + H]+ calcd for C14H12ClN2O2 275.0582, found 275.0665.
O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.66–7.60 (3H, m, HAr), 7.53 (1H, d, J = 9.0 Hz, HAr), 7.21 (1H, t, J = 8.5 Hz, HAr), 7.14 (1H, d, J = 8.5 Hz, HAr), 3.96 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 155.6, 149.8, 147.4, 138.8, 126.3 (2C), 122.1 (2C), 121.2 (2C), 120.1, 115.8, 112.7, 56.9. LC-MS (m/z) [M + H]+ calcd for C14H12ClN2O2 275.0582, found 275.0665.
6-Chloro-2-(3-nitrophenyl)-1H-benzo[d]imidazole (1m). Yellow solid, mp 243–244 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.46 (1H, s, –NH–), 8.99 (1H, s, HAr), 8.59 (1H, d, J = 7.5 Hz, HAr), 8.34 (1H, dd, J = 8.5, 1.5 Hz, HAr), 7.86 (1H, t, J = 8.0 Hz, HAr), 7.70 (1H, s, HAr), 7.66 (1H, d, J = 8.5 Hz, HAr), 7.27 (1H, dd, J = 8.5, 1.5 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 150.4, 148.3, 132.6, 131.2, 130.7, 127.0 (2C), 124.5 (2C), 123.0 (2C), 121.0 (2C). LC-MS (m/z) [M − H]− calcd for C13H7ClN3O2 272.0232, found 272.0206; [M + H]+ calcd for C13H9ClN3O2 274.0378, found 274.0318.
6-Chloro-2-(4-nitrophenyl)-1H-benzo[d]imidazole (1n). Yellow solid, mp 260–262 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.47 (1H, s, –NH–), 8.50–8.40 (4H, m, HAr), 7.72–7.68 (2H, m, HAr), 7.30 (1H, d, J = 8.5 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 148.1, 135.5, 134.0 (2C), 127.6 (2C), 124.3 (2C), 123.8 (2C), 118.8, 113.2, 111.5. LC-MS (m/z) [M − H]− calcd for C13H7ClN3O2 272.0232, found 272.0104; [M + H]+ calcd for C13H9ClN3O2 274.0378, found 273.9934.
4-(6-Chloro-1H-benzo[d]imidazole-2-yl)-N,N-dimethylaniline (1o). White solid, mp 246–248 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 12.71 (1H, s, –NH–), 7.99 (2H, d, J = 9.0 Hz, HAr), 7.54–7.49 (2H, m, HAr), 7.15 (1H, dd, J = 8.5, 1.5 Hz, HAr), 6.84 (2H, d, J = 9.0 Hz, HAr), 3.00 (6H, s, –N(CH3)2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.7, 151.4, 134.9 (2C), 127.7 (2C), 125.6 (2C), 121.4 (2C), 116.7 (2C), 111.8, 39.7. LC-MS (m/z) [M − H]− calcd for C15H13ClN3 270.0803, found 270.0948; [M + H]+ calcd for C15H15ClN3 272.0949, found 272.0909.
2-(Benzo[d][1,3]dioxol-5-yl)-6-chloro-1H-benzo[d]imidazole (1p). White solid, mp 211–213 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 12.92 (1H, s, –NH–), 7.71 (1H, d, J = 8.0 Hz, HAr), 7.66 (1H, s, HAr), 7.65–7.50 (2H, m, HAr), 7.20 (1H, d, J = 8.5 Hz, HAr), 7.10 (1H, d, J = 8.0 Hz, HAr), 6.13 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 149.1, 147.9 (2C), 123.7 (3C), 121.2 (2C), 108.8 (2C), 106.6 (2C), 101.7, 89.2. LC-MS (m/z) [M − H]− calcd for C14H8ClN2O2 271.0280, found 271.0257; [M + H]+ calcd for C14H10ClN2O2 273.0425, found 273.0423.
6-Chloro-2-(furan-2-yl)-1H-benzo[d]imidazole (1q). White solid, mp 175–177 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.11 (1H, s, –NH–), 7.96 (1H, d, J = 1.0 Hz, HAr), 7.80–7.40 (2H, m, HAr), 7.23–7.20 (2H, m, HAr), 6.74–6.72 (1H, m, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 145.0, 142.4, 135.0, 133.1, 126.8, 122.6, 122.2, 119.9, 118.1, 112.6, 111.1. LC-MS (m/z) [M − H]− calcd for C11H6ClN2O 217.0174, found 217.0118; [M + H]+ calcd for C11H8ClN2O 219.0320, found 219.0366.
2-(2-Chlorophenyl)-6-nitro-1H-benzo[d]imidazole (2a). Yellow solid, mp 173–175 °C. IR (ν, cm−1): 1593.2 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1435.0 (C
N), 1435.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1336.7 (N
C), 1336.7 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 737.2 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.42 (2H, s, –NH–), 8.61 (1H, s, HAr), 8.46 (1H, s, HAr), 8.20–8.14 (2H, m, HAr), 7.98–7.76 (4H, m, HAr), 7.71–7.70 (2H, d, J = 8.0 Hz, HAr), 7.63–7.60 (2H, m, HAr), 7.58–7.55 (2H, m, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.8, 147.6, 146.2, 140.3, 137.5, 132.9, 130.1, 129.6, 126.4, 121.9, 120.8, 118.7, 114.0. LC-MS (m/z) [M + H]+ calcd for C13H9ClN3O2 274.0378, found 274.0365.
O), 737.2 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.42 (2H, s, –NH–), 8.61 (1H, s, HAr), 8.46 (1H, s, HAr), 8.20–8.14 (2H, m, HAr), 7.98–7.76 (4H, m, HAr), 7.71–7.70 (2H, d, J = 8.0 Hz, HAr), 7.63–7.60 (2H, m, HAr), 7.58–7.55 (2H, m, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.8, 147.6, 146.2, 140.3, 137.5, 132.9, 130.1, 129.6, 126.4, 121.9, 120.8, 118.7, 114.0. LC-MS (m/z) [M + H]+ calcd for C13H9ClN3O2 274.0378, found 274.0365.
2-(4-Chlorophenyl)-6-nitro-1H-benzo[d]imidazole (2b). Yellow solid, mp 309–311 °C. IR (ν, cm−1): 1597.1 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1442.8 (C
N), 1442.8 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1284.6 (N
C), 1284.6 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 733.0 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.66 (1H, s, –NH–), 8.48 (1H, s, HAr), 8.24–8.21 (2H, m, HAr), 8.14 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.78 (1H, d, J = 8.5 Hz, HAr), 7.70–7.68 (2H, m, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.1, 146.4, 144.2, 139.7, 138.3, 134.5, 128.5 (2C), 126.2 (2C), 122.8, 119.0, 116.6. LC-MS (m/z) [M + H]+ calcd for C13H9ClN3O2 274.0378, found 274.0344.
O), 733.0 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.66 (1H, s, –NH–), 8.48 (1H, s, HAr), 8.24–8.21 (2H, m, HAr), 8.14 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.78 (1H, d, J = 8.5 Hz, HAr), 7.70–7.68 (2H, m, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.1, 146.4, 144.2, 139.7, 138.3, 134.5, 128.5 (2C), 126.2 (2C), 122.8, 119.0, 116.6. LC-MS (m/z) [M + H]+ calcd for C13H9ClN3O2 274.0378, found 274.0344.
2-(2,4-Dichlorophenyl)-6-nitro-1H-benzo[d]imidazole (2c). Yellow solid, mp 245–246 °C. IR (ν, cm−1): 1624.1 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1435.0 (C
N), 1435.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1339.3 (N
C), 1339.3 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 733.0 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.45 (1H, s, –NH–), 8.54 (1H, s, HAr), 8.16 (1H, dd, J = 8.5, 1.5 Hz, HAr), 7.97 (1H, d, J = 8.5 Hz, HAr), 7.87 (1H, d, J = 2.0 Hz, HAr), 7.81 (1H, d, J = 9.0 Hz, HAr), 7.65 (1H, dd, J = 8.5, 2.0 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.5, 142.9, 135.9, 133.4 (2C), 132.7 (3C), 130.0, 127.9, 127.7, 118.1 (2C). LC-MS (m/z) [M − H]− calcd for C13H6Cl2N3O2 305.9843, found 305.9880.
O), 733.0 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.45 (1H, s, –NH–), 8.54 (1H, s, HAr), 8.16 (1H, dd, J = 8.5, 1.5 Hz, HAr), 7.97 (1H, d, J = 8.5 Hz, HAr), 7.87 (1H, d, J = 2.0 Hz, HAr), 7.81 (1H, d, J = 9.0 Hz, HAr), 7.65 (1H, dd, J = 8.5, 2.0 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.5, 142.9, 135.9, 133.4 (2C), 132.7 (3C), 130.0, 127.9, 127.7, 118.1 (2C). LC-MS (m/z) [M − H]− calcd for C13H6Cl2N3O2 305.9843, found 305.9880.
2-(3,4-Dichlorophenyl)-6-nitro-1H-benzo[d]imidazole (2d). Yellow solid, mp 302–304 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.81 (1H, s, –NH–), 8.51 (1H, s, HAr), 8.44 (1H, d, J = 2.0 Hz, HAr), 8.19 (1H, dd, J = 8.5, 2.0 Hz, HAr), 8.16 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.89 (1H, d, J = 8.5 Hz, HAr), 7.81 (1H, d, J = 9.0 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.4, 148.2, 145.3, 141.5, 136.9, 133.6, 132.5, 129.0, 125.8, 121.6, 120.2, 118.9, 113.8. LC-MS (m/z) [M − H]− calcd for C13H6Cl2N3O2 305.9843, found 305.9795.
2-(2-Chloro-6-fluorophenyl)-6-nitro-1H-benzo[d]imidazole (2e). Yellow solid, mp 183–184 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.59 (1H, s, HAr), 8.19 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.84 (1H, d, J = 9.0 Hz, HAr), 7.69 (1H, q, J = 8.5 Hz, HAr), 7.58 (1H, d, J = 8.0 Hz, HAr), 7.50 (1H, t, J = 8.5 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 161.5, 159.5, 148.2, 143.0, 134.1, 134.0, 133.3, 133.2, 126.02, 125.99, 118.8, 118.6, 118.1, 115.1, 115.0. LC-MS (m/z) [M − H]− calcd for C13H6ClFN3O2 290.0138, found 290.0016; [M + H]+ calcd for C13H8ClFN3O2 292.0284, found 292.0235.
2-(3,4-Dimethoxyphenyl)-6-nitro-1H-benzo[d]imidazole (2f). Yellow solid, mp 126–128 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.44 (1H, d, J = 2.0 Hz, HAr), 8.17 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.73 (1H, d, J = 2.0 Hz, HAr), 7.70 (1H, dd, J = 8.0, 2.0 Hz, HAr), 7.66 (1H, d, J = 8.5 Hz, HAr), 7.13 (1H, d, J = 8.0 Hz, HAr), 3.98 (3H, s, –OCH3), 3.93 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 157.8, 153.5, 151.0, 145.0, 122.3 (2C), 121.7, 119.5 (2C), 115.2, 112.3 (2C), 111.5, 56.6, 56.5. LC-MS (m/z) [M − H]− calcd for C15H12N3O4 298.0833, found 298.0710.
2-(4-Ethoxyphenyl)-6-nitro-1H-benzo[d]imidazole (2g). Brown solid, mp 283–285 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.30 (1H, s, –NH–), 8.41 (1H, s, HAr), 8.14 (2H, d, J = 8.5 Hz, HAr), 8.10 (1H, d, J = 8.5 Hz, HAr), 7.71 (1H, d, J = 8.0 Hz, HAr), 7.12 (2H, t, J = 8.5 Hz, HAr), 4.13 (2H, q, J = 7.0 Hz, –CH2-), 1.36 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 160.8, 142.5 (2C), 128.7 (2C), 121.2 (3C), 117.7 (3C), 115.0 (2C), 63.4, 14.5. LC-MS (m/z) [M − H]− calcd for C15H12N3O3 282.0884, found 282.0950; [M + H]+ calcd for C15H14N3O3 284.1030, found 284.1096.
2-Ethoxy-4-(6-nitro-1H-benzo[d]imidazole-2-yl)phenol (2h). Yellow solid, mp 170–172 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.46 (1H, d, J = 2.0 Hz, HAr), 8.18 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.73 (1H, d, J = 2.0 Hz, HAr), 7.66 (1H, d, J = 8.5 Hz, HAr), 7.61 (1H, dd, J = 8.5, 2.0 Hz, HAr), 6.99 (1H, d, J = 8.0 Hz, HAr), 4.25 (2H, q, J = 7.0 Hz, –CH2–), 1.51 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.5, 148.8, 144.9, 121.9 (2C), 121.3 (2C), 119.3 (2C), 116.9 (2C), 112.8 (2C), 65.8, 15.1. LC-MS (m/z) [M − H]− calcd for C15H12N3O4 298.0833, found 298.0721.
2-(4-Fluorophenyl)-6-nitro-1H-benzo[d]imidazole (1i). Yellow solid, mp 219–221 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.60 (1H, s, –NH–), 8.47 (1H, s, HAr), 8.26 (2H, d, J = 9.0 Hz, HAr), 8.13 (1H, dd, J = 9.0, 1.5 Hz, HAr), 7.76 (1H, s, HAr), 7.45 (2H, t, J = 9.0 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 164.7, 162.7, 142.7 (2C), 129.5 (3C), 129.4, 125.6 (2C), 116.3 (2C), 116.2. LC-MS (m/z) [M − H]− calcd for C13H7FN3O2 256.0528, found 256.0454; [M + H]+ calcd for C13H9FN3O2 258.0673, found 258.0654.
3-(6-Nitro-1H-benzo[d]imidazole-2-yl)phenol (2j). Red solid, mp 307–309 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 12.89 (1H, s, –NH–), 9.78 (1H, s, –OH), 8.11–8.02 (4H, m, HAr), 7.42 (1H, t, J = 8.0 Hz, HAr), 7.34 (1H, d, J = 7.0 Hz, HAr), 6.97 (1H, dd, J = 8.0, 1.5 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 157.5, 152.9, 144.5, 135.1, 133.9, 130.7, 126.3, 122.9, 119.6, 117.5, 113.5, 112.8, 110.7. LC-MS (m/z) [M − H]− calcd for C13H8N3O3 254.0571, found 254.0576; [M + H]+ calcd for C13H10N3O3 256.0717, found 256.0738.
2-(3-Methoxyphenyl)-6-nitro-1H-benzo[d]imidazole (2k). Yellow solid, mp 234–236 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.02 (1H, s, –NH–), 8.46 (1H, s, HAr), 8.11 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.75 (1H, d, J = 8.5 Hz, HAr), 7.72–7.52 (3H, m, HAr), 7.09 (1H, dd, J = 8.5, 1.5 Hz, HAr), 3.85 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 159.5, 152.1, 142.1, 135.8, 133.6, 130.6, 126.9, 122.5, 120.2, 118.7, 116.5, 112.9, 111.5, 55.4. LC-MS (m/z) [M − H]− calcd for C14H10N3O3 268.0728, found 268.0814; [M + H]+ calcd for C14H12N3O3 270.0873, found 270.0749.
2-Methoxy-5-(6-nitro-1H-benzo[d]imidazole-2-yl)phenol (2l). Yellow solid, mp 263–265 °C. IR (ν, cm−1): 1600.1 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1436.9 (C
N), 1436.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1333.7 (N
C), 1333.7 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.50 (1H, s, HAr), 8.22–8.19 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.71 (1H, d, J = 9.0 Hz, HAr), 7.65–7.61 (2H, m, HAr), 7.14 (1H, d, J = 8.5 Hz, HAr), 3.98 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.7, 149.5, 147.6, 144.1, 126.3, 122.0 (2C), 120.9, 120.2 (2C), 115.9, 112.8 (2C), 56.4. LC-MS (m/z) [M + H]+ calcd for C14H12N3O4 286.0822, found 286.0871.
O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.50 (1H, s, HAr), 8.22–8.19 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.71 (1H, d, J = 9.0 Hz, HAr), 7.65–7.61 (2H, m, HAr), 7.14 (1H, d, J = 8.5 Hz, HAr), 3.98 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.7, 149.5, 147.6, 144.1, 126.3, 122.0 (2C), 120.9, 120.2 (2C), 115.9, 112.8 (2C), 56.4. LC-MS (m/z) [M + H]+ calcd for C14H12N3O4 286.0822, found 286.0871.
6-Nitro-2-(3-nitrophenyl)-1H-benzo[d]imidazole (2m). Yellow solid, mp 290–292 °C. IR (ν, cm−1): 1518.0 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1446.6 (C
N), 1446.6 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1342.5 (N
C), 1342.5 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.92 (1H, s, –NH–), 9.00 (1H, t, J = 2.0 Hz, HAr), 8.62 (1H, d, J = 8.0 Hz, HAr), 8.50 (1H, s, HAr), 8.36 (1H, dd, J = 8.5, 1.5 Hz, HAr), 8.14 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.88 (1H, t, J = 8.0 Hz, HAr), 7.80 (1H, d, J = 9.0 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.5, 148.3 (2C), 143.0, 133.0, 130.9 (2C), 130.5, 125.2, 121.4 (2C), 118.4 (2C). LC-MS (m/z) [M − H]− calcd for C13H7N4O4 283.0473, found 283.0440; [M + H]+ calcd for C13H9N4O4 285.0618, found 285.0601.
O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.92 (1H, s, –NH–), 9.00 (1H, t, J = 2.0 Hz, HAr), 8.62 (1H, d, J = 8.0 Hz, HAr), 8.50 (1H, s, HAr), 8.36 (1H, dd, J = 8.5, 1.5 Hz, HAr), 8.14 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.88 (1H, t, J = 8.0 Hz, HAr), 7.80 (1H, d, J = 9.0 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.5, 148.3 (2C), 143.0, 133.0, 130.9 (2C), 130.5, 125.2, 121.4 (2C), 118.4 (2C). LC-MS (m/z) [M − H]− calcd for C13H7N4O4 283.0473, found 283.0440; [M + H]+ calcd for C13H9N4O4 285.0618, found 285.0601.
6-Nitro-2-(4-nitrophenyl)-1H-benzo[d]imidazole (2n). Yellow solid, mp 285–286 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.36 (1H, s, –NH–), 8.50 (1H, s, HAr), 8.43 (2H, d, J = 8.0 Hz, HAr), 8.16 (2H, d, J = 8.0 Hz, HAr), 8.04 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.71 (1H, d, J = 8.5 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 148.7, 143.6 (2C), 135.8, 134.2, 127.5 (2C), 124.4 (2C), 123.6, 118.9, 113.3, 111.7. LC-MS (m/z) [M − H]− calcd for C13H7N4O4 283.0473, found 283.0411; [M + H]+ calcd for C13H9N4O4 285.0618, found 285.0670.
N,N-Dimethyl-4-(6-nitro-1H-benzo[d]imidazole-2-yl)aniline (2o). Red solid, mp 212–214 °C. IR (ν, cm−1): 1606.7 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1494.8 (C
N), 1494.8 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1330.9 (N
C), 1330.9 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.19 (1H, s, –NH–), 8.42–8.26 (1H, m, HAr), 8.07 (1H, d, J = 8.5 Hz, HAr), 8.03 (2H, d, J = 8.5 Hz, HAr), 7.69–7.60 (1H, m, HAr), 6.86 (2H, d, J = 9.0 Hz, HAr), 3.02 (6H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.1, 151.3, 127.6 (2C), 126.5 (2C), 122.4, 117.1 (3C), 112.5 (3C), 39.6. LC-MS (m/z) [M − H]− calcd for C15H13N4O2 281.1044, found 281.0968; [M + H]+ calcd for C15H15N4O2 283.1190, found 283.1166.
O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.19 (1H, s, –NH–), 8.42–8.26 (1H, m, HAr), 8.07 (1H, d, J = 8.5 Hz, HAr), 8.03 (2H, d, J = 8.5 Hz, HAr), 7.69–7.60 (1H, m, HAr), 6.86 (2H, d, J = 9.0 Hz, HAr), 3.02 (6H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.1, 151.3, 127.6 (2C), 126.5 (2C), 122.4, 117.1 (3C), 112.5 (3C), 39.6. LC-MS (m/z) [M − H]− calcd for C15H13N4O2 281.1044, found 281.0968; [M + H]+ calcd for C15H15N4O2 283.1190, found 283.1166.
2-(Benzo[d][1,3]dioxol-5-yl)-6-nitro-1H-benzo[d]imidazole (2p). Yellow solid, mp 208–210 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 12.85 (1H, s, –NH–), 8.41 (1H, s, HAr), 8.07 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.68 (1H, d, J = 8.5 Hz, HAr), 7.74 (1H, d, J = 8.0 Hz, HAr), 7.66–7.51 (1H, m, HAr), 7.10 (1H, d, J = 8.0 Hz, HAr), 6.15 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 150.2, 148.1 (3C), 143.6, 123.9 (2C), 121.5 (2C), 109.5 (2C), 106.8, 101.9, 89.4. LC-MS (m/z) [M − H]− calcd for C14H8N3O4 282.0520, found 282.0568; [M + H]+ calcd for C14H10N3O4 284.0666, found 284.0653.
2-(Furan-2-yl)-6-nitro-1H-benzo[d]imidazole (2q). Yellow solid, mp 228–230 °C. 1H NMR (500 MHz, DMSO-d6, δ ppm): 13.62 (1H, s, –NH–), 8.44 (1H, s, HAr), 8.13 (1H, dd, J = 8.5, 2.0 Hz, HAr), 8.03 (1H, d, J = 1.0 Hz, HAr), 7.72 (1H, s, HAr), 7.35 (1H, d, J = 3.5 Hz, HAr), 6.79 (1H, dd, J = 3.5, 2.0 Hz, HAr). 13C NMR (125 MHz, DMSO-d6, δ ppm): 146.9, 145.6 (2C), 145.2 (3C), 119.7 (2C), 113.9, 113.6 (2C). LC-MS (m/z) [M − H]− calcd for C11H6N3O3 228.0415, found 228.0409.
The reflux method. A mixture of 6-substituted 1H-benzimidazole derivatives 1–2 (1 mmol) and potassium carbonate (1 mmol) in acetonitrile (10 mL) was added to substituted halides (1.2 mmol) at 80 °C. The reaction mixture was then heated for 12–24 h and monitored by TLC. After cooling, the reaction crude was poured on a mixture of ice/water to give a solid that was filtered off in a Büchner funnel. The resulting solid was purified by column chromatography on silica gel using hexane/ethyl acetate as eluent. Reaction yields ranged within 26–43%.
The microwave-assisted method. A mixture of 6-substituted 1H-benzimidazole derivatives 1–2 (1 mmol), potassium carbonate (1 mmol), acetonitrile (10 mL), and substituted halides (1.2 mmol) was placed in a microwave oven and irradiated at a power of 300 W for 20–60 min at 80 °C. After cooling, the reaction crude was poured on a mixture of ice/water to give a solid that was filtered off in a Büchner funnel. The resulting solid was purified by column chromatography on silica gel using hexane/ethyl acetate as eluent. Reaction yields ranged within 40–50%.
1-Allyl-6-chloro-2-(2-chlorophenyl)-1H-benzo[d]imidazole (3a). White solid, mp 91–92 °C. IR (ν, cm−1): 1610.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1452.4 (C
N), 1452.4 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 761.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.79 (1H, d, J = 1.5 Hz, HAr), 7.73 (1H, d, J = 8.5 Hz, HAr), 7.69 (1H, d, J = 8.5 Hz, HAr), 7.64–7.60 (2H, m, HAr), 7.53 (1H, d, J = 8.0 Hz, HAr), 7.35 (1H, dd, J = 8.5, 2.0 Hz, HAr), 5.85–5.79 (1H, m, –CH
C), 761.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.79 (1H, d, J = 1.5 Hz, HAr), 7.73 (1H, d, J = 8.5 Hz, HAr), 7.69 (1H, d, J = 8.5 Hz, HAr), 7.64–7.60 (2H, m, HAr), 7.53 (1H, d, J = 8.0 Hz, HAr), 7.35 (1H, dd, J = 8.5, 2.0 Hz, HAr), 5.85–5.79 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.09 (1H, d, J = 10.5 Hz,
), 5.09 (1H, d, J = 10.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.87 (1H, d, J = 17.0 Hz,
CH2), 4.87 (1H, d, J = 17.0 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.70 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.8, 143.3, 141.2, 135.3, 132.9, 132.1, 129.6, 127.3, 126.5, 122.8, 120.7, 118.8, 117.2, 112.5, 111.0, 46.2. LC-MS (m/z) [M + H]+ calcd for C16H13Cl2N2 303.0450, found 303.09.
CH2), 4.70 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.8, 143.3, 141.2, 135.3, 132.9, 132.1, 129.6, 127.3, 126.5, 122.8, 120.7, 118.8, 117.2, 112.5, 111.0, 46.2. LC-MS (m/z) [M + H]+ calcd for C16H13Cl2N2 303.0450, found 303.09.
1-Allyl-6-chloro-2-(4-chlorophenyl)-1H-benzo[d]imidazole (3b). White solid, mp 95–97 °C. IR (ν, cm−1): 1608.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1456.3 (C
N), 1456.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 790.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.90 (2H, d, J = 8.5 Hz, HAr), 7.77 (1H, d, J = 1.5 Hz, HAr), 7.71 (1H, d, J = 8.5 Hz, HAr), 7.58 (2H, d, J = 9.0 Hz, HAr), 7.32 (1H, dd, J = 8.5, 2.0 Hz, HAr), 6.08–6.01 (1H, m, –CH
C), 790.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.90 (2H, d, J = 8.5 Hz, HAr), 7.77 (1H, d, J = 1.5 Hz, HAr), 7.71 (1H, d, J = 8.5 Hz, HAr), 7.58 (2H, d, J = 9.0 Hz, HAr), 7.32 (1H, dd, J = 8.5, 2.0 Hz, HAr), 6.08–6.01 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.20 (1H, d, J = 10.5 Hz,
), 5.20 (1H, d, J = 10.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.94 (1H, d, J = 17.0 Hz,
CH2), 4.94 (1H, d, J = 17.0 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.87 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.3, 143.3, 141.2, 136.6, 135.0, 133.1, 130.7 (2C), 128.9 (2C), 127.2, 122.8, 120.5, 118.6, 112.4, 46.7. LC-MS (m/z) [M + H]+ calcd for C16H13Cl2N2 303.0450, found 303.19.
CH2), 4.87 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.3, 143.3, 141.2, 136.6, 135.0, 133.1, 130.7 (2C), 128.9 (2C), 127.2, 122.8, 120.5, 118.6, 112.4, 46.7. LC-MS (m/z) [M + H]+ calcd for C16H13Cl2N2 303.0450, found 303.19.
1-Allyl-6-chloro-2-(2,4-dichlorophenyl)-1H-benzo[d]imidazole (3c). White solid, mp 79–81 °C. IR (ν, cm−1): 1609.5 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1450.5 (C
N), 1450.5 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 790.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.88 (1H, s, HAr), 7.79 (1H, d, J = 1.5 Hz, HAr), 7.74–7.70 (3H, m, HAr), 7.33 (1H, dd, J = 8.5, 1.5 Hz, HAr), 5.85–5.80 (1H, m, –CH
C), 790.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.88 (1H, s, HAr), 7.79 (1H, d, J = 1.5 Hz, HAr), 7.74–7.70 (3H, m, HAr), 7.33 (1H, dd, J = 8.5, 1.5 Hz, HAr), 5.85–5.80 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.09 (1H, d, J = 10.5 Hz,
), 5.09 (1H, d, J = 10.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.87 (1H, dd, J = 17.0, 1.5 Hz,
CH2), 4.87 (1H, dd, J = 17.0, 1.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.72 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 150.8, 143.2, 141.2, 135.9, 134.2, 133.5, 132.4, 129.4, 128.1, 127.7, 126.7, 123.0, 120.8, 118.9, 112.6, 46.3. LC-MS (m/z) [M + H]+ calcd for C16H12Cl3N2 337.0061, found 336.17.
CH2), 4.72 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 150.8, 143.2, 141.2, 135.9, 134.2, 133.5, 132.4, 129.4, 128.1, 127.7, 126.7, 123.0, 120.8, 118.9, 112.6, 46.3. LC-MS (m/z) [M + H]+ calcd for C16H12Cl3N2 337.0061, found 336.17.
1-Allyl-6-chloro-2-(3,4-dichlorophenyl)-1H-benzo[d]imidazole (3d). White solid, mp 96–98 °C. IR (ν, cm−1): 1610.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1458.2 (C
N), 1458.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 792.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.01 (1H, d, J = 2.0 Hz, HAr), 7.85 (1H, d, J = 8.5 Hz, HAr), 7.78 (1H, d, J = 2.0 Hz, HAr), 7.74 (1H, d, J = 8.5 Hz, HAr), 7.62 (1H, d, J = 8.5 Hz, HAr), 7.35 (1H, dd, J = 8.5, 2.0 Hz, HAr), 6.05–6.01 (1H, m, –CH
C), 792.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.01 (1H, d, J = 2.0 Hz, HAr), 7.85 (1H, d, J = 8.5 Hz, HAr), 7.78 (1H, d, J = 2.0 Hz, HAr), 7.74 (1H, d, J = 8.5 Hz, HAr), 7.62 (1H, d, J = 8.5 Hz, HAr), 7.35 (1H, dd, J = 8.5, 2.0 Hz, HAr), 6.05–6.01 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.21 (1H, d, J = 10.5 Hz,
), 5.21 (1H, d, J = 10.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.98 (1H, dd, J = 17.0, 1.5 Hz,
CH2), 4.98 (1H, dd, J = 17.0, 1.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.88 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.9, 143.1, 141.1, 136.6, 134.7, 133.1, 131.6, 130.6, 128.9, 127.5, 126.9, 123.1, 120.7, 118.7, 112.5, 46.8. LC-MS (m/z) [M + H]+ calcd for C16H12Cl3N2 337.0061, found 337.11.
CH2), 4.88 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.9, 143.1, 141.1, 136.6, 134.7, 133.1, 131.6, 130.6, 128.9, 127.5, 126.9, 123.1, 120.7, 118.7, 112.5, 46.8. LC-MS (m/z) [M + H]+ calcd for C16H12Cl3N2 337.0061, found 337.11.
1-Allyl-6-chloro-2-(3,4-dimethoxyphenyl)-1H-benzo[d]imidazole (3e). White solid, mp 108–110 °C. IR (ν, cm−1): 1608.9 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1459.3 (C
N), 1459.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1248.0 (C–O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.74 (1H, d, J = 1.5 Hz, HAr), 7.69 (1H, d, J = 8.5 Hz, HAr), 7.53 (1H, d, J = 8.5 Hz, HAr), 7.33–7.25 (2H, m, HAr), 7.14 (1H, d, J = 8.5 Hz, HAr), 6.13–6.07 (1H, m, –CH
C), 1248.0 (C–O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.74 (1H, d, J = 1.5 Hz, HAr), 7.69 (1H, d, J = 8.5 Hz, HAr), 7.53 (1H, d, J = 8.5 Hz, HAr), 7.33–7.25 (2H, m, HAr), 7.14 (1H, d, J = 8.5 Hz, HAr), 6.13–6.07 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.24 (1H, d, J = 10.0 Hz,
), 5.24 (1H, d, J = 10.0 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.92 (1H, dd, J = 17.0, 1.5 Hz,
CH2), 4.92 (1H, dd, J = 17.0, 1.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.90 (2H, s, –CH2–), 3.85 (3H, s, –CH3), 3.82 (3H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.5, 150.4, 148.7, 143.4, 141.3, 136.7, 134.7, 133.3, 126.7, 122.3, 121.8, 120.2, 118.3, 112.3, 111.6, 55.6, 55.3, 46.8. LC-MS (m/z) [M + H]+ calcd for C18H18ClN2O2 329.1051, found 329.22.
CH2), 4.90 (2H, s, –CH2–), 3.85 (3H, s, –CH3), 3.82 (3H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.5, 150.4, 148.7, 143.4, 141.3, 136.7, 134.7, 133.3, 126.7, 122.3, 121.8, 120.2, 118.3, 112.3, 111.6, 55.6, 55.3, 46.8. LC-MS (m/z) [M + H]+ calcd for C18H18ClN2O2 329.1051, found 329.22.
1-Allyl-6-chloro-2-(4-ethoxyphenyl)-1H-benzo[d]imidazole (3f). White solid, mp 92–94 °C. IR (ν, cm−1): 1610.5 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1462.0 (C
N), 1462.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1244.1 (C–O), 790.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.72 (1H, d, J = 1.5 Hz, HAr), 7.70 (2H, d, J = 8.5 Hz, HAr), 7.53 (1H, d, J = 8.5 Hz, HAr), 7.28 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.10 (2H, d, J = 9.0 Hz, HAr), 6.11–6.03 (1H, m, –CH
C), 1244.1 (C–O), 790.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.72 (1H, d, J = 1.5 Hz, HAr), 7.70 (2H, d, J = 8.5 Hz, HAr), 7.53 (1H, d, J = 8.5 Hz, HAr), 7.28 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.10 (2H, d, J = 9.0 Hz, HAr), 6.11–6.03 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.20 (1H, d, J = 10.5 Hz,
), 5.20 (1H, d, J = 10.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.92 (2H, s, –CH2–), 4.88 (1H, d, J = 17.0 Hz,
CH2), 4.92 (2H, s, –CH2–), 4.88 (1H, d, J = 17.0 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.12 (2H, q, J = 7.0 Hz, –CH2–), 1.37 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 159.9, 154.5, 143.4, 134.7, 133.2, 130.4 (2C), 126.5, 122.3, 121.6, 118.3, 116.7, 114.7 (2C), 112.2, 63.3, 46.8, 14.5. LC-MS (m/z) [M + H]+ calcd for C18H18ClN2O 313.1102, found 313.24.
CH2), 4.12 (2H, q, J = 7.0 Hz, –CH2–), 1.37 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 159.9, 154.5, 143.4, 134.7, 133.2, 130.4 (2C), 126.5, 122.3, 121.6, 118.3, 116.7, 114.7 (2C), 112.2, 63.3, 46.8, 14.5. LC-MS (m/z) [M + H]+ calcd for C18H18ClN2O 313.1102, found 313.24.
1-Allyl-6-chloro-2-(4-fluorophenyl)-1H-benzo[d]imidazole (3g). White solid, mp 106–108 °C. IR (ν, cm−1): 1606.7 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1458.4 (C
N), 1458.4 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1224.8 (C–F), 736.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.82 (2H, d, J = 8.5 Hz, HAr), 7.76 (1H, d, J = 2.0 Hz, HAr), 7.70 (1H, d, J = 8.5 Hz, HAr), 7.41 (2H, d, J = 8.5 Hz, HAr), 7.32 (1H, dd, J = 8.5, 2.0 Hz, HAr), 6.08–6.00 (1H, m, –CH
C), 1224.8 (C–F), 736.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.82 (2H, d, J = 8.5 Hz, HAr), 7.76 (1H, d, J = 2.0 Hz, HAr), 7.70 (1H, d, J = 8.5 Hz, HAr), 7.41 (2H, d, J = 8.5 Hz, HAr), 7.32 (1H, dd, J = 8.5, 2.0 Hz, HAr), 6.08–6.00 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.20 (1H, d, J = 10.5 Hz,
), 5.20 (1H, d, J = 10.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.92 (1H, dd, J = 17.0, 1.5 Hz,
CH2), 4.92 (1H, dd, J = 17.0, 1.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.87 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 164.0, 153.5, 143.3, 136.5, 133.1, 131.4, 127.0 (2C), 122.7, 120.4, 118.5, 116.7 (2C), 115.9, 112.4, 46.7. LC-MS (m/z) [M + H]+ calcd for C16H13ClFN2 287.0746, found 287.0679.
CH2), 4.87 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 164.0, 153.5, 143.3, 136.5, 133.1, 131.4, 127.0 (2C), 122.7, 120.4, 118.5, 116.7 (2C), 115.9, 112.4, 46.7. LC-MS (m/z) [M + H]+ calcd for C16H13ClFN2 287.0746, found 287.0679.
1-Allyl-6-chloro-2-(3-nitrophenyl)-1H-benzo[d]imidazole (3h). Yellow solid, mp 130–132 °C. IR (ν, cm−1): 1612.5 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1533.4 (C
N), 1533.4 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1350.2 (N
C), 1350.2 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 707.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.58 (1H, t, J = 2.0 Hz, HAr), 8.41 (1H, d, J = 9.0 Hz, HAr), 8.24 (1H, d, J = 9.0 Hz, HAr), 7.90–7.86 (1H, m, HAr), 7.83 (1H, d, J = 2.0 Hz, HAr), 7.77 (1H, d, J = 9.0 Hz, HAr), 7.37 (1H, dd, J = 9.0, 2.0 Hz, HAr), 6.14–6.06 (1H, m, –CH
O), 707.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.58 (1H, t, J = 2.0 Hz, HAr), 8.41 (1H, d, J = 9.0 Hz, HAr), 8.24 (1H, d, J = 9.0 Hz, HAr), 7.90–7.86 (1H, m, HAr), 7.83 (1H, d, J = 2.0 Hz, HAr), 7.77 (1H, d, J = 9.0 Hz, HAr), 7.37 (1H, dd, J = 9.0, 2.0 Hz, HAr), 6.14–6.06 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.25 (1H, dd, J = 10.5, 1.0 Hz,
), 5.25 (1H, dd, J = 10.5, 1.0 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.02 (2H, s, –CH2–), 4.93 (1H, dd, J = 17.0, 1.0 Hz,
CH2), 5.02 (2H, s, –CH2–), 4.93 (1H, dd, J = 17.0, 1.0 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.0, 147.9, 143.2, 141.1, 136.7, 135.1, 133.0, 131.0, 127.7, 124.6, 123.5, 122.9, 120.8, 118.8, 112.6, 46.9. LC-MS (m/z) [M + H]+ calcd for C16H13ClN3O2 314.0691, found 314.18.
CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.0, 147.9, 143.2, 141.1, 136.7, 135.1, 133.0, 131.0, 127.7, 124.6, 123.5, 122.9, 120.8, 118.8, 112.6, 46.9. LC-MS (m/z) [M + H]+ calcd for C16H13ClN3O2 314.0691, found 314.18.
1-Allyl-6-chloro-2-(4-nitrophenyl)-1H-benzo[d]imidazole (3i). Yellow solid, mp 140–142 °C. IR (ν, cm−1): 1600.9 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1460.1 (C
N), 1460.1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1347.6 (N
C), 1347.6 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 707.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.41 (2H, d, J = 9.0 Hz, HAr), 8.07 (2H, d, J = 9.0 Hz, HAr), 7.83 (1H, d, J = 2.0 Hz, HAr), 7.65 (1H, d, J = 8.5 Hz, HAr), 7.37 (1H, dd, J = 8.5, 2.0 Hz, HAr), 6.09–6.02 (1H, m, –CH
O), 707.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.41 (2H, d, J = 9.0 Hz, HAr), 8.07 (2H, d, J = 9.0 Hz, HAr), 7.83 (1H, d, J = 2.0 Hz, HAr), 7.65 (1H, d, J = 8.5 Hz, HAr), 7.37 (1H, dd, J = 8.5, 2.0 Hz, HAr), 6.09–6.02 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.30 (1H, dd, J = 10.5, 1.0 Hz,
), 5.30 (1H, dd, J = 10.5, 1.0 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.01 (2H, s, –CH2–), 4.89 (1H, dd, J = 17.0, 1.5 Hz,
CH2), 5.01 (2H, s, –CH2–), 4.89 (1H, dd, J = 17.0, 1.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.2, 148.1, 143.3, 141.2, 136.8, 135.6, 133.0, 130.3, 127.8 (2C), 123.9 (2C), 120.9, 118.9, 112.7, 46.9. LC-MS (m/z) [M + H]+ calcd for C16H13ClN3O2 314.0691, found 314.16.
CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.2, 148.1, 143.3, 141.2, 136.8, 135.6, 133.0, 130.3, 127.8 (2C), 123.9 (2C), 120.9, 118.9, 112.7, 46.9. LC-MS (m/z) [M + H]+ calcd for C16H13ClN3O2 314.0691, found 314.16.
1-Benzyl-6-chloro-2-(2-chlorophenyl)-1H-benzo[d]imidazole (3j). White solid, mp 83–84 °C. IR (ν, cm−1): 1606.9 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1454.3 (C
N), 1454.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 723.3 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.79 (1H, d, J = 1.5 Hz, HAr), 7.74 (1H, d, J = 9.0 Hz, HAr), 7.67 (1H, d, J = 8.5 Hz, HAr), 7.62–7.57 (2H, m, HAr), 7.55 (1H, d, J = 9.0 Hz, HAr), 7.30 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.24–7.21 (3H, m, HAr), 6.93 (2H, dd, J = 8.5, 2.0 Hz, HAr), 5.32 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.2, 141.3, 136.0, 135.5, 133.6, 133.1, 132.3, 129.8, 129.1, 128.6 (2C), 127.7, 126.7 (2C), 123.1, 122.7, 120.9, 119.0, 112.7, 47.4. LC-MS (m/z) [M + H]+ calcd for C20H15Cl2N2 353.0607, found 353.03.
C), 723.3 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.79 (1H, d, J = 1.5 Hz, HAr), 7.74 (1H, d, J = 9.0 Hz, HAr), 7.67 (1H, d, J = 8.5 Hz, HAr), 7.62–7.57 (2H, m, HAr), 7.55 (1H, d, J = 9.0 Hz, HAr), 7.30 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.24–7.21 (3H, m, HAr), 6.93 (2H, dd, J = 8.5, 2.0 Hz, HAr), 5.32 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.2, 141.3, 136.0, 135.5, 133.6, 133.1, 132.3, 129.8, 129.1, 128.6 (2C), 127.7, 126.7 (2C), 123.1, 122.7, 120.9, 119.0, 112.7, 47.4. LC-MS (m/z) [M + H]+ calcd for C20H15Cl2N2 353.0607, found 353.03.
1-Benzyl-6-chloro-2-(4-chlorophenyl)-1H-benzo[d]imidazole (3k). White solid, mp 98–101 °C. IR (ν, cm−1): 1606.8 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1454.5 (C
N), 1454.5 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 723.3 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.80 (1H, d, J = 2.0 Hz, HAr), 7.76–7.73 (3H, m, HAr), 7.59 (2H, d, J = 8.5 Hz, HAr), 7.31 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.29–7.24 (3H, m, HAr), 6.97 (2H, dd, J = 8.5, 2.0 Hz, HAr), 5.61 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.6, 143.4, 141.3, 136.7, 135.0, 134.9, 130.8, 128.9 (2C), 128.5 (2C), 127.6 (2C), 126.8 (2C), 123.0, 120.6, 118.7, 112.6, 47.6. LC-MS (m/z) [M + H]+ calcd for C20H15Cl2N2 353.0607, found 353.08.
C), 723.3 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.80 (1H, d, J = 2.0 Hz, HAr), 7.76–7.73 (3H, m, HAr), 7.59 (2H, d, J = 8.5 Hz, HAr), 7.31 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.29–7.24 (3H, m, HAr), 6.97 (2H, dd, J = 8.5, 2.0 Hz, HAr), 5.61 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.6, 143.4, 141.3, 136.7, 135.0, 134.9, 130.8, 128.9 (2C), 128.5 (2C), 127.6 (2C), 126.8 (2C), 123.0, 120.6, 118.7, 112.6, 47.6. LC-MS (m/z) [M + H]+ calcd for C20H15Cl2N2 353.0607, found 353.08.
1-Benzyl-6-chloro-2-(2,4-dichlorophenyl)-1H-benzo[d]imidazole (3l). White solid, mp 119–121 °C. IR (ν, cm−1): 1645.3 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1452.4 (C
N), 1452.4 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 794.6 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.85 (1H, d, J = 2.0 Hz, HAr), 7.81 (1H, d, J = 1.5 Hz, HAr), 7.75 (1H, d, J = 8.5 Hz, HAr), 7.63 (1H, dd, J = 8.5 Hz, HAr), 7.58 (1H, dd, J = 8.5, 1.5 Hz, HAr), 7.31 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.26–7.21 (3H, m, HAr), 6.95 (2H, dd, J = 8.5, 2.0 Hz, HAr), 5.35 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.1, 143.4, 141.3, 136.0, 135.9, 135.6, 134.3, 133.6, 129.4 (2C), 128.3, 127.7, 126.8 (2C), 123.2, 122.7, 120.9, 119.0, 112.7, 47.4. LC-MS (m/z) [M + H]+ calcd for C20H14Cl3N2 387.0217, found 386.98.
C), 794.6 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.85 (1H, d, J = 2.0 Hz, HAr), 7.81 (1H, d, J = 1.5 Hz, HAr), 7.75 (1H, d, J = 8.5 Hz, HAr), 7.63 (1H, dd, J = 8.5 Hz, HAr), 7.58 (1H, dd, J = 8.5, 1.5 Hz, HAr), 7.31 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.26–7.21 (3H, m, HAr), 6.95 (2H, dd, J = 8.5, 2.0 Hz, HAr), 5.35 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.1, 143.4, 141.3, 136.0, 135.9, 135.6, 134.3, 133.6, 129.4 (2C), 128.3, 127.7, 126.8 (2C), 123.2, 122.7, 120.9, 119.0, 112.7, 47.4. LC-MS (m/z) [M + H]+ calcd for C20H14Cl3N2 387.0217, found 386.98.
1-Benzyl-6-chloro-2-(3,4-dichlorophenyl)-1H-benzo[d]imidazole (3m). White solid, mp 143–145 °C. IR (ν, cm−1): 1601.8 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1452.4 (C
N), 1452.4 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 702.5 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.80–7.74 (2H, m, HAr), 7.70–7.69 (2H, m, HAr), 7.56 (1H, d, J = 9.0 Hz, HAr), 7.30 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.28–7.24 (3H, m, HAr), 6.99 (2H, d, J = 7.5 Hz, HAr), 5.61 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.3, 143.3, 141.2, 136.9, 134.9, 133.1, 131.7, 130.8, 130.2, 129.1 (2C), 127.7, 127.1 (2C), 126.1, 123.4, 120.8, 118.9, 112.7, 47.7. LC-MS (m/z) [M + H]+ calcd for C20H14Cl3N2 387.0217, found 386.89.
C), 702.5 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.80–7.74 (2H, m, HAr), 7.70–7.69 (2H, m, HAr), 7.56 (1H, d, J = 9.0 Hz, HAr), 7.30 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.28–7.24 (3H, m, HAr), 6.99 (2H, d, J = 7.5 Hz, HAr), 5.61 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.3, 143.3, 141.2, 136.9, 134.9, 133.1, 131.7, 130.8, 130.2, 129.1 (2C), 127.7, 127.1 (2C), 126.1, 123.4, 120.8, 118.9, 112.7, 47.7. LC-MS (m/z) [M + H]+ calcd for C20H14Cl3N2 387.0217, found 386.89.
1-Benzyl-6-chloro-2-(3,4-dimethoxyphenyl)-1H-benzo[d]imidazole (3n). White solid, mp 157–159 °C. IR (ν, cm−1): 1610.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1494.8 (C
N), 1494.8 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1253.7 (C–O), 815.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.71 (1H, d, J = 8.5 Hz, HAr), 7.62 (1H, d, J = 2.0 Hz, HAr), 7.34–7.31 (2H, m, HAr), 7.28–7.25 (3H, m, HAr), 7.23 (1H, d, J = 2.0 Hz, HAr), 7.08 (1H, d, J = 8.5 Hz, HAr), 7.02 (2H, d, J = 7.0 Hz, HAr), 5.61 (2H, s, –CH2–), 3.81 (3H, s, –OCH3), 3.65 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.4, 150.3, 148.6, 141.3, 136.9, 136.8, 128.8 (2C), 127.5, 126.9 (2C), 125.9, 122.5, 121.8, 121.7, 120.3, 112.3, 111.7, 110.8, 55.6, 55.3, 47.6. The NOESY correlation including: δH 7.71 with δH 7.28–7.25; δH 7.62 with δH 5.61; δH 7.34–7.31 with δH 7.02; δH 7.28–7.25 with δH 7.08, 5.61 and 3.81–3.65; δH 7.08 with δH 3.81; δH 7.02 with δH 5.61. LC-MS (m/z) [M + H]+ calcd for C22H20ClN2O2 379.1208, found 379.13.
C), 1253.7 (C–O), 815.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.71 (1H, d, J = 8.5 Hz, HAr), 7.62 (1H, d, J = 2.0 Hz, HAr), 7.34–7.31 (2H, m, HAr), 7.28–7.25 (3H, m, HAr), 7.23 (1H, d, J = 2.0 Hz, HAr), 7.08 (1H, d, J = 8.5 Hz, HAr), 7.02 (2H, d, J = 7.0 Hz, HAr), 5.61 (2H, s, –CH2–), 3.81 (3H, s, –OCH3), 3.65 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.4, 150.3, 148.6, 141.3, 136.9, 136.8, 128.8 (2C), 127.5, 126.9 (2C), 125.9, 122.5, 121.8, 121.7, 120.3, 112.3, 111.7, 110.8, 55.6, 55.3, 47.6. The NOESY correlation including: δH 7.71 with δH 7.28–7.25; δH 7.62 with δH 5.61; δH 7.34–7.31 with δH 7.02; δH 7.28–7.25 with δH 7.08, 5.61 and 3.81–3.65; δH 7.08 with δH 3.81; δH 7.02 with δH 5.61. LC-MS (m/z) [M + H]+ calcd for C22H20ClN2O2 379.1208, found 379.13.
1-Benzyl-6-chloro-2-(4-ethoxyphenyl)-1H-benzo[d]imidazole (3o). White solid, mp 128–129 °C. IR (ν, cm−1): 1611.4 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1462.0 (C
N), 1462.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1249.9 (C–O), 794.7 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.75 (1H, d, J = 1.5 Hz, HAr), 7.70 (1H, d, J = 8.5 Hz, HAr), 7.65 (2H, d, J = 8.5 Hz, HAr), 7.32 (1H, d, J = 9.0 Hz, HAr), 7.29–7.23 (3H, m, HAr), 7.05 (2H, d, J = 8.5 Hz, HAr), 6.99 (2H, d, J = 7.5 Hz, HAr), 5.58 (2H, s, –CH2–), 4.09 (2H, q, J = 7.0 Hz, –CH2–), 1.35 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 159.9, 154.8, 143.5, 136.7, 134.7, 130.5 (2C), 128.8 (2C), 127.5, 126.8 (2C), 122.4, 121.6, 120.3, 118.4, 114.7 (2C), 112.3, 63.3, 47.6, 14.5. LC-MS (m/z) [M + H]+ calcd for C22H20ClN2O 363.1259, found 363.18.
C), 1249.9 (C–O), 794.7 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.75 (1H, d, J = 1.5 Hz, HAr), 7.70 (1H, d, J = 8.5 Hz, HAr), 7.65 (2H, d, J = 8.5 Hz, HAr), 7.32 (1H, d, J = 9.0 Hz, HAr), 7.29–7.23 (3H, m, HAr), 7.05 (2H, d, J = 8.5 Hz, HAr), 6.99 (2H, d, J = 7.5 Hz, HAr), 5.58 (2H, s, –CH2–), 4.09 (2H, q, J = 7.0 Hz, –CH2–), 1.35 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 159.9, 154.8, 143.5, 136.7, 134.7, 130.5 (2C), 128.8 (2C), 127.5, 126.8 (2C), 122.4, 121.6, 120.3, 118.4, 114.7 (2C), 112.3, 63.3, 47.6, 14.5. LC-MS (m/z) [M + H]+ calcd for C22H20ClN2O 363.1259, found 363.18.
1-Benzyl-6-chloro-2-(4-fluorophenyl)-1H-benzo[d]imidazole (3p). White solid, mp 117–118 °C. IR (ν, cm−1): 1606.7 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1475.2 (C
N), 1475.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1226.7 (C–F), 794.7 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.73 (2H, d, J = 8.5 Hz, HAr), 7.66 (1H, d, J = 2.0 Hz, HAr), 7.52 (1H, d, J = 8.5 Hz, HAr), 7.37 (2H, d, J = 8.5 Hz, HAr), 7.30 (1H, d, J = 8.5 Hz, HAr), 7.28–7.24 (3H, m, HAr), 6.98 (2H, d, J = 8.0 Hz, HAr), 5.59 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 171.5, 162.9, 152.9, 146.2, 145.9, 144.1, 140.9, 138.3, 137.0, 136.2, 135.7, 132.3, 130.0 (2C), 128.1 (2C), 125.5 (2C), 122.0, 120.5 (2C), 57.1. LC-MS (m/z) [M + H]+ calcd for C20H15ClFN2 337.0902, found 337.0900.
C), 1226.7 (C–F), 794.7 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.73 (2H, d, J = 8.5 Hz, HAr), 7.66 (1H, d, J = 2.0 Hz, HAr), 7.52 (1H, d, J = 8.5 Hz, HAr), 7.37 (2H, d, J = 8.5 Hz, HAr), 7.30 (1H, d, J = 8.5 Hz, HAr), 7.28–7.24 (3H, m, HAr), 6.98 (2H, d, J = 8.0 Hz, HAr), 5.59 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 171.5, 162.9, 152.9, 146.2, 145.9, 144.1, 140.9, 138.3, 137.0, 136.2, 135.7, 132.3, 130.0 (2C), 128.1 (2C), 125.5 (2C), 122.0, 120.5 (2C), 57.1. LC-MS (m/z) [M + H]+ calcd for C20H15ClFN2 337.0902, found 337.0900.
1-Benzyl-6-chloro-2-(3-nitrophenyl)-1H-benzo[d]imidazole (3q). Yellow solid, mp 143–145 °C. IR (ν, cm−1): 1603.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1510.3 (C
N), 1510.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 727.2 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.50 (1H, d, J = 2.0 Hz, HAr), 8.36 (1H, d, J = 9.0 Hz, HAr), 8.17 (1H, d, J = 9.0 Hz, HAr), 7.86–7.79 (1H, m, HAr), 7.75 (1H, d, J = 2.0 Hz, HAr), 7.61 (1H, d, J = 9.0 Hz, HAr), 7.35 (1H, d, J = 8.5 Hz, HAr), 7.30–7.23 (3H, m, HAr), 7.01 (2H, d, J = 8.0 Hz, HAr), 5.67 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.3, 147.9, 143.3, 141.2, 137.0, 135.2, 131.1, 130.5, 128.8 (2C), 127.8, 127.1 (2C), 126.1, 124.6, 123.7, 120.9, 119.0, 112.8, 47.8. LC-MS (m/z) [M + H]+ calcd for C20H15ClN3O2 364.0847, found 364.14.
C), 727.2 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.50 (1H, d, J = 2.0 Hz, HAr), 8.36 (1H, d, J = 9.0 Hz, HAr), 8.17 (1H, d, J = 9.0 Hz, HAr), 7.86–7.79 (1H, m, HAr), 7.75 (1H, d, J = 2.0 Hz, HAr), 7.61 (1H, d, J = 9.0 Hz, HAr), 7.35 (1H, d, J = 8.5 Hz, HAr), 7.30–7.23 (3H, m, HAr), 7.01 (2H, d, J = 8.0 Hz, HAr), 5.67 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.3, 147.9, 143.3, 141.2, 137.0, 135.2, 131.1, 130.5, 128.8 (2C), 127.8, 127.1 (2C), 126.1, 124.6, 123.7, 120.9, 119.0, 112.8, 47.8. LC-MS (m/z) [M + H]+ calcd for C20H15ClN3O2 364.0847, found 364.14.
1-Benzyl-6-chloro-2-(4-nitrophenyl)-1H-benzo[d]imidazole (3r). Yellow solid, mp 186–188 °C. IR (ν, cm−1): 1601.4 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1518.9 (C
C), 1518.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1348.1 (N
N), 1348.1 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 732.7 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.36 (2H, d, J = 8.0 Hz, HAr), 8.04 (2H, d, J = 8.0 Hz, HAr), 7.76 (1H, s, HAr), 7.61 (1H, d, J = 9.0 Hz, HAr), 7.34 (1H, t, J = 8.5 Hz, HAr), 7.29–7.22 (3H, m, HAr), 6.98 (2H, d, J = 8.5 Hz, HAr), 5.68 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.5, 148.1, 143.4, 137.7, 136.3, 135.8, 130.4, 128.8 (2C), 127.9 (2C), 126.1 (2C), 123.9 (2C), 123.1, 121.0, 119.0, 112.9, 47.6. LC-MS (m/z) [M + H]+ calcd for C20H15ClN3O2 364.0847, found 364.0811.
O), 732.7 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.36 (2H, d, J = 8.0 Hz, HAr), 8.04 (2H, d, J = 8.0 Hz, HAr), 7.76 (1H, s, HAr), 7.61 (1H, d, J = 9.0 Hz, HAr), 7.34 (1H, t, J = 8.5 Hz, HAr), 7.29–7.22 (3H, m, HAr), 6.98 (2H, d, J = 8.5 Hz, HAr), 5.68 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.5, 148.1, 143.4, 137.7, 136.3, 135.8, 130.4, 128.8 (2C), 127.9 (2C), 126.1 (2C), 123.9 (2C), 123.1, 121.0, 119.0, 112.9, 47.6. LC-MS (m/z) [M + H]+ calcd for C20H15ClN3O2 364.0847, found 364.0811.
4-(1-Benzyl-6-chloro-1H-benzo[d]imidazole-2-yl)-N,N-dimethylaniline (3s). White solid, mp 184–186 °C. IR (ν, cm−1): 1607.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1456.3 (C
N), 1456.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 796.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.66–7.50 (4H, m, HAr), 7.33 (1H, t, J = 8.5 Hz, HAr), 7.30–7.21 (3H, m, HAr), 7.01 (2H, d, J = 7.5 Hz, HAr), 6.78 (2H, d, J = 8.5 Hz, HAr), 5.58 (2H, s, –CH2–), 2.96 (6H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.8, 150.3, 148.6, 143.5, 136.8, 134.9, 128.8 (2C), 127.5 (2C), 126.6 (2C), 125.9, 122.5, 121.8, 118.4, 112.3 (2C), 111.7, 47.7, 39.0 (2C). LC-MS (m/z) [M + H]+ calcd for C22H21ClN3 362.1419, found 361.19.
C), 796.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.66–7.50 (4H, m, HAr), 7.33 (1H, t, J = 8.5 Hz, HAr), 7.30–7.21 (3H, m, HAr), 7.01 (2H, d, J = 7.5 Hz, HAr), 6.78 (2H, d, J = 8.5 Hz, HAr), 5.58 (2H, s, –CH2–), 2.96 (6H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.8, 150.3, 148.6, 143.5, 136.8, 134.9, 128.8 (2C), 127.5 (2C), 126.6 (2C), 125.9, 122.5, 121.8, 118.4, 112.3 (2C), 111.7, 47.7, 39.0 (2C). LC-MS (m/z) [M + H]+ calcd for C22H21ClN3 362.1419, found 361.19.
6-Chloro-1-(2-chlorobenzyl)-2-(4-chlorophenyl)-1H-benzo[d]imidazole (3t). White solid, mp 140–142 °C. IR (ν, cm−1): 1610.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1465.9 (C
N), 1465.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 752.2 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.80 (1H, d, J = 2.0 Hz, HAr), 7.76 (1H, d, J = 8.5 Hz, HAr), 7.67 (2H, d, J = 8.5 Hz, HAr), 7.60 (2H, d, J = 8.5 Hz, HAr), 7.48 (1H, dd, J = 8.0, 1.5 Hz, HAr), 7.32–7.27 (2H, m, HAr), 7.20 (1H, d, J = 8.5 Hz, HAr), 6.65 (1H, d, J = 8.0 Hz, HAr), 5.61 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.8, 143.4, 135.1, 134.8, 133.6, 131.5, 130.7, 130.6, 129.8, 129.5 (2C), 129.0 (2C), 127.8, 127.3, 123.2, 120.8, 118.9, 112.5, 46.1. LC-MS (m/z) [M + H]+ calcd for C20H14Cl3N2 387.0217, found 386.90.
C), 752.2 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.80 (1H, d, J = 2.0 Hz, HAr), 7.76 (1H, d, J = 8.5 Hz, HAr), 7.67 (2H, d, J = 8.5 Hz, HAr), 7.60 (2H, d, J = 8.5 Hz, HAr), 7.48 (1H, dd, J = 8.0, 1.5 Hz, HAr), 7.32–7.27 (2H, m, HAr), 7.20 (1H, d, J = 8.5 Hz, HAr), 6.65 (1H, d, J = 8.0 Hz, HAr), 5.61 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.8, 143.4, 135.1, 134.8, 133.6, 131.5, 130.7, 130.6, 129.8, 129.5 (2C), 129.0 (2C), 127.8, 127.3, 123.2, 120.8, 118.9, 112.5, 46.1. LC-MS (m/z) [M + H]+ calcd for C20H14Cl3N2 387.0217, found 386.90.
6-Chloro-1-(4-chlorobenzyl)-2-(3,4-dichlorophenyl)-1H-benzo[d]imidazole (3u). White solid, mp 168–169 °C. IR (ν, cm−1): 1610.3 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1458.7 (C
N), 1458.7 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 754.1 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.94 (1H, d, J = 2.0 Hz, HAr), 7.82–7.66 (3H, m, HAr), 7.57 (1H, d, J = 9.0 Hz, HAr), 7.37–7.30 (3H, m, HAr), 7.02–6.99 (2H, m, HAr), 5.62 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.5, 142.9, 135.3, 134.9, 133.4, 131.7, 130.8, 129.9, 129.6, 129.2 (2C), 128.7 (2C), 127.6, 127.0, 123.1, 120.6, 118.5, 112.4, 46.8. LC-MS (m/z) [M + H]+ calcd for C20H12Cl4N2 420.9827, found 420.9731.
C), 754.1 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.94 (1H, d, J = 2.0 Hz, HAr), 7.82–7.66 (3H, m, HAr), 7.57 (1H, d, J = 9.0 Hz, HAr), 7.37–7.30 (3H, m, HAr), 7.02–6.99 (2H, m, HAr), 5.62 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.5, 142.9, 135.3, 134.9, 133.4, 131.7, 130.8, 129.9, 129.6, 129.2 (2C), 128.7 (2C), 127.6, 127.0, 123.1, 120.6, 118.5, 112.4, 46.8. LC-MS (m/z) [M + H]+ calcd for C20H12Cl4N2 420.9827, found 420.9731.
6-Chloro-1-(4-chlorobenzyl)-2-(furan-2-yl)-1H-benzo[d]imidazole (3v). White solid, mp 129–131 °C. IR (ν, cm−1): 1612.4 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1459.3 (C
N), 1459.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 753.2 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.94 (1H, d, J = 2.0 Hz, HAr), 7.75 (1H, d, J = 2.0 Hz, HAr), 7.63 (1H, d, J = 8.5 Hz, HAr), 7.36 (2H, d, J = 8.5 Hz, HAr), 7.30 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.19 (1H, d, J = 3.0 Hz, HAr), 7.11 (2H, d, J = 8.5 Hz, HAr), 6.73 (1H, dd, J = 4.0, 1.5 Hz, HAr), 5.80 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.8, 148.3, 143.6, 141.0, 136.8, 135.6, 134.9, 132.9, 130.2 (2C), 127.0 (2C), 123.4, 123.0, 120.9, 116.9, 111.2, 46.7. LC-MS (m/z) [M − H]− calcd for C18H11Cl2N2O 341.0254, found 341.0219; [M + H]+ calcd for C18H13Cl2N2O 343.0399, found 343.0349.
C), 753.2 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.94 (1H, d, J = 2.0 Hz, HAr), 7.75 (1H, d, J = 2.0 Hz, HAr), 7.63 (1H, d, J = 8.5 Hz, HAr), 7.36 (2H, d, J = 8.5 Hz, HAr), 7.30 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.19 (1H, d, J = 3.0 Hz, HAr), 7.11 (2H, d, J = 8.5 Hz, HAr), 6.73 (1H, dd, J = 4.0, 1.5 Hz, HAr), 5.80 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 151.8, 148.3, 143.6, 141.0, 136.8, 135.6, 134.9, 132.9, 130.2 (2C), 127.0 (2C), 123.4, 123.0, 120.9, 116.9, 111.2, 46.7. LC-MS (m/z) [M − H]− calcd for C18H11Cl2N2O 341.0254, found 341.0219; [M + H]+ calcd for C18H13Cl2N2O 343.0399, found 343.0349.
5-Chloro-1-(4-chlorobenzyl)-2-(4-chlorophenyl)-1H-benzo[d]imidazole (3w). White solid, mp 165–167 °C. IR (ν, cm−1): 1600.9 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1465.9 (C
N), 1465.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 796.6 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.80 (1H, d, J = 2.0 Hz, HAr), 7.73 (2H, d, J = 9.0 Hz, HAr), 7.60 (2H, d, J = 8.5 Hz, HAr), 7.53 (1H, d, J = 9.0 Hz, HAr), 7.35 (2H, d, J = 8.5 Hz, HAr), 7.30 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.00 (2H, d, J = 9.0 Hz, HAr), 5.60 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.5, 143.4, 135.5, 135.0, 134.7, 132.2, 130.8, 128.9 (2C), 128.8 (2C), 128.4 (2C), 128.0 (2C), 126.9, 123.1, 118.8, 112.5, 48.0. The NOESY correlation including: δH 7.73 with δH 7.60 and 5.60; δH 7.53 with δH 7.30 and 5.60; δH 7.35 with δH 7.00; δH 7.00 with δH 5.60. LC-MS (m/z) [M + H]+ calcd for C20H14Cl3N2 387.0217, found 386.92.
C), 796.6 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.80 (1H, d, J = 2.0 Hz, HAr), 7.73 (2H, d, J = 9.0 Hz, HAr), 7.60 (2H, d, J = 8.5 Hz, HAr), 7.53 (1H, d, J = 9.0 Hz, HAr), 7.35 (2H, d, J = 8.5 Hz, HAr), 7.30 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.00 (2H, d, J = 9.0 Hz, HAr), 5.60 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.5, 143.4, 135.5, 135.0, 134.7, 132.2, 130.8, 128.9 (2C), 128.8 (2C), 128.4 (2C), 128.0 (2C), 126.9, 123.1, 118.8, 112.5, 48.0. The NOESY correlation including: δH 7.73 with δH 7.60 and 5.60; δH 7.53 with δH 7.30 and 5.60; δH 7.35 with δH 7.00; δH 7.00 with δH 5.60. LC-MS (m/z) [M + H]+ calcd for C20H14Cl3N2 387.0217, found 386.92.
1-Benzyl-5-chloro-2-(3,4-dimethoxyphenyl)-1H-benzo[d]imidazole (3x). White solid, mp 146–148 °C. IR (ν, cm−1): 1604.8 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1487.1 (C
N), 1487.1 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1222.9 (C–O), 790.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.74 (1H, d, J = 2.0 Hz, HAr), 7.49 (1H, d, J = 8.5 Hz, HAr), 7.33–7.23 (6H, m, HAr), 7.09 (1H, d, J = 8.5 Hz, HAr), 7.02 (2H, d, J = 7.0 Hz, HAr), 5.61 (2H, s, –CH2-), 3.81–3.66 (6H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.8, 150.3, 148.6, 143.5, 136.8, 134.9, 128.8 (2C), 127.5 (2C), 126.6, 125.9, 122.5, 121.8, 121.7, 118.4, 112.3, 112.2, 111.7, 55.6, 55.3, 47.7. The NOESY correlation including: δH 7.49 with δH 7.28–7.23 and 5.61; δH 7.33–7.30 with δH 7.02; δH 7.28–7.23 with δH 7.09, 5.61 and 3.81–3.66; δH 7.09 with δH 3.81–3.66; δH 7.02 with δH 5.61. LC-MS (m/z) [M + H]+ calcd for C22H20ClN2O2 379.1208, found 379.14.
C), 1222.9 (C–O), 790.8 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 7.74 (1H, d, J = 2.0 Hz, HAr), 7.49 (1H, d, J = 8.5 Hz, HAr), 7.33–7.23 (6H, m, HAr), 7.09 (1H, d, J = 8.5 Hz, HAr), 7.02 (2H, d, J = 7.0 Hz, HAr), 5.61 (2H, s, –CH2-), 3.81–3.66 (6H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.8, 150.3, 148.6, 143.5, 136.8, 134.9, 128.8 (2C), 127.5 (2C), 126.6, 125.9, 122.5, 121.8, 121.7, 118.4, 112.3, 112.2, 111.7, 55.6, 55.3, 47.7. The NOESY correlation including: δH 7.49 with δH 7.28–7.23 and 5.61; δH 7.33–7.30 with δH 7.02; δH 7.28–7.23 with δH 7.09, 5.61 and 3.81–3.66; δH 7.09 with δH 3.81–3.66; δH 7.02 with δH 5.61. LC-MS (m/z) [M + H]+ calcd for C22H20ClN2O2 379.1208, found 379.14.
1-Allyl-2-(2,4-dichlorophenyl)-6-nitro-1H-benzo[d]imidazole (4a). Yellow solid, mp 150–152 °C. IR (ν, cm−1): 1516.1 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1438.9 (C
N), 1438.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1332.8 (N
C), 1332.8 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 734.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.62 (1H, d, J = 2.0 Hz, HAr), 8.24 (1H, d, J = 9.0, 2.5 Hz, HAr), 8.07 (1H, d, J = 2.0 Hz, HAr), 7.92 (1H, d, J = 9.0 Hz, HAr), 7.88 (1H, dd, J = 8.5, 1.0 Hz, HAr), 7.81 (1H, d, J = 8.5 Hz, HAr), 6.13–6.04 (1H, m, –CH
O), 734.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.62 (1H, d, J = 2.0 Hz, HAr), 8.24 (1H, d, J = 9.0, 2.5 Hz, HAr), 8.07 (1H, d, J = 2.0 Hz, HAr), 7.92 (1H, d, J = 9.0 Hz, HAr), 7.88 (1H, dd, J = 8.5, 1.0 Hz, HAr), 7.81 (1H, d, J = 8.5 Hz, HAr), 6.13–6.04 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.23 (1H, d, J = 9.5 Hz,
), 5.23 (1H, d, J = 9.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.06 (2H, s, –CH2–), 4.92 (1H, d, J = 17.5 Hz,
CH2), 5.06 (2H, s, –CH2–), 4.92 (1H, d, J = 17.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 155.5, 146.7, 143.3, 141.5, 140.2, 135.3, 133.6, 131.7, 130.9, 129.5, 119.8, 118.5, 117.1, 115.5, 111.8, 47.1. LC-MS (m/z) [M + H]+ calcd for C16H12Cl2N3O2 348.0301, found 348.0136.
CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 155.5, 146.7, 143.3, 141.5, 140.2, 135.3, 133.6, 131.7, 130.9, 129.5, 119.8, 118.5, 117.1, 115.5, 111.8, 47.1. LC-MS (m/z) [M + H]+ calcd for C16H12Cl2N3O2 348.0301, found 348.0136.
1-Allyl-2-(3,4-dichlorophenyl)-6-nitro-1H-benzo[d]imidazole (4b). Yellow solid, mp 190–192 °C. IR (ν, cm−1): 1518.0 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1438.9 (C
N), 1438.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1334.7 (N
C), 1334.7 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 742.6 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.61 (1H, d, J = 2.0 Hz, HAr), 8.23 (1H, dd, J = 9.0, 2.5 Hz, HAr), 8.07 (1H, d, J = 2.0 Hz, HAr), 7.92 (1H, d, J = 9.0 Hz, HAr), 7.88 (1H, d, J = 8.5 Hz, HAr), 7.80 (1H, d, J = 8.5 Hz, HAr), 6.13–6.04 (1H, m, –CH
O), 742.6 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.61 (1H, d, J = 2.0 Hz, HAr), 8.23 (1H, dd, J = 9.0, 2.5 Hz, HAr), 8.07 (1H, d, J = 2.0 Hz, HAr), 7.92 (1H, d, J = 9.0 Hz, HAr), 7.88 (1H, d, J = 8.5 Hz, HAr), 7.80 (1H, d, J = 8.5 Hz, HAr), 6.13–6.04 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.23 (1H, d, J = 10.5 Hz,
), 5.23 (1H, d, J = 10.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.10 (2H, s, –CH2–), 4.90 (1H, d, J = 17.5 Hz,
CH2), 5.10 (2H, s, –CH2–), 4.90 (1H, d, J = 17.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 155.5, 146.7, 143.3, 141.5, 140.2, 135.3, 133.7, 131.8, 130.9, 129.5, 119.8, 118.5, 117.1, 115.5, 111.9, 47.1. LC-MS (m/z) [M + H]+ calcd for C16H12Cl2N3O2 348.0301, found 348.0306.
CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 155.5, 146.7, 143.3, 141.5, 140.2, 135.3, 133.7, 131.8, 130.9, 129.5, 119.8, 118.5, 117.1, 115.5, 111.9, 47.1. LC-MS (m/z) [M + H]+ calcd for C16H12Cl2N3O2 348.0301, found 348.0306.
1-Allyl-2-(3,4-dimethoxyphenyl)-6-nitro-1H-benzo[d]imidazole (4c). Yellow solid, mp 144–146 °C. IR (ν, cm−1): 1516.2 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1437.0 (C
N), 1437.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1332.8 (N
C), 1332.8 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.57 (1H, d, J = 2.0 Hz, HAr), 8.20 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.92 (1H, d, J = 9.0 Hz, HAr), 7.87 (1H, d, J = 8.5 Hz, HAr), 7.40 (1H, d, J = 2.0 Hz, HAr), 7.17 (1H, dd, J = 8.5, 2.0 Hz, HAr), 6.19–6.09 (1H, m, –CH
O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.57 (1H, d, J = 2.0 Hz, HAr), 8.20 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.92 (1H, d, J = 9.0 Hz, HAr), 7.87 (1H, d, J = 8.5 Hz, HAr), 7.40 (1H, d, J = 2.0 Hz, HAr), 7.17 (1H, dd, J = 8.5, 2.0 Hz, HAr), 6.19–6.09 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.27 (1H, d, J = 10.5 Hz,
), 5.27 (1H, d, J = 10.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.10 (2H, s, –CH2–), 4.92 (1H, d, J = 17.0 Hz,
CH2), 5.10 (2H, s, –CH2–), 4.92 (1H, d, J = 17.0 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 3.86 (3H, s, –OCH3), 3.83 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 158.1, 150.8, 150.7, 148.7, 143.0, 140.4, 135.3, 133.2, 122.0, 121.1, 119.1, 117.9, 114.9, 112.3, 111.3, 55.6, 55.3, 47.1. LC-MS (m/z) [M + H]+ calcd for C18H18N3O4 340.1292, found 340.1264.
CH2), 3.86 (3H, s, –OCH3), 3.83 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 158.1, 150.8, 150.7, 148.7, 143.0, 140.4, 135.3, 133.2, 122.0, 121.1, 119.1, 117.9, 114.9, 112.3, 111.3, 55.6, 55.3, 47.1. LC-MS (m/z) [M + H]+ calcd for C18H18N3O4 340.1292, found 340.1264.
1-Allyl-2-(4-ethoxyphenyl)-6-nitro-1H-benzo[d]imidazole (4d). Yellow solid, mp 129–131 °C. IR (ν, cm−1): 1610.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1510.3 (C
N), 1510.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1330.9 (N
C), 1330.9 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1253.7 (C–O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.55 (1H, d, J = 2.5 Hz, HAr), 8.18 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.85 (1H, d, J = 9.0 Hz, HAr), 7.75 (2H, d, J = 8.5 Hz, HAr), 7.12 (2H, d, J = 8.5 Hz, HAr), 6.15–6.06 (1H, m, –CH
O), 1253.7 (C–O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.55 (1H, d, J = 2.5 Hz, HAr), 8.18 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.85 (1H, d, J = 9.0 Hz, HAr), 7.75 (2H, d, J = 8.5 Hz, HAr), 7.12 (2H, d, J = 8.5 Hz, HAr), 6.15–6.06 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.24 (1H, d, J = 10.5 Hz,
), 5.24 (1H, d, J = 10.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.09 (2H, s, –CH2–), 4.90 (1H, d, J = 17.5 Hz,
CH2), 5.09 (2H, s, –CH2–), 4.90 (1H, d, J = 17.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.13 (2H, q, J = 7.0 Hz, –CH2–), 1.38 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 160.4, 158.0, 147.2, 142.5, 140.3, 133.1, 130.7 (2C), 120.9, 119.1, 117.9, 116.8, 114.9 (2C), 111.3, 63.4, 47.0, 14.5. LC-MS (m/z) [M + H]+ calcd for C18H18N3O3 324.1343, found 324.1301.
CH2), 4.13 (2H, q, J = 7.0 Hz, –CH2–), 1.38 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 160.4, 158.0, 147.2, 142.5, 140.3, 133.1, 130.7 (2C), 120.9, 119.1, 117.9, 116.8, 114.9 (2C), 111.3, 63.4, 47.0, 14.5. LC-MS (m/z) [M + H]+ calcd for C18H18N3O3 324.1343, found 324.1301.
1-Allyl-2-(4-fluorophenyl)-6-nitro-1H-benzo[d]imidazole (4e). Yellow solid, mp 211–213 °C. IR (ν, cm−1): 1506.4 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1448.5 (C
N), 1448.5 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1331.6 (N
C), 1331.6 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.59 (1H, d, J = 1.5 Hz, HAr), 8.21 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.88 (2H, d, J = 8.5 Hz, HAr), 7.78 (1H, d, J = 8.5 Hz, HAr), 7.44 (2H, d, J = 8.5 Hz, HAr), 6.12–6.04 (1H, m, –CH
O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.59 (1H, d, J = 1.5 Hz, HAr), 8.21 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.88 (2H, d, J = 8.5 Hz, HAr), 7.78 (1H, d, J = 8.5 Hz, HAr), 7.44 (2H, d, J = 8.5 Hz, HAr), 6.12–6.04 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.23 (1H, d, J = 10.5 Hz,
), 5.23 (1H, d, J = 10.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.05 (2H, s, –CH2–), 4.90 (1H, d, J = 17.5 Hz,
CH2), 5.05 (2H, s, –CH2–), 4.90 (1H, d, J = 17.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 162.4, 157.1, 146.9, 142.9, 135.2, 133.0, 131.7 (2C), 125.5, 119.5, 118.2, 117.0, 116.9, 116.2 (2C), 115.3, 111.7, 47.0. LC-MS (m/z) [M + H]+ calcd for C16H13FN3O2 298.0986, found 298.0972.
CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 162.4, 157.1, 146.9, 142.9, 135.2, 133.0, 131.7 (2C), 125.5, 119.5, 118.2, 117.0, 116.9, 116.2 (2C), 115.3, 111.7, 47.0. LC-MS (m/z) [M + H]+ calcd for C16H13FN3O2 298.0986, found 298.0972.
4-(1-Allyl-6-nitro-1H-benzo[d]imidazole-2-yl)-N,N-dimethylaniline (4f). Red solid, mp 170–172 °C. IR (ν, cm−1): 1611.7 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1458.2 (C
N), 1458.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1323.2 (N
C), 1323.2 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.51 (1H, d, J = 2.0 Hz, HAr), 8.14 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.80 (1H, d, J = 9.0 Hz, HAr), 7.67 (2H, d, J = 8.5 Hz, HAr), 6.86 (2H, d, J = 8.5 Hz, HAr), 6.18–6.09 (1H, m, –CH
O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.51 (1H, d, J = 2.0 Hz, HAr), 8.14 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.80 (1H, d, J = 9.0 Hz, HAr), 7.67 (2H, d, J = 8.5 Hz, HAr), 6.86 (2H, d, J = 8.5 Hz, HAr), 6.18–6.09 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.24 (1H, d, J = 10.5 Hz,
), 5.24 (1H, d, J = 10.5 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 5.04 (2H, m, –CH2–), 4.93 (1H, d, J = 17.0 Hz,
CH2), 5.04 (2H, m, –CH2–), 4.93 (1H, d, J = 17.0 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 3.01 (6H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 158.9, 151.6, 147.6, 142.9, 140.6, 135.5, 130.1 (2C), 118.5, 116.7, 115.3, 114.4, 111.7 (2C), 111.0, 47.1, 41.5 (2C). LC-MS (m/z) [M + H]+ calcd for C18H19N4O2 323.1503, found 323.1437.
CH2), 3.01 (6H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 158.9, 151.6, 147.6, 142.9, 140.6, 135.5, 130.1 (2C), 118.5, 116.7, 115.3, 114.4, 111.7 (2C), 111.0, 47.1, 41.5 (2C). LC-MS (m/z) [M + H]+ calcd for C18H19N4O2 323.1503, found 323.1437.
1-(4-Chlorobenzyl)-2-(3,4-dichlorophenyl)-6-nitro-1H-benzo[d]imidazole (4g). Yellow solid, mp 190–192 °C. IR (ν, cm−1): 1516.1 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1438.9 (C
N), 1438.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1332.8 (N
C), 1332.8 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 734.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.61 (1H, d, J = 2.5 Hz, HAr), 8.20 (1H, dd, J = 9.0, 2.5 Hz, HAr), 8.00 (1H, d, J = 2.0 Hz, HAr), 7.96 (1H, d, J = 9.0 Hz, HAr), 7.83 (1H, d, J = 8.5 Hz, HAr), 7.73 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.37–7.35 (2H, m, HAr), 7.04 (2H, d, J = 8.5 Hz, HAr), 5.78 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.9, 142.4, 135.8, 134.2, 133.0, 130.7, 129.4, 131.1, 129.5, 129.0 (2C), 128.2 (2C), 126.6, 127.1, 122.8, 120.5, 118.2, 112.1, 46.9. LC-MS (m/z) [M − H]− calcd for C20H11Cl3N3O2 429.9922, found 429.9852.
O), 734.9 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.61 (1H, d, J = 2.5 Hz, HAr), 8.20 (1H, dd, J = 9.0, 2.5 Hz, HAr), 8.00 (1H, d, J = 2.0 Hz, HAr), 7.96 (1H, d, J = 9.0 Hz, HAr), 7.83 (1H, d, J = 8.5 Hz, HAr), 7.73 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.37–7.35 (2H, m, HAr), 7.04 (2H, d, J = 8.5 Hz, HAr), 5.78 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.9, 142.4, 135.8, 134.2, 133.0, 130.7, 129.4, 131.1, 129.5, 129.0 (2C), 128.2 (2C), 126.6, 127.1, 122.8, 120.5, 118.2, 112.1, 46.9. LC-MS (m/z) [M − H]− calcd for C20H11Cl3N3O2 429.9922, found 429.9852.
1-(4-Chlorobenzyl)-2-(3,4-dimethoxyphenyl)-6-nitro-1H-benzo[d]imidazole (4h). Yellow solid, mp 145–146 °C. IR (ν, cm−1): 1517.8 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1450.4 (C
N), 1450.4 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1334.7 (N
C), 1334.7 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 740.2 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.60 (1H, d, J = 2.0 Hz, HAr), 8.17 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.72 (1H, d, J = 9.0 Hz, HAr), 7.39–7.37 (2H, m, HAr), 7.29 (2H, m, HAr), 7.12 (1H, d, J = 9.0 Hz, HAr), 7.06 (2H, d, J = 8.5 Hz, HAr), 5.69 (2H, s, –CH2–), 3.82 (3H, s, –OCH3), 3.70 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.7, 150.4, 148.9, 142.1, 137.2, 136.5, 128.9 (2C), 127.6 (2C), 127.0, 126.1, 123.5, 122.5, 121.4, 120.8, 112.9, 111.6, 110.5, 55.7, 55.5, 47.8. LC-MS (m/z) [M − H]− calcd for C22H17ClN3O4 422.0913, found 422.0811; [M + H]+ calcd for C22H19ClN3O4 424.1059, found 424.1087.
O), 740.2 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.60 (1H, d, J = 2.0 Hz, HAr), 8.17 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.72 (1H, d, J = 9.0 Hz, HAr), 7.39–7.37 (2H, m, HAr), 7.29 (2H, m, HAr), 7.12 (1H, d, J = 9.0 Hz, HAr), 7.06 (2H, d, J = 8.5 Hz, HAr), 5.69 (2H, s, –CH2–), 3.82 (3H, s, –OCH3), 3.70 (3H, s, –OCH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 154.7, 150.4, 148.9, 142.1, 137.2, 136.5, 128.9 (2C), 127.6 (2C), 127.0, 126.1, 123.5, 122.5, 121.4, 120.8, 112.9, 111.6, 110.5, 55.7, 55.5, 47.8. LC-MS (m/z) [M − H]− calcd for C22H17ClN3O4 422.0913, found 422.0811; [M + H]+ calcd for C22H19ClN3O4 424.1059, found 424.1087.
4-(1-(4-Chlorobenzyl)-6-nitro-1H-benzo[d]imidazole-2-yl)-2-ethoxyphenol (4i). Yellow solid, mp 150–152 °C. IR (ν, cm−1): 1519.0 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1451.6 (C
N), 1451.6 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1334.5 (N
C), 1334.5 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 739.5 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.66 (1H, d, J = 2.0 Hz, HAr), 8.18 (1H, dd, J = 8.0, 2.5 Hz, HAr), 7.75 (1H, d, J = 8.0 Hz, HAr), 7.46 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.37 (2H, d, J = 8.5 Hz, HAr), 7.26 (1H, d, J = 2.0 Hz, HAr), 7.18 (1H, d, J = 9.0 Hz, HAr), 7.03 (2H, d, J = 8.5 Hz, HAr), 5.68 (2H, s, –CH2–), 3.95 (2H, q, J = 7.0 Hz, –CH2–), 1.27 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.6, 151.2, 149.1, 143.6, 137.4, 136.3, 129.1, 127.8 (2C), 127.0 (2C), 126.5, 123.2, 122.7, 121.6, 120.4, 112.5, 111.8, 110.9, 55.6, 47.7, 14.8. LC-MS (m/z) [M − H]− calcd for C22H17ClN3O4 422.0913, found 422.0915.
O), 739.5 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.66 (1H, d, J = 2.0 Hz, HAr), 8.18 (1H, dd, J = 8.0, 2.5 Hz, HAr), 7.75 (1H, d, J = 8.0 Hz, HAr), 7.46 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.37 (2H, d, J = 8.5 Hz, HAr), 7.26 (1H, d, J = 2.0 Hz, HAr), 7.18 (1H, d, J = 9.0 Hz, HAr), 7.03 (2H, d, J = 8.5 Hz, HAr), 5.68 (2H, s, –CH2–), 3.95 (2H, q, J = 7.0 Hz, –CH2–), 1.27 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 153.6, 151.2, 149.1, 143.6, 137.4, 136.3, 129.1, 127.8 (2C), 127.0 (2C), 126.5, 123.2, 122.7, 121.6, 120.4, 112.5, 111.8, 110.9, 55.6, 47.7, 14.8. LC-MS (m/z) [M − H]− calcd for C22H17ClN3O4 422.0913, found 422.0915.
3-(1-(4-Chlorobenzyl)-6-nitro-1H-benzo[d]imidazole-2-yl)phenol (4j). Yellow solid, mp 212–213 °C. IR (ν, cm−1): 1522.4 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1454.8 (C
N), 1454.8 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1351.7 (N
C), 1351.7 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 737.1 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 9.86 (1H, s, –OH), 8.62 (1H, d, J = 2.0 Hz, HAr), 8.18 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.70 (1H, d, J = 9.0 Hz, HAr), 7.37 (2H, d, J = 8.5 Hz, HAr), 7.35 (1H, d, J = 8.0 Hz, HAr), 7.14 (1H, d, J = 1.5 Hz, HAr), 7.13 (1H, d, J = 8.0 Hz, HAr), 7.03 (2H, d, J = 8.5 Hz, HAr), 6.98 (1H, dd, J = 9.0, 1.5 Hz, HAr), 5.66 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 157.9, 152.7, 142.5, 135.6, 134.5, 133.1, 131.3, 130.6, 129.8, 129.1, 128.5 (2C), 127.5 (2C), 126.8, 123.2, 120.8, 118.4, 111.7, 46.8. LC-MS (m/z) [M − H]− calcd for C20H13ClN3O3 378.0651, found 378.0595.
O), 737.1 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 9.86 (1H, s, –OH), 8.62 (1H, d, J = 2.0 Hz, HAr), 8.18 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.70 (1H, d, J = 9.0 Hz, HAr), 7.37 (2H, d, J = 8.5 Hz, HAr), 7.35 (1H, d, J = 8.0 Hz, HAr), 7.14 (1H, d, J = 1.5 Hz, HAr), 7.13 (1H, d, J = 8.0 Hz, HAr), 7.03 (2H, d, J = 8.5 Hz, HAr), 6.98 (1H, dd, J = 9.0, 1.5 Hz, HAr), 5.66 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 157.9, 152.7, 142.5, 135.6, 134.5, 133.1, 131.3, 130.6, 129.8, 129.1, 128.5 (2C), 127.5 (2C), 126.8, 123.2, 120.8, 118.4, 111.7, 46.8. LC-MS (m/z) [M − H]− calcd for C20H13ClN3O3 378.0651, found 378.0595.
4-(1-(4-Chlorobenzyl)-6-nitro-1H-benzo[d]imidazole-2-yl)-N,N-dimethylaniline (4k). Red solid, mp 169–171 °C. IR (ν, cm−1): 1536.9 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1457.3 (C
N), 1457.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1352.5 (N
C), 1352.5 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 740.6 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.54 (1H, d, J = 2.5 Hz, HAr), 8.12 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.64 (1H, d, J = 9.0 Hz, HAr), 7.60 (2H, d, J = 9.0 Hz, HAr), 7.39 (2H, d, J = 9.0 Hz, HAr), 7.05 (2H, d, J = 8.5 Hz, HAr), 6.81 (2H, d, J = 9.0 Hz, HAr), 5.68 (2H, s, –CH2–), 2.98 (6H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 155.0, 150.9, 141.5, 136.6, 126.4, 129.5 (2C), 128.4 (2C), 127.1 (2C), 126.0, 125.6, 121.8, 119.5, 116.2, 111.4 (2C), 110.1, 47.4, 39.3 (2C). LC-MS (m/z) [M + H]+ calcd for C22H20ClN4O2 407.1269, found 407.1198.
O), 740.6 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.54 (1H, d, J = 2.5 Hz, HAr), 8.12 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.64 (1H, d, J = 9.0 Hz, HAr), 7.60 (2H, d, J = 9.0 Hz, HAr), 7.39 (2H, d, J = 9.0 Hz, HAr), 7.05 (2H, d, J = 8.5 Hz, HAr), 6.81 (2H, d, J = 9.0 Hz, HAr), 5.68 (2H, s, –CH2–), 2.98 (6H, s, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 155.0, 150.9, 141.5, 136.6, 126.4, 129.5 (2C), 128.4 (2C), 127.1 (2C), 126.0, 125.6, 121.8, 119.5, 116.2, 111.4 (2C), 110.1, 47.4, 39.3 (2C). LC-MS (m/z) [M + H]+ calcd for C22H20ClN4O2 407.1269, found 407.1198.
Ethyl 2-(2-(4-chlorophenyl)-6-nitro-1H-benzo[d]imidazole-1-yl)acetate (4l). Yellow solid, mp 272–274 °C. IR (ν, cm−1): 1737.9 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1518.0 (C
O), 1518.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1448.5 (C
N), 1448.5 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1329.0 (N
C), 1329.0 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 733.0 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.78 (1H, d, J = 2.0 Hz, HAr), 8.25 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.92 (1H, d, J = 9.0 Hz, HAr), 7.78 (2H, d, J = 8.5 Hz, HAr), 7.68 (2H, d, J = 8.5 Hz, HAr), 5.44 (2H, s, –CH2–), 4.11 (2H, q, J = 7.0 Hz, –CH2–), 1.12 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 166.8, 152.3, 145.9, 144.5, 139.6, 138.4, 133.8, 128.7 (2C), 126.1 (2C), 122.6, 119.1, 116.5, 62.5, 47.8, 14.6. LC-MS (m/z) [M + H]+ calcd for C17H15ClN3O4 360.0746, found 360.0747.
O), 733.0 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.78 (1H, d, J = 2.0 Hz, HAr), 8.25 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.92 (1H, d, J = 9.0 Hz, HAr), 7.78 (2H, d, J = 8.5 Hz, HAr), 7.68 (2H, d, J = 8.5 Hz, HAr), 5.44 (2H, s, –CH2–), 4.11 (2H, q, J = 7.0 Hz, –CH2–), 1.12 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 166.8, 152.3, 145.9, 144.5, 139.6, 138.4, 133.8, 128.7 (2C), 126.1 (2C), 122.6, 119.1, 116.5, 62.5, 47.8, 14.6. LC-MS (m/z) [M + H]+ calcd for C17H15ClN3O4 360.0746, found 360.0747.
Ethyl 2-(2-(4-(dimethylamino)phenyl)-6-nitro-1H-benzo[d]imidazole-1-yl)acetate (4m). Yellow solid, mp 162–163 °C. IR (ν, cm−1): 1741.7 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1606.7 (C
O), 1606.7 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1434.6 (C
N), 1434.6 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1325.1 (N
C), 1325.1 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.78 (1H, d, J = 2.0 Hz, HAr), 8.24 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.91 (1H, d, J = 9.0 Hz, HAr), 7.77 (2H, d, J = 8.5 Hz, HAr), 7.69 (2H, d, J = 8.5 Hz, HAr), 5.38 (2H, s, –CH2–), 4.12 (2H, q, J = 7.0 Hz, –CH2–), 1.12 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 167.0, 152.7, 150.8, 143.2, 139.5, 138.1, 132.6, 127.5 (2C), 124.9, 121.6, 117.2, 112.4 (2C), 62.3, 47.6, 39.5 (2C), 14.5. LC-MS (m/z) [M − H]− calcd for C19H19N4O4 367.1412, found 367.1332; [M + H]+ calcd for C19H21N4O4 369.1557, found 369.1457.
O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.78 (1H, d, J = 2.0 Hz, HAr), 8.24 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.91 (1H, d, J = 9.0 Hz, HAr), 7.77 (2H, d, J = 8.5 Hz, HAr), 7.69 (2H, d, J = 8.5 Hz, HAr), 5.38 (2H, s, –CH2–), 4.12 (2H, q, J = 7.0 Hz, –CH2–), 1.12 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 167.0, 152.7, 150.8, 143.2, 139.5, 138.1, 132.6, 127.5 (2C), 124.9, 121.6, 117.2, 112.4 (2C), 62.3, 47.6, 39.5 (2C), 14.5. LC-MS (m/z) [M − H]− calcd for C19H19N4O4 367.1412, found 367.1332; [M + H]+ calcd for C19H21N4O4 369.1557, found 369.1457.
Ethyl 2-(2-(4-fluorophenyl)-6-nitro-1H-benzo[d]imidazole-1-yl)acetate (4n). Yellow solid, mp 178–179 °C. IR (ν, cm−1): 1737.9 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1519.9 (C
O), 1519.9 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1375.3 (C
N), 1375.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1217.1 (N
C), 1217.1 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1153.4 (C–F). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.80 (1H, d, J = 2.5 Hz, HAr), 8.25 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.93 (1H, d, J = 9.0 Hz, HAr), 7.82 (2H, d, J = 8.5 Hz, HAr), 7.46 (2H, d, J = 8.5 Hz, HAr), 5.45 (2H, s, –CH2–), 4.11 (2H, q, J = 7.0 Hz, –CH2–), 1.12 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 166.9, 164.6, 162.5, 146.8, 142.5, 129.7, 129.2 (2C), 125.5, 119.6, 118.0, 117.2, 116.5, 116.4 (2C), 115.2, 62.2, 47.3, 14.6. LC-MS (m/z) [M − H]− calcd for C17H13FN3O4 342.0896, found 342.0811; [M + H]+ calcd for C17H15FN3O4 344.1041, found 344.0937.
O), 1153.4 (C–F). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.80 (1H, d, J = 2.5 Hz, HAr), 8.25 (1H, dd, J = 9.0, 2.5 Hz, HAr), 7.93 (1H, d, J = 9.0 Hz, HAr), 7.82 (2H, d, J = 8.5 Hz, HAr), 7.46 (2H, d, J = 8.5 Hz, HAr), 5.45 (2H, s, –CH2–), 4.11 (2H, q, J = 7.0 Hz, –CH2–), 1.12 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 166.9, 164.6, 162.5, 146.8, 142.5, 129.7, 129.2 (2C), 125.5, 119.6, 118.0, 117.2, 116.5, 116.4 (2C), 115.2, 62.2, 47.3, 14.6. LC-MS (m/z) [M − H]− calcd for C17H13FN3O4 342.0896, found 342.0811; [M + H]+ calcd for C17H15FN3O4 344.1041, found 344.0937.
Ethyl 2-(2-(3-(2-ethoxy-2-oxoethoxy)-4-methoxyphenyl)-6-nitro-1H-benzo[d]imidazole-1-yl)acetate (4o). Yellow solid, mp 117–118 °C. IR (ν, cm−1): 1766.8 and 1737.9 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1603.2 (C
O), 1603.2 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1439.8 (C
N), 1439.8 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1330.9 (N
C), 1330.9 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1251.8 (C–O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.75 (1H, d, J = 2.0 Hz, HAr), 8.22 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.88 (1H, d, J = 9.0 Hz, HAr), 7.37 (1H, dd, J = 8.0, 2.0 Hz, HAr), 7.28 (1H, d, J = 2.0 Hz, HAr), 7.23 (1H, d, J = 8.0 Hz, HAr), 5.42 (2H, s, –CH2–), 5.34 (2H, s, –CH2–), 4.20–4.12 (4H, m, –CH2–), 3.89 (3H, s, –OCH3), 1.24–1.15 (6H, m, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 169.1, 166.8, 153.8, 149.6, 147.5, 144.3, 135.5, 129.5, 126.4, 122.1, 121.0, 120.3, 118.2, 115.8, 112.5, 65.6, 61.3, 61.0, 56.5, 47.4, 14.5, 14.3. LC-MS (m/z) [M − H]− calcd for C22H22N3O8 456.1412, found 456.1298; [M + H]+ calcd for C22H24N3O8 458.1558, found 458.1457.
O), 1251.8 (C–O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.75 (1H, d, J = 2.0 Hz, HAr), 8.22 (1H, dd, J = 9.0, 2.0 Hz, HAr), 7.88 (1H, d, J = 9.0 Hz, HAr), 7.37 (1H, dd, J = 8.0, 2.0 Hz, HAr), 7.28 (1H, d, J = 2.0 Hz, HAr), 7.23 (1H, d, J = 8.0 Hz, HAr), 5.42 (2H, s, –CH2–), 5.34 (2H, s, –CH2–), 4.20–4.12 (4H, m, –CH2–), 3.89 (3H, s, –OCH3), 1.24–1.15 (6H, m, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 169.1, 166.8, 153.8, 149.6, 147.5, 144.3, 135.5, 129.5, 126.4, 122.1, 121.0, 120.3, 118.2, 115.8, 112.5, 65.6, 61.3, 61.0, 56.5, 47.4, 14.5, 14.3. LC-MS (m/z) [M − H]− calcd for C22H22N3O8 456.1412, found 456.1298; [M + H]+ calcd for C22H24N3O8 458.1558, found 458.1457.
1-Allyl-2-(furan-2-yl)-6-nitro-1H-benzo[d]imidazole (4p). Brown solid, mp 168–170 °C. IR (ν, cm−1): 1606.7 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1510.3 (C
N), 1510.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1351.8 (N
C), 1351.8 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1282.6 (C–O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.60 (1H, d, J = 2.0 Hz, HAr), 8.19 (1H, dd, J = 9.0, 2.0 Hz, HAr), 8.06 (1H, d, J = 1.0 Hz, HAr), 7.83 (1H, d, J = 9.0 Hz, HAr), 7.37 (1H, d, J = 3.5 Hz, HAr), 6.81 (1H, d, J = 3.0 Hz, HAr), 6.13–6.07 (1H, m, –CH
O), 1282.6 (C–O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.60 (1H, d, J = 2.0 Hz, HAr), 8.19 (1H, dd, J = 9.0, 2.0 Hz, HAr), 8.06 (1H, d, J = 1.0 Hz, HAr), 7.83 (1H, d, J = 9.0 Hz, HAr), 7.37 (1H, d, J = 3.5 Hz, HAr), 6.81 (1H, d, J = 3.0 Hz, HAr), 6.13–6.07 (1H, m, –CH![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) ), 5.30 (2H, s, –CH2–), 5.18 (1H, d, J = 10.0 Hz,
), 5.30 (2H, s, –CH2–), 5.18 (1H, d, J = 10.0 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2), 4.93 (1H, d, J = 17.0 Hz,
CH2), 4.93 (1H, d, J = 17.0 Hz, ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 148.2, 147.2, 143.7, 141.7, 134.9, 132.9, 119.2, 116.9, 115.0, 114.4, 112.5, 111.2, 107.6, 47.0. LC-MS (m/z) [M + H]+ calcd for C14H12N3O3 270.0873, found 270.0815.
CH2). 13C NMR (125 MHz, DMSO-d6, δ ppm): 148.2, 147.2, 143.7, 141.7, 134.9, 132.9, 119.2, 116.9, 115.0, 114.4, 112.5, 111.2, 107.6, 47.0. LC-MS (m/z) [M + H]+ calcd for C14H12N3O3 270.0873, found 270.0815.
1-(4-Chlorobenzyl)-2-(furan-2-yl)-6-nitro-1H-benzo[d]imidazole (4q). Yellow solid, mp 168–170 °C. IR (ν, cm−1): 1532.4 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1455.7 (C
N), 1455.7 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1349.8 (N
C), 1349.8 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 742.0 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.57 (1H, d, J = 2.0 Hz, HAr), 8.19 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.99 (1H, d, J = 1.5 Hz, HAr), 7.85 (1H, d, J = 9.0 Hz, HAr), 7.37 (2H, d, J = 8.5 Hz, HAr), 7.30 (1H, dd, J = 4.0, 1.0 Hz, HAr), 7.13 (2H, d, J = 8.5 Hz, HAr), 6.76 (1H, dd, J = 3.5, 2.0 Hz, HAr), 5.90 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.2, 148.5, 143.8, 141.3, 136.7, 135.8, 135.0, 133.2, 130.4 (2C), 127.2 (2C), 123.5, 123.1, 121.2, 116.8, 111.5, 46.8. LC-MS (m/z) [M − H]− calcd for C18H11ClN3O3 352.0494, found 352.0427; [M + H]+ calcd for C18H13ClN3O3 354.0640, found 354.0699.
O), 742.0 (C–Cl). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.57 (1H, d, J = 2.0 Hz, HAr), 8.19 (1H, dd, J = 8.5, 2.0 Hz, HAr), 7.99 (1H, d, J = 1.5 Hz, HAr), 7.85 (1H, d, J = 9.0 Hz, HAr), 7.37 (2H, d, J = 8.5 Hz, HAr), 7.30 (1H, dd, J = 4.0, 1.0 Hz, HAr), 7.13 (2H, d, J = 8.5 Hz, HAr), 6.76 (1H, dd, J = 3.5, 2.0 Hz, HAr), 5.90 (2H, s, –CH2–). 13C NMR (125 MHz, DMSO-d6, δ ppm): 152.2, 148.5, 143.8, 141.3, 136.7, 135.8, 135.0, 133.2, 130.4 (2C), 127.2 (2C), 123.5, 123.1, 121.2, 116.8, 111.5, 46.8. LC-MS (m/z) [M − H]− calcd for C18H11ClN3O3 352.0494, found 352.0427; [M + H]+ calcd for C18H13ClN3O3 354.0640, found 354.0699.
Ethyl 2-(2-(furan-2-yl)-6-nitro-1H-benzo[d]imidazole-1-yl)acetate (4r). Brown solid, mp 136–137 °C. IR (ν, cm−1): 1745.6 (C
![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O), 1508.3 (C
O), 1508.3 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1435.0 (C
N), 1435.0 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1329.0 (N
C), 1329.0 (N![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.54 (1H, d, J = 1.5 Hz, HAr), 8.21 (1H, dd, J = 9.0, 2.5 Hz, HAr), 8.02 (1H, s, HAr), 7.95 (1H, d, J = 9.0 Hz, HAr), 7.36 (1H, d, J = 3.5 Hz, HAr), 6.80 (1H, d, J = 2.0 Hz, HAr), 5.56 (2H, s, –CH2–), 4.18 (2H, q, J = 7.0 Hz, –CH2–), 1.19 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 166.5, 147.5, 145.8, 144.9, 135.6, 133.4, 130.5, 127.3, 122.2, 119.8, 113.7, 113.5, 61.6, 47.8, 14.5. LC-MS (m/z) [M + H]+ calcd for C15H14N3O5 316.0928, found 316.0814.
O). 1H NMR (500 MHz, DMSO-d6, δ ppm): 8.54 (1H, d, J = 1.5 Hz, HAr), 8.21 (1H, dd, J = 9.0, 2.5 Hz, HAr), 8.02 (1H, s, HAr), 7.95 (1H, d, J = 9.0 Hz, HAr), 7.36 (1H, d, J = 3.5 Hz, HAr), 6.80 (1H, d, J = 2.0 Hz, HAr), 5.56 (2H, s, –CH2–), 4.18 (2H, q, J = 7.0 Hz, –CH2–), 1.19 (3H, t, J = 7.0 Hz, –CH3). 13C NMR (125 MHz, DMSO-d6, δ ppm): 166.5, 147.5, 145.8, 144.9, 135.6, 133.4, 130.5, 127.3, 122.2, 119.8, 113.7, 113.5, 61.6, 47.8, 14.5. LC-MS (m/z) [M + H]+ calcd for C15H14N3O5 316.0928, found 316.0814.
4.3. In vitro antibacterial and antifungal activities
The minimum inhibitory concentration of the test compounds (MIC) was determined by the micro-broth dilution technique using nutrient broth.28 All bacterial strains were maintained on nutrient agar medium at ±37 °C, and fungal strains were maintained on potato dextrose agar at ±25 °C. Serial twofold dilutions ranging from 1024 to 2 μg mL−1 were prepared in media. The inoculum was prepared using a 4–6 h old broth culture of each bacteria and fungi and diluted in broth media to give a final concentration of 5 × 105 CFU mL−1 in the test tray. The trays were covered and placed in plastic bags to prevent evaporation and are incubated at 35 °C for 18–20 h with the bacteria, and the fungal culture was incubated at 25 °C for 72 h. All determinations were done in triplicates. Ciprofloxacin and fluconazole were used as the positive control for antibacterial and antifungal activities, respectively. The MIC was defined as the lowest concentration of the compound giving complete inhibition of visible growth.4.4. In vitro anticancer activity
The cytotoxic activity of the synthesized compounds was evaluated against the human hepatocyte carcinoma cell line (HepG2), human breast adenocarcinoma cell line (MDA-MB-231), human breast cancer cell line (MCF7), human rhabdomyosarcoma cell line (RMS), and colon carcinoma cell line (C26) using the methyl thiazolyl tetrazolium (MTT) method conducted according to the MTT assay protocol. Paclitaxel was used as the positive control. The assay detects the reduction of yellow tetrazolium (MTT) by metabolically active cells to be purple formazan measured using spectrophotometry.34The cells lines were seeded into 96-well plates at a density of 5000 cells per well, replenished with growth media consisting of Eagle's Minimum Essential Medium (EMEM), 10% Fetal Calf Serum (FCS), 2 mM L-glutamine, 100 IU mL−1 penicillin, 100 μg mL−1 streptomycin. The cells were incubated at 37 °C in 5% CO2 for 24 h. Then, a series of concentrations of the tested compounds and paclitaxel in DMSO was added to each well of the plate and incubated for 48 h. After that, 10 μL fresh solution of MTT reagent was added to each well, and the plate was incubated in a CO2 incubator at 37 °C for 4 h. After the purple precipitate was obtained, the cells were dissolved in ethanol and their optical density was recorded at 570 nm. The experiment was performed on 6 wells for a concentration of the test sample and conducted in parallel with the control DMSO at the same concentration. The percent of proliferation inhibition was calculated using the following formula:
At = absorption of test compound, Ab = absorption of blank, Ac = absorption of control.
4.5. ADME-Tox predictions
The physicochemical properties were calculated using ChemBio3D (ChemBioOffice Ultra 18.0 suite). In silico prediction of the ADME properties (absorption, distribution, metabolism, and excretion) and the toxicity risks (mutagenicity, tumorigenicity, irritation, and reproduction) was performed using ADMETlab 2.0 descriptors algorithm protocol and SwissADME web tool.454.6. In silico molecular docking studies
The structure of ligand molecules and the standards were drawn in ChemBioDraw Ultra 19.0. The energy of each molecule was minimized using ChemBio3D Ultra 19.0. The ligand molecules with minimized energy were then used as input for AutoDock Vina, in order to carry out the docking simulation. The ligand molecules with minimized energy were then used as input for AutoDock Vina, in order to carry out the docking simulation.29 Protein molecules of dihydrofolate reductase, N-myristoyl transferase, gyrase B, vascular endothelial growth factor receptor 2, fibroblast growth factor receptor 1, and histone deacetylase 6 were retrieved from the protein data bank (Table 6). These protein molecules were retrieved from the protein data bank. The receptors were removed all the water molecules and added only polar hydrogen and Kollman charges. The Graphical User Interface program BMGL Tools was used to set the grid box for docking simulations. The compounds or commercial drugs were docked with the target in order to determine the docking parameters with the help of Grid-based ligand docking. Auto Dock Vina was compiled and run under Windows 10.0 Professional operating system. Discovery Studio 2021 was used to deduce the pictorial representation of the interaction between the ligands and the target protein.| Entry | Target | Symbol | PDB ID | Organism | Expression system |
|---|---|---|---|---|---|
| 1 | Dihydrofolate reductase | DHFR-B | 3FYV | Staphylococcus aureus | Escherichia coli |
| 2 | Dihydrofolate reductase | DHFR-F | 4HOF | Candida albicans | Escherichia coli BL21 (DE3) |
| 3 | N-Myristoyl transferase | NMT | 1IYL | Candida albicans | Escherichia coli |
| 4 | Gyrase B | GyrB | 4URM | Staphylococcus aureus | Escherichia coli BL21 (DE3) |
| 5 | Vascular endothelial growth factor receptor 2 | VEGFR-2 | 5EW3 | Homo sapiens | Spodoptera frugiperda |
| 6 | Fibroblast growth factor receptor 1 | FGFR-1 | 5A46 | Homo sapiens | Escherichia coli |
| 7 | Histone deacetylase 6 | HDAC6 | 5EEF | Danio rerio | Escherichia coli BL21 (DE3) |
4.7. Statistical analysis
All values are expressed in Mean ± SEM (Standard Error of Mean). The difference in IC50 value between tested compounds and paclitaxel was analyzed by one-way analysis of variance (ANOVA) with Tukey's Honestly Significant Difference (Tukey HSD) post hoc test using Minitab 18.0 software. The results were considered statistically significant if p < 0.05. The chart is drawn using Microsoft Excel 2019 software.Author contributions
Em Canh Pham: conceptualization, methodology, investigation, data curation, supervision, writing-original draft preparation, writing – review & editing. Tuong Vi Thi Le: investigation, software. Tuyen Ngoc Truong: data curation, supervision, writing-original draft preparation, writing – review & editing.Conflicts of interest
The authors declare that they have no known competing financial interests or personal relationships that could have appeared to influence the work reported in this paper.References
- M. F. Brana, J. M. Castellano, G. Keilhauer, A. Machuca, Y. Martin, C. Redondo, E. Schlick and N. Walker, Anticancer Drug Des., 1994, 9, 527–538 CAS.
- H. M. Refaat, Eur. J. Med. Chem., 2010, 45, 2949–2956 CrossRef CAS PubMed.
- B. T. Buu Hue, H. T. Kim Quy, W. K. Oh, V. D. Duy, C. N. Tram Yen, T. T. Kim Cuc, P. C. Em, T. P. Thao, T. T. Loan and M. V. Hieu, Tetrahedron Lett., 2016, 57, 887–891 CrossRef.
- T. H. Kim Chi, T. N. Hong An, T. N. Cam Thu and T. H. Kim Dung, RSC Adv., 2020, 10, 20543 RSC.
- M. Tunçbilek, T. Kiper and N. Altanlar, Eur. J. Med. Chem., 2009, 44, 1024–1033 CrossRef PubMed.
- H. Z. Zhang, G. L. Damu, G. X. Cai and C. H. Zhou, Eur. J. Med. Chem., 2013, 64, 329–344 CrossRef CAS PubMed.
- S. Malasala, M. N. Ahmad, R. Akunuri, M. Shukla, G. Kaul, A. Dasgupta, Y. V. Madhavi, S. Chopra and S. Nanduri, Eur. J. Med. Chem., 2021, 212, 112996 CrossRef CAS PubMed.
- K. C. Achar, K. M. Hosamani and H. R. Seetharamareddy, Eur. J. Med. Chem., 2010, 45, 2048–2054 CrossRef CAS PubMed.
- N. Pribut, A. E. Basson, W. A. L. van Otterlo, D. C. Liotta and S. C. Pelly, ACS Med. Chem. Lett., 2019, 10, 196–202 CrossRef CAS PubMed.
- M. C. Sharma, D. V. Kohli and S. Sharma, Int. J. Drug Delivery, 2010, 2, 228–237 CrossRef CAS.
- D. C. Cole, J. L. Gross, T. A. Comery, S. Aschmies, W. D. Hirst, C. Kelley, J. I. Kim, K. Kubek, X. Ning, B. J. Platt, A. J. Robichaud, W. R. Solvibile, J. R. Stock, G. Tawa, M. J. Williams and J. W. Ellingboe, Bioorg. Med. Chem. Lett., 2010, 20, 1237–1240 CrossRef CAS PubMed.
- G. Surineni, Y. Gao, M. Hussain, Z. Liu, Z. Lu, C. Chhotaray, M. M. Islam, H. M. A. Hameed and T. Zhang, Medchemcomm, 2019, 10, 49–60 RSC.
- L. Welage and R. Berardi, J. Am. Pharm. Assoc., 2000, 40, 52 CAS.
- B. G. Mohamed, A. A. Abdel-Alim and M. A. Hussein, Acta Pharm., 2006, 56, 31–48 CAS.
- M. Ouattara, D. Sissouma, M. W. Koné, H. E. Menan, S. A. Touré and L. Ouattara, Trop. J. Pharm. Res., 2011, 10, 767–775 CrossRef CAS.
- D. Valdez-Padilla, S. Rodriguez-Morales, A. Hernandez-Campos, F. Hernandez-Luis, L. Yepez-Mulia, A. Tapia-Contreras and R. Castillo, Bioorg. Med. Chem., 2009, 17, 1724–1730 CrossRef CAS PubMed.
- N. Bharti, M. T. Shailendra, G. Garza, D. E. Cruz-Vega, J. Catro-Garza, K. Saleem, F. Naqvi, M. R. Maurya and A. Azam, Bioorg. Med. Chem. Lett., 2002, 12, 869–871 CrossRef CAS PubMed.
- B. Bhrigu, N. Siddiqui, D. Pathak, M. S. Alam, R. Ali and B. Azad, Acta Pol. Pharm., 2012, 69, 53–62 CAS.
- J. Valdez, R. Cedillo, A. Hernández-Campos, L. Yépez, F. Hernández-Luis, G. Navarrete-Vázquez, A. Tapia, R. Cortés, M. Hernández and R. Castillo, Bioorg. Med. Chem. Lett., 2002, 12, 2221–2224 CrossRef CAS.
- S. Tahlan, K. Ramasamy, S. M. Lim, S. A. A. Shah, V. Mani and B. Narasimhan, BMC Chem., 2019, 13, 12 CrossRef PubMed.
- D. Hao, J. D. Rizzo, S. Stringer, R. V. Moore, J. Marty, D. L. Dexter, G. L. Mangold, J. B. Camden, D. D. Von Hoff and S. D. Weitman, Invest. New Drugs, 2002, 20, 261–270 CrossRef CAS PubMed.
- H. Joensuu, J. Y. Blay, A. Comandone, J. Martin-Broto, E. Fumagalli, G. Grignani, X. G. Del Muro, A. Adenis, C. Valverde, A. L. Pousa, O. Bouché, A. Italiano, S. Bauer, C. Barone, C. Weiss, S. Crippa, M. Camozzi, R. Castellana and A. Le Cesne, Br. J. Cancer, 2017, 117, 1278–1285 CrossRef CAS PubMed.
- C. Y. Hsieh, P. W. Ko, Y. J. Chang, M. Kapoor, Y. C. Liang, H. H. Lin, J. C. Horng and M. H. Hsu, Molecules, 2019, 24, 3259 CrossRef PubMed.
- G. Mariappan, R. Hazarika, F. Alam, R. Karki, U. Patangia and S. Nath, Arabian J. Chem., 2015, 8, 715–719 CrossRef CAS.
- P. Boggu, E. Venkateswararao, M. Manickam, D. Kwak, Y. Kim and S. H. Jung, Bioorg. Med. Chem., 2016, 24, 1872–1878 CrossRef CAS PubMed.
- D. R. Gund, A. P. Tripathi and S. D. Vaidya, Eur. J. Chem., 2017, 8, 149–154 CrossRef CAS.
- E. M. E. Dokla, N. S. Abutaleb, S. N. Milik, D. Li, K. El-Baz, M. W. Shalaby, R. Al-Karaki, M. Nasr, C. D. Klein, K. A. M. Abouzid and M. N. Seleem, Eur. J. Med. Chem., 2020, 186, 111850 CrossRef CAS PubMed.
- P. C. Em, L. T. Tuong Vi, T. P. Long, T. N. Huong-Giang, N. B. L. Khanh and N. T. Tuyen, Arabian J. Chem., 2022, 15, 103682 CrossRef.
- P. C. Em, V. V. Lenh, V. N. Cuong, N. D. Ngoc Thoi, L. T. Tuong Vi and N. T. Tuyen, Biomed. Pharmacother., 2022, 146, 112611 CrossRef PubMed.
- P. Singla, V. Luxami and K. Paul, J. Photochem. Photobiol., B, 2017, 168, 156–164 CrossRef CAS PubMed.
- S. B. Nabuurs, M. Wagener and J. de Vlieg, J. Med. Chem., 2007, 50, 6507–6518 CrossRef CAS PubMed.
- S. M. Rizvi, S. Shakil and M. Haneef, EXCLI J., 2013, 12, 831–857 Search PubMed.
- N. El-Hachem, B. Haibe-Kains, A. Khalil, F. H. Kobeissy and G. Nemer, Methods Mol. Biol., 2017, 1598, 391–403 CrossRef CAS PubMed.
- P. C. Em, T. N. Tuyen, D. H. Nguyen, V. D. Duy and D. T. Hong Tuoi, Med. Chem., 2022, 18, 558–573 CrossRef.
- M. Congreve, R. Carr, C. Murray and H. Jhoti, Drug Discovery Today, 2003, 8, 876–877 CrossRef PubMed.
- C. A. Lipinski, Drug Discovery Today: Technol., 2004, 1, 337–341 CrossRef CAS PubMed.
- M. M. Morcoss, E. S. M. N. Abdelhafez, R. A. Ibrahem, H. M. Abdel-Rahman, M. Abdel-Aziz and D. A. Abou El-Ella, Bioorg. Chem., 2020, 101, 103956 CrossRef CAS PubMed.
- Y. Keng Yoon, M. Ashraf Ali, T. S. Choon, R. Ismail, A. Chee Wei, R. Suresh Kumar, H. Osman and F. Beevi, BioMed Res. Int., 2013, 2013, 926309 Search PubMed.
- L. Shi, T. T. Wu, Z. Wang, J. Y. Xue and Y. G. Xu, Bioorg. Med. Chem., 2014, 22, 4735–4744 CrossRef CAS PubMed.
- L. V. Nayak, B. Nagaseshadri, M. V. P. S. Vishnuvardhan and A. Kamal, Bioorg. Med. Chem. Lett., 2016, 26, 3313–3317 CrossRef PubMed.
- Y. K. Yoon, M. A. Ali, A. C. Wei, A. N. Shirazi, K. Parang and T. S. Choon, Eur. J. Med. Chem., 2014, 83, 448–454 CrossRef CAS PubMed.
- J. Zhang, D. Yao, Y. Jiang, J. Huang, S. Yang and J. Wang, Bioorg. Chem., 2017, 72, 168–181 CrossRef CAS PubMed.
- N. S. Goud, S. M. Ghouse, J. Vishnu, D. Komal, V. Talla, R. Alvala, J. Pranay, J. Kumar, I. A. Qureshi and M. Alvala, Bioorg. Chem., 2019, 89, 103016 CrossRef CAS PubMed.
- U. Das, S. Kumar, J. R. Dimmock and R. K. Sharma, Curr. Cancer Drug Targets, 2012, 12, 667–692 CrossRef CAS PubMed.
- G. Xiong, Z. Wu, J. Yi, L. Fu, Z. Yang, C. Hsieh, M. Yin, X. Zeng, C. Wu, A. Lu, X. Chen, T. Hou and D. Cao, Nucleic Acids Res., 2021, 49, W5–W14 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available: ADMET and Docking Information, and NMR spectra. See https://doi.org/10.1039/d2ra03491c |
| This journal is © The Royal Society of Chemistry 2022 |