Open Access Article
Open Access ArticleAnti-SARS-CoV-2 and cytotoxic activity of two marine alkaloids from green alga Caulerpa cylindracea Sonder in the Dardanelles†
Ebru Erol a,
Muge Didem Orhan
a,
Muge Didem Orhan b,
Timucin Avsar
b,
Timucin Avsar bc,
Atilla Akdemir
bc,
Atilla Akdemir d,
Emine Sukran Okudan
d,
Emine Sukran Okudan e,
Gulbahar Ozge Alim Toraman
e,
Gulbahar Ozge Alim Toraman f and
Gulacti Topcu
f and
Gulacti Topcu *fg
*fg
aDept. of Analytical Chemistry, Faculty of Pharmacy, Bezmialem Vakif University, Istanbul, Turkey
bBahcesehir University, Health Sciences Institute, Neuroscience Laboratory, Istanbul, Turkey
cBahcesehir University, School of Medicine, Department of Medical Biology, Istanbul, Turkey
dComputer-Aided Drug Discovery Lab., Dept. of Pharmacology, Faculty of Pharmacy, Bezmialem Vakif University, Istanbul, Turkey
eFaculty of Aquatic Sciences and Fisheries, Akdeniz University, Antalya, Turkey
fDept. of Pharmacognosy, Faculty of Pharmacy, Bezmialem Vakif University, Istanbul, Turkey
gDrug Application & Research Center, Bezmialem Vakif University, Istanbul, Turkey
First published on 19th October 2022
Abstract
Caulerpa cylindracea Sonder is a green alga belonging to the Caulerpaceae family. This is the first chemical investigation of C. cylindracea in the Dardanelles which resulted in the isolation of four compounds, caulerpin (1), monomethyl caulerpinate (2), beta-sitosterol (3), and palmitic acid (4). Their structures were elucidated by spectroscopic analyses including 1D- and 2D NMR and mass. The isolated compounds 1 and 2 were tested against the SARS-CoV-2 viral targets spike protein and main protease (3CL) enzyme, and both compounds significantly inhibit the interaction of spike protein and ACE2, while the main protease activity was not significantly reduced. Docking studies suggested that compounds 1 and 2 may bind to the ACE2 binding pocket on spike, and compound 2 may also bind to an allosteric site on spike. As such, these compounds may inhibit the spike–ACE2 complex formation competitively and/or allosterically and have the potential to be used against SARS-CoV-2 virus infection. In addition, compounds 1 and 2 showed at least two-fold higher cytotoxicity against breast cancer cell lines MCF7 and MDA-MB-231 compared to the CCD fibroblast control cell line.
1 Introduction
Severe Acute Respiratory Syndrome Coronavirus 2 (SARS-CoV-2) emerged in China at the end of 2019 and has spread all over the world, causing serious health risks to both humans and animals. Up to now, the total number of cases in the world is over 595 million, while the number of deaths is 6.4 million according to Worldometer Online, https://www.worldometers.info/coronavirus/, (accessed August 2022). The U.S. Food and Drug Administration (FDA) approved the first mRNA-based vaccine (Pfizer-BioNTech Covid-19) on August 23, 2021. This vaccine and other vaccines have emergency use authorization in the world, and are rejected by a certain majority due to short-term side effects such as low-grade fever or pain or redness at the injection site or uncertainty of long-term side effects. In addition, no specific approved anti-viral drug is yet available for the treatment of the disease except FDA-approved drug remdesivir (Veklury). Considering all of this, drug discovery studies, particularly for antiviral agents, have become even more important, especially in natural products, which have low toxicity and fewer adverse effects. The entry of SARS-CoV-2 into host cells depends on the combination of its spike glycoprotein (S protein) and angiotensin-converting enzyme II (ACE2) on the human cell membranes.1 Interfering or blocking the binding of S protein and ACE2 in cells will be beneficial in drug discovery against COVID-19. In recent years, many research groups have worked on this target to discover lead compounds. Therefore, several publications of new anti-viral agents derived from synthetic and natural sources including repurposing studies have been made and are still going on to be published.2–4Seaweeds have been consumed as food by humans for centuries, and the present worldwide market is worth more than $6 billion per year, with an annual volume of around 12 million tonnes in 2018. Macroalgae are prevalent in coastal ecosystems and provide a source of bioactive metabolites with a variety of potential and noteworthy biological activities, capable of impacting the abundance, distribution, and survival of marine organisms.5
Seaweeds are classified into three major groups, including Chlorophyta (green algae), Rhodophyta (red algae), and Ochrophyta (brown algae) based on their color. It is estimated that 1800 green, and about 1800 brown macroalgae, and 6200 red macroalgae are found in the marine environment.6 Caulerpaceae is one of the green alga families, and represented by a unique genus “Caulerpa”. The genus Caulerpa has 101 species, with 40 varieties and 67 forms in southern Australia.7 They are typically grown in shallow-water tropical and subtropical marine habitats. Seaweeds are well-known for their polysaccharides, minerals, and vitamins.8 Marine algae have also been identified as a possible source of antioxidants9,10 as well as readily available food, particularly in coastal populations.11–13 The fatty acids and enzymatic and non-enzymatic antioxidant capabilities of Caulerpa species have been studied by some researchers.14–16
Caulerpa cylindracea Sonder is a species of seaweed that is native to the Australian coast in a large area extending from around Perth North into the Mid-West and Pilbara to the Kimberley region of the Western Australia. However, C. cylindracea was first introduced to the Mediterranean Sea in the early 1990s, where it has spread extensively and is considered an invasive species. Although this species has been explored in terms of taxonomy, invasiveness, and ecological features, as well as bioactivities and applications,17 it has not been studied in terms of isolation of its secondary metabolites.
In the present study, the green alga Caulerpa cylindracea Sonder was collected in the North Aegean Sea (Dardanelles), and investigated for secondary metabolites for the first time. The four pure compounds caulerpin (1), monomethyl caulerpinate (2), beta-sitosterol (3) and palmitic acid (4) were isolated and their structures were elucidated by 1D- and 2D-NMR and HRMS techniques.
Caulerpin is a red pigment, which was first isolated from a green alga Caulerpa spp., and characterized as a phenazine derivative in 1970,18 the chemical structure was later reassigned.19 It was then isolated from several Caulerpa species, such as Caulerpa peltata,20 C. ashmeadii,21 C. racemosa,22 C. serrulata,23 C. prolifera,24 C. lentillifera,25 C. microphysa,25 C. sertularioides,25 C. taxifolia,26 and C. lamourouxii.27 Caulerpin was also isolated from other alga genera: Halimeda incrossate, Hypnea concornis, Caloglossa leprieuri,28 Chondria armata,29 Laurencia majuscule,28 and from a few brown alga Spatoglossum asperum,30 and Sargassum platycarpum.31 However, we couldn't isolate caulerpin or derivatives in Laurencia obtusa collected from North Aegean Sea in our previous study.32 A number of bioactivity studies have been carried out with caulerpin, including cytotoxic, antitumor, anticancer, antilarvicidal, anticorrosion, antitubercular, antimicrobial, antiviral, antiherpes, antidiabetic, anti-inflammator, spasmolytic, antinociceptive, and plant growth regulatory activities.28,33–35
Searches for combination therapies were carried out with a molecular docking approach, and many potential drugs were investigated.1 There are several antiviral studies on caulerpin. In an earlier investigation, caulerpin was found to be an alternative agent against herpes simplex virus type-1.36 In another study, caulerpin exhibited strong anti-viral activity (EC50 = 0.8 μM) against Chikungunya virus.37 Caulerpin is also being studied as a possible SARS-CoV-2 protein inhibitor. Results of a recent study based on in silico techniques, caulerpin has the highest binding affinity among all examined receptors when compared to other bioactive compounds.31,38
Thus, we have investigated isolated two compounds from C. cylindracea against the SARS-CoV-2 viral targets spike protein and main protease (3CL) enzyme along with their docking studies.
In this study, the cytotoxic activity of the extract of C. cylindracea, and two isolates caulerpin and monomethyl caulerpinate were also investigated against the aggressive breast cancer cell line MDA-MB-231 for the first time besides breast cell line MCF-7. In fact, in a previous studies, caulerpin was evaluated by Li et al. against a panel of human cancer cell lines, and IC50 values were found to be 4.67 (human leukemia: K562), 4.20 (human lung: A549), 1.95 (human cervical: HeLa), 4.04 (human colon: HT29), 3.71 (human breast: SK-BR-3), and 0.72 (human liver: Huh7) μM,39 but not on MDA-MB-231 and MCF-7 breast cell lines.
2 Results and discussion
2.1 Structure elucidation of the isolated compounds
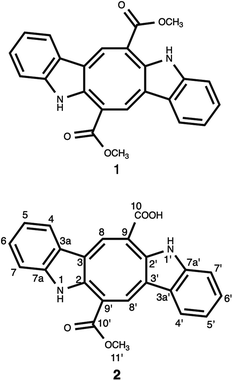
Chemical investigation of the CH2Cl2–MeOH extract of C. cylindracea afforded four known (1–4) compounds.Compound 1 was isolated as an orange-red amorphous solid needles; mp. 315.5–317 °C. Its molecular formula was determined to be C24H17N2O4 corresponding to an [M–H]− ion peak at m/z 397.11945 (calcd for C24H17N2O4, 397.11880) in the HR-MS spectrum with the combination of 1H and 13C-NMR experimental data (ESI†). The 13C NMR (BB) experiment implied the presence of twelve carbon atoms suggesting that this pigment is a dimer, exhibited only 11 signals owing to C-2 symmetry which verified by the Orbitrap high resolution mass spectrometer.
The 13C NMR spectrum (125 MHz, CDCl3, Table 1) of compound 1 showed the presence of a methyl ester carbonyl in the molecule with two signals resonated at δ 166.69 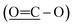 and at δ 52.55
and at δ 52.55 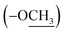 . Aromatic carbons were observed at δ 142.82, 137.72, 132.87, 128.16, 125.48, 123.40, 120.75, 118.07, 112.49 and 111.55. The 1H-NMR data indicated the presence of two methyl signals at δ 3.90 (6H, s). The chemical shift value at δ 9.20 (2H, s) is attributed the presence of N–H groups. A chemical shift observed at δ 8.06 is assigned to the two proton signals as singlets of an eight-member ring. A pair of doublet of triplet at δ 7.31 (2H, dt, 8.1, 0.9 Hz, 1H), and δ 7.43 (2H, dq, 8.0, 0.8 Hz, 1H), and a signal corresponding to two protons at δ 7.10 (2H, ddd, 8.0, 7.0, 1.0 Hz), and δ 7.18 (2H, ddd, 8.1, 7.0, 1.1 Hz) proposed the bis-indole skeleton of the structure, based on all NMR (Table 1), and mass results and literature data,31,40,41 structure of the isolated compound 1 was found to be identical to caulerpin (Fig. 1).
. Aromatic carbons were observed at δ 142.82, 137.72, 132.87, 128.16, 125.48, 123.40, 120.75, 118.07, 112.49 and 111.55. The 1H-NMR data indicated the presence of two methyl signals at δ 3.90 (6H, s). The chemical shift value at δ 9.20 (2H, s) is attributed the presence of N–H groups. A chemical shift observed at δ 8.06 is assigned to the two proton signals as singlets of an eight-member ring. A pair of doublet of triplet at δ 7.31 (2H, dt, 8.1, 0.9 Hz, 1H), and δ 7.43 (2H, dq, 8.0, 0.8 Hz, 1H), and a signal corresponding to two protons at δ 7.10 (2H, ddd, 8.0, 7.0, 1.0 Hz), and δ 7.18 (2H, ddd, 8.1, 7.0, 1.1 Hz) proposed the bis-indole skeleton of the structure, based on all NMR (Table 1), and mass results and literature data,31,40,41 structure of the isolated compound 1 was found to be identical to caulerpin (Fig. 1).
| Pos. | Caulerpin (1) (in CDCl3) | Monomethyl caulerpinate (2) (in CD3OD) | |||
|---|---|---|---|---|---|
| δC | δH,mult. (J Hz) | δC | δH,mult. (J Hz) | HMBC (H→C) | |
| 1 (N) | — | 9.20 s (2H) | — | — | — |
| 2 | 132.87 | 8.06 s (2H) | 131.60 | — | — |
| 3 | 112.49 | — | 112.64 | — | — |
| 3a | 128.16 | — | 127.99 | — | — |
| 4 | 118.07 | 7.43 dt (2H, 8.0, 0.8) | 116.99 | 7.32 dt (1H, 7.9, 1.1) | C5, C7, C7a |
| 5 | 120.75 | 7.10 ddd (2H, 8.0, 7.0, 1.0) | 121.82 | 6.94 dtd (1H, 7.9, 7.0, 1.0) | C3a, C7 |
| 6 | 123.40 | 7.18 ddd (2H, 8.1, 7.0, 1.1) | 119.03 | 7.02 dddd (1H, 8.2, 7.2, 2.4, 1.2) | C4, C7a |
| 7 | 111.55 | 7.31 dt (2H, 8.1, 0.9 Hz) | 110.69 | 7.22 dt (1H, 8.1, 0.9) | C6, C3a |
| 7a | 137.72 | — | 137.66 | — | — |
| 8 | 142.82 | 8.06 s (2H) | 135.11 | 7.76 s (1H) | C10, C2′, C2, C3 |
| 9 | 125.48 | — | 133.78 | — | — |
| 10 | 166.68 | — | 173.22 | — | — |
| 11 | 52.55 | 3.90 s (6H) | — | — | — |
| 1′ (N) | — | — | — | ||
| 2′ | 136.77 | — | — | ||
| 3′ | 110.59 | — | — | ||
| 3a′ | 128.39 | — | — | ||
| 4′ | 117.52 | 7.41 dt (1H, 7.9, 1.0) | C5′, C7a′ | ||
| 5′ | 121.82 | 6.94 dtd (1H, 7.9, 7.0, 1.0) | C3a′, C7′ | ||
| 6′ | 119.49 | 7.02 ddd (1H, 8.1, 7.0, 1.1) | C4′, C7a′ | ||
| 7′ | 111.06 | 7.25 dt (1H, 8.1, 0.9) | C6′, C3a′ | ||
| 7a′ | 138.02 | — | — | ||
| 8′ | 142.88 | 8.12 s (1H) | C10′, C2′, C2, C9′ | ||
| 9′ | 125.13 | — | — | ||
| 10′ | 167.18 | — | — | ||
| 11′ | 51.21 | 3.80 s (3H) | C10′ | ||
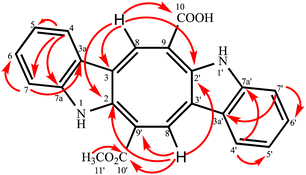
Compound 2 was isolated as an orange-red amorphous solid; mp. 160–162 °C (decomposition). Its molecular ion peak observed at m/z (rel. int.) 384.11128 (26) in the HRESI-MS which corresponded to a molecular formula C23H16N2O4. But, [M–H]− peak of compound 2 were observed at m/z 383.10364 (100) as the main ion (calcd for C23H15N2O4, 383.10320) in the HRESI-MS spectrum. The fragment ions were observed at m/z (rel. int.) 369.08786 [M–CH3]+ (16), 353.34222 [M–OCH3]+ (2), 325.18411 [M–OCOCH3]+ (6). Its 1D- and 2D-NMR spectra were very similar to those of compound 1. However, the main difference of compound 2 from caulerpin (1) was observed in the 1H- and 13C-NMR spectra (Table 1) due to the lack of one of the methyl signals (ESI†). While in caulerpin, the molecule has a symmetry axis, in compound 2 this symmetry axis was not present anymore due to one of the ester groups converted into acid group. Therefore, acid carbonyl-C was observed at δ 173.22 while ester carbonyl resonated at δ 167.18. In addition, the proton signals H-8 and H-8′ were differed appearing at δ 7.76 and 8.12, each showed three-bond away correlations with C-2 and C-2′ in the HMBC (Table 1, Fig. 2) experiment. In addition, the rest of C-signals of compound 2 were observed separately, except a C signal resonated at δ 121.82 for two carbon atoms (C-5, 5′). As a result, totally 22C signals resonated for 23 carbon atoms consisting of 10 aromatic methine C's, 10 quaternary C's, 2 carbonyl C's (an acid and an ester), and a methoxy carbon.
Thus, structure of compound 2 was elucidated as monomethyl caulerpinate considering 1D- and 2D-NMR and HRMS spectroscopic analyses, isolated from a marine green-alga Caulerpa cylindracea. In fact, it was first isolated from C. racemosa,22 but, the previous 1H-NMR data were not given sufficiently, even no 13C-NMR data were present. The 1H-NMR and 13C-NMR spectra of monomethyl caulerpinate (2) were taken in MeOD-d4, for the first time, herein.
2.2 Caulerpin and monomethyl caulerpinate inhibited spike binding but not main protease activity
The purpose of main protease inhibition assay was to assess whether caulerpin and mono methyl caulerpinate inhibit the main protease enzyme which is important in replication of virus.Caulerpin did not show any significant enzyme inhibition at any of concentrations. The all concentrations were tested two separate studies at triplicates, 10 nM of caulerpin also prevented the fluorescence measurement. The reason might be due to autofluorescence characteristics of caulerpin that can mask the fluorescence at 460 nm (Fig. 4a). Monomethyl caulerpinate caused slight decrease in main protease activity at higher concentrations. Maximum tested concentration 100 μM of mono methyl caulerpinate reduced 22.4% enzyme activity, SARS-CoV-2 virus (Fig. 4b). Viral spike and human ACE2 protein which was not significant. Additionally, no other concentrations significantly reduced the main protease enzyme activity of binding were tested to understand whether caulerpin and monomethyl caulerpinate can inhibit the interaction between these proteins. Both of the molecules significantly inhibited the spike–ACE2 binding (p < 0.001). Half maximal inhibitory concentration (IC50) of caulerpin was 87.5 nM and IC50 of monomethyl caulerpinate was 190.2 nM (S17, S18) maximum inhibition at 100 μM concentration for caulerpin was 59% (Fig. 4a) and 66% (Fig. 4b) for monomethyl caulerpinate compound. Overall, it can be stated that caulerpin and mono methyl caulerpinate can significantly inhibit the spike2–ACE2 binding, but not main protease activity. Therefore, both compounds can be used to prevent SARS-CoV-2 infection rather than directly targeting virus replication.
2.3 Molecular modelling studies
Caulerpin and monomethyl caulerpinate were docked into two different crystal structures of the SARS-CoV-2 spike protein receptor binding domain.42The study revealed that both compounds could interact with the pocket of the spike protein with which ACE2 also interacts and as such interfere with the binding of the latter (Fig. 3). Caulerpin forms a hydrogen bond with Tyr449 and π–π stacking with Tyr505. On the other hand, monomethyl caulerpinate forms more extensive interactions with the binding pocket, including hydrogen bonds with the sidechains of Arg403 and Tyr505 and the backbone of Gly496. In addition, the anionic carboxylic acid tail of the ligand forms electrostatic interactions with Arg403 and π–π stacking with Tyr505 are formed.
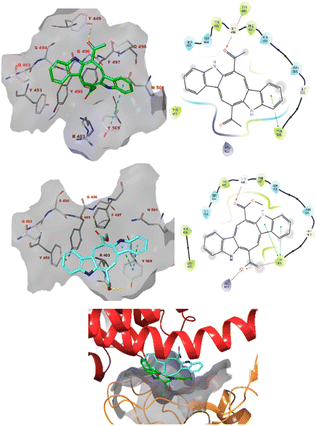 | ||
| Fig. 3 The docked pose caulerpin (top) and monomethyl caulerpinate (middle) with the SARS-CoV-2 spike protein (pdb 6m0j). The ligand interacts with the pocket on spike to which ACE2 also binds (bottom). Hydrogen bonds are indicated in yellow dashed (3D) or purple solid (2D plots) lines. π–π stackings are shown in blue dashed (3D) and green solid (2D plots) lines. | ||
Furthermore, monomethyl caulerpinate was able to interact with the pocket of the spike protein where the cyclic peptide inhibitor was bound in the 7l4z cocrystal structure (Fig. 5). The ligand forms hydrogen bonds with the sidechains of Arg357 and Thr393 and with the backbones of Phe392, Asn394 and Val395.
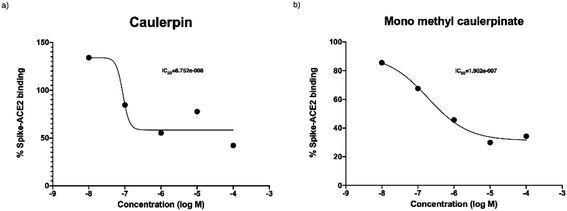 | ||
| Fig. 4 Spike and ACE2 binding activity of (a) caulerpin and (b) monomethyl caulerpinate at different concentrations. | ||
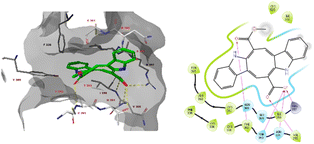 | ||
| Fig. 5 The docked pose monomethyl caulerpinate with the SARS-CoV-2 spike protein (pdb 7l4z). Hydrogen bonds are indicated in yellow dashed (3D) or purple solid (2D plots) lines. π–π stackings are shown in blue dashed (3D) and green solid (2D plots) lines. | ||
The modelling studies suggest that especially monomethyl caulerpinate and possibly also caulerpin may bind to different sites on the SARS-CoV-2 spike protein and interfere with the formation of the spike–ACE2 complex which is necessary to viral cell entry.
2.4 Strong cytotoxic activity profile
It is worthy that the IC50 values of the alga extract of C. cylindracea showed no toxic effect in the healthy fibroblast cell line (CCD-1079Sk) as well as in cancer cell lines, which indicates that the possible use as food of this Caulerpa species (Table 2). On the other hand, the isolated two bis-indole alkaloids from C. cylindracea showed strong activity both in the MDA-MB-231 and the MCF-7 breast cell lines. Moreover, monomethyl caulerpinate showed high IC50 value (424.6 ± 0.47 μg mL−1) on the healthy cell lines, particularly after 24 hours incubation that indicated healthy cells remain viable, almost two-fold higher than the standard drug doxorubicin. Thus, monomethyl caulerpinate exhibited almost six times higher viability compared to that of caulerpin. In this work, two bis-indole alkaloids isolated from C. cylindracea tested for the first time against both the MCF-7 and the aggressive MDA-MB-231 cell lines. These results indicate that both compounds have a high potential to be lead drugs in the treatment of aggressive breast cancer.| Cell lines | ||||||
|---|---|---|---|---|---|---|
| CCD-1079Sk | MDA-MB-231 | MCF7 | ||||
| 24 h | 48 h | 24 h | 48 h | 24 h | 48 h | |
| a IC50 value is calculated as μg mL−1 for only the extract, for the others given as μM. | ||||||
| C. cylindracea extracta | 465.87 ± 1.87 | 295.91 ± 0.25 | 369.12 ± 1.18 | 245.45 ± 0.24 | 346.58 ± 2.56 | 217.64 ± 0.65 |
| Caulerpin | 77.44 ± 0.12 | 36.2 ± 0.69 | 11.85 ± 1.03 | 14.68 ± 0.51 | 15.68 ± 0.38 | 13.82 ± 0.72 |
| Monomethyl caulerpinate | 424.6 ± 0.47 | 53.97 ± 0.86 | 13.96 ± 0.52 | 20.59 ± 0.92 | 16.82 ± 1.12 | 17.28 ± 1.32 |
| Doxorubicin | 284.5 ± 0.04 | — | 62.8 ± 0.06 | — | 42.39 ± 0.08 | — |
3 Experimental
3.1 General experimental procedures
NMR data were acquired using Bruker Magnet System 500'54 Ascend 500 NMR spectrometer (1H-NMR 500 MHz, 13C-NMR 125 MHz). HRESI-MS analyses were carried out by Thermo Orbitrap Q-Exactive mass spectrometer at Drug Application and Research Center, Bezmialem Vakif University, Istanbul. Melting points were recorded on a STUART SMP40. All the NMR solvents were obtained from Merck. Antiviral activity was analyzed by using multimode microplate reader (HIDEX Sense) for SARS-CoV-2 assays (BPS Biosciences, MBP-tagged Assay Kit #79955-2, ACE2 Inhibitor Screening Assay Kit, #79931).3.2 Alga material and extraction procedures
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, 2.4 L × 10 days) at room temperature. The solution was concentrated to give an extract EEL-CC (8.62 g), by using a rotary evaporator under reduced pressure at 38–40 °C (yield 3.83%).
1, 2.4 L × 10 days) at room temperature. The solution was concentrated to give an extract EEL-CC (8.62 g), by using a rotary evaporator under reduced pressure at 38–40 °C (yield 3.83%).The extract (EEL-CC, 8.0 g) was applied to the column chromatography (CC) over a silica gel column (70 cm × 5 cm), [silica gel pore size; 60 Å (70–230 mesh)]. The elution was started using hexane, followed by increasing 25% polarity of CH2Cl2, then after 100% CH2Cl2 elution, polarity increased using acetone as a gradient, and then MeOH was used as final gradient to afford totally 23 fractions (EEL-CC-1—EEL-CC-23). Fr. EEL-CC-8 (377.5 mg) was subjected to a silica gel column (70 cm × 2.2 cm) eluting with a solvent mixture [hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) CH2Cl2 (1
CH2Cl2 (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4)] to isolate compound 1 (caulerpin, 70 mg). Frs. EEL-CC-18 and EEL-CC-19 were combined (440.0 mg), then they were uploaded to a silica gel column (80 cm × 2.2 cm) and eluted with CHCl3
4)] to isolate compound 1 (caulerpin, 70 mg). Frs. EEL-CC-18 and EEL-CC-19 were combined (440.0 mg), then they were uploaded to a silica gel column (80 cm × 2.2 cm) and eluted with CHCl3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) MeOH (9
MeOH (9![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give 33 fractions. Fr. EEL-CC-18–29 was re-chromatographed on a Sephadex LH-20 column (60 cm × 2 cm) using 100% MeOH to afford compound 2 (monomethyl caulerpinate, 9.5 mg). Furthermore, Fr. EEL-CC-7 (918.6 mg) was subjected to a silica gel column (80 cm × 2.2 cm) and eluted with a mixture [hexane
1) to give 33 fractions. Fr. EEL-CC-18–29 was re-chromatographed on a Sephadex LH-20 column (60 cm × 2 cm) using 100% MeOH to afford compound 2 (monomethyl caulerpinate, 9.5 mg). Furthermore, Fr. EEL-CC-7 (918.6 mg) was subjected to a silica gel column (80 cm × 2.2 cm) and eluted with a mixture [hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) diethyl ether (3
diethyl ether (3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1)] to isolate compound 3 (β-sitosterol, 75 mg). Fr. EEL-CC-14–4 (108.2 mg) was uploaded to a silica gel column (50 cm × 2.3 cm) and eluted with hexane
1)] to isolate compound 3 (β-sitosterol, 75 mg). Fr. EEL-CC-14–4 (108.2 mg) was uploaded to a silica gel column (50 cm × 2.3 cm) and eluted with hexane![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) acetone (4
acetone (4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1) to give compound 4 (palmitic acid, 30 mg).
1) to give compound 4 (palmitic acid, 30 mg).
3.3 Anti-viral activity
3.4 Docking studies against the receptor binding domain of SARS-CoV-2 spike protein
The crystal structures of SARS-CoV-2 spike protein in complex with ACE2 (pdb 6m0j) and a cyclic peptide inhibitor (pdb 7l4z) were obtained from the RCSB Protein Data Bank. Subsequently, the structure was prepared using the protein preparation tool of Schrödinger (v2022-3, Schrödinger, Inc., New York, USA). All water and buffer molecules were omitted. Subunit A was retained and all other subunits, if present, were omitted. Subsequently, hydrogen atoms were added (pH 7.4), and the system was minimized using the OPLS4 forcefield.The three-dimensional structures of caulerpin and mono methyl caulerpinate were prepared using the LigPrep tool of Schrödinger, protonated according to pH 7.4 and minimized with the OPLS4 forcefield. Subsequently, all ligands were docked into the binding sites of the target enzymes. The binding sites have been assigned as all residues at the spike–ACE2 interaction interface (pdb 6m0j) or within 5 Å of the cocrystallized ligand (pdb 7l4z). Docking was performed using the Glide tool of Schrödinger with the SP settings. The three highest scoring poses were obtained for each ligand and the poses were subsequently minimized using the Prime tool and MM-GBSA forcefield. To this end, the ligand and all residues within 5.5 Å were unrestrained.
3.5 Viability/cytotoxic activity
In vitro cytotoxic activities were determined by measuring cell proliferation at 6 concentrations and two incubation times (24, 48 h) using the MTT (3-(4,5-dimethylthiazol-2-yl)-2,3-diphenyl-tetrazolium).Cytotoxic activity was performed against three different mammalian cell lines; the healthy cell line (CCD fibroblast) and two the cytotoxic breast cell lines; MCF7 and MDA-MB-231. A 96-well plate was seeded with 5000 cells per well (50% cell occupancy) in Dulbecco's modified Eagle's (DMEM) medium containing 10% FBS, L-glutamine, 100 units per mL penicillin or 100 μg mL−1 streptomycin. They were allowed to adhere to the surface by keeping them at 37 °C overnight. The next day, the medium was aspirated and 100 μL of the new medium was added and the compounds prepared at six different concentrations were added to the cells. After incubation for different times, 10 μL of MTT was added and incubated 5% CO2 for 3 hours in the dark and at 37 °C. After incubation, the absorption at 570 nm was measured with a spectrophotometer. These tests were repeated 3 times for each concentration, the data were compared and plotted according to the concentration, and the IC50 values were determined.43
4 Conclusions
Caulerpin and mono methyl caulerpinate were isolated from the green alga Caulerpa cylindracea species for the first time. Both bis-indole alkaloids, but especially monomethyl caulerpinate, show strong cytotoxic effects against breast cancer cell lines MDA-MB-231 and MCF-7. In addition, both alkaloids inhibit the formation of the spike–ACE2 complex in in vitro assays, and the possible binding interactions of both compounds have been investigated by docking studies revealing possible allosteric and competitive interference with ACE2 binding to spike. Thus, these compounds may be further investigated as possible leads to prevent SARS-CoV-2 infection.Author contributions
Ebru Erol: extraction, isolation and structural elucidation of the compounds, methodology, investigation, writing – original draft. Muge Didem Orhan and Timucin Avsar: testing of antiviral activity, writing – original draft. Atilla Akdemir: molecular modelling studies. Emine Sukran Okudan: identification of the alga. Gulbahar O. Alim Toraman: testing of cytotoxicity activity. Gulacti Topcu: resources, structural elucidation of the compounds, writing – original draft, conceptualization, data curation.Conflicts of interest
There are no conflicts to declare.Acknowledgements
This work was supported by a TUBITAK (The Scientific and Technological Research Council of Turkey) project (project number: TUBITAK 117Z892).References
- H. R. A. El-Mageed, D. A. Abdelrheem, S. A. Ahmed, A. A. Rahman, K. N. M. Elsayed, S. A. Ahmed, A. A. EL-Bassuony and H. S. Mohamed, Struct. Chem., 2021, 32, 1415–1430 CrossRef CAS PubMed.
- N. M. Tam, M. Q. Pham, H. T. Nguyen, N. D. Hong, N. K. Hien, D. T. Quang, H. T. Thu Phung and S. T. Ngo, RSC Adv., 2021, 11, 22206–22213 RSC.
- A. I. Elshamy, T. A. Mohamed, M. A. A. Ibrahim, M. A. M. Atia, T. Yoneyama, A. Umeyama and M. E. F. Hegazy, RSC Adv., 2021, 11, 20151–20163 RSC.
- G. Topcu, H. SenoL, G. O. Alim Toraman and V. M. Altan, Bezmialem Sci., 2020, 8, 131–139 CrossRef.
- L. J. Villarreal-Gómez, I. E. Soria-Mercado, G. Guerra-Rivas and N. E. Ayala-Sánchez, Rev. Biol. Mar. Oceanogr., 2010, 45, 267–275 CrossRef.
- M. I. Rushdi, I. A. M. Abdel-Rahman, E. Z. Attia, W. M. Abdelraheem, H. Saber, H. A. Madkoure, E. Amin, H. M. Hassan, U. R. Abdelmohsen and S. Afr, J. Bot., 2020, 226–241 CAS.
- G. S. Belton, S. G. A. Draisma, W. F. Prud'homme van Reine, J. M. Huisman and C. F. D. Gurgel, Phycologia, 2019, 58, 234–253 CrossRef CAS.
- K. Senthilkumar and S. K. Kim, Adv. Food Nutr. Res., 2014, 72, 195–213 CAS.
- K. Chakraborty, D. Joseph, M. Joy and V. K. Raola, Food Chem., 2016, 212, 778–788 CrossRef CAS PubMed.
- K. Hardouin, G. Bedoux, A. S. Burlot, C. Donnay-Moreno, J. P. Bergé, P. Nyvall-Collén and N. Bourgougnon, Algal Res., 2016, 16, 233–239 CrossRef.
- S. Ghosh, T. Sarkar, S. Pati, Z. A. Kari, H. A. Edinur and R. Chakraborty, Front. Mar. Sci., 2022, 9, 585–598 Search PubMed.
- Y. J. Yang, S. J. Nam, G. Kong and M. K. Kim, Br. J. Nutr., 2010, 103, 1345–1353 CrossRef CAS PubMed.
- Y. Matsumura, Asia Pac. J. Clin. Nutr., 2001, 10, 40–47 CrossRef.
- J. Terrados and J. A. Lopez-Jimenez, Biochem. Mol. Biol. Int., 1996, 39, 863–869 CAS.
- M. Blažina, L. Iveša and M. Najdek, Eur. J. Phycol., 2009, 44, 183–189 CrossRef.
- M. Kumar, V. Gupta, P. Kumari, C. R. K. Reddy and B. Jha, J. Food Compos. Anal., 2011, 24, 270–278 CrossRef CAS.
- L. Stabili, S. Fraschetti, M. I. Acquaviva, R. A. Cavallo, Sa. A. De Pascali, F. P. Fanizzi, C. Gerardi, M. Narracci and L. Rizzo, Mar. Drugs, 2016, 14, 210 CrossRef PubMed.
- G. Aguilar-Santos, J. Chem. Soc. C, 1970, 842–843 RSC.
- B. C. Maiti, R. H. Thomson and M. Mahendran, J. Chem. Res., Synop., 1978, 4, 126–127 Search PubMed.
- R. J. Capon, E. L. Ghisalberti and P. R. Jefferies, Phytochemistry, 1983, 22, 1465–1467 CrossRef CAS.
- V. J. Paul, M. M. Littler, D. S. Littler and W. Fenical, J. Chem. Ecol., 1987, 13, 1171–1185 CrossRef CAS PubMed.
- A. S. R. Anjaneyulu, C. V. S. Prakash and U. V Mallavadhani, Phytochemistry, 1991, 30, 3041–3042 CrossRef CAS.
- J. Y. Su, Y. Zhu, L. M. Zeng and X. H. Xu, J. Nat. Prod., 1997, 60, 1043–1044 CrossRef CAS.
- L. D. Napoli, S. Magno, L. Mayol and E. Novellino, Experientia, Birkhauser Verlag, CH-4010 Basel, Switzerland, 1983, vol. 39, pp. 141–143 Search PubMed.
- C. S. Vairappan, Asian J. Microbiol., Biotechnol. Environ. Sci., 2004, 6, 197–201 CAS.
- S. C. Mao, Y. W. Guo and X. Shen, Bioorg. Med. Chem. Lett., 2006, 16, 2947–2950 CrossRef CAS PubMed.
- V. Kamath and A. Pai, Int. J. Green Pharm., 2018, 12, S46–S50 CAS.
- K. C. Güven, A. Percot and E. Sezik, Mar. Drugs, 2010, 8, 269–284 CrossRef PubMed.
- M. B. Govenkar and S. Wahidulla, Phytochemistry, 2000, 54, 979–981 CrossRef CAS PubMed.
- J. Ara, V. Sultana, A. Tariq, D. Taj, M. Azam and V. U. Ahmed, J. Chem. Soc. Pak., 2019, 41, 379–382 CAS.
- D. A. Abdelrheem, H. R. Abd El-Mageed, H. S. Mohamed, A. A. Rahman, K. N. M. Elsayed and S. A. Ahmed, J. Biomol. Struct. Dyn., 2021, 39, 5137–5147 CrossRef CAS PubMed.
- G. Topcu, Z. Aydogmus, S. Imre, A. C. Gören, J. M. Pezzuto, J. A. Clement and D. G. I. Kingston, J. Nat. Prod., 2003, 66, 1505–1508 CrossRef CAS PubMed.
- J. Lunagariya, P. Bhadja, S. Zhong, R. Vekariya and S. Xu, Mini-Rev. Med. Chem., 2017, 19, 751–761 CrossRef PubMed.
- A. M. M. Lucena, C. R. M. Souza, J. T. Jales, P. M. M. Guedes, G. E. C. de Miranda, A. M. A. de Moura, J. X. Araujo-Junior, G. J. Nascimento, K. C. Scortecci, B. V. O. Santos and J. T. Souto, Mar. Drugs, 2018, 16, 1–18 CrossRef PubMed.
- É. T. De Souza, D. P. De Lira, A. C. De Queiroz, D. J. C. Da Silva, A. B. De Aquino, E. A. Campessato Mella, V. P. Lorenzo, G. E. C. De Miranda, J. X. De Araújo-Júnior, M. C. De Oliveira Chaves, J. M. Barbosa-Filho, P. F. De Athayde-Filho, B. V. De Oliveira Santos and M. S. Alexandre-Moreira, Mar. Drugs, 2009, 7, 689–704 CrossRef PubMed.
- N. Regina, P. Vieira, M. S. Ribeiro, R. C. Villaça, W. Ferreira, A. M. Pinto, V. L. Teixeira, C. Cirne-santos and C. N. P. Izabel, Rev. Bras. Farmacogn., 2011, 22, 861–867 Search PubMed.
- I. C. P. P. Priscilla, O. Esteves, M. C. de Oliveira, C. de Souza Barros, C. C. Cirne-Santos and V. T. Laneuvlille, Nat. Prod. Commun., 2019, 14, 1–6 Search PubMed.
- D. A. Abdelrheem, S. A. Ahmed, H. R. Abd El-Mageed, H. S. Mohamed, A. A. Rahman, K. N. M. Elsayed and S. A. Ahmed, J. Environ. Sci. Health, Part A: Toxic/Hazard. Subst. Environ. Eng., 2020, 55, 1373–1386 CrossRef CAS PubMed.
- H. Li, X. Liao, Y. Sun, R. Zhou, W. Long, L. Li, L. Gu and S. Xu, ChemistrySelect, 2018, 3, 12406–12409 CrossRef CAS.
- C. I. Canché Chay, R. G. Cansino, C. I. Espitia Pinzón, R. O. Torres-Ochoa and R. Martínez, Mar. Drugs, 2014, 12, 1757–1772 CrossRef PubMed.
- A. M. M. Lucena, C. R. M. Souza, J. T. Jales, P. M. M. Guedes, G. E. C. de Miranda, A. M. A. de Moura, J. X. Araújo-Júnior, G. J. Nascimento, K. C. Scortecci, B. V. O. Santos and J. T. Souto, Mar. Drugs, 2018, 16, 318 CrossRef PubMed.
- S. Durdagi, T. Avsar, M. D. Orhan, M. Serhatli, B. K. Balcioglu, H. U. Ozturk, A. Kayabolen, Y. Cetin, S. Aydinlik, T. Bagci-Onder, S. Tekin, H. Demirci, M. Guzel, A. Akdemir, S. Calis, L. Oktay, I. Tolu, Y. E. Butun, E. Erdemoglu, A. Olkan, N. Tokay, Ş. Işık, A. Ozcan, E. Acar, S. Buyukkilic and Y. Yumak, Mol. Ther., 2022, 30, 963–974 CrossRef CAS PubMed.
- T. Mosmann, J. Immunol. Methods, 1983, 65, 55–63 CrossRef CAS PubMed.
Footnote |
| † Electronic supplementary information (ESI) available. See DOI: https://doi.org/10.1039/d2ra03358e |
| This journal is © The Royal Society of Chemistry 2022 |