Open Access Article
Open Access ArticleGreen synthesis of biocompatible core–shell (Au–Ag) and hybrid (Au–ZnO and Ag–ZnO) bimetallic nanoparticles and evaluation of their potential antibacterial, antidiabetic, antiglycation and anticancer activities
Sumaira Anjum *a,
Khadija Nawaza,
Bushra Ahmadb,
Christophe Hano
*a,
Khadija Nawaza,
Bushra Ahmadb,
Christophe Hano c and
Bilal Haider Abbasi
c and
Bilal Haider Abbasi d
d
aDepartment of Biotechnology, Kinnaird College for Women, 92-Jail Road, Lahore-54000, Pakistan. E-mail: sumaira.anjum@kinnaird.edu.pk; Tel: +92-3006957038
bDepartment of Biochemistry, Shaheed Benzair Bhutto Women University, Peshwar-25120, Pakistan
cLaboratoire de Biologie des Ligneux et des Grandes Cultures, INRAE USC1328, University of Orleans, 45067 Orléans Cedex 2, France
dDepartment of Biotechnology, Quaid-i-Azam University, Islamabad-45320, Pakistan
First published on 23rd August 2022
Abstract
The fabrication of bimetallic nanoparticles (BNPs) using plant extracts is applauded since it is an environmentally and biologically safe method. In this research, Manilkara zapota leaf extract was utilized to bioreduce metal ions for the production of therapeutically important core–shell Au–Ag and hybrid (Au–ZnO and Ag–ZnO) BNPs. The phytochemical profiling of the leaf extract in terms of total phenolic and flavonoid content is attributed to its high free radical scavenging activity. FTIR data also supported the involvement of these phytochemicals (polyphenols, flavonoids, aromatic compounds and alkynes) in the synthesis of BNPs. Whereas, TEM and XRD showed the formation of small sized (16.57 nm) spherical shaped core–shell Au–Ag BNPs and ZnO nano-needles with spherical AuNPs (48.32 nm) and ZnO nano-rods with spherical AgNP (19.64 nm) hybrid BNPs. The biological activities of BNPs reinforced the fact that they show enhanced therapeutic efficacy as compared to their monometallic components. All BNPs showed comparable antibacterial activities as compared to standard tetracycline discs. While small sized Au–Ag BNPs were most effective in killing human hepato-cellular carcinoma cells (HepG2) in terms of lowest cell viability, highest intracellular ROS/RNS production, loss of mitochondrial membrane potential, induction of caspase-3 gene expression and enhanced caspase-3/7 activity. BNPs also effectively inhibited advanced glycation end products and carbohydrate digesting enzymes which can be used as a nano-medicine for aging and diabetes. The most important finding was the permissible biocompatibility of these BNPs towards brine shrimp larvae and human RBCs, which suggests their environmental and biological safety. This research study gives us insight into the promise of using a green route to synthesize commercially important BNPs with enhanced therapeutic efficacy as compared to conventional treatment options.
1 Introduction
Bimetallic nanoparticles (BNPs) are more attractive from a scientific and technological viewpoint as compared to monometallic nanoparticles (NPs) due to their superior characteristics. BNPs are composed of two different metals with enhanced electronic, plasmonic, catalytic, optical, thermal, magnetic and biological properties due to a synergistic effect.1 The synthesis of BNPs via a green route is gaining particular attention as NPs are produced without any harsh synthesis conditions or using expensive and hazardous chemicals.2 Moreover, plant mediated synthesis of BNPs has an advantage of producing biocompatible BNPs as they are capped by therapeutically important plant secondary metabolites.3 Among different types of BNPs, gold (Au) is the most biologically prepared bimetallic nanostructure reported so far. Particularly Au–Ag nano-systems have received an enormous amount of attention. Given that, in our research we have synthesized other important BNPs including gold–zinc oxide bimetallic NPs (Au–ZnO BNPs), gold–silver bimetallic NPs (Au–Ag BNPs) and silver–zinc oxide bimetallic NPs (Ag–ZnO BNPs). These BNPs were green synthesized from Manilkara zapota leaf extract (LE) which is valued for its medicinal and nutritional properties. M. zapota is known to possess a rich content of vitamin, minerals, bioactive phenols and flavonoids,4 therefore making it a superb candidate for the green synthesis of BNPs as these metabolites contribute in the bio-reduction, capping and stabilizing of BNPs.Advancement in the synthesis of BNPs has paved way to their diversified applications especially in biomedicine. One of the widely explored therapeutic application of BNPs include their anti-microbial action towards Gram positive and negative multi-drug resistant microbes. The BNPs are emerging as an alternate to antibiotics which have become ineffective due to their inappropriate and inadequate use.5 Secondly, BNPs especially Au–Ag NPs have been reported as important agents for cancer diagnosis and therapeutics as they show increased efficiency and greater stability.6 BNPs have demonstrated a potential to treat a wide range of cancers including ovarian, lung, oral, breast and many others.7–9 Other promising application of BNPs include their anti-diabetic and anti-aging properties.10,11 The advantage of using green BNPs is their target specificity and biocompatibility due to phytochemical capping.12 In this research, the therapeutic efficacy of green synthesized BNPs was evaluated including anti-bacterial activity against Bacillus subtilis, Pseudomonas aeruginosa and Pseudomonas fluorescens; anti-cancerous activity against HepG2 cells in terms of cell viability, intracellular ROS/RNS production, mitochondrial membrane potential and caspase-3 gene expression as well as anti-glycation, anti-diabetic and biocompatibility with brine shrimp larvae and human RBCs. According to our knowledge, this is first report in which a variety of BNPs have been synthesized from a single plant source and comparatively evaluated for their various biological activities, especially the hybrid Au–ZnO BNPs.
2 Materials and methods
2.1. Manilkara zapota leaves extract preparation
Leaves of M. zapota were plucked afresh and verified by the Department of Botany, Kinnaird College for Women, Lahore. 10 g leaves were dry weighed and washed with tap water as well as distilled water thrice. The washed leaves were set to boil in 400 mL distilled water on burner to reduce its volume up to 100 mL. The mixture was grounded with pestle and mortar and filtered with Whatman's filter paper. The filtrate LE was refrigerated at 4 °C.2.2. Phytochemical analysis of Manilkara zapota
Total phenolic contents (TPC) of M. zapota LE were measured by Folin–Ciocalteu's method13 while total flavonoid contents (TFC) were estimated by aluminum chloride colorimetric protocol following Khan et al.13 For TPC, 1 mL LE or gallic acid standard (50–1000 μg mL−1), 0.5 mL Folin–Ciocalteu's reagent and 5 mL distilled water were taken in a test-tube. The mixture was properly shaken and incubated at room temperature (RT) for 5 min, and 1.5 mL of 20% sodium carbonate and distilled water were added to make total volume 5 mL. A deep blue color of mixture developed. Lastly, the mixture was incubated for 2 h at RT and absorbance of known standard was measured at 750 nm using a spectrophotometer (Analytik Jena, Specord 200 Plus, Jena, Germany). Lastly, the TPC of LE was estimated from a gallic acid standard curve (y = 0.00004x + 0.0089, R2 = 0.9953) and expressed as mg g−1 of gallic acid equivalent in milligrams per gram (mg GAE per g) of dry extract.For TFC, 1 mL M. zapota extract/quercetin standard (25–200 μg mL−1), 0.2 mL of 1 M potassium acetate, 5.6 mL distilled water and 0.2 mL of 10% (w/v) AlCl3 were taken in a test tube, shaken and incubated for 30 min at RT. Absorbance of standard was measured at 415 nm by spectrophotometer. Lastly, the TFC of LE was determined from a quercetin standard curve (y = 0.0057x + 0.0127, R2 = 0.9973) and expressed as quercetin equivalent (QE) per gram DW.
TPC and TFC were measured by following equation;
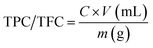 | (1) |
Free radical scavenging activity (FRSA) was evaluated to measure antioxidant potential of M. zapota LE using DPPH (2,2-diphenyl-1-picrylhydrazyl) according to the protocol of Anjum et al.14 Briefly, 0.5 mL LE and 4.5 mL DPPH (3.2 mg/100 mL methanol) were mixed in a test tube and incubated for 1 h at RT. The absorbance of mixture and standard were measured by spectrophotometer at 517 nm. FRSA was expressed as a percentage of discoloration of DPPH calculated using the equation as follows;
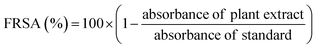 | (2) |
2.3. Synthesis of bimetallic nanoparticles from M. zapota
Au–Ag BNPs were prepared using protocol as mentioned by Elemike et al.15 after some changes. To synthesize Au–Ag BNPs (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5), 10 mL of LE and 50 mL of 1 mM silver nitrate solution (SNS) were mixed together for 5 min followed by addition of 50 mL of 1 mM chloroauric acid. A purple color of solution developed. The solution was constantly stirred for 2 h on magnetic stirrer.
5), 10 mL of LE and 50 mL of 1 mM silver nitrate solution (SNS) were mixed together for 5 min followed by addition of 50 mL of 1 mM chloroauric acid. A purple color of solution developed. The solution was constantly stirred for 2 h on magnetic stirrer.
Au–ZnO BNPs (0.1/0.5) were synthesized from the method of Pandiyan et al.16 1 mL LE and 50 mL of 1 mM chloroauric acid mixture was heated for 5 min at 60 °C followed by adding 50 mL of 0.5 M zinc acetate stock solution. Whereas, Ag–ZnO BNPs (0.1/0.1) were prepared as stated by Sumbal et al.17 with some changes. 1 mL LE was heated for 5 min at 60 °C with 50 mL of 0.1 M zinc acetate. After 5 min, 5 mL of 0.1 M silver nitrate for Ag–ZnO BNPs was added and pH was adjusted to 12 with dropwise addition of 2 M NaOH. The mixture was incubated at 60 °C for 2 h and stirred constantly.
The progress and synthesis of all BNPs was confirmed from UV-visible spectrophotometer. Lastly, the solutions of all BNPs were centrifuged at 10![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 15 min, supernatant discarded and pellets were re-suspended in distilled water. This step was repeated thrice to properly wash BNPs. Finally the BNPs were dried at 40 °C overnight and subjected to characterization and biological activities.
000 rpm for 15 min, supernatant discarded and pellets were re-suspended in distilled water. This step was repeated thrice to properly wash BNPs. Finally the BNPs were dried at 40 °C overnight and subjected to characterization and biological activities.
2.4. Characterization of BNPs
BNPs were characterized for their physico-chemical properties from various techniques including UV-visible spectroscopy, Transmission Electron Microscopy (TEM), X-ray Diffraction (XRD), Fourier-Transform Infrared Spectroscopy (FTIR) and Dynamic Light Scattering (DLS).Green synthesis of BNPs was monitored by recording their UV-vis spectra by spectrophotometer. FTIR analysis was carried out as stated by Tungmunnithum et al.18 using Brucker (Palaiseau, France) V70 interferometer. The wave numbers were measured in the range 400–4500 cm−1.
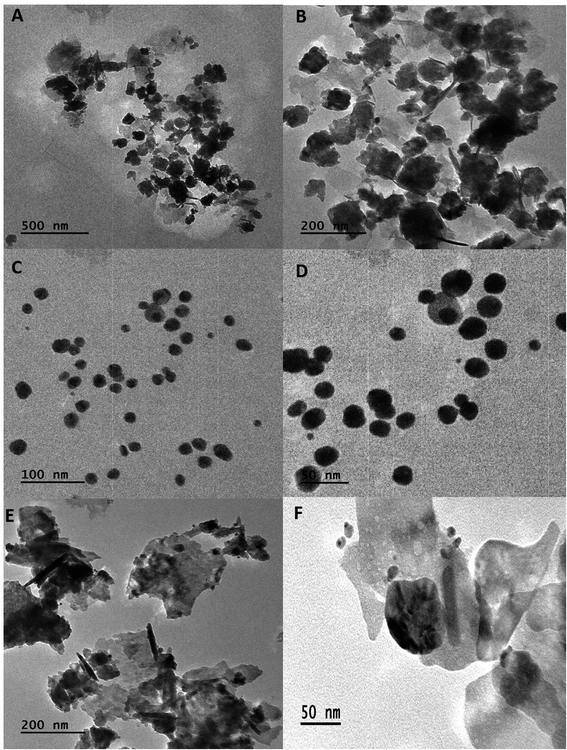
The shape of BNPs was evaluated from TEM (JEM-2100, JEOL Ltd., Tokyo, Japan) by dissolving 1 mg BNPs in 1 mL of 50% ethanol and sonicated for 25 min to ensure proper mixing. 10 μL BNPs were dropped onto a para-film and a grid was placed over it for 5–10 min. The following day BNPs were observed under TEM.19
The crystallinity of BNPs was evaluated by XRD using an X-ray diffractometer (Shimadzu-Model, XRD6000). Furthermore, the size of BNPs was determined by the Debye–Scherrer equation.20
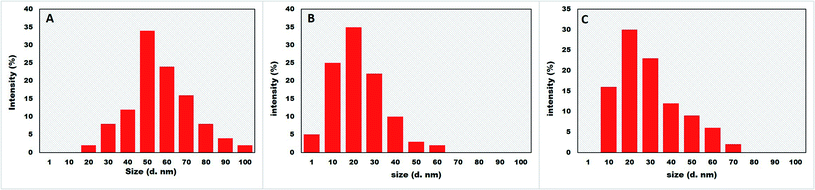
DLS was applied to measure the size distribution of BNPs with the help of Zetasizer (Malvern, NanoZSP, UK) as reported by Sohail et al.21
2.5. Antibacterial activity of BNPs
The potential of BNPs as antibacterial agents was assessed against Bacillus subtilis, Pseudomonas aeruginosa and Pseudomonas fluorescens by well-diffusion method as described by Abbasi et al.22 The stock cultures of B. subtilis, P. aeruginosa and P. fluorescens were revived on nutrient agar followed by swabbing a single colony of bacterial strains on Mueller Hinton Agar (MHA). 15 μL of LE as a negative control while positive controls 1 mM chloroauric acid–silver nitrate solution (CAA–SNS), 0.1 M chloroauric acid solution–0.5 M zinc acetate (CAA–ZAS) and 0.1 M silver nitrate–zinc acetate solution (SN–ZAS) were added. 15 μL 10 mg mL−1 BNPs and 10 μg per disc tetracyclin antibiotic discs (Thermo Fisher Scientific™ Oxoid™) were added to the wells on MHA plates. The plates were incubated at 37 °C overnight and the zones of inhibition in millimeter (mm) were measured.2.6. Biocompatibility of BNPs
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm for 10 min. 100 μL supernatant was taken in a 96-well plate and BioTek ELX800 Absorbance Microplate Reader (BioTek Instruments, France) was used to measure the release of hemoglobin at 540 nm. Triton X-100 (positive) and DMSO (negative) acted as controls. The results were expressed as % hemolysis measured by the below stated formula:24
000 rpm for 10 min. 100 μL supernatant was taken in a 96-well plate and BioTek ELX800 Absorbance Microplate Reader (BioTek Instruments, France) was used to measure the release of hemoglobin at 540 nm. Triton X-100 (positive) and DMSO (negative) acted as controls. The results were expressed as % hemolysis measured by the below stated formula:24
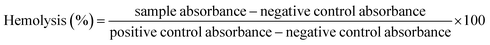 | (3) |
2.7. Anti-diabetic and anti-glycation activities of BNPs
Anti-diabetic activity of BNPs was determined in terms of inhibition of α-glucosidase and α-amylase as reported by Hano et al.25 α-Amylase inhibition by BNPs was evaluated by using porcine pancreatic α-amylase (Sigma Aldrich). For α-glucosidase inhibition assay, 1 μL rat intestinal fluid having 4-nitrophenyl-α-D-glucopyranoside (5 mM, 4NPG; Sigma, France) was used by a chromogenic method as described in (ref. 26). The antidiabetic activity was reported as % inhibition of both enzymes.The anti-glycation activity of BNPs was measured as % inhibition of AGEs as mentioned by Shah et al.26 BNPs were mixed with 20 mg mL−1 bovine serum albumin (Sigma Aldrich) prepared in phosphate buffer (0.1 M; pH 7.4), 1 mL of phosphate buffer containing sodium azide 0.02% (w/v) and glucose solution (0.5 M; Sigma Aldrich) were incubated for 5 days at 37 °C in dark. The formation of fluorescent AGEs monitored by VersaFluor fluorometer (Bio-Rad, France) set at 410 nm emission and 330 nm excitation wavelengths and % inhibition was calculated.
2.8. Anti-cancerous activities of BNPs
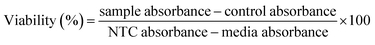 | (4) |
Apo-ONE Homogeneous Caspase-3/7 Assay kit (Promega) helped to evaluate in vitro caspase-3/7 activity by following manufacturer's instructions. All the data was measured in triplicates.
2.9. Data analysis
Mean and standard deviation (SD) of all data was calculated with the help of SPSS (Windows Version 7.5.1, SPSS Inc., Chicago, IL, USA). The data was expressed as mean ± SD.3 Results and discussion
3.1. Phytochemical analysis of Manilkara zapota
Phenolic compounds form an important constituent of plants as they bear a hydroxyl group which impart them the free radical scavenging potential.29 Whereas flavonoids are a sub-group of polyphenols which have important pharmacological role as free radical scavengers.30 Recently, metabolic profiling of M. zapota leaves detected the presence of secondary metabolites including flavonoids and phenolic compounds.31 The phytochemical analysis of M. zapota LE was carried out in this study which revealed TPC and TFC to be 67.16 ± 2.31 mg GAE per g DW and 18.27 ± 0.95 mg QE per g DW respectively. Previous study also shows enriched phenolic and flavonoid content present in aqueous LE of M. zapota reporting TPC 106.19 ± 1.99 mg GAE per g and TFC to be 37.04 ± 0.37 mg QE per g.32 Since the ability of plant extract to fabricate stable NPs depends on their phytochemical profile, here the enriched flavonoid and phenolic content present in M. zapota LE show their potential to effectively synthesize BNPs.33A single assay cannot be the representative of the antioxidant potential of plant extract. Therefore we have also performed the DPPH assay in which when DPPH is mixed with the test substance, reduced form of DPPH i.e. diphenypicrylhydrazine forms with a loss of violet color.34 The FRSA of M. zapota LE was 80.04 ± 1.28%, which correlates with the flavonoid and phenolic contents present in the LE. Kaneria and Chanda also reported good FRSA (65.41%) of aqueous extract of M. zapota which was comparable to other solvent extracts.32 The plant extract exhibits strong antioxidant activity attributed to its phytochemical profile which means it can chelate metal ions and reduce it into BNPs effectively.
3.2. Characterization of BNPs
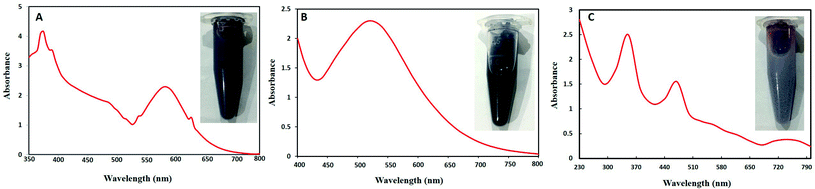
Visually brown to deep purple color development indicated the synthesis of Au–Ag BNPs. Weng et al.36 also reported purple color formation of the mixture of Au–Ag BNPs during synthesis by using Deinococcus radiodurans protein extract. UV-visible spectroscopic analysis of Au–Ag BNPs was recorded as shown in Fig. 1B. Wavelength at 530 nm was observed which is in agreement with previous results for 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 Au–Ag BNPs where single sharp peak centered at 559 nm was observed.37 Moreover, single peak indicates that Au–Ag BNPs are in an alloy form and not a heterogeneous mixture of individual NPs. Likewise, soybean leaf extract-mediated Au–Ag BNPs also showed a single peak at 533 nm.38
1 Au–Ag BNPs where single sharp peak centered at 559 nm was observed.37 Moreover, single peak indicates that Au–Ag BNPs are in an alloy form and not a heterogeneous mixture of individual NPs. Likewise, soybean leaf extract-mediated Au–Ag BNPs also showed a single peak at 533 nm.38
Fig. 1C shows a dark brown color of reaction mixture of Ag–ZnO BNPs as well as absorption peaks at 350 nm for ZnONPs and at 470 nm for AgNPs. Sumbal et al.17 also reported brown colored mixture of ZnO–AgNPs when prepared at ratio 0.5![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0.5 (v/v). These peaks are characteristic of pure zero valent ZnO and AgNPs confirming that prepared BNPs are composed of Ag–ZnO. Previously, peaks at 355 nm and 450 nm were observed for bimetallic ZnO–AgNPs from UV-visible spectroscopy.39 Moreover, two characteristic SPR band between 400–480 nm and 350 and 380 nm were reported for bimetallic Ag–ZnONPs by Anjum et al.40 when synthesized from Morus macroura leaf extract.
0.5 (v/v). These peaks are characteristic of pure zero valent ZnO and AgNPs confirming that prepared BNPs are composed of Ag–ZnO. Previously, peaks at 355 nm and 450 nm were observed for bimetallic ZnO–AgNPs from UV-visible spectroscopy.39 Moreover, two characteristic SPR band between 400–480 nm and 350 and 380 nm were reported for bimetallic Ag–ZnONPs by Anjum et al.40 when synthesized from Morus macroura leaf extract.
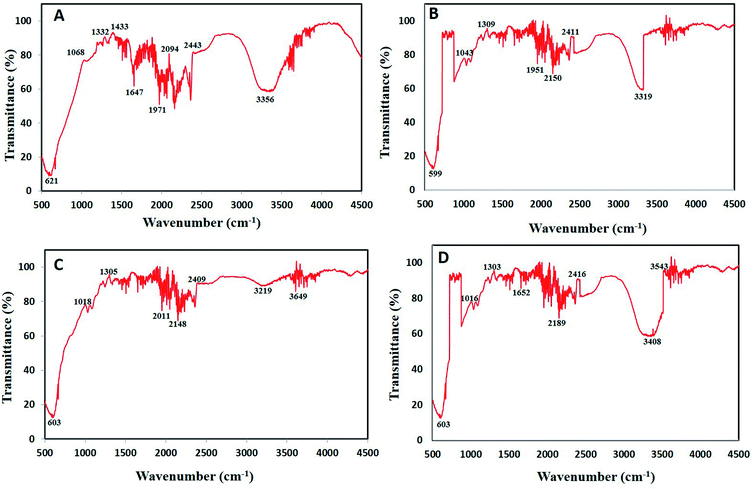
![[triple bond, length as m-dash]](https://www.rsc.org/images/entities/char_e002.gif) C stretching of alkynes.43 Wavenumber at 599–621 cm−1 represents
C stretching of alkynes.43 Wavenumber at 599–621 cm−1 represents ![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C–H bend of alkenes.44 Lastly, strong peaks between 1068–1305 cm−1 represents C–O stretch of carboxylic acids and alcohols while wavenumbers 1647–1652 cm−1 represents C–C stretch of aromatic compounds.45,46
C–H bend of alkenes.44 Lastly, strong peaks between 1068–1305 cm−1 represents C–O stretch of carboxylic acids and alcohols while wavenumbers 1647–1652 cm−1 represents C–C stretch of aromatic compounds.45,46
The present data recommends the involvement of various phytochemicals such as polyphenols, flavonoids, aromatic compounds, carboxylic acid, alkenes and alkynes in the process of bioreduction as well as capping of BNPs. Similarly, Kuppusamy et al.47 also demonstrated via FTIR analysis that functional groups like amine, alkane, phenol and alcohol present in Commelina nudiflora plant extract played vital role in Au–Ag NPs alloy synthesis. These antioxidant phytochemicals play role during synthesis by donating hydrogen atom/electrons to reduce the metal salt into zero valent state which nucleate to form NPs.48 Lastly, phytochemicals act as stabilizing agents which cap the NPs. These bio-stabilized NPs are more biocompatible having better biological activities.49
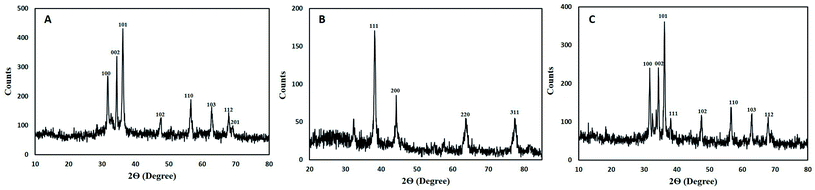
XRD pattern of core–shell Au–Ag BNPs showed FCC structure as peaks at 2θ were 38.3°, 44.25°, 64.05° and 77.75° which corresponds to the 111, 200, 220 and 311 planes, respectively (Fig. 4B). Additionally the reflections in XRD diffractogram resembles to the monometallic Au and Ag NPs without any peak of impurity. The crystalline structure of Au–Ag BNPs is analogous to the one stated by Kumari et al.60 when BNPs were green synthesized from pomegranate juice. Similar crystalline structure of bimetallic Au–AgNPs was reported previously from XRD data.61 The LE of M. zapota promoted the formation of smaller sized Au–Ag BNPs 16.57 nm as calculated by Debye Scherrer formula using XRD data. Biogenic synthesis of Ag–Au BNPs from a polymer kondagogu resulted in the formation of 21.6 nm sized NPs.62
The crystalline structure of hybrid Ag–ZnO BNPs was examined from XRD pattern as shown in Fig. 4C in which major and minor peaks observed at 2θ were 31.8°, 34.3°, 36.1°, 38.2°, 47.65°, 56.7°, 62.7° and 67.85°. These peaks correspond to the reflection planes 100, 002, 101, 111, 102, 110, 103 and 112 of hexagonal (ZnO) and FCC (Ag) structure nature, respectively. Moreover the average diameter of Ag–ZnO BNPs was 19.64 nm while the identified peaks indicate the purity of BNPs. Zare et al.63 showed the formation of very small 5 nm sized Ag–ZnO nanocomposites having similar crystallinity synthesized by using T. vulgaris extract. Moreover, minor peaks for Ag while major peaks for ZnO were observed in XRD pattern for plate and spherical shaped bimetallic Ag–ZnO NPs prepared from Silybum marianum.64
3.3. Biological activities of bimetallic NPs
| Strain type | Sample type | Zone of inhibition (mm) (15 μL per well) Ag–Au BNPs | Sample type | Zone of inhibition (mm) (15 μL per well) Au–ZnO BNPs | Sample type | Zone of inhibition (mm) (15 μL per well) Ag–ZnO BNPs |
|---|---|---|---|---|---|---|
| a LE represent to leaf extract, CAA–SNS to chloroauric acid–silver nitrate solution, CAA–ZAS to chloroauric acid–zinc acetate solution and SN–ZAS to silver nitrate–zinc acetate solution. | ||||||
| Bacillus subtilis | LE | 5 ± 0.24 | LE | 6 ± 0.28 | LE | 5 ± 0.24 |
| CAA–SNS | 11 ± 0.86 | CAA–ZAS | 16 ± 1.46 | SN–ZAS | 10 ± 0.78 | |
| Ag–Au BNPs | 11.5 ± 1.02 | Au–ZnO BNPs | 12 ± 1.26 | Ag–ZnO BNPs | 12 ± 0.79 | |
| Antibiotic disc | 15 ± 1.46 | Antibiotic disc | 13 ± 1.01 | Antibiotic disc | 12.5 ± 1.21 | |
| Pseudomonas aeruginosa | LE | 5 ± 0.24 | LE | 5 ± 0.24 | LE | 5 ± 0.24 |
| CAA–SNS | 6.5 ± 0.30 | CAA–ZAS | 13.5 ± 1.21 | SN–ZAS | 9.5 ± 0.40 | |
| Ag–Au BNPs | 7.5 ± 0.42 | Au–ZnO BNPs | 15.5 ± 1.36 | Ag–ZnO BNPs | 12.5 ± 1.32 | |
| Antibiotic disc | 8 ± 0.41 | Antibiotic disc | 7.5 ± 0.36 | Antibiotic disc | 11 ± 1.12 | |
| Pseudomonas flourescens | LE | 5 ± 0.24 | LE | 5 ± 0.24 | LE | 5 ± 0.24 |
| CAA–SNS | 9.5 ± 0.44 | CAA–ZAS | 8.5 ± 0.46 | SN–ZAS | 8.5 ± 0.40 | |
| Ag–Au BNPs | 8.5 ± 0.36 | Au–ZnO BNPs | 11 ± 0.42 | Ag–ZnO BNPs | 9.5 ± 1.12 | |
| Antibiotic disc | 8 ± 0.38 | Antibiotic disc | 8.5 ± 0.41 | Antibiotic disc | 9 ± 0.48 | |
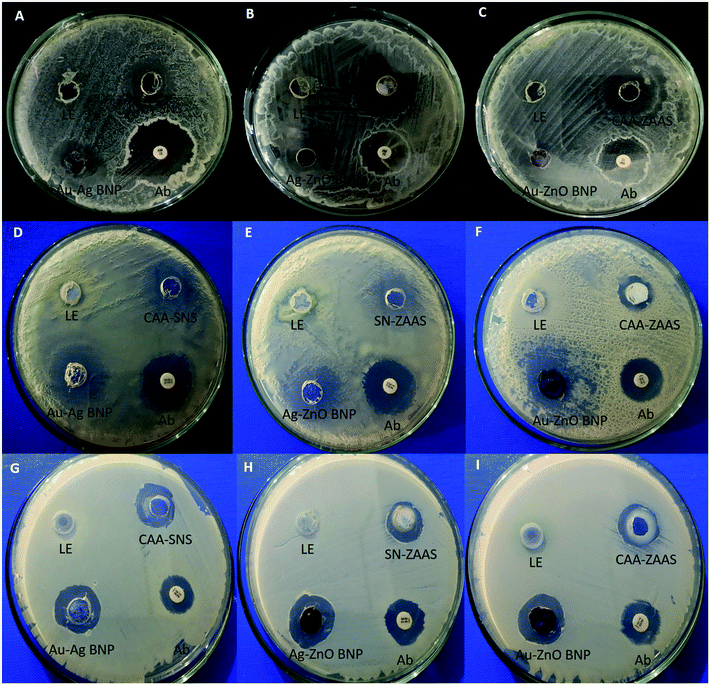 | ||
| Fig. 6 Antibacterial activity of bimetallic NPs.5 (A–C); B. subtilis (D–F); P. aeruginosa (G–I) P. flourescens. Note: LE represent to leaf extract, Ab to tetracycline antibiotic disc, CAA–SNS to chloroauric acid–silver nitrate solution, CAA–ZAS to chloroauric acid–zinc acetate solution and SN–ZAS to silver nitrate–zinc acetate solution. | ||
Antibacterial activity of BNPs against P. aeruginosa were shown in Fig. 6D–F. The zone of inhibitions were measured to be Au–ZnO BNPs 15.5 mm, Ag–ZnO BNPs 12.5 mm and Au–Ag BNPs 8.5 mm. Their antibacterial activity is better than tetracycline disc which formed zone of inhibition 8–9 mm. Whereas Fig. 6G–I shows that Au–ZnO BNPs formed the highest zone of inhibition 11 mm, Ag–ZnO BNPs 9.5 mm and least by Au– Ag BNPs 8.5 mm against P. flourescens. Tetracycline discs formed inhibition zone of around 8–9 mm. Bimetallic Ag–ZnO nanostructure, having star-shaped ZnONPs and spherical, rod and plate shaped AgNPs showed better antibacterial activity against P. aeruginosa in comparison with pure ZnONPs.71
Here it is observed that in all bacterial strains, Gram positive or negative, the ZnO nano-rods combined with spherical AuNPs and AgNPs have shown superior antibacterial activity as compared to spherical Au–Ag BNPs. This suggest morphology based antibacterial activity of BNPs due to higher penetrative ability and surface area of nano-rods as compared to nano-spheres. This further reinforces the fact that catalytic, optical, magnetic and biological activities of BNPs are improved to a greater extent in comparison to monometallic NPs.3 Similar observation was made by Alshareef et al.72 that truncated octahedral Ag–Au BNPs were more effective in killing Enterococcus faecium and Escherichia coli in comparison with spherical AgNPs.
 | ||
| Fig. 7 Biocompatibility studies (A) toxicity of BNPs against brine shrimp larvae (B) % hemolysis of human RBCs by BNPs. | ||
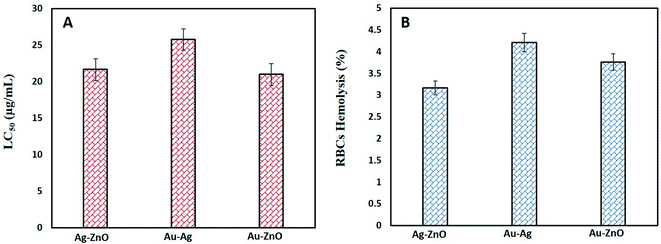
Biocompatibility with human RBCs was also assessed to evaluate their % hemolysis as shown in Fig. 7B. All bimetallic NPs were found only slightly hemolytic as Ag–ZnO BNPs were 3.16 ± 0.18%, Au–Ag BNPs 4.22 ± 0.34 and 3.76 ± 0.53% hemolytic. Magnetite/silver NPs showed negligible hemolysis 1% at concentration up to 200 μg mL−1 while increase in hemolysis was observed with increase in concentration.75 Another type of Ag–Pd NPs were also found biocompatible with RBCs up to its maximum dose of 200 μg mL−1.76 Our BNPs are also safe for human use as an alternate drug since 5% hemolysis is acceptable for biomaterials.77
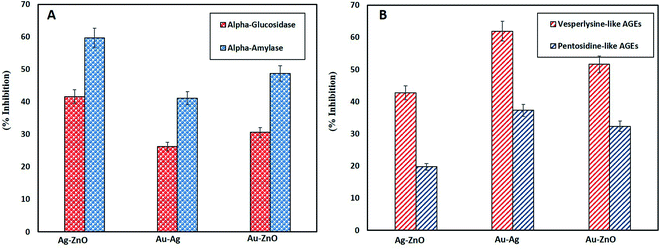
Besides antidiabetic activity, anti-glycation activity is also a remarkable ability of NPs. Glycation is a process of generating glucose related products under oxidative stress called as advanced glycation end products (AGEs) which end up in disrupting cell membrane and eventually apoptosis. The damaging effects of AGEs are associated with age and diabetes related diseases.83 Therefore, in this study the efficacy of green synthesized BNPs in inhibiting vesperlysine-like AGEs and pentosidine-like AGEs was evaluated as anti-aging and anti-inflammatory tools. Fig. 8B reports highest vesperlysine-like AGEs and pentosidine-like AGEs was inhibited by Au–Ag BNPs as 61.93 ± 1.10% and 37.30 ± 0.60%, respectively. Au–ZnO BNPs resulted in 51.6 ± 0.60% and 32.35 ± 0.49% while Ag–ZnO BNPs in 42.76 ± 1.66% and 19.77 ± 0.89% inhibition of vesperlysine-like AGEs and pentosidine-like AGEs, respectively. Previous reports also shed light on the anti-glycation activities of AgNPs, ZnONPs and Ag–ZnO BNPs.26,40,84 Here we have observed that spherical shaped smaller sized Au–Ag BNPs were better anti-glycation agents as compared to hybrid BNPs. The probable explanation for better activities of Au–Ag BNPs is that enzyme–NPs interaction vary depending on their size and shape. It is reported that curved NPs surface provides more flexibility to the adsorbed enzyme as compared to the planar NPs surface.85 Furthermore, smaller size means greater surface area of NPs. A loss of enzyme activity results due to conformational changes in the active site of enzyme when enzyme interacts with NPs surface.86 However further research is required in this domain to explore the underlying mechanism of antidiabetic and AGEs inhibition by NPs.
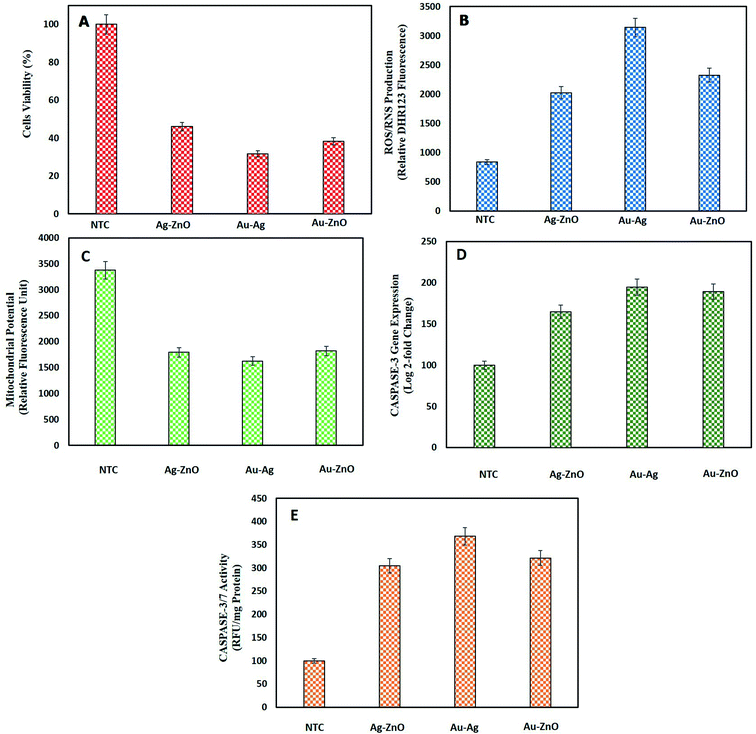
3.3.4.1. Cell viability assay of HepG2 cells by MTT. MTT assay was performed to find out the % of viable HepG2 cells when treated with BNPs. Fig. 9A shows a substantial loss of cell viability by Au–Ag BNPs (31.7 ± 3.46%), Au–ZnO BNPs (38.37 ± 1.60%) and Ag–ZnO BNPs (46.02 ± 3.46%) as compared to NTC which were 100% viable. The large surface area of small sized Au–Ag BNPs were more cytotoxic towards HepG2 cells which was also confirmed from previous reports.87 Previously bimetallic ZnO–Ag NPs synthesized via laser ablation method were found cytotoxic towards HeLa and HCT116 cancer cells at higher concentration as compared to their monometallic NPs.88 Pandiyan et al.16 reported exceptional anticancerous potential of Ag–Au/ZnO NPs against HeLa cell line which proves that bi-metallic Ag and Au doping enhanced the anticancerous efficacy of ZnONPs.
3.3.4.2. Intracellular ROS/RNS production. Disproportional increase in intracellular ROS/RNS level can induce cancer cell cycle arrest and cell death.89 Consequent escalation in intracellular ROS/RNS production in HepG2 cells in response to green synthesized BNPs is reported in Fig. 9B. In coherence with lowest cell viability, highest ROS/RNS were produced from Au–Ag BNPs 3143 ± 88.77 relative DHR123 fluorescence unit (RFU-DHR123). Furthermore, Au–ZnO BNPs resulted in 2325.3 ± 184.58 RFU-DHR123 and Ag–ZnO BNPs in 2025.28 ± 88.77 RFU-DHR123. The NTC produced normal level of intracellular ROS/RNS of 835 ± 80.17 RFU-DHR123. Sivamaruthi et al.76 also verified ROS production in A549 lung cancer cells when exposed to bimetallic Ag–Pd NPs synthesized from Terminalia chebula. These findings shed light on the increase in intracellular stress by BNPs as a way to fight cancer.
3.3.4.3. Mitochondrial membrane potential. Injury to mitochondria is important in deciding the fate of cell especially apoptosis i.e. programmed cell death. Mitochondrial injury includes a loss of mitochondrial membrane permeability, swelling of mitochondria, lysis of mitochondria and release of apoptotic proteins.90 The loss of MMP in response to NPs is a key anti-cancerous strategy to kill cancer cells. In this study, notable loss of MMP in HepG2 cells was observed when incubated with green synthesized BNPs as shown in Fig. 9C. Au–Ag BNPs resulted in maximum loss of fluorescence 1626 ± 61.78 RFU representing the highest loss of MMP as compared to other synthesized BNPs. Ag–ZnO BNPs and Au–ZnO BNPs resulted in 1796.3 ± 61.09 RFU and 1820 ± 36.72 RFU, respectively as compared to NTC 3374.94 ± 105.31 RFU. 1947.40 ± 60.91 RFU loss of MMP was recorded when HepG2 cells were treated with biogenic ZnONPs while we report higher loss of MMP from bimetallic NPs.13 These findings shows that bimetallic NPs are highly effective in disrupting MMP especially core–shell Au–Ag BNPs owing to their smaller size.
3.3.4.4. Caspase-3 gene expression and caspase-3/7 activity. Caspases are enzyme involved in the programmed cell death which is an outcome of the disruption of outer mitochondrial membrane pore and loss of MMP.91 We also assessed the increase in caspase-3 gene expression as a result of BNPs treatment of HepG2 cells as a measure of anti-cancerous potential of green NPs. Fig. 9D shows induction of high level of caspase-3 gene expression by BNPs as compared to NTC (normalized to 100% (i.e., 100 ± 1.81% in log
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2 fold change basis)). Highest caspase-3 gene expression was induced by smaller sized spherical shaped Au–Ag BNPs 194.67 ± 4.51%. Au–ZnO BNPs resulted in 189.2 ± 8.50 and Ag–ZnO BNPs in 164.76 ± 11.06% caspase-3 gene expression. Here we can state that Au based bimetallic NPs show better cytotoxic properties due to their excellent optical features and biochemical consistency thereby enhancing the properties as bimetallic NPs. Katifelis et al.92 reported that bimetallic Au–Ag alloy synthesized via chemical reduction activated caspase cascade in mice with tumor.
2 fold change basis)). Highest caspase-3 gene expression was induced by smaller sized spherical shaped Au–Ag BNPs 194.67 ± 4.51%. Au–ZnO BNPs resulted in 189.2 ± 8.50 and Ag–ZnO BNPs in 164.76 ± 11.06% caspase-3 gene expression. Here we can state that Au based bimetallic NPs show better cytotoxic properties due to their excellent optical features and biochemical consistency thereby enhancing the properties as bimetallic NPs. Katifelis et al.92 reported that bimetallic Au–Ag alloy synthesized via chemical reduction activated caspase cascade in mice with tumor.The consequent increase in caspase-3/7 activity was also observed correlating with caspase-3 gene expression. Au–Ag BNPs resulted in 368.23 ± 9.71, Au–ZnO BNPs 321.67 ± 15.72 and Ag–ZnO BNPs 304.67 ± 9.07 RFU per mg protein caspase-3/7 activity. The control showed 100 ± 6.61 RFU per mg protein caspase-3/7 activity. Baharara et al.93 reported the induction of apoptosis through enhanced caspase-3/9 activity in HeLa cells by Zataria multiflora extract synthesized AuNPs. Sambale et al.94 revealed high caspase 3/7 activity in A-549 cells cultivated with 10 ppm AgNPs indicating the cell death by apoptosis. These biocompatible BNPs synthesized from natural extract show anti-cancer activities which can be used as alternative to chemotherapeutic agents.
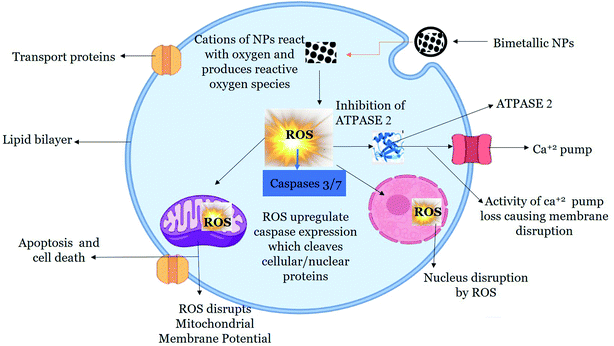
Once internalized, NPs mediate cytotoxicity through several proposed mechanism. ROS and free radical generation is the key mechanism of NPs toxicity which induces oxidative stress.99 In our study, we found enhanced level of ROS production in HepG2 cells when exposed to 200 μg mL−1 of green synthesized BNPs. The accumulation of ROS alters the health of cells by inhibiting Atpase-2 enzyme, which leads to the influx of extracellular Ca2+ ions.100 Increase in intracellular Ca2+ ions particularly leads to mitochondrial dysfunction, DNA damage and fragmentation.101 Mitochondria is the powerhouse as well as arsenal in cells, where cell death by apoptosis and non-apoptosis mechanism is triggered which results in disruption of electron transport, adenosine triphosphate production, oxidative phosphorylation, release of caspase protease and alteration in cellular reduction–oxidation potential.102 The increase in intracellular ROS level opens mitochondrial transition pore which decreases MMP, initiates caspase cascade which eventually leads to cell death.103 Caspases are intracellular proteases which are activated in a sequential manner and results in the formation of apoptotic bodies. Caspases 3 and 7 are effector caspases which executes apoptosis amongst other caspases.104 The outcome of caspases include formation of apoptotic bodies, expression of ligands for phagocytic cell receptors and uptake of the apoptotic bodies by phagocytic cells. In our study, we found that BNPs effectively enhanced ROS level in HepG2 cells, which caused a loss of MMP. Furthermore, BNPs enhanced the activity of caspase-3 and caspase-7 which eventually led to cancer cells death evident from lower cell viability in HepG2 cells treated with BNPs as compared to NTCs. A possible mechanism is proposed in Fig. 10.
4 Conclusions
The plant based synthesis of BNPs provide a new scope for NPs synthesis being a simple and eco-friendly method. In our study, different BNPs including Au–ZnO, Au–Ag and Ag–ZnO were synthesized from M. zapota LE which possess rich content of bio-reducing and capping phytochemicals such as phenolic and flavonoid compounds. Characterization of NPs showed the size of Au–ZnO BNPs was 48.32 nm, Au–Ag BNPs 16.57 nm and Ag–ZnO BNPs 19.64 nm while a variety of shapes were formed. These green synthesized BNPs showed biocompatibility with human RBCs as well as brine shrimp larvae, therefore showing their biological and environmental safety. Furthermore, the BNPs particularly Au–Ag BNPs showed significant therapeutic activities including anti-cancerous against HepG2 cells, anti-bacterial, anti-diabetic and anti-glycations. We can conclude that the green synthesized BNPs have a potential to be used as future nano-medicine. Lastly future research should focus on developing green synthesis protocol which is scalable for mass production.Ethical approval
The study was approved by the Bioethical committee of Kinnaird College for Women, Lahore, Pakistan.Author contributions
Conceptualization, S. A and B. H. A.; methodology, K. N., S. A. and B. A.; validation, S. A., C. H., and B. H. A.; formal analysis, C. H., B. H. A. and S. A.; data curation, B. A., C. H., B. H. A. and S. A.; writing—original draft preparation, K. N., and S. A.; writing—review and editing, S. A., B. H. A. and C. H.; supervision, S. A. and C. H.; project administration, S. A.; funding acquisition, S. A. and C. H. All authors have read and agreed to the published version of the manuscript.Conflicts of interest
Authors declare no conflict of interest.References
- G. Sharma, A. Kumar, S. Sharma, M. Naushad, R. Prakash Dwivedi, Z. A. Alothman and G. T. Mola, J. King Saud Univ., Sci., 2019, 31, 257–269 CrossRef.
- R. M. C. R. Ramos and M. D. Regulacio, ACS Omega, 2021, 6, 7212–7228 CrossRef CAS PubMed.
- A. Behera, B. Mittu, S. Padhi, N. Patra and J. Singh, in Multifunctional Hybrid Nanomaterials for Sustainable Agri-Food and Ecosystems, ed. K. A. Abd-Elsalam, Elsevier, 2020, pp. 639–682, DOI:10.1016/B978-0-12-821354-4.00025-X.
- A. Tulloch, A. Goldson-Barnaby, D. Bailey and S. Gupte, Int. J. Fruit Sci., 2020, 20, S1–S7 CrossRef.
- A. L. Padilla-Cruz, J. A. Garza-Cervantes, X. G. Vasto-Anzaldo, G. García-Rivas, A. León-Buitimea and J. R. Morones-Ramírez, Sci. Rep., 2021, 11, 5351 CrossRef CAS PubMed.
- Y. Cao, H. A. Dhahad, M. A. El-Shorbagy, H. Q. Alijani, M. Zakeri, A. Heydari, E. Bahonar, M. Slouf, M. Khatami, M. Naderifar, S. Iravani, S. Khatami and F. F. Dehkordi, Sci. Rep., 2021, 11, 23479 CrossRef CAS PubMed.
- S. Gupta, H. Hemlata and K. Tejavath, Beilstein Arch., 2020, 1, 95 Search PubMed.
- K. Sathya, R. Saravanathamizhan and G. Baskar, Mol. Biol. Rep., 2018, 45, 1397–1404 CrossRef CAS PubMed.
- M.-T. Tsai, Y.-S. Sun, M. Keerthi, A. K. Panda, U. Dhawan, Y.-H. Chang, C.-F. Lai, M. Hsiao, H.-Y. Wang and R.-J. Chung, Nanomaterials, 2022, 12, 61 CrossRef CAS PubMed.
- J. Ashraf, M. Ansari, S. Fatma, S. Abdullah, J. Iqbal, A. Madkhali, H. Hamali, S. Ahmad, A. Jerah, V. Echeverria, G. Barreto and G. Ashraf, Mol. Neurobiol., 2018, 55, 7438–7452 CrossRef CAS PubMed.
- F. Mohammadi Arvanag, A. Bayrami, A. Habibi-Yangjeh and S. Rahim Pouran, Mater. Sci. Eng., C, 2019, 97, 397–405 CrossRef CAS PubMed.
- M. Bhardwaj, P. Yadav, S. Dalal and S. K. Kataria, Biomed. Pharmacother., 2020, 127, 110198 CrossRef CAS PubMed.
- A. K. Khan, S. Renouard, S. Drouet, J.-P. Blondeau, I. Anjum, C. Hano, B. H. Abbasi and S. Anjum, Pharmaceutics, 2021, 13, 1977 CrossRef CAS PubMed.
- S. Anjum, B. H. Abbasi and C. Hano, Plant Cell, Tissue Organ Cult., 2017, 129, 73–87 CrossRef CAS.
- E. E. Elemike, D. C. Onwudiwe, O. E. Fayemi and T. L. Botha, Appl. Phys. A: Mater. Sci. Process., 2019, 125, 1–12 CrossRef CAS.
- N. Pandiyan, B. Murugesan, M. Arumugam, J. Sonamuthu, S. Samayanan and S. Mahalingam, J. Photochem. Photobiol., B, 2019, 198, 111559 CrossRef CAS PubMed.
- Sumbal, A. Nadeem, S. Naz, J. S. Ali, A. Mannan and M. Zia, Biotechnol. Rep., 2019, 22, e00338 CrossRef CAS PubMed.
- D. Tungmunnithum, S. Renouard, S. Drouet, J.-P. Blondeau and C. Hano, Plants, 2020, 9, 921 CrossRef CAS PubMed.
- S. Faisal, H. Jan, S. A. Shah, S. Shah, A. Khan, M. T. Akbar, M. Rizwan, F. Jan, Wajidullah and N. Akhtar, ACS Omega, 2021, 6, 9709–9722 CrossRef CAS PubMed.
- S. Fakhari, M. Jamzad and H. Kabiri Fard, Green Chem. Lett. Rev., 2019, 12, 19–24 CrossRef CAS.
- M. F. Sohail, M. Rehman, S. Z. Hussain, Z.-E. Huma, G. Shahnaz, O. S. Qureshi, Q. Khalid, S. Mirza, I. Hussain and T. J. Webster, J. Drug Delivery Sci. Technol., 2020, 59, 101911 CrossRef.
- B. H. Abbasi, S. Anjum and C. Hano, RSC Adv., 2017, 7, 15931–15943 RSC.
- M. Ahmed, H. Fatima, M. Qasim and B. Gul, BMC Complementary Altern. Med., 2017, 17, 1–16 CrossRef PubMed.
- A. T. Khalil, M. Ovais, I. Ullah, M. Ali, Z. K. Shinwari, S. Khamlich and M. Maaza, Nanomedicine, 2017, 12, 1767–1789 CrossRef CAS PubMed.
- C. Hano, S. Renouard, R. Molinié, C. Corbin, E. Barakzoy, J. Doussot, F. Lamblin and E. Lainé, Bioorg. Med. Chem. Lett., 2013, 23, 3007–3012 CrossRef CAS PubMed.
- M. Shah, S. Nawaz, H. Jan, N. Uddin, A. Ali, S. Anjum, N. Giglioli-Guivarc'h, C. Hano and B. H. Abbasi, Mater. Sci. Eng., C, 2020, 112, 110889 CrossRef CAS PubMed.
- B. H. Abbasi, M. Shah, S. S. Hashmi, M. Nazir, S. Naz, W. Ahmad, I. U. Khan and C. Hano, Nanomaterials, 2019, 9, 1171 CrossRef CAS PubMed.
- M. Nazir, D. Tungmunnithum, S. Bose, S. Drouet, L. Garros, N. Giglioli-Guivarc’h, B. H. Abbasi and C. Hano, J. Agric. Food Chem., 2019, 67, 1847–1859 CrossRef CAS PubMed.
- N. Babbar, H. S. Oberoi, S. K. Sandhu and V. K. Bhargav, J. Food Sci. Technol., 2014, 51, 2568–2575 CrossRef CAS PubMed.
- L. H. Yao, Y.-M. Jiang, J. Shi, F. Tomas-Barberan, N. Datta, R. Singanusong and S. Chen, Plant Foods Hum. Nutr., 2004, 59, 113–122 CrossRef CAS PubMed.
- S. Islam, M. B. Alam, H.-J. Ann, J.-H. Park, S.-H. Lee and S. Kim, Int. J. Mol. Sci., 2021, 22, 132 CrossRef CAS.
- M. Kaneria and S. Chanda, Asian Pac. J. Trop. Biomed., 2012, 2, S1526–S1533 CrossRef.
- S. Bawazeer, A. Rauf, S. U. A. Shah, A. M. Shawky, Y. S. Al-Awthan, O. S. Bahattab, G. Uddin, J. Sabir and M. A. El-Esawi, Green Process. Synth., 2021, 10, 85–94 CrossRef CAS.
- P. Molyneux, Songklanakarin J. Sci. Technol., 2004, 26, 211–219 CAS.
- M. Khan, S. K. Behera, P. Paul, B. Das, M. Suar, R. Jayabalan, D. Fawcett, G. E. J. Poinern, S. K. Tripathy and A. Mishra, Med. Microbiol. Immunol., 2019, 208, 609–629 CrossRef CAS PubMed.
- Y. Weng, J. Li, X. Ding, B. Wang, S. Dai, Y. Zhou, R. Pang, Y. Zhao, H. Xu and B. Tian, Int. J. Nanomed., 2020, 15, 1823 CrossRef CAS PubMed.
- K. Gopinath, S. Kumaraguru, K. Bhakyaraj, S. Mohan, K. S. Venkatesh, M. Esakkirajan, P. Kaleeswarran, N. S. Alharbi, S. Kadaikunnan, M. Govindarajan, G. Benelli and A. Arumugam, Microb. Pathog., 2016, 101, 1–11 CrossRef CAS PubMed.
- D. Alti, M. Veeramohan Rao, D. N. Rao, R. Maurya and S. K. Kalangi, ACS Omega, 2020, 5, 16238–16245 CrossRef CAS PubMed.
- Z. Zaheer and S. M. Albukhari, Arab. J. Chem., 2020, 13, 7921–7938 CrossRef CAS.
- S. Anjum, A. K. Khan, A. Qamar, N. Fatima, S. Drouet, S. Renouard, J. P. Blondeau, B. H. Abbasi and C. Hano, Int. J. Mol. Sci., 2021, 22, 11294 CrossRef CAS PubMed.
- T. Petit and L. Puskar, Diamond Relat. Mater., 2018, 89, 52–66 CrossRef CAS.
- S. Anjum and B. H. Abbasi, Int. J. Nanomed., 2016, 11, 715 CrossRef CAS PubMed.
- R. Geetha, T. Ashokkumar, S. Tamilselvan, K. Govindaraju, M. Sadiq and G. Singaravelu, Cancer Nanotechnol., 2013, 4, 91–98 CrossRef PubMed.
- M. Roni, K. Murugan, C. Panneerselvam, J. Subramaniam and J.-S. Hwang, Parasitol. Res., 2013, 112, 981–990 CrossRef PubMed.
- S. Some, O. Bulut, K. Biswas, A. Kumar, A. Roy, I. K. Sen, A. Mandal, O. L. Franco, İ. A. İnce and K. Neog, Sci. Rep., 2019, 9, 1–13 CrossRef CAS PubMed.
- S. Vennila and S. S. Jesurani, Int. J. ChemTech Res., 2017, 10, 271–275 CAS.
- P. Kuppusamy, S. Ilavenil, S. Srigopalram, D. H. Kim, N. Govindan, G. P. Maniam, M. M. Yusoff and K. C. Choi, J. Inorg. Organomet. Polym. Mater., 2017, 27, 562–568 CrossRef CAS.
- B. Kim, W. C. Song, S. Y. Park and G. Park, Catalysts, 2021, 11, 347 CrossRef CAS.
- R. Javed, M. Zia, S. Naz, S. O. Aisida, N. u. Ain and Q. Ao, J. Nanobiotechnol., 2020, 18, 172 CrossRef PubMed.
- A. M. Mostafa and E. A. Mwafy, J. Mater. Res. Technol., 2020, 9, 3241–3248 CrossRef CAS.
- Y. Sun, Y. Sun, T. Zhang, G. Chen, F. Zhang, D. Liu, W. Cai, Y. Li, X. Yang and C. Li, Nanoscale, 2016, 8, 10774–10782 RSC.
- I. S. Saputra, D. O. B. Apriandanu, Y. Yulizar, E. Maryanti, Y. N. Permana, S. Suhartati and Sudirman, Opt. Mater., 2021, 121, 111628 CrossRef CAS.
- E. Csapó, A. Oszkó, E. Varga, Á. Juhász, N. Buzás, L. Kőrösi, A. Majzik and I. Dékány, Colloids Surf., A, 2012, 415, 281–287 CrossRef.
- S. Ganaie, T. Abbasi and S. Abbasi, J. Exp. Nanosci., 2016, 11, 395–417 CrossRef CAS.
- R. K. Sali, M. S. Pujar, S. Patil and A. H. Sidarai, Adv. Mater. Lett., 2021, 12, 1–7 CrossRef.
- S. Slathia, T. Gupta and R. P. Chauhan, Phys. B, 2021, 621, 413287 CrossRef CAS.
- F. Pragati, S. Gangopadhyay and S. Pande, RSC Adv., 2014, 4, 294962 Search PubMed.
- R. Biswas, B. Banerjee, M. Saha, I. Ahmed, S. Mete, R. A. Patil, Y.-R. Ma and K. K. Haldar, J. Phys. Chem. C, 2021, 125, 6619–6631 CrossRef CAS.
- M. K. Choudhary, J. Kataria and S. Sharma, ACS Appl. Nano Mater., 2018, 1, 1870–1878 CrossRef CAS.
- M. Meena Kumari, J. Jacob and D. Philip, Spectrochim. Acta, Part A, 2015, 137, 185–192 CrossRef CAS PubMed.
- N. Berahim, W. J. Basirun, B. F. Leo and M. R. Johan, Catalysts, 2018, 8, 412 CrossRef.
- S. Velpula, S. R. Beedu and K. Rupula, Int. J. Biol. Macromol., 2021, 190, 159–169 CrossRef CAS PubMed.
- M. Zare, K. Namratha, S. Alghamdi, Y. H. E. Mohammad, A. Hezam, M. Zare, Q. A. Drmosh, K. Byrappa, B. N. Chandrashekar, S. Ramakrishna and X. Zhang, Sci. Rep., 2019, 9, 8303 CrossRef PubMed.
- S. Hameed, A. T. Khalil, M. Ali, M. Numan, S. Khamlich, Z. K. Shinwari and M. Maaza, Nanomedicine, 2019, 14, 655–673 CrossRef CAS PubMed.
- P. M. Carvalho, M. R. Felício, N. C. Santos, S. Gonçalves and M. M. Domingues, Front. Chem., 2018, 6, 237 CrossRef PubMed.
- M. I. Khan, S. K. Behera, P. Paul, B. Das, M. Suar, R. Jayabalan, D. Fawcett, G. E. J. Poinern, S. K. Tripathy and A. Mishra, Med. Microbiol. Immunol., 2019, 208, 609–629 CrossRef CAS PubMed.
- X. Jiang, X. Fan, W. Xu, R. Zhang and G. Wu, ACS Biomater. Sci. Eng., 2019, 6, 680–689 CrossRef PubMed.
- Z. Noohpisheh, H. Amiri, S. Farhadi and A. Mohammadi-gholami, Spectrochim. Acta, Part A, 2020, 240, 118595 CrossRef CAS PubMed.
- T. Mazhar, V. Shrivastava and R. S. Tomar, J. Pharm. Sci. Res., 2017, 9, 102 CAS.
- K. Bankura, D. Maity, M. M. R. Mollick, D. Mondal, B. Bhowmick, I. Roy, T. Midya, J. Sarkar, D. Rana and K. Acharya, Carbohydr. Polym., 2014, 107, 151–157 CrossRef CAS PubMed.
- G. R. S. Andrade, C. C. Nascimento, Z. M. Lima, E. Teixeira-Neto, L. P. Costa and I. F. Gimenez, Appl. Surf. Sci., 2017, 399, 573–582 CrossRef CAS.
- A. Alshareef, K. Laird and R. B. M. Cross, Appl. Surf. Sci., 2017, 424, 310–315 CrossRef CAS.
- S. Hnamte, K. Kaviyarasu and B. Siddhardha, in Model Organisms to Study Biological Activities and Toxicity of Nanoparticles, Springer, 2020, pp. 401–415 Search PubMed.
- S. Krishnan Sundarrajan and L. Pottail, Appl. Nanosci., 2021, 11, 971–981 CrossRef CAS.
- C. M. Ramírez-Acosta, J. Cifuentes, J. C. Cruz and L. H. Reyes, Nanomaterials, 2020, 10, 1857 CrossRef PubMed.
- B. S. Sivamaruthi, V. S. Ramkumar, G. Archunan, C. Chaiyasut and N. Suganthy, J. Drug Delivery Sci. Technol., 2019, 51, 139–151 CrossRef CAS.
- M. Zare, K. Namratha, M. Thakur and K. Byrappa, Mater. Res. Bull., 2019, 109, 49–59 CrossRef CAS.
- U. Etxeberria, A. L. de la Garza, J. Campión, J. A. Martínez and F. I. Milagro, Expert Opin. Ther. Targets, 2012, 16, 269–297 CrossRef CAS PubMed.
- V. Malapermal, J. N. Mbatha, R. M. Gengan and K. Anand, Adv. Mater. Lett., 2015, 6, 1050–1057 CrossRef CAS.
- P. Robkhob, S. Ghosh, J. Bellare, D. Jamdade, I. M. Tang and S. Thongmee, J. Trace Elem. Med. Biol., 2020, 58, 126448 CrossRef CAS PubMed.
- M. Kazmi, S. Zaib, A. Ibrar, S. T. Amjad, Z. Shafique, S. Mehsud, A. Saeed, J. Iqbal and I. Khan, Bioorg. Chem., 2018, 77, 190–202 CrossRef CAS PubMed.
- S. Poovitha and M. Parani, BMC Complementary Altern. Med., 2016, 16, 185 CrossRef PubMed.
- P. Gkogkolou and M. Böhm, Derm.-Endocrinol., 2012, 4, 259–270 CrossRef CAS PubMed.
- J. M. Ashraf, M. A. Ansari, S. Fatma, S. Abdullah, J. Iqbal, A. Madkhali, A. H. Hamali, S. Ahmad, A. Jerah and V. Echeverria, Mol. Neurobiol., 2018, 55, 7438–7452 CrossRef CAS PubMed.
- A. Verma and F. Stellacci, Small, 2010, 6, 12–21 CrossRef CAS PubMed.
- S. R. Saptarshi, A. Duschl and A. L. Lopata, J. Nanobiotechnol., 2013, 11, 26 CrossRef CAS PubMed.
- D. Sahu, G. Kannan, M. Tailang and R. Vijayaraghavan, J. Nanosci., 2016, 2016, 1–9 CrossRef.
- K. A. Elsayed, M. Alomari, Q. A. Drmosh, M. Alheshibri, A. Al Baroot, T. S. Kayed, A. A. Manda and A. L. Al-Alotaibi, Alexandria Eng. J., 2022, 61, 1449–1457 CrossRef.
- G.-Y. Liou and P. Storz, Free Radical Res., 2010, 44, 479–496 CrossRef CAS PubMed.
- S. Hussain, in Nanotoxicity, Springer, 2019, pp. 123–131 Search PubMed.
- Q. Tang, H. Xia, W. Liang, X. Huo and X. Wei, J. Photochem. Photobiol., B, 2020, 202, 111698 CrossRef CAS PubMed.
- H. Katifelis, I. Mukha, P. Bouziotis, N. Vityuk, C. Tsoukalas, A. C. Lazaris, A. Lyberopoulou, G. E. Theodoropoulos, E. P. Efstathopoulos and M. Gazouli, Int. J. Nanomed., 2020, 15, 6019 CrossRef CAS PubMed.
- J. Baharara, T. Ramezani, A. Divsalar, M. Mousavi and A. Seyedarabi, Avicenna J. Med. Biotechnol., 2016, 8, 75 Search PubMed.
- F. Sambale, S. Wagner, F. Stahl, R. Khaydarov, T. Scheper and D. Bahnemann, J. Nanomater., 2015, 16(1), 1–6 Search PubMed.
- L. Le Marchand, Biomed. Pharmacother., 2002, 56, 296–301 CrossRef CAS.
- H. Chen, L. Luo, S. Fan, Y. Xiong, Y. Ling and S. Peng, J. Pharm. Pharmacol., 2021, 73, 221–232 CrossRef PubMed.
- N. Oh and J.-H. Park, Int. J. Nanomed., 2014, 9, 51 Search PubMed.
- F. Gulbagça, A. Aygun, E. E. Altuner, M. Bekmezci, T. Gur, F. Sen, H. Karimi-Maleh, N. Zare, F. Karimi and Y. Vasseghian, Chem. Eng. Res. Des., 2022, 180, 254–264 CrossRef.
- K. A. Elsayed, M. Alomari, Q. Drmosh, M. Alheshibri, A. Al Baroot, T. Kayed, A. A. Manda and A. L. Al-Alotaibi, Alexandria Eng. J., 2022, 61, 1449–1457 CrossRef.
- S. Zhu, L. Gong, Y. Li, H. Xu, Z. Gu and Y. Zhao, Adv. Sci., 2019, 6, 1802289 CrossRef PubMed.
- K.-N. Yu, T.-J. Yoon, A. Minai-Tehrani, J.-E. Kim, S. J. Park, M. S. Jeong, S.-W. Ha, J.-K. Lee, J. S. Kim and M.-H. Cho, Toxicol. in Vitro, 2013, 27, 1187–1195 CrossRef CAS PubMed.
- B. Javed, M. Ikram, F. Farooq, T. Sultana, Z.-u.-R. Mashwani and N. I. Raja, Appl. Microbiol. Biotechnol., 2021, 105, 2261–2275 CrossRef CAS PubMed.
- G. Chen, S.-Y. Li, H. T. Malik, Y.-G. Ma, H. Xu and L.-K. Sun, Biotechnol. Lett., 2016, 38, 1269–1276 CrossRef CAS PubMed.
- F. Mobaraki, M. Momeni, M. E. T. Yazdi, Z. Meshkat, M. S. Toosi and S. M. Hosseini, Process Biochem., 2021, 111, 167–177 CrossRef CAS.
| This journal is © The Royal Society of Chemistry 2022 |