Open Access Article
Open Access ArticleGerberdriasins A–F, six undescribed coumarin derivatives from Gerbera anandria (Linn) Sch-Bip and their protective effects on scopolamine-induced injury in PC12 cells†
Zhi-li Wu‡ ab,
Ze-shi Sun‡a,
Jia-yu Lia,
Yong-xun Yangc,
Xian-peng Zua,
Hui-liang Li*a and
Wei-dong Zhang
ab,
Ze-shi Sun‡a,
Jia-yu Lia,
Yong-xun Yangc,
Xian-peng Zua,
Hui-liang Li*a and
Wei-dong Zhang *ab
*ab
aSchool of Pharmacy, Second Military Medical University, Shanghai 200433, P. R. China. E-mail: faranli@hotmail.com; wdzhangy@hotmail.com
bSchool of Traditional Chinese Pharmacy, China Pharmaceutical University, Nanjing, Jiangsu 211198, P. R. China
cSchool of Animal Science, Xichang College, Xichang, Sichuan 615000, P. R. China
First published on 20th July 2022
Abstract
A chemical investigation on the herb Gerbera anandria (Linn) Sch-Bip led to the isolation and identification of six previously undescribed coumarin derivatives, named Gerberdriasins A–F (1–6). Structurally, their chemical structures and absolute configurations were determined by nuclear magnetic resonance (1D and 2D NMR), high resolution electrospray ionization mass spectroscopy (HR-ESI-MS), experimental and quantum mechanical nuclear magnetic resonance (QM-NMR) methods, Mosher's method and calculated electronic circular dichroism (ECD) experiments. The biological activity of the obtained compounds showed that they displayed significant neuroprotective effects against scopolamine-induced injury in PC12 cells at the concentrations 12.5, 25.0 and 50.0 nM. Further study demonstrated that 1 could inhibit cell apoptosis, decrease malondialdehyde (MDA) levels and increase superoxide dismutase (SOD) activity in scopolamine-treated PC12 cells.
Introduction
The large genus Gerbera belonging to the family Asteraceae comprises about 80 species. Many of them exist in Africa and Asia, and twenty of them are widely distributed in most South-western Asian countries, especially Yunnan Province in China.1–4 In addition, they had been widely used as Traditional Chinese Medicine (TCM) to treat enteritis, coughs, lung heat, dysentery, etc.5–7 Previous phytochemical investigations of this genus Gerbera yielded coumarins, terpenoids, sterols, flavonoids and phenylacetones.8–13 Some of them exhibited a wide array of bioactivities, including anti-bacterial, anti-inflammatory, anti-coagulant and antitumor activities.14–16 To date, research on the phytochemistry and pharmacological activity of Gerbera anandria (Linn) Sch-Bip has rarely been reported. In this study, six undescribed coumarin derivatives, Gerberdriasins A–F (1–6), were isolated from the herb Gerbera anandria (Linn) Sch-Bip. In bioactivity assays, all compounds exhibited neuroprotective effects on scopolamine-induced injury in PC12 cells. The isolation, structural elucidation and neuroprotective activities of these compounds (Fig. 1) are described in this paper.Experimental
General experimental procedure
Column chromatography (CC): silica gel H (10–40 μm) and silica gel (200–300 mesh) (Marine Chemical Factory, Qingdao, P. R. China); MCI gel CHP–20P: (Daiso, Co., Japan) and RP-C18 gel (40–63 μm; Daiso, Co., Japan) were used for column chromatography; Sephadex LH-20 (Pharmacia Fine Chemicals, Piscataway, NJ, USA). TLC: silica gel plates, visualization by spraying with 10% H2SO4 in EtOH and Dragendorff's reagent. Semi-preparative HPLC: Agilent 1260 series (Agilent Technologies, US) with a Zorbax SB-C18 (5 μM, 9.4 mm × 25 cm) column. NMR Spectra: Bruker Avance III-500 spectrometer (Bruker, Switzerland). MS: Agilent MSD-Trap-XCT (for ESI) and Agilent-6520 Q-TOF mass spectrometer (for HR-ESI). IR: Thermo Scientific Nicolet 6700 (Thermo Scientific, US). UV spectra: Agilent 1260 series DAD detector (Agilent Technologies, US). CD spectrum: Brighttime Chirascan (Applied Photophysics Ltd, UK). Optical rotation: Rudolph Autopo V (Rudolph Research Analytical, Hackettstown, NJ).Plant material
The herb of Gerbera anandria (Linn) Sch-Bip was collected from Yunnan province of China in October 2019, and identified by Prof. Bao-kang Huang (Department of pharmacognosy, Second Military Medical University, Shanghai, China). A voucher specimen (no. 201910-VS) is deposited in the department of pharmacognosy, Second Military Medical University.Extraction and isolation
The dried herb of Gerbera anandria (Linn) Sch-Bip (50.0 kg) was extracted by maceration with 95% methanol overnight at room temperature (3 × 60 L). After remove of solvent, the methanol extract (8.60 kg) was partitioned between water and petroleum ether (PE)/ethyl acetate (EtOAc) (3 times with 10 L each). PE extract (2.15 Kg) was segmented by Silica gel H (10–40 μm) column chromatography (PE/EtOAc, 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 to 0
1 to 0![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 v/v) to give 21 fractions (Fr. 1–21). Fraction 18 (5.5 g) was purified through MCI column chromatography (MeOH/H2O, 20
1 v/v) to give 21 fractions (Fr. 1–21). Fraction 18 (5.5 g) was purified through MCI column chromatography (MeOH/H2O, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 to 100
80 to 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 0 v/v) to give 9 subfractions (Fr. 18.1–18.9). Then, subfraction 18.5 was isolated by ODS (CH3CN/H2O, 20
0 v/v) to give 9 subfractions (Fr. 18.1–18.9). Then, subfraction 18.5 was isolated by ODS (CH3CN/H2O, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 to 50
80 to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 v/v) to afford 7 subfractions (Fr. 18.5.1–18.5.7). Subfractions 18.5.3 and 18.5.5 were further separated using semi-preparative RP-C18 HPLC (CH3CN/H2O, 55
50 v/v) to afford 7 subfractions (Fr. 18.5.1–18.5.7). Subfractions 18.5.3 and 18.5.5 were further separated using semi-preparative RP-C18 HPLC (CH3CN/H2O, 55![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 45 v/v 1.0 mL min−1) to produce compounds 1 (9.3 mg), 2 (8.5 mg) and 3 (7.6 mg). Subfraction 18.6 was also subjected to ODS column chromatography (CH3CN/H2O, 20
45 v/v 1.0 mL min−1) to produce compounds 1 (9.3 mg), 2 (8.5 mg) and 3 (7.6 mg). Subfraction 18.6 was also subjected to ODS column chromatography (CH3CN/H2O, 20![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 80 to 50
80 to 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 v/v) to afford 6 subfractions (Fr. 18.6.1–18.6.5). Subfraction 18.6.2 was purified by semi-preparative RP-C18 HPLC (CH3CN/H2O, 50
50 v/v) to afford 6 subfractions (Fr. 18.6.1–18.6.5). Subfraction 18.6.2 was purified by semi-preparative RP-C18 HPLC (CH3CN/H2O, 50![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 50 v/v 1.0 mL min−1) to afforded compounds 5 (10.3 mg) and 6 (7.9 mg). Subfraction 18.7 was separated by a Sephadex LH-20 column eluted with MeOH/H2O (70
50 v/v 1.0 mL min−1) to afforded compounds 5 (10.3 mg) and 6 (7.9 mg). Subfraction 18.7 was separated by a Sephadex LH-20 column eluted with MeOH/H2O (70![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 30, v/v) to yield 5 subfractions (Fr. 18.7.1–18.7.5), and subfraction 18.7.4 was separated through semi-preparative RP-C18 HPLC (CH3CN/H2O, 65
30, v/v) to yield 5 subfractions (Fr. 18.7.1–18.7.5), and subfraction 18.7.4 was separated through semi-preparative RP-C18 HPLC (CH3CN/H2O, 65![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 35 v/v 1.0 mL min−1) to get compound 4 (6.5 mg). Above all, compounds 1–6 were obtained (Fig. 1).
35 v/v 1.0 mL min−1) to get compound 4 (6.5 mg). Above all, compounds 1–6 were obtained (Fig. 1).
Compound characterizations of 1–11
| No. | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| a NMR Data were measured at 125 MHz in CD3OD for 1–6. | ||||||
| 2 | 160.8 | 160.7 | 160.8 | 161.5 | 166.4 | 166.3 |
| 3 | 108.5 | 108.4 | 108.5 | 105.4 | 99.0 | 98.9 |
| 4 | 167.5 | 167.5 | 167.5 | 166.9 | 167.8 | 167.8 |
| 5 | 136.5 | 136.4 | 136.5 | 136.5 | 138.5 | 138.5 |
| 6 | 126.3 | 126.3 | 126.3 | 126.3 | 116.5 | 116.5 |
| 7 | 131.8 | 131.7 | 131.8 | 131.7 | 133.4 | 133.4 |
| 8 | 114.3 | 114.2 | 114.3 | 114.3 | 112.6 | 112.6 |
| 9 | 155.8 | 155.8 | 155.8 | 155.7 | 149.1 | 149.1 |
| 10 | 111.7 | 111.7 | 111.7 | 111.8 | 118.9 | 118.9 |
| 11 | 20.0 | 20.0 | 20.0 | 20.0 | 80.9 | 80.9 |
| 12 | 45.9 | 46.0 | 46.0 | 47.0 | 22.3 | 22.2 |
| 13 | 89.5 | 89.4 | 89.4 | 94.7 | 120.4 | 120.4 |
| 14 | 34.7 | 34.7 | 34.6 | 26.7 | 136.3 | 135.9 |
| 15 | 24.4 | 24.5 | 24.5 | 37.3 | 36.4 | 35.3 |
| 16 | 79.5 | 79.1 | 79.6 | 137.8 | 29.1 | 32.8 |
| 17 | 71.3 | 71.3 | 71.4 | 124.1 | 77.4 | 74.6 |
| 18 | 24.1 | 24.1 | 24.0 | 57.9 | 72.3 | 147.4 |
| 19 | 24.9 | 25.0 | 25.0 | 14.7 | 24.2 | 110.0 |
| 20 | 18.0 | 18.1 | 18.1 | 21.3 | 23.5 | 16.1 |
| 21 | 14.5 | 14.4 | 14.4 | 28.4 | 14.9 | 14.8 |
| 1′ | 173.8 | 166.3 | 166.6 | |||
| 2′ | 33.8 | 120.5 | 122.7 | |||
| 3′ | 21.1 | 146.8 | 146.6 | |||
| 4′ | 38.1 | 38.0 | 41.3 | |||
| 5′ | 66.7 | 66.7 | 66.0 | |||
| 6′ | 22.1 | 21.9 | 21.9 | |||
| No. | 1 | 2 | 3 | 4 | 5 | 6 |
|---|---|---|---|---|---|---|
| a NMR data were measured at 500 MHz in CD3OD for 1–6. | ||||||
| 6 | 7.13, d, (7.5) | 7.10, d, (7.5) | 7.12, d, (7.5) | 7.10, d, (7.5) | 7.27, d, (7.4) | 7.28, d, (7.4) |
| 7 | 7.48, tlike, (8.1) | 7.45, tlike, (8.3) | 7.47, tlike, (8.2) | 7.46, tlike, (7.8) | 7.60, tlike, (7.6) | 7.60, tlike, (7.6) |
| 8 | 7.20, d, (8.4) | 7.18, d, (8.3) | 7.20, d, (8.4) | 7.20, d, (8.3) | 7.15, d, (8.2) | 7.15, d, (8.2) |
| 11 | 2.67, s | 2.66, s | 2.67, s | 2.65, s | 5.85, s | 5.84, s |
| 12 | 3.07, dd, (8.3, 3.8) | 3.17, d, (7.3) | 3.16, d, (7.2) | |||
| 13 | 4.91, dd, (13.3, 6.7) | 4.89, dd, (13.0, 6.5) | 4.91, dd, (13.3, 6.7) | 5.33, m | 5.31, m | |
| 14 | 1.82, 1.69, m | 1.85, 1.67, m | 1.85, 1.67, m | 2.06, 1.77, m | ||
| 15 | 1.64, m | 1.67, m | 1.67, m | 2.18, 2.06, m | 2.25, 2.04, m | 2.00, m |
| 16 | 4.79, m | 4.82, m | 4.83, m | 1.72, 1.34, m | 1.60, m | |
| 17 | 5.41, m | 3.22 | 3.95, t, (6.8) | |||
| 18 | 1.12, s | 1.12, s | 1.13, s | 4.08, d, (6.7) | ||
| 19 | 1.13, s | 1.13, s | 1.13, s | 1.71, s | 1.13, s | 4.86, 4.76, s |
| 20 | 1.27, s | 1.26, s | 1.26, s | 1.60, s | 1.10, s | 1.67, s |
| 21 | 1.48, d, (6.6) | 1.46, d, (6.6) | 1.46, d, (6.7) | 1.56, s | 1.77, s | 1.76, s |
| 2′ | 2.40, m | 5.92, d, (11.4) | 5.94, d, (15.7) | |||
| 3′ | 1.76, 1.68, m | 6.40, m | 7.04, m | |||
| 4′ | 1.46, m | 2.79, m | 2.36, m | |||
| 5′ | 3.73, m | 3.86, m | 3.89, m | |||
| 6′ | 1.15, d, (6.2) | 1.17, d, (6.3) | 1.19, d, (6.3) | |||
ECD calculations
In general, the conformational analyses were generated on the basis of the OPLS3 force field. Then, the generated conformers of compounds 1–4 whose energy threshold less than 3.0 kcal mol−1 were subjected to optimization with DFT calculation at the B3LYP/6-31G (d, p) level in the gas phase through the Gaussian 09 software. ECD calculations were performed using the TD-DFT methodology at the b3lyp/6–311 + g (d, p) level in methanol. Finally, the ECD spectrum was obtained by the Boltzmann-calculated and the SpecDis software. The ECD curves were drawn using the Origin Pro 8 program (Origin Lab Corporation, Northampton, MA, USA).Preparation of the (S)-MTPA and (R)-MTPA esters of 1–3 and 5
To each compounds (each 1.5 mg) in pyridine-d5 (130 μl) were separately added (R)-(−)-MTPA (5 μL) and (S)-(+)-MTPA (5 μL) at room temperature, followed by stirring at 40 °C for 8 h. Then the reaction was monitored through HPLC following the four time points: 0, 2, 4, and 8 h, and the reaction was found to be completed at 8 h. Finally, we transferred them into a 1.7 mm NMR tube, respectively.(S)-MTPA ester of 1 (1a). 1H NMR (pyridine-d5, 500 MHz): δH: 1.30 (3H, H3-6′), 1.51 (1H, H-4′a), 2.13 (1H, H-3′a), 1.41 (3H, H3-18), 1.42 (3H, H3-19).
(R)-MTPA ester of 1 (1b). 1H NMR (pyridine-d5, 500 MHz): δH: 1.32 (3H, H3-6′), 1.45 (1H, H-4′a), 2.07 (1H, H-3′a), 1.39 (3H, H3-18), 1.40 (3H, H3-19).
(S)-MTPA ester of 2 (2a). 1H NMR (pyridine-d5, 500 MHz): δH: 1.28 (3H, H3-6′), 2.06 (1H, H-4′a), 6.11 (1H, H-3′), 5.42 (1H, H-2′), 1.42 (3H, H3-18), 1.42 (3H, H3-19).
(R)-MTPA ester of 2 (2b). 1H NMR (pyridine-d5, 500 MHz): δH: 1.31 (3H, H3-6′), 2.01 (1H, H-4′a), 6.01 (1H, H-3′), 5.36 (1H, H-2′), 1.39 (3H, H3-18), 1.39 (3H, H3-19).
(S)-MTPA ester of 3 (3a). 1H NMR (pyridine-d5, 500 MHz): δH: 1.31 (3H, H3-6′), 2.03 (1H, H-4′a), 6.04 (1H, H-3′), 5.35 (1H, H-2′), 1.23 (3H, H3-18), 1.24 (3H, H3-19).
(R)-MTPA ester of 3 (3b). 1H NMR (pyridine-d5, 500 MHz): δH: 1.28 (3H, H3-6′), 2.06 (1H, H-4′a), 6.18 (1H, H-3′), 5.43 (1H, H-2′), 1.27 (3H, H3-18), 1.27 (3H, H3-19).
(S)-MTPA ester of 5 (5a). 1H NMR (pyridine-d5, 500 MHz): δH: 1.31 (3H, H3-19), 1.36 (3H, H3-20), 2.11 (1H, H-16a).
(R)-MTPA ester of 5 (5b). 1H NMR (pyridine-d5, 500 MHz): δH: 1.43 (3H, H3-19), 1.45 (3H, H3-20), 2.06 (1H, H-16a).
Cell culture and cell viability assay
The PC12 cells were purchased from Shanghai Institute of Biochemistry and Cell Biology, Chinese Academy of Sciences (Shanghai, China). The cells were cultured in Dulbecco's modified eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS),100 U mL−1 penicillin and 100 μg mL−1 streptomycin at 37 °C in a humidified atmosphere under 5% CO2.Cell viability was determined by the CCK-8 assay. In brief, PC12 cells were seed in 96-well plates at a density of 5 × 103 cells per well for 24 h. Then the cells were incubated with scopolamine for an additional 24 h and the compounds with various concentrations were pretreated for 12 h before treated with scopolamine. After treatment, each well with 10 μL CCK-8 reagent and incubated at 37 °C for 30 min in the dark. Afterward, the optical OD-value was measured at 450 nm through a microplate reader, respectively (BioTek Instruments, Inc).
Cell apoptosis assay
For apoptosis assay, the treated cells were washed with PBS and stained with PI followed by Annexin V–FITC at room temperature for 20 min, then, the samples were examined by flow cytometry (BD Biosciences, San Jose, CA, USA).MDA and SOD assay
Malondialdehyde (MDA) and superoxide dismutase (SOD) assay were determined by the commercial assay kits (Beyotime Institute of Biotechnology, Nantong, China) based on manufacture instructions. At the same time, protein concentrations were measured on the basis of the BCA protein assay kit (Beyotime Institute of Biotechnology, Nantong, China). Finally, all the samples were detected according to the manufacturer's protocol.Statistical analysis
The statistical analysis of the values was analyzed according to Prism 8 (GraphPad, San Diego, CA, USA) software. P < 0.05 was considered to indicate statistical significance. One-way analysis of variance (ANOVA) and Tukey post hoc test were used for the dates of three separate studies. And, all values were expressed as mean ± standard deviation (SD).Results and discussion
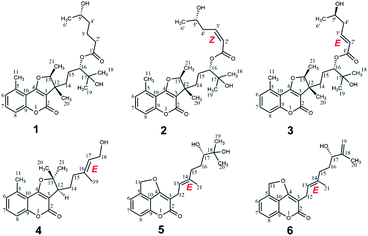
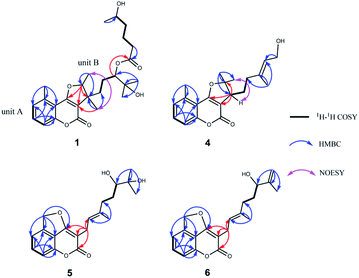
Compound 1 was isolated as colorless oil. Its molecular formula was established as C26H36O7 based on the positive HRESIMS ion peak at m/z 483.2351 ([M + Na]+, calcd for C26H36O7Na, 483.2353), suggesting 9 indices of hydrogen deficiency. The IR spectrum showed the absorption bands 3430 cm−1 and 1720 cm−1, which confirmed to the presence of the hydroxyl group and conjugated ester-carbonyl group.The 1H NMR spectra data (Table 1) showed three aromatic olefinic protons: δH 7.13 (1H, d, J = 7.5 Hz, H-6); 7.48 (1H, tlike, J = 8.1 Hz, H-7); 7.20 (1H, d, J = 8.4 Hz, H-8), three oxygenated methines: δH 4.91 (1H, dd, J = 13.3, 6.7 Hz, H-13); 4.79 (1H, H-16); 3.73 (1H, H-5′), six methyl peaks: δH 2.67 (3H, s, H3-11); 1.12 (3H, s, H3-18); 1.13 (3H, s, H3-19); 1.27 (3H, s, H3-20); 1.48 (3H, d, J = 6.6 Hz, H3-21) and 1.15 (3H, d, J = 6.2 Hz, H3-6′). The 13C NMR and DEPT spectra data (Table 2) indicated 26 carbons, including six methyls (δC 14.5, 18.0, 20.0, 22.1, 24.1, 24.9), five methylenes (δC 21.1, 24.4, 33.8, 34.7, 38.1), six methines (including 3 sp2 carbons at δC 114.3, 126.3, 131.8, 3 sp3 carbons at δC 66.7, 79.5, 89.5), nine quaternary carbons (2 ester carbonyls at δC 160.8, 173.8, 5 sp2 carbons at δC 108.5, 111.7, 136.5, 155.8, 167.5, 2 sp3 carbons at δC 45.9, 71.3). Meanwhile, the presence of a 1,2,3-trisubstituted methylbenzene ring was determined by the NMR features as follows: δH 7.13 (H-6) to δC 126.3 (C-6), δH 7.48 (H-7) to δC 131.8 (C-7), and δH 7.20 (H-8) to δC 114.3 (C-8), which together with an aromatic methyl (δH 2.67, H3-11, δC 20.0, CH3-11) and a conjugated system (a conjugated ester at δC 160.8, two olefinic carbons at δC 167.5 and δC 108.5) determined that unit A should be a 5-methylcoumarin moiety.17,18 Furthermore, two spin-coupling systems: H2-14/H2–15H-16 and H2-2′/H2-3′/H2-4′/H-5′ as revealed by the 1H–1H COSY spectra and the HMBC cross-peaks from H3-18 to C-16, C-17 and C-19, from H3-20 to C-12 and C-14, from H3-21 to C-12 and C-13, as well as from H2-2′ to C-1′ (an ester carbonyl at δC 173.8), from H3-6′ to C-5′ established the structure of unit B (the remaining sixteen carbons) could be a monoterpene moiety and six carbon short-chain fatty acid ester (Fig. 2). Meanwhile, they should be linked directly via C-16–O–C-1′ bond based on the HMBC correlation from H-16 (δH 4.79) to C-1′ (δC 173.8). Finally, the connectivity of unit A and unit B, via a furan ring, was deduced by the HMBC correlations from H-13 (δH 4.91) to C-3 (δC 108.5), C-4 (δC 167.5), C-12 (δC 45.9) and from H3-20 (δH 1.27) to C-3 (δC 108.5) and C-14 (δC 34.7). Thus, the planar structure of 1 was determined.
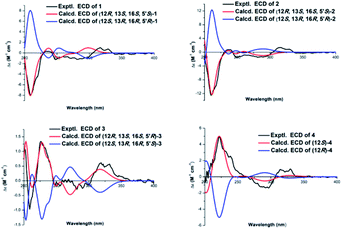
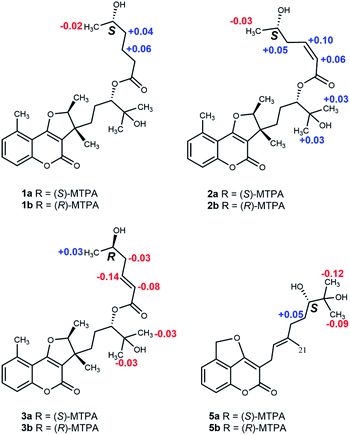
The relative configuration was deduced from its NOESY spectrum. The cross-peaks from H3-21 to H3-20 indicated these protons were oriented on the same direction and assigned as β-oriented (Fig. 2). In order to further confirm its absolution configuration, the method of ECD calculation was applied. As seen from the Fig. 3, the calculated ECD curve of (12R, 13S, 16S, 5′S)-1 was in good agreement with the experimental ECD spectrum of 1. In addition, the observed ΔδH(S–R) values of the (S)- and (R)-MTPA esters established the absolute configuration of C-5′ in 1 as S (Fig. 4), which was consistent with the ECD results. Thus, the structure of 1 was defined and named gerberdriasin A.
Compound 2, isolated as colorless oil, possessed a molecular formula of C26H34O7 as established by the ion peak at m/z 481.2201 ([M + Na]+, calcd for C26H34O7Na, 481.2197) in the positive HRESIMS spectrum, corresponding to 10 indices of hydrogen deficiency. Overall comparison of 1D NMR data of 2 (Tables 1 and 2) with those of 1 revealed that they share the similar skeleton with the exception of the presence of a double bond (δH 5.92, 6.40, H-2′, H-3′; δC 120.5, 146.8, C-2′, C-3′) at the C-2′ and C-3′ positions in the six carbon short-chain fatty acid ester in 2. This deduction was supported by the 2 Da less than 1, the one more unsaturation and the 1H–1H COSY correlations from H-2′ (δH 5.92) to H-3′ (δH 6.40) H2-4′ (δH 2.79) and H-5′ (δH 3.86), as well as the HMBC correlations from H-2′ and H-3′ both to C-1′ (δC 166.3), C-4′ (δC 38.0). Additionally, the C-2′/C-3′ double bond should be Z-configured due to the coupling constants of J2′/3′ = 11.4 Hz and the absence of the NOESY correlation from H-2′ to H2-4′. Meanwhile, compound 2 shared the same relative configurations with 1 through the analysis of its NOESY data. The absolute configuration of 2 was assigned as 12R, 13S, 16S, 5′S based on the good qualitative agreement between the experimental and calculated ECD spectra (Fig. 3). Additionally, the absolute configuration of C-5′ in 2 was also S determined by the ΔδH(S–R) results (Fig. 4), which also was fitted well with the ECD results. Finally, the structure of 2 was determined and named Gerberdriasin B.
Compound 3 was isolated as colorless oil and assigned the same molecular formula as that of 2 according to the HRESIMS data: m/z 481.2200 ([M + Na]+, calcd for C26H34O7Na, 481.2197). Detail comparison of the 1D and 2D NMR spectroscopic data (1H, 13C, DEPT, 1H–1H COSY and HMBC) of 2 and 3 showed that they possessed the same 2D structure. The differences between them on NMR data were the C-4′ and H2-4′, the chemical shift of C-4′ was down field shifted from δC 38.0 in 2 to δC 41.3 in 3, at the same time, an obvious up field shifted of H2-4′ from δH 2.79 in 2 to δH 2.36 in 3. The de-shielding of C-4′ (Δδ +3.3) and the shielding of H2-4′ (Δδ −0.43) in 3 were clearly implied the configurational change of C-5′. This deduction was further evidenced by the ΔδH(S–R) results (Fig. 4). In addition, correlations between H-2′ and H2-4′ and the large coupling constants of J2′/3′ = 15.7 Hz indicated that the C-2′/C-3′ double bond was E-configured. By means of ECD calculation, the experimental ECD spectrum of 3 fitted well with the calculated spectrum of (12R, 13S, 16S, 5′R)-3 (Fig. 3). Therefore, its structure was established and named Gerberdriasin C.
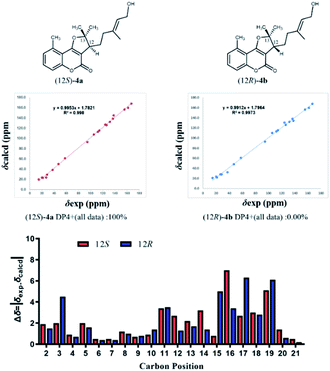
Compound 4, isolated as colorless oil, displayed a molecular formula of C20H24O4 as defined by the positive HRESIMS ion peak at m/z 351.1569 ([M + Na]+, calcd for C20H24O4Na, 351.1567), with 9 degrees of unsaturation. A detailed analysis of the 1D NMR spectra of 4 (Tables 1 and 2) indicated that 4 also had a 5-methylcoumarin moiety. The major difference was the side chain monoterpene moiety, as supported by the 1H–1H COSY correlations from H-12 to H2-14 and H2-15 and the HMBC correlations from H3-19 (δH 1.71) to C-15 (δC 37.3), C-16 (δC 137.8) and C-17 (δC 124.1); from H2-18 (δH 4.08) to C-17 (δC 124.1) and from H3-20 (δH 1.60) to CH3-21 (δC 28.4), C-12 (δC 47.0) and C-13 (δC 94.7) (Fig. 2). Meanwhile, the two moieties were also linked at C-3 and C-4 through a furan ring by the HMBC correlations from H-12 (δH 3.07) to C-3 (δC 105.4), C-4 (δC 166.9) and C-13 (δC 94.7) and the remaining one unsaturation (Fig. 2). The relative configuration of 4 was established by analyzing its NOESY data. Correlations from H2-15 to H-17 determined that the C-16/C-17 double bond might be E conformation. And, the NOESY correlations from H3-21 to H-12 suggested a same orientation of these protons. To further confirm the relative configuration of C-12, geometrical optimizations at TDDFT level for two isomers (12S*)-4a and (12R*)-4b were undertaken. Then, the NMR calculations performed through the QM-NMR method at the mPW1PW91/6-31G(d) level by the GIAO approach.19,20 Further DP4+ analyses based on the 13C NMR data indicated (12S*)-4a (R2 = 0.9980) was the correct structure with 100% probability (Fig. 5) Furthermore, the absolute configuration of 4 (12S) was determined by comparative analysis of calculated and experimental ECD spectra (Fig. 3). Finally, the structure of 4 was defined and named Gerberdriasin D.
Compound 5 was isolated as colorless oil. Its molecular formula was confirmed to be C20H24O5 based on the positive HRESIMS data: 367.1514 ([M + Na]+, calcd for C20H24O5Na, 367.1516). Analyzing the 1H and 13C NMR spectroscopic data (Tables 1 and 2) in detail established that compound 5 consisted of a coumarin moiety and a monoterpene moiety. Explicit structural information was derived from 2D NMR spectroscopic data (1H–1H COSY and HMBC) (Fig. 2). The 1H–1H COSY correlations of H-6/H-7/H-8 and the HMBC correlations from H2-11 (δH 5.85) to C-5, C-6 and C-10, from H-8 to C-6, C-9 and C-10 together with a conjugated system (a conjugated ester at δC 166.4, two olefinic carbons at δC 167.8 and δC 99.0), a characteristic of the 5-methylol-4-hydroxycoumarin moiety was composed. Furthermore, a furan ring newly generated fused with the coumarin moiety at C-4, C-10 and C-5 through C4–O–C11 bond. This conclusion was revealed by the HMBC cross-peaks from H2-11 (δH 5.85) to C-4 (δC 167.8), C-5 (δC 138.5) and C-10 (δC 118.9). Another the spin-coupling system, H2-15/H2-16/H-17, in the 1H–1H COSY spectrum, as well as the HMBC correlations from H3-19 to C-17, C-18 and C-20, from H3-21 to C-13, C-14 and C-15, and from H2-12 to C-13 revealed the presence of a monoterpene moiety. Meanwhile, the HMBC correlations from H2-12 (δH 3.17) to C-3 (δC 99.0), C-2 (δC 166.4), C-4 (δC 167.8) and C-13 (δC 120.4) defined that this monoterpene moiety was attached to the 5-methylol-4-hydroxycoumarin moiety via a C3–C12 single bond. Meanwhile, the C-13/C-14 double bond was confirmed to be E conformation based on the NOESY correlations between H-13 and H2-15. Additionally, based on the Mosher's method, the 17S configuration in 5 was established (Fig. 4). Therefore, its structure was depicted and named Gerberdriasin E.
Compound 6 was assigned a molecular formula of C20H22O4 as established by the [M + Na]+ ion peak m/z 349.1407 (calcd for C20H22O4Na, 349.1410) in the positive HRESIMS data. Its 1H and 13C NMR data (Tables 1 and 2) were similar as those of 5 except that the appearance of an exocyclic Δ18(19) double bond in 6 instead of an oxygenated quaternary carbon (C-18, δC 72.3) and a methyl (CH3-19, δC 24.2) in 5. This characteristic was verified by the less than 18 Da between their molecular weight, one more unsaturation as well as the HMBC correlations from H2-19 (δH 4.86, 4.76) to C-17 (δC 74.6), C-18 (δC 147.4), C-20 (δC 16.1) (Fig. 2). Similar as 5, it also tended to have an E double bond at C-13 and C-14 positions according to the NOESY data. Hence, the structure of 6 was deduced and named Gerberdriasin F.
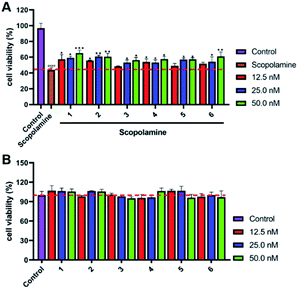
All the isolated compounds (1–6) were tested for their protective effects on scopolamine-induced PC12 cells injury by the CCK8 assay. As shown in Fig. 6A and B, compounds 1–6 showed varying degrees of protection against scopolamine-damaged PC12 cells with no significant cytotoxicity at dose concentrations of 12.5, 25.0 and 50.0 nM. Furthermore, compound 1 showed stronger activities than others. Thus, we chose 1 as the material to further study neuroprotective effects.
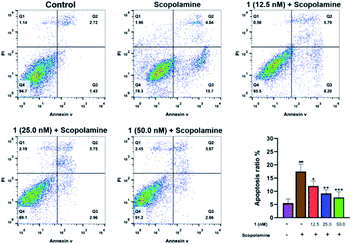
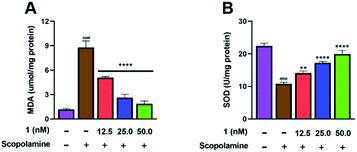
Flow cytometry analysis was performed to investigate whether pre-treatment with 1 could reduce apoptosis in scopolamine-induced PC12 cells. As shown in Fig. 7, after treated with scopolamine, the apoptosis rate of PC12 cells was significantly increased (4.15% → 19.74%). However, the percentage of apoptotic cells was reduced to 13.99%, 8.70%, and 6.33% after pre-treatment with 1 at 12.5, 25.0 and 50.0 nM, respectively. Meanwhile, to examine whether 1 could enhance anti-oxidant activity, the MDA and SOD assay of PC12 cells were carried out. As shown in Fig. 8A, treatment with 1 prior to scopolamine evidently reduced the level of MDA, compare with the scopolamine group. Additionally, in comparison with the scopolamine group, pre-treatment with 1 significantly increased the SOD levels as shown in Fig. 8B. These above results exhibited that 1 could prevent apoptosis and improve anti-oxidant activity in scopolamine-induced PC12 cells.
Conclusions
In conclusion, Gerberdriasin A–F (1–6), six new coumarin derivatives were isolated and elucidated from the herb of Gerbera anandria (Linn) Sch-Bip. Their chemical structures including absolute configurations were identified by the analyses of their 1D and 2D NMR, HRESIMS spectra, Mosher's method, QM-NMR method and ECD calculation. In addition, all the isolated compounds revealed the neuroprotective effects on scopolamine-induced injury in PC12 cells. Meanwhile, pre-treatment with 1 significantly ameliorated apoptosis and oxidative stress in scopolamine-injured PC12 cells. Taken together, this research provided a series of rare coumarin derivatives as a candidate molecule for the treatment of neurodegenerative disease.Conflicts of interest
The authors declare no conflicts of interest in this paper.Acknowledgements
This study was supported by the National Natural Science Foundation of China (82141203, 82004003, 82004215, 82003624); National Key R&D Program of China (2019YFC1711000); Innovation Team and Talents Cultivation Program of National Administration of Traditional Chinese Medicine (ZYYCXTDD-202004); Shanghai Sailing Program (20YF1459000), Shanghai Municipal Health Commission Project (20204Y0326) and Science and Technology Commission of Shanghai Municipality (20YF1458700).References
- W. Zheng, X. D. Xu and J. Wen, Econ. Bot., 2017, 71, 380–386 CrossRef.
- F. Li, S. C. Li and Q. L. Shan, Hort Science, 2019, 54, 1164–1167 Search PubMed.
- P. Sorina and G. Mihaela, Rom. Biotechnol. Lett., 2020, 25, 1635–1640 CrossRef.
- H. A. Oketch-Rabah, E. Lemmich, S. F. Dossaji, G. Theander, E. Olsen, C. Cornett, A. Kharazmi and S. Christensen, J. Nat. Prod., 1997, 60, 458–461 CrossRef CAS PubMed.
- F. He, M. Wang, M. H. Guo, M. Zhao, Y. H. Bai and C. J. Zhao, Molecules, 2014, 19, 4046–4057 CrossRef PubMed.
- B. Krishna, C. Ana and S. Y. Xiao, BMC Plant Biol., 2020, 20539 Search PubMed.
- C. Debasis and K. Subodh, Acta Physiol. Plant., 2008, 30, 325–331 CrossRef.
- S. Z. Liu, J. Q. Feng, J. Wu and W. M. Zhao, Helv. Chim. Acta, 2010, 93, 2026–2029 CrossRef CAS.
- T. Li, X. Ma, F. Daniil, L. Kjaerulfet, K. Frydenvang, S. Coriani, P. R. Hansen, K. T. Kongstad and D. Staerk, Molecules, 2020, 25, 1706 CrossRef CAS PubMed.
- Y. Qiang, Y. J. Chen, Y. Li, J. Zhao and K. Gao, Planta Med., 2011, 77, 175–178 CrossRef CAS PubMed.
- Y. Xiao, Y. Ding, J. B. Li and T. Nohara, Chem. Pharm. Bull., 2004, 52, 1362–1364 CrossRef CAS PubMed.
- Y. J. Chen, Y. Li, J. J. Chen and K. Gao, Helv. Chim. Acta, 2007, 90, 176–182 CrossRef CAS.
- Y. Xiao, J. B. Li and Y. Ding, Chin. Tradit. Herb. Drugs, 2003, 34, 109–111 CAS.
- C. M. Zhong, Y. Tang, B. Pang, X. K. Li, Y. P. Yang, J. Deng, C. Y. Feng, L. F. Li, G. P. Ren, Y. P. Wang, J. Z. Peng, S. L. Sun, S. Liang and X. J. Wang, Hortic. Res., 2020, 7, 78 CrossRef CAS PubMed.
- S. Nagumo, T. Inoue and T. T. Nagai, Chem. Pharm. Bull., 1989, 37, 2621–2623 CrossRef CAS.
- S. S. Hassan, M. Ishaq, W. D. Zhang and H. Z. Jin, Curr. Pharm. Des., 2021, 27, 2605–2614 CrossRef PubMed.
- Y. Qiang, Y. J. Chen, Y. Li, J. Zhao and K. Cao, Planta Med., 2011, 77, 175–178 CrossRef CAS PubMed.
- L. Lei, Y. B. Xue, Z. Liu, S. S. Peng, Y. He, Y. Zhang, R. Fang, J. Y. Wang, Z. W. Luo, G. M. Yao, J. W. Zhang, G. Zhang, H. P. Song and Y. H. Zhang, Sci. Rep., 2014, 5–13544 Search PubMed.
- J. Hao, T. X. Zhou, Y. R. Ma, J. T. Deng, H. T. Cheng, Q. Wang, Q. X. Lin, X. Z. Yang and H. Y. Choi, Front. Chem., 2121, 9, 717904 CrossRef PubMed.
- S. M. Shen, Z. Y. Zhang, L. G. Yao, J. R. Wang, Y. W. Guo and X. W. Li, Chin. J. Chem., 2020, 40, 235–246 CrossRef.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra03166c |
| ‡ These authors are co-first authors. |
| This journal is © The Royal Society of Chemistry 2022 |