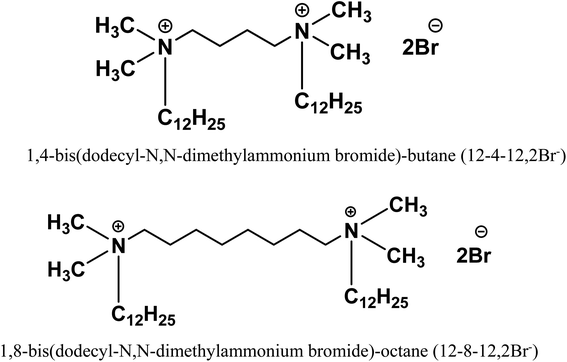
Open Access Article
Open Access ArticleRefolding of denatured gold nanoparticles-conjugated bovine serum albumin through formation of catanions between gemini surfactant and sodium dodecyl sulphate†
Rishika Aggrawal‡
,
Sayantan Halder‡,
Shalini Dyagala and
Subit K. Saha *
*
Department of Chemistry, Birla Institute of Technology & Science (BITS) Pilani, Hyderabad Campus, Hyderabad, Telangana 500078, India. E-mail: sksaha@hyderabad.bits-pilani.ac.in; sksaha@pilani.bits-pilani.ac.in; subitksaha@gmail.com; Tel: +91-40-66303643
First published on 27th May 2022
Abstract
The present work elucidates binding interactions of sodium dodecyl sulphate (SDS) with the conjugated gold nanoparticles (AuNPs)-bovine serum albumin (BSA), unfolded by each of two gemini surfactants, 1,4-bis(dodecyl-N,N-dimethylammonium bromide)-butane (12-4-12,2Br−) or 1,8-bis(dodecyl-N,N-dimethylammonium bromide)-octane (12-8-12,2Br−). Initially, at a low concentration of SDS there is a relaxation of bioconjugates from their compressed form due to the formation of catanions between SDS and gemini surfactants. On moving towards higher concentrations of SDS, these relaxed unfolded bioconjugates renature by removal of residual bound gemini surfactants. Mixed assemblies of SDS and gemini surfactants formed during refolding of bioconjugates are characterized by DLS and FESEM measurements. A step-by-step process of refolding observed for these denatured protein bioconjugates is exactly the inverse of their unfolding phenomenon. Parameters concerning nanometal surface energy transfer (NSET) and Förster's resonance energy transfer (FRET) phenomenon were employed to develop a binding isotherm. Moreover, there remains an inverse relationship between α-helix and β-turns of bioconjugates during the refolding process. Significantly, in the presence of 12-8-12,2Br−, SDS induces more refolding as compared to that for 12-4-12,2Br−. Bioconjugation shows an effect on the secondary structures of refolded BSA, which has been explored in detail through various studies such as Fourier transform infrared spectroscopy, fluorescence, and circular dichroism (CD). Therefore, this approach vividly describes the refolding of denatured bioconjugates, exploring structural information regarding various catanions formed during the process that would help in understanding distance-dependent optical biomolecular detection methodologies and physicochemical properties.
1 Introduction
Proteins adopt a specific three dimensional structure which is utilized in several biological functions.1,2 Improper folding of protein or aggregation of protein are the preliminary causes of serious human degenerative diseases such as cancer, Alzheimer's, amyotrophic lateral sclerosis, etc.3,4 Thus, the prevention of protein aggregation and the refolding of protein have always been attractive topics of progressing interest.The renaturation of a multi-domain protein is a very complicated process. There is independent refolding witnessed in each domain of the protein, and the degree of renaturation is governed by the chaotrope used for this phenomenon.5,6 Different chemical agents like poly(ethylene glycol), sugars, salts, surfactants, β-cyclodextrin (β-CD), etc. including molecular chaperones have been implemented to prevent unwanted aggregations in proteins, thereby, effectively causing refolding of the denatured proteins back to their native conformations.3,7–10
BSA, one of the most aqueous soluble proteins is one of the extensively explored plasma proteins.4,11–14 It can bind with a variety of compounds such as fatty acids, bile acids, many drugs, surfactants etc.4,11,15–20 The surfactants, especially the cationic surfactants, are very well known protein denaturing agents.11,18,21 The proteins can interact with surfactants in its various forms such as monomer, micelles, mixed micelles, vesicles etc.14 However, some reports describe the refolding of BSA in the presence of SDS. BSA has been reported to get thermally stabilized in the monomeric binding region by SDS, which in the submicellar region unfolds the protein.22 The renaturation of urea-mediated unfolded BSA, in presence of a small amount of SDS, was demonstrated by Moriyama et al.23 Lu et al.24 reported insignificant structural changes of BSA on addition of an equimolar mixture of an anionic surfactant, sodium decylsulfonate and a cationic surfactant, decyltriethylammonium bromide. Strong electrostatic attraction operating between oppositely charged headgroups as well as hydrophobic interactions between the tails of cationic and anionic surfactants leads to the formation of mixed micelles (catanions).25 A strong synergism exists between the oppositely charged cationic and anionic surfactants. As a result, mixed aggregates are formed in the bulk accompanied by weaker interactions of these mixed aggregates with the protein, suggesting that this mixture can be used as an artificial chaperone.24 Often there is precipitation of the equimolarly mixed cationic–anionic surfactants with occurrence of turbidity at lower concentrations,26,27 which is frequently a hurdle to overcome. But the interactions involved bear significant experimental and theoretical importance. When the molar ratio of the mixture of cationic and anionic surfactant, exceeds 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1, catanions are formed. These catanionic micelles often behave as refolding agents for unfolded proteins. Kumari et al.4 have investigated the formation of the mixed micellar systems between gemini surfactants, 12-n-12,2Br− (n = 3, 6, 8, 12) and SDS in order to refold back the gemini-induced denatured BSA into its native conformation. When protein gets refolded, simultaneously some big particles formed in the system are aggregates of SDS and gemini surfactants. Literature says even in the absence of a protein, the aggregates like rod-like micelles, vesicles, lamellar micelles etc. are formed.28–32 Reports are available for the formation of these structures even at a very low concentration of catanions.33
1, catanions are formed. These catanionic micelles often behave as refolding agents for unfolded proteins. Kumari et al.4 have investigated the formation of the mixed micellar systems between gemini surfactants, 12-n-12,2Br− (n = 3, 6, 8, 12) and SDS in order to refold back the gemini-induced denatured BSA into its native conformation. When protein gets refolded, simultaneously some big particles formed in the system are aggregates of SDS and gemini surfactants. Literature says even in the absence of a protein, the aggregates like rod-like micelles, vesicles, lamellar micelles etc. are formed.28–32 Reports are available for the formation of these structures even at a very low concentration of catanions.33
Interactions between protein and nanoparticles (NPs) have garnered immense significance in biosensors, delivery of proteins, and in several medicinal applications.34–36 “Bioconjugation” usually refers to NPs covalently attached to protein.37–40 However, besides these covalent interactions, several other interactions such as hydrophobic,34,41,42 electrostatic,34,41 and coordination binding43 also have a significant part in the conjugation of NPs with BSA. The conjugated nanoparticles (NPs)-BSA have been investigated by various groups.44–47 Shang et al.37 have broadly elucidated the changes in conformations of conjugated (AuNPs)-BSA with pH changes utilizing different spectroscopic techniques. Förster resonance energy transfer (FRET), a non-radiative energy transfer process, can successfully measure the separation distance within 20–90 Å.48,49 It has been observed in many nanocomposite systems that metal nanostructures quench the fluorescence of the nearby fluorophores.50,51 This dipole-to-metal surface excitation energy transfer is called nanometal surface energy transfer (NSET).52 NSET considerably differs from FRET because the requirement of a spectral overlap between donor emission and the acceptor absorption is not mandatory in the former process.53,54 Sen and group38 have vividly shown the variations in conformations of BSA at different pH using the NSET mechanism involving AuNPs.
This study attempts to understand the interactions of sodium dodecyl sulphate (SDS) with conjugated AuNPs-BSA, unfolded by two different cationic gemini surfactants, 12-4-12,2Br−and 12-8-12,2Br− [Scheme 1]. Earlier, the unfolding of the conjugated AuNPs-BSA has been carried out by these two cationic gemini surfactants and demonstrated by the FRET/NSET method.13 After the consolidation of the gemini surfactant-BSA binding isotherm, the refolding study of unfolded bioconjugated protein using SDS through the formation of catanion/mixed assemblies with gemini surfactant has been performed by the FRET and NSET methods. On interacting with cationic gemini surfactant molecules, SDS molecules form catanion due to the oppositely charged headgroups. On addition of SDS to the BSA–gemini complex solution, it extracts gemini surfactant molecules leading to catanion formation. Subsequently, BSA molecules would palliate themselves from the gemini surfactants and regain their native form. FRET parameters such as the energy transfer efficiency (ET), Förster's distance (R0) (in FRET mechanism), distance (d0) (in NSET mechanism), and the actual distance between AuNPs and BSA Trp residues, r and d from FRET and NSET mechanisms, respectively were calculated at the consecutive stages of protein refolding upon gradually adding SDS. The two gemini surfactants, distinctly differing in the spacer chain length, play significant roles in the refolding of protein. It has been explained here. The variations in different elements of the secondary structures of protein like α-helix, β-sheets, turns, and random coil, were observed using CD and FTIR spectroscopy to elucidate the structural changes during the refolding process of unfolded conjugated AuNPs-BSA. In addition, CD and FTIR spectroscopy was also used to highlight the effect of conjugation with AuNPs on the secondary of BSA refolded by SDS. Dynamic light scattering (DLS) size distribution and FESEM images have supported the formation of mixed assemblies of SDS and gemini surfactant in absence and presence of bioconjugates.
It is well established in our previous report that the protein's helicity is majorly retained even after bioconjugation in its native as well as unfolded states.13 The ionic surfactants have a strong affinity to bind to the proteins stepwise. Thus, a detailed picture of the protein–surfactant interactions is presented here. Henceforth, bioconjugation of protein with AuNPs and the emphasis upon the catanion-induced renaturation of the denatured bioconjugates would pave the way to some interesting protein research. It may be applicable for devising suitable bioconjugated nanomaterials in near future.
2 Experimental section
2.1 Materials
Gemini surfactants, namely, 1,4-bis(dodecyl-N,N-dimethylammonium bromide)-butane (12-4-12,2Br−) and 1,8-bis(dodecyl-N,N-dimethylammonium bromide)-octane (12-8-12,2Br−) were synthesized according to reported methods.55–58 A mixture of ethyl acetate and methanol was used to recrystallize the synthesized gemini surfactants. The molecular structure was verified by FT-IR and 1H NMR measurements.55,59 Chloroauric acid (HAuCl4·3H2O) was used as a good nanoparticle precursor and was procured from Merck. Sodium borohydride (NaBH4), sodium dodecyl sulphate (SDS), bovine serum albumin (BSA), and quinine sulphate were obtained from Merck as well and were used as it is. Sulfuric acid (H2SO4) was purchased from SDFCL, India. Sodium hydroxide (NaOH) and 4-(2-hydroxyethyl)-1-piperazineethanesulfonic acid (HEPES) buffer were obtained from SRL, India.2.2 Methods
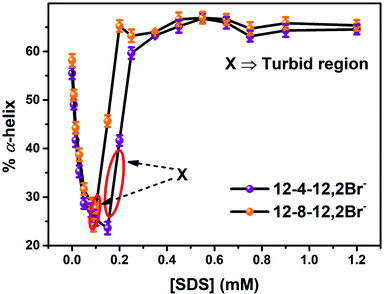
Conjugated AuNPs-BSA was synthesized according to the method available in the literature that has been described in our previous work as well.13,38 The pH of HEPES buffer (∼10 mM) solution prepared with Milli-Q water was maintained as 7.4 using dilute solutions of H2SO4 and NaOH. This HEPES buffer solution was used to prepare the stock solutions of the surfactants, SDS (∼25 mM), 12-4-12,2Br− and 12-8-12,2Br− (each having a concentration of 50 mM). The final pH of each stock solution was once again adjusted at 7.4 using dilute solutions of H2SO4 and NaOH in Milli-Q water. The stock solutions were degassed with pure N2 for around 20 minutes before further use. To carry out the refolding study, the concentration of conjugated AuNPs-BSA was kept fixed where the concentration of BSA and AuNPs was maintained as 10 μM and 0.0124 μM, respectively. The final concentration of the gemini surfactants was kept at 0.2 mM (lying in the saturative binding region of BSA–surfactant interactions), while the concentration of SDS was gradually increased from 0–1.2 mM. The final volume for each solution was kept as 2 mL. It is noteworthy that turbidity is formed at a certain range of composition of mixture of gemini surfactant and SDS in presence of bioconjugates in HEPES buffer (∼10 mM) at pH 7.4. These ranges are avoided in collecting any experimental data. The concentration ranges of SDS at which turbidity is formed are between 0.16 to 0.19 mM for 12-4-12,2Br− and between 0.09 to 0.1 mM for 12-8-12,2Br−. None of the data falls in these ranges is reported here. These ranges are indicated in figures as well.The relative fluorescence quantum yields of different BSA (QD) solutions devoid of AuNPs, were determined. Quinine sulphate solution in 0.1 N H2SO4 with quantum yield of 0.55 was taken as the standard solution.60
The absorption measurements were carried out with a JASCO (model V-650) UV-Vis spectrophotometer. FluoroLog-TM (Horiba Scientific) spectrofluorimeter was used to carry out all the steady-state fluorescence measurements. A quartz cuvette with a Teflon stopper and path length of 1 cm was used for all the measurements. A 2 nm slit width was maintained for both excitation and emission with a uniform scan rate for all fluorescence measurements. The corrected fluorescence spectra were recorded to maintain a uniform output from the xenon lamp. The same fluorescence spectrofluorimeter was used for recording the three-dimensional fluorescence measurements of the various BSA systems where slit widths were maintained at 1 nm for both the excitation as well as emission fluorescence measurements. Excitation and emission were scanned at increasing intervals of 5 and 1 nm per scanning, respectively. While the excitation wavelength ranged between 250 and 350 nm, the range of emission wavelength was 250 to 600 nm. It is noteworthy that in case of the fluorescence measurements, the sample's concentration is precautiously maintained and a negligible change in absorbance value at the wavelength of excitation as determined from the absorption spectra excludes any possible inner filter effect. The far – UV circular dichroism (CD) spectra of relevant samples were recorded on Jasco – J 1500 CD spectropolarimeter in 190–260 nm wavelength range, maintaining a scan speed of 50 nm min−1 and a spectral bandwidth of 2.5 nm for each spectrum. A quartz cuvette with a path length of 0.1 cm was used to record the CD measurements. The background correction was done by subtracting the buffer spectrum from each spectrum recorded for all BSA systems. The sizes of the AuNPs were estimated with the help of dynamic light scattering (DLS) and scanning transmission electron microscopy (STEM) measurements. Bright-field STEM images were taken using a field emission scanning electron microscope (FE-SEM, FEI-Apreo S), maintaining a magnification of ‘3![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000
000![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 00×’ along with a horizontal field width (HFW) of 1.38 μm. Its instrumental details are given elsewhere.13,20 The FE-SEM images were recorded by the same instrument, working at 20 kV, fitted with an E–T detector for determining the structural changes of AuNPs-conjugated BSA on unfolding and refolding. FE-SEM images of mixed assemblies of SDS–gemini surfactants have also been recorded. The FE-SEM samples were prepared by drop casting 10 μL of the selected solutions on the silicon wafers and were dried in vacuum, while the STEM samples were prepared by drop casting 5 μL of the required solutions on the carbon-coated Cu grid. Energy-dispersive X-ray fluorescence (ED-XRF) spectra of the different AuNPs-conjugated BSA systems were recorded on a Panalytical Epsilon-1 (M-788) instrument possessing a SDD5 detector. The required samples were poured on a polypropylene-based thin-film plastic placed within a cup, and used for measurements. The parameters of the ED-XRF spectrum were maintained at a potential of 50 kV and an electric current of 100 μA. Zetasizer Nano-ZS 90 from Malvern Instruments Ltd was used for DLS measurements.13 The Milli-Q water was first filtered with 0.22 μm pore size filter paper (Millipore) and that was used to prepare the buffer solution. This buffer solution filtered with the same filter paper was then used to prepare BSA, gemini surfactant and SDS solutions. After that except BSA solutions, the stock solutions of gemini surfactant and SDS each were filtered with the same Millipore filter paper. A laser light of wavelength 632.8 nm and a scattering angle of 173° were used for the measurement of DLS size distributions. The corresponding G function was utilized to judge the size distribution. Note 1† of the ESI presents details of method of determination of hydrodynamic radius. It is true that in case of XRF, STEM and SEM, the morphologies obtained in solid phase may be different from that in suspension. That is why DLS measurements have been carried out. However, results do not show very significant difference between solid phase and liquid phase morphologies. A combined pH and conductometer from Eutech Instruments Pte. Ltd, (Model – PC 510) was used to measure the pH of the buffer and all stock solution. A Jasco FTIR-4200 spectrometer was used to record the FT-IR spectra. The sample preparation for FT-IR measurements, its parameters and instrumental details have been reported earlier.13 Each measurement was recorded three times and the corresponding standard deviations obtained are given in the relevant figures and tables accordingly. The temperature was kept fixed at 298.15 ± 1 K for all measurements.
00×’ along with a horizontal field width (HFW) of 1.38 μm. Its instrumental details are given elsewhere.13,20 The FE-SEM images were recorded by the same instrument, working at 20 kV, fitted with an E–T detector for determining the structural changes of AuNPs-conjugated BSA on unfolding and refolding. FE-SEM images of mixed assemblies of SDS–gemini surfactants have also been recorded. The FE-SEM samples were prepared by drop casting 10 μL of the selected solutions on the silicon wafers and were dried in vacuum, while the STEM samples were prepared by drop casting 5 μL of the required solutions on the carbon-coated Cu grid. Energy-dispersive X-ray fluorescence (ED-XRF) spectra of the different AuNPs-conjugated BSA systems were recorded on a Panalytical Epsilon-1 (M-788) instrument possessing a SDD5 detector. The required samples were poured on a polypropylene-based thin-film plastic placed within a cup, and used for measurements. The parameters of the ED-XRF spectrum were maintained at a potential of 50 kV and an electric current of 100 μA. Zetasizer Nano-ZS 90 from Malvern Instruments Ltd was used for DLS measurements.13 The Milli-Q water was first filtered with 0.22 μm pore size filter paper (Millipore) and that was used to prepare the buffer solution. This buffer solution filtered with the same filter paper was then used to prepare BSA, gemini surfactant and SDS solutions. After that except BSA solutions, the stock solutions of gemini surfactant and SDS each were filtered with the same Millipore filter paper. A laser light of wavelength 632.8 nm and a scattering angle of 173° were used for the measurement of DLS size distributions. The corresponding G function was utilized to judge the size distribution. Note 1† of the ESI presents details of method of determination of hydrodynamic radius. It is true that in case of XRF, STEM and SEM, the morphologies obtained in solid phase may be different from that in suspension. That is why DLS measurements have been carried out. However, results do not show very significant difference between solid phase and liquid phase morphologies. A combined pH and conductometer from Eutech Instruments Pte. Ltd, (Model – PC 510) was used to measure the pH of the buffer and all stock solution. A Jasco FTIR-4200 spectrometer was used to record the FT-IR spectra. The sample preparation for FT-IR measurements, its parameters and instrumental details have been reported earlier.13 Each measurement was recorded three times and the corresponding standard deviations obtained are given in the relevant figures and tables accordingly. The temperature was kept fixed at 298.15 ± 1 K for all measurements.
Different parameters such as ET, R0, and r were calculated to quantitatively analyze the FRET phenomenon.18,61 Details about FRET mechanism and determination of its parameters is provided in Note 2† of the ESI.
A decrease in fluorescence lifetime along with steady-state fluorescence quenching in the presence of NPs has been reported by many groups.38,49,62–64 The NSET process is responsible for these consequences. Details about NSET mechanism and corresponding calculation of its parameters are given in Note 3† of the ESI.
3 Results and discussion
3.1 Interaction between AuNPs-conjugated BSA (bioconjugates) and gemini surfactants: denaturation of bioconjugates
Refolding of the unfolded conjugated AuNPs-BSA has been carried out at a concentration of 0.2 mM each of gemini surfactant, 12-4-12,2Br− and 12-8-12,2Br− based on our previous reports.13 Fig. S1† shows the binding isotherm to display binding between the conjugated AuNPs-BSA and gemini surfactants based on changes in fluorescence intensity and α-helix% with an increase in surfactant concentration. One can see that a minimum appears at a concentration of gemini surfactant close to 0.06 mM. This concentration is the critical aggregation concentration (cac) of a gemini surfactant in presence of AuNPs (0.0124 μM)-conjugated BSA (10 μM) in HEPES buffer (∼10 mM) at pH 7.4 which was determined by conductivity measurements (Fig. S2†) reported earlier.13 The concentration at which a clear break-point noted is ∼0.06 mM which is the cac of the gemini surfactant. A reduced aggregation concentration of a gemini surfactant in the present system as compared to pure surfactant supports the fact that gemini surfactants interact with the bioconjugates.65 It can be seen from Fig. S1† that at a concentration below cac which is pre-aggregation region of a gemini surfactant, specific as well as non-cooperative bindings take place, and above this concentration in post-aggregation region cooperative and massive bindings of surfactants occur.13 At 0.2 mM concentration of a gemini surfactant, the binding of micelles with the bioconjugate reaches almost saturation. As reported earlier, in this state protein remains in a compressed form due to the presence of hydrophobic microdomain of micelles showing significantly high % of α-helical structures (Fig. S1†).13,66–683.2 STEM images of conjugated AuNPs-BSA
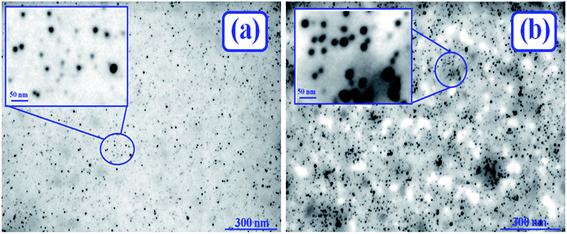
AuNPs (0.0124 μM) conjugated with BSA (10 μM) was synthesized following the method available in the literature.38 For the systematic justification of the synthesized AuNPs conjugated with folded and unfolded BSA, the STEM images have been recorded and published earlier.13 AuNPs conjugated with BSA proteins that are initially unfolded by 0.2 mM of 12-4-12,2Br− and 12-8-12,2Br− each and then refolded by SDS in HEPES buffer (∼10 mM) at pH 7.4 have been characterized by recoding STEM images, and are displayed in Fig. 1(a) and (b), respectively. As mentioned above, the STEM images of AuNPs conjugated with native and unfolded BSA are documented elsewhere.13 AuNPs of spherical shape with an average diameter of 5.0 ± 0.5 nm are noted for folded and refolded BSA and also for unfolded BSA. These sizes and shapes accord well with the literature report.38 Fig. 1 insets show enlarged view of AuNPs-conjugated with BSA. These insets are obtained by cropping the selected portion of images.3.3 Energy Dispersive X-ray Fluorescence (ED-XRF) spectra of AuNPs-conjugated BSA
The ED-XRF spectra were recorded to account for the preparation of conjugated AuNPs in folded, unfolded, and refolded states of BSA. The ED-XRF spectra for conjugated AuNPs in folded and unfolded states of BSA (containing 0.2 mM each of 12-4-12,2Br− and 12-8-12,2Br−) have already been reported.13 Fig. S3† displays the ED-XRF spectra of conjugated AuNPs-BSA refolded by 0.35 mM SDS (denatured by 0.2 mM each of 12-4-12,2Br− and 12-8-12,2Br−) in HEPES buffer. The corresponding table (Table S1†), emphasizes the proper binding between the protein and AuNPs by calculation of the ratio of S and Au as determined from the spectra, bearing well agreement with literature reports.13,69,70 The three forms of S-containing species in BSA namely, cysteine, methionine, and cystine are found to recognize the AuNPs to form Au–thiol co-ordination bonds. It suggests that BSA is conjugated with AuNPs due to formation of Au–S bonds as well as electrostatic interactions.71 The peak of Br arises from the bromide counter-ions of the gemini surfactants.3.4 UV-Visible absorption study of conjugated AuNPs-BSA
The UV-Visible absorption spectra of BSA, conjugated AuNPs-BSA (folded state), 0.2 mM gemini surfactant-induced unfolded states, and their corresponding refolded counterparts containing 0.35 mM of SDS and 0.2 mM each of 12-4-12,2Br− and 12-8-12,2Br−, are displayed in Fig. S4(a) and (b),† respectively. The absorption spectra for the native as well as AuNPs-conjugated BSA, as shown here, are also reported in our earlier work.13 Besides, their corresponding unfolded states have already been reported there.13 An absorption spectrum of pure BSA shows two absorption peak maxima one at 210 nm and other at 279 nm. The absorption peak maximum at 210 nm characterizes the π → π* transition of the of C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) O group in polypeptide backbone structure while the absorption peak maximum at 279 nm signifies the n → π* transition of aromatic amino acids present in BSA protein, i.e., Phenylalanine (Phe), Tyrosine (Tyr), and Tryptophan (Trp).11,72 The surface plasmon resonance (SPR) band for refolded conjugated AuNPs-BSA was also observed at 525 nm, which is the same as for folded and unfolded states for both surfactant systems. This SPR band's position, which is characteristic of AuNPs, agrees with the earlier reports.13,37,73–75 As discussed earlier, the presence of numerous amino acids as well as disulfide bonds along with one free thiol in the cysteine residues in BSA may be responsible for the AuNPs conjugation through thiolate linkage and/or weak covalent bonds with alkylamines. Thus, all the above characterizations along with the characteristic SPR band justify the conjugation of AuNPs with the protein. Reports have shown that different kinds of interactions between moieties in protein and AuNPs are responsible for their conjugation with each other.37,38,43,76,77
O group in polypeptide backbone structure while the absorption peak maximum at 279 nm signifies the n → π* transition of aromatic amino acids present in BSA protein, i.e., Phenylalanine (Phe), Tyrosine (Tyr), and Tryptophan (Trp).11,72 The surface plasmon resonance (SPR) band for refolded conjugated AuNPs-BSA was also observed at 525 nm, which is the same as for folded and unfolded states for both surfactant systems. This SPR band's position, which is characteristic of AuNPs, agrees with the earlier reports.13,37,73–75 As discussed earlier, the presence of numerous amino acids as well as disulfide bonds along with one free thiol in the cysteine residues in BSA may be responsible for the AuNPs conjugation through thiolate linkage and/or weak covalent bonds with alkylamines. Thus, all the above characterizations along with the characteristic SPR band justify the conjugation of AuNPs with the protein. Reports have shown that different kinds of interactions between moieties in protein and AuNPs are responsible for their conjugation with each other.37,38,43,76,77
3.5 CD spectral study of conjugated AuNPs-BSA
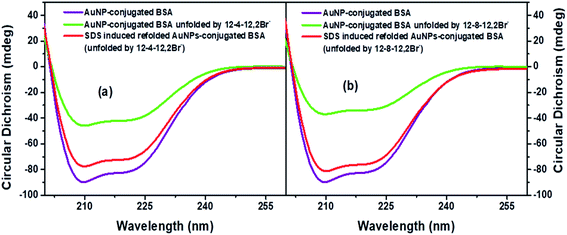
Far-UV CD spectra were recorded to study the secondary/tertiary structural changes of BSA upon conjugating with AuNPs, as reported by Shang et al.37 In our earlier study, we have reported the impact of AuNPs on the secondary structures of BSA partially unfolded by each of 12-4-12,2Br− and 12-8-12,2Br−. In the present work, the effect of conjugation on secondary structures of the protein refolded by 0.35 mM of SDS has been studied. The far UV CD spectra of conjugated AuNPs-BSA in folded and refolded states in the presence of 12-4-12,2Br− and 12-8-12,2Br−, have been shown in Fig. 2(a) and (b), respectively. The spectra for conjugated AuNPs-BSA unfolded by 0.06 mM each of 12-4-12,2Br− and 12-8-12,2Br−, respectively, have also been displayed in the same figures. The spectra corresponding to the AuNPs-conjugated BSA as well as their unfolded states are displayed in our previous report too.13 Two distinct negative minima at 208 and 222 nm seen in the UV region are observed in the CD spectrum of conjugated AuNPs-refolded BSA, which is the same as that for BSA in its native form.4 The Best Sel software (online version)78,79 was used to analyze all the CD data. The % α-helix of native BSA was calculated in order to validate the data obtained after computing from the software.78,79 The value obtained was 62.3% which corroborates well with the literature.66,80 As reported, the secondary structures of BSA are reduced after conjugation with AuNPs, and further reduced upon unfolding. The % α-helix in folded conjugated AuNPs-BSA, and after getting denatured by 12-4-12,2Br− and 12-8-12,2Br− (0.06 mM) decrease to 59.8, 30.8 and 23.8%, respectively.13 This shows that BSA is unfolded more by 12-8-12,2Br−. Although the denaturation studies of BSA have been carried out in 0.2 mM each of a gemini surfactant for other studies, 0.06 mM each of a gemini surfactant has been used for recording CD spectrum. The reasons for choosing this concentration are explained in an earlier report.13 After refolding the conjugated AuNPs-BSA with 0.35 mM of SDS, the % α-helix increases to 54.4 and 56.4 for 12-4-12,2Br− and 12-8-12,2Br−, respectively. The determined values lie very close to that for conjugated AuNPs-BSA in the folded state, and thus the % refolding can be determined as ∼91.0 and ∼94.3 for 12-4-12,2Br− and 12-8-12,2Br−, respectively.However, the refolding process is more in the case of 12-8-12,2Br− than 12-4-12,2Br−. As the spacer elongates, it tends to bend towards the tails, thus increasing the hydrophobicity. Hence, the SDS interacts more efficiently with a gemini surfactant with a lengthier spacer, enhancing the formation of catanionic micelles or mixed assemblies and easily stripping off the gemini surfactant. The effect of AuNPs on the refolded BSA with respect to its unconjugated state is also evidenced by the slight decrease in the % α-helix on conjugation. The % α-helix observed in case of SDS induced refolded BSA denatured by 12-4-12,2Br− and 12-8-12,2Br− are 57.2 and 59.0, respectively. Thus, the decrement in α-helical contents can be seen as ∼2.5% and ∼2.6–2.8% for folded and refolded BSA, respectively, in the presence of AuNPs. Therefore, the secondary structures of BSA stay almost unaffected even on AuNPs conjugation, which is the main criterion for the preparation of conjugated AuNPs-BSA.81
3.6 Steady-state fluorescence study of conjugated AuNPs-BSA
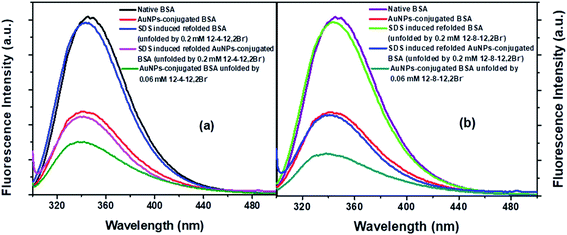
Steady-state fluorescence spectra of native BSA, its counterpart conjugated AuNPs-BSA, unfolded conjugated AuNPs-BSA, and refolded BSA in native form and upon conjugation are displayed by Fig. 3(a) and (b) for 12-4-12,2Br− and 12-8-12,2Br−, respectively.13 The spectra displaying the native and AuNPs-conjugated BSA as well as the unfolded bioconjugates are depicted in our earlier report also.13 The excitation wavelength was chosen as 295 nm specifically for Trp residues of BSA. The emission peak for native BSA was at 345 nm, while that for SDS induced refolded BSA with 0.06 mM each of 12-4-12,2Br− and 12-8-12,2Br− was observed at 342 and 343 nm, respectively. After introducing SDS into the system, a minor blue shift of 2 to 3 nm is observed. The SDS molecules interact with the protein surfactant complex and provide a more hydrophobic environment around the Trp residues causing the blue shift. A possibility of minute changes in the protein's tertiary structures may cause the environment around the Trp residues to become less polar, which cannot be ignored.In the case of the conjugated AuNPs-BSA, the fluorescence intensity is substantially quenched, while the peak maximum is observed at 340 nm. On unfolding the conjugated AuNPs-BSA by 0.06 mM each of the two gemini surfactants, peak maximum undergoes a blue shift with respect to the conjugated folded state. However, after adding 0.35 mM of SDS, the fluorescence intensity is regained. The corresponding peak maximum is observed at 341 and 340 nm for 12-4-12,2Br− and 12-8-12,2Br−, respectively, suggesting that the protein is refolded. Although the fluorescence intensity for folded and refolded conjugated AuNPs-BSA is nearly in the same range yet, there is a slight difference in the peak maximum of around 1 to 3 nm.
The fluorescence intensity of the Trp residues gets quenched in the folded and refolded states of BSA upon getting conjugated, as compared to its unconjugated versions, which is detailed in Note 4.†
After the conjugation of BSA with AuNPs, the radiative rate constants and oscillator strength change slightly. The latter's values lie in the range of 1.57 to 3.99%, as observed from Table S2.† The percentage quenching of Trp fluorescence lies in the range of 52.93 to 60.02%, which is higher than the corresponding systems' oscillator strength. Hence, it can be stated that energy transfer from BSA Trp moieties to AuNPs is the leading cause behind the quenching of Trp fluorescence in the presence of AuNPs. Also, the residence of AuNPs in close proximity of Trp residues is verified, which goes by the earlier reports.38,82,83
3.7 Refolding of unfolded conjugated AuNPs-BSA by SDS
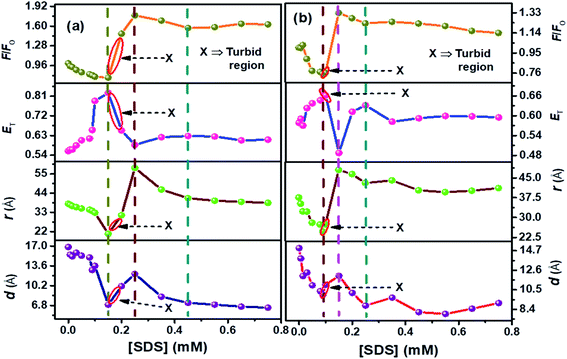 | ||
| Fig. 4 Plots compiled for F/F0, (ET), (r) (FRET phenomenon), (d) (NSET phenomenon) for SDS-prompted refolding of denatured conjugated AuNPs-BSA by (a) 12-4-12,2Br−, and (b) 12-8-12,2Br−. [BSA] = 10 μM, [AuNPs] = 0.0124 μM, [gemini surfactant] = 0.2 mM, λex = 295 nm for fluorescence studies. The errors related to the FRET and NSET parameters are given in Table S3.† | ||
It can be seen from Fig. 4(a) and (b) that ET follows an inverse relationship with r and d for both the surfactant systems and the changes in F/F0 are in good agreement with the trends of these parameters. With the initial addition of SDS molecules, the protein surfactant complex gets dissociated, and the AuNPs approach near the Trp residues. Thus the distance between the donor and acceptor moieties decreases. At the same time, the energy transfer is enhanced with the increasing concentration of SDS. It can be seen from Fig. 4(a) and (b) that the values of F/F0 and distance (r and d) start increasing while ET starts decreasing on going beyond the [SDS] = 0.15 mM and 0.09 mM for 12-4-12,2Br− and 12-8-12,2Br−, respectively. As the SDS concentration increases, the protein renaturation process commences with the removal of remaining gemini surfactant molecules from the protein chain. Therefore, the AuNPs experience difficulties staying in close vicinity of BSA Trp residues, unlike in the case of unfolded conjugated AuNPs-BSA. Hence, the d and r increase with a subsequent decrease in ET as the SDS concentration increases.
Furthermore, analysis of FRET/NSET parameters brings several fascinating facts to light. As per our earlier reports, it is found that the values of r and d are higher for gemini surfactant with a higher spacer length as it forms bigger micelles.13 Possibly, the donor–acceptor distance increases as the micellar size grows during the gradual binding between the micelles and the protein chain. On the onset of refolding, at 0.35 mM of SDS, the values of r and d attained are 46.04 Å, 44.04 Å, and 8.30 Å, 9.66 Å, for 12-4-12,2Br−, and 12-8-12,2Br−, respectively. Corresponding ‘ET’ achieved are 0.62 and 0.58 for 12-4-12,2Br−, and 12-8-12,2Br−, respectively. Beyond this concentration, the protein gets denatured by the SDS molecules.86,87 Interestingly, the SDS molecules can extract the gemini molecules from the BSA–gemini aggregates with maximum efficiency at a molar ratio of gemini![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) SDS as 1
SDS as 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 2. But, it is difficult to study the extraction process at this molar ratio of gemini and SDS due to precipitation. Hence, the study shows that AuNPs can be successfully used as a tool to monitor the protein's conformational changes during its denaturation and renaturation process.
2. But, it is difficult to study the extraction process at this molar ratio of gemini and SDS due to precipitation. Hence, the study shows that AuNPs can be successfully used as a tool to monitor the protein's conformational changes during its denaturation and renaturation process.
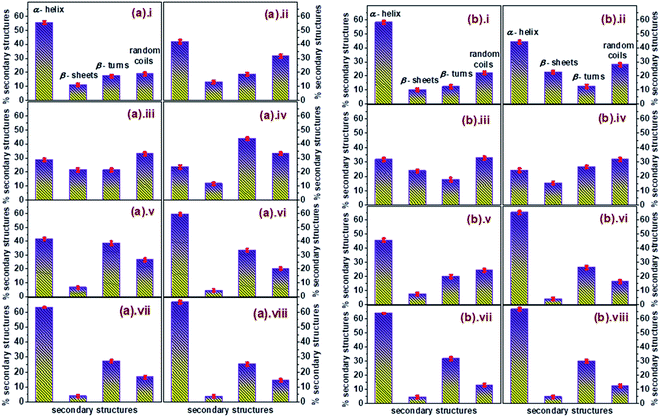
The percentage of different elements of the secondary structures like α-helix, β-sheets, β-turns, and random coils were calculated to get a comprehensive illustration of variations in the secondary structure of unfolded conjugated AuNPs-BSA during its refolding process. The calculated data has been depicted in Fig. 6 in a histogram format which clearly describes the relationship of the percentage of α-helix content with each of the other secondary structures during the refolding of conjugated AuNPs- BSA in the presence of SDS. α-Helix is in an inverse relationship with both β-turns and random coils; however, it bears an arbitrary relation with β-sheets. An enhancement in β-turns and random coils with a decrease in α-helix is observed before the refolding process starts. However, as the refolding process initiates, the α-helix increases with a considerable reduction in β-turns and random coils.
Fig. 7(a)–(d) depict the changes in the FTIR spectra observed for native, unfolded, and refolded BSA in the absence, and those in the presence of AuNPs, respectively. However, parts (a) and (c) of Fig. 7 are represented for changes in FTIR spectra for the case of 12-4-12,2Br−, while parts (b) and (d) of Fig. 7 stands for 12-8-12,2Br−.
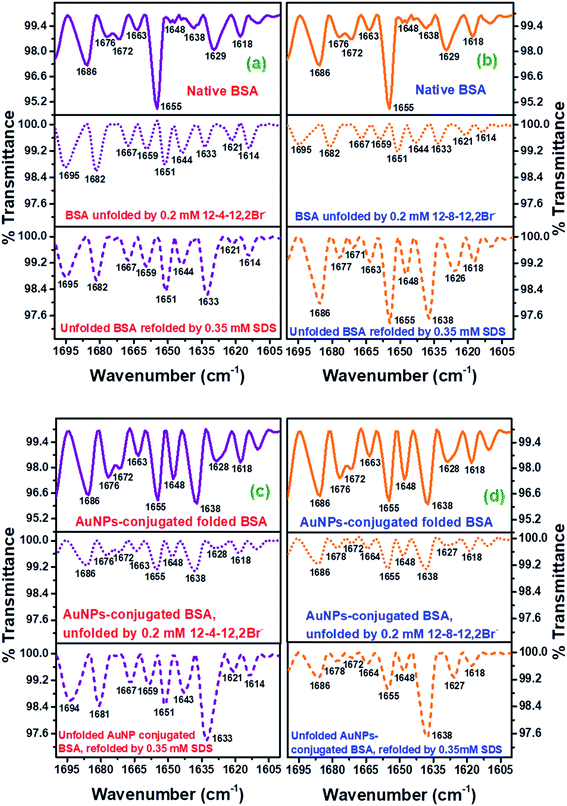 | ||
| Fig. 7 FTIR spectra corresponding to the amide I bands for peptide backbone of the native BSA, BSA denatured by a gemini surfactant, and SDS-prompted refolded BSA, denatured by a gemini surfactant, for gemini surfactant (a) 12-4-12,2Br−, (b) 12-8-12,2Br−, and conjugated AuNPs-BSA, conjugated AuNPs-BSA denatured by a gemini surfactant, and SDS-prompted refolded conjugated AuNPs-BSA, denatured by a gemini surfactant, for gemini surfactant (c) 12-4-12,2Br−, and (d) 12-8-12,2Br−. Standard deviations related to the wavenumbers are depicted in the Table S4.† | ||
The α-helical content of BSA is represented by the band at 1655 cm−1. In Fig. 7(a) and (b), this peak splits into two new peaks at 1651 and 1659 cm−1 in the unfolded state possessing lower intensity than in the native state. These new peaks contribute to helical content but are weak, supporting that the protein chains get compressed, which is inferred from the CD and fluorescence experimental data. This also displays that some α-helical content is lost upon the unfolding of the protein. A new band at 1681 cm−1 also supports the same phenomenon. The β-sheets are indicated by the bands at 1618, 1629, and at 1638 cm−1, while the turns are indicated by the bands at 1672, 1676, and 1686 cm−1 for the secondary structures of the protein. These bands gain intensities upon unfolding of the protein, which again states the protein getting denatured. Now, with the addition of SDS to the unfolded BSA systems, the refolding of protein happens with regain in the intensities of the peaks at 1651 and 1659 cm−1, responsible for the recovery of the α-helical content. While the intensities of bands contributing to the β-sheets and turns relapse. This supports the renaturation of protein by SDS. Interestingly, the peak at 1655 cm−1 reappeared again in the refolded BSA (containing 12-8-12,2Br−), resembling to that in the native BSA and the intensity of this peak is greater to the 1651 cm−1 of the unfolded protein. This result again strengthens the fact that surfactant 12-8-12,2Br− is removed off more easily than 12-4-12,2Br− by SDS.
Focusing on all the conjugated systems of BSA represented Fig. 7(c) and (d), it can be seen that the intensity of the band at 1655 cm−1 is decreased compared to the unconjugated BSA systems; however, the other secondary structures' band intensities increase which is due to conjugation with AuNPs.13,37 Similar to the case of unconjugated systems, the refolded conjugated systems experience an increment in the band intensities corresponding to the regain in α-helical content at 1651 and 1659 cm−1 for 12-4-12,2Br−, and 1655 cm−1 for 12-8-12,2Br−, respectively. It is noteworthy that for 12-4-12,2Br−, loss of α-helical content in the unfolded conjugated AuNPs-BSA is denoted by a decrement in the band intensity at 1655 cm−1. However, α-helical content regain in the refolded conjugated AuNPs-BSA is depicted by increment in the band intensities at 1651 and 1659 cm−1, which also attributes to α-helix. Whereas, in the case of 12-8-12,2Br−, α-helical loss and regain in unfolded and refolded conjugated AuNPs-BSA, respectively, is seen by their corresponding decrement and increment of band intensity at 1655 cm−1 only, which is a characteristic band for α-helix. Though the regain in the intensities of the peaks responsible for α-helical content is less for refolded conjugated AuNPs-BSA systems than its unconjugated counterparts, the refolding of unfolded conjugated AuNPs-BSA is clearly evidenced. Significantly, the refolding of conjugated AuNPs-BSA in the system containing 12-8-12,2Br− is more than that for 12-4-12,2Br−, which is similar to observed for unconjugated systems due to its intensity regain in the characteristic band for α-helix at 1655 cm−1 than 1651 and 1659 cm−1. The appearance of few bands other than for α-helix correspond to other secondary structural elements, maybe because of the loss of helical form in the presence of AuNPs at the expense of β-sheets, turns, etc.
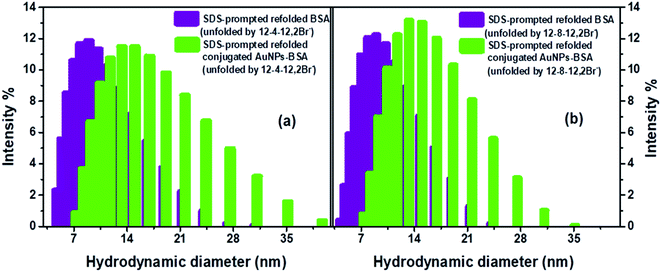
DLS measurement have also been carried out to get detailed information regarding the structural changes of AuNPs-conjugated BSA complexed with 0.2 mM of gemini surfactants along with different other aggregates formed in the solution upon addition of varying concentrations of SDS. Fig. S8† represents the changes in hydrodynamic diameter of conjugated AuNPs-BSA denatured by 0.2 mM of gemini surfactants, 12-4-12,2Br− and 12-8-12,2Br− each, with increasing concentration of SDS in HEPES buffer (∼10 mM) at pH 7.4. It can be seen that initially the sizes of the bioconjugate–surfactant complexes increase up to ∼60 nm after addition of 0.15 mM and 0.09 mM of SDS in presence of 12-4-12,2Br− and 12-8-12,2Br− each. After that the sizes are decreased before they reach a minimum value of size of a particle of ∼12 nm. Interestingly, the two concentrations of SDS mentioned above match well with the concentrations at which refolding of bioconjugates start as discussed based on fluorescence data (Fig. 4) and CD data (Fig. 5).
Formations of comparatively bigger-sized particles (mixed assemblies) upon addition of SDS to a system containing bioconjugate and gemini surfactant are also evident from DLS measurements. Fig. S9(a)† shows the sizes of these particles in presence of conjugated AuNPs (0.0124 μM)-BSA (10 μM) and 0.2 mM of 12-4-12,2Br− and 12-8-12,2Br− each at different concentrations of SDS. In order to investigate whether any mixed assemblies of surfactants, SDS and gemini surfactants form in absence of bioconjugates, DLS experiments have been performed (Fig. S9(b)†). Sizes of all particles obtained from DLS measurements along with the proposed shapes are tabulated in Tables S6(a) and (b).† The hydrodynamic diameter of micelles of 12-4-12,2Br− and 12-8-12,2Br− are found to be 1.5 and 1.2 nm, respectively in absence of bioconjugates. In our earlier study, we have found particles of similar sizes.56 Hu et al.92 have reported the hydrodynamic diameter of 1.64 nm for micelles in 10 mM solution of 14-4-14,2Br−. The hydrodynamic diameters noted by us are corroborating well with that reported by Hu et al.92 for similar gemini surfactant except for different tail length. The data thus support the authenticity of our results. The representative DLS size distributions for mixed assemblies in presence and absence of BSA bioconjugates are given as Fig. S10(a) and (b).† One can see from the data in Tables S6(a) and (b)† that bigger-sized assemblies formed in presence and in absence of bioconjugates are almost same. These particles' sizes become as high as ∼800 nm at [SDS] = 0.05 mM, after that it decreases to ∼400 nm at [SDS] = 0.1 mM, then once again it increases to ∼500–600 nm at 0.2 mM of SDS and become almost constant thereafter. On the other hand, the sizes of smaller particles those are complexes of bioconjugates and gemini surfactants, decrease with increasing SDS concentration. These results rule out the possibility of interactions of mixed assembles of SDS and gemini surfactants with bioconjugates. Aswal and co-workers26 have reported the formation of various assemblies of mixed anionic SDS and cationic DTAB surfactants in absence and presence of BSA. They have found interactions of mixed surfactant-assemblies with the protein forming different structures upon addition of different mole fractions of SDS and DTAB. These observations are different from that in the present study could be because in the present work pure SDS is added to the complexes of bioconjugates and gemini surfactants. In their study, Aswal's group26 has noticed that in the absence of BSA, with increasing mole fraction of either surfactant, there are formations of assemblies such as ellipsoidal and rod-like micelles, and vesicles and then at equimolar mixture there is an evidence of formation of multi-lamellar vesicles. Our group also has reported before the formation of various structures in a solution containing SDS and different gemini surfactants.4 There are many other groups who have reported the formation of catanions/mixed assemblies like rod-like micelles, vesicles, lamellar micelles etc. even in the absence of a protein.29–33 Keeping these results in mind and FESEM images discussed below we have also proposed the shapes of mixed surfactant assemblies formed after addition of different concentrations of SDS and given in Tables S6(a) and (b).† With increasing concentration of SDS, the different structures formed are proposed to be rod-like micelles, bilayer vesicles, and rod-like micelles/vesicles. The sizes of mixed assemblies of SDS and gemini surfactants noted in the present study are bigger than that in case of SDS and DTAB reported by Aswal and co-workers.26 This is because gemini surfactants are more surface active and hydrophobic than their conventional counterparts.93–95 It is noteworthy that the width of a distribution corresponding to the mixed assemblies is more than that for biconjugates could be because chances of getting particles of different sizes are more for the former case.
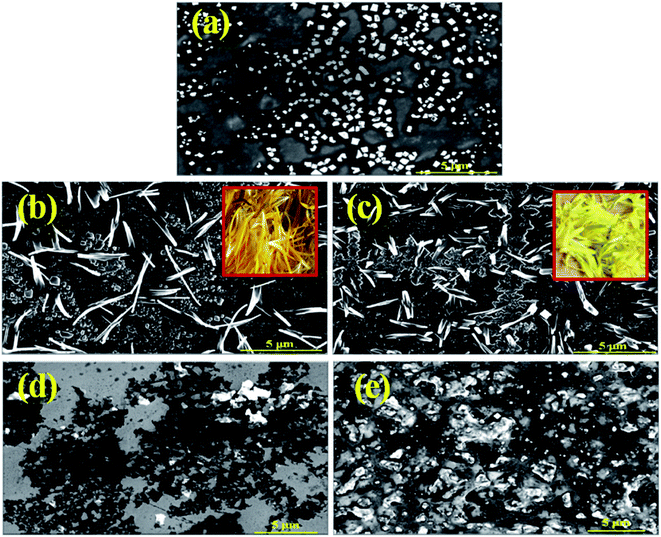
3.7.7.1 FESEM images of AuNPs-conjugated BSA in folded, unfolded and refolded form. In order to support unfolding of bioconjugates by 0.2 mM of a gemini surfactant followed by refolding by the addition of SDS in HEPES buffer (∼10 mM) at pH 7.4, FESEM images have been recorded. FE-SEM images (Fig. 9), recorded on a magnified scale, depict the morphological changes of the conjugated AuNPs-BSA in all the three states viz. folded, unfolded and refolded. Square-like flakes can be seen for folded conjugated AuNPs-BSA in Fig. 9(a). However, structures similar to dispersed petals of a dandelion flower (inset of Fig. 9(b) and (c)) can be seen for the unfolded states as represented in Fig. 9(b) and (c), for 12-4-12,2Br− and 12-8-12,2Br−, respectively. Upon addition of SDS to the unfolded states, in addition to different other structures, some square flakes, can be observed in Fig. 9(d) and (e) for the refolded conjugated AuNPs-BSA in presence of 12-4-12,2Br− and 12-8-12,2Br−, respectively. Some other assemblies noted in Fig. 9(d) and (e) are absent in Fig. 9(a). These structures could be the mixed assemblies of SDS and gemini surfactants as evident from the DLS data as well. Results thereby indicate that the AuNPs-conjugated BSA molecules tend to relieve themselves from interacting with the molecules of gemini surfactant as a result of formation of mixed assemblies (catanion) between gemini surfactant and SDS, and therefore get refolded. These images somewhat demonstrate that the conjugated AuNPs-BSA in the folded, as well as refolded forms, partly resemble each other; however, those in the unfolded ones are quite different.
3.7.7.2 FESEM images of mixed assemblies of SDS and gemini surfactants. Further to support the fact that mixed assemblies of SDS and gemini surfactants are formed, the FESEM images have been recorded for the samples containing 0.2 mM of a gemini surfactant and varying concentrations of SDS in HEPES buffer (∼10 mM) at pH 7.4. Fig. S11(i)(a–e)† represents the structures of mixed assemblies at 0.03, 0.09, 0.35 and 0.55 mM of SDS. Similarly, Fig. S11(ii)(a–e)† denotes the above systems in case of 12-8-12,2Br−. While at a lower concentration range of SDS (0.03–0.09 mM) bilayer vesicles are formed, but at a higher concentration range of SDS (0.35–0.55 mM), the system seems to have mixed structures such as rod-like micelles and vesicles, etc. as evident from the DLS results as well (Fig. S9(b)†).
The formation of mixed assemblies of SDS and gemini surfactants even in presence of AuNPs-conjugated BSA can be found from Fig. 9(d) and (e) in Section 3.7.7.1. As discussed above in addition to square flake-like structures for refolded bioconjugates, other structures those could be mixed assemblies of SDS and gemini surfactants are also noted.
4 Conclusions
The SDS-prompted refolding of conjugated AuNPs-BSA denatured by two gemini surfactants, 12-4-12,2Br− and 12-8-12,2Br− each, has been elucidated here. Techniques like ED-XRF, STEM, and DLS have been used to support the conjugation of AuNPs to BSA. Since both the NSET and FRET phenomena are applicable to describe the energy transfer occurring between Trp residues of BSA (donor) and AuNPs (acceptor), both methods were used to probe the refolding of conjugated AuNPs-BSA by calculating their corresponding parameters obtained from fluorescence measurements. Other than this, CD, FT-IR, DLS and FESEM measurements were also used to understand the refolding mechanism. The refolding phenomenon follows an inverse order in correspondence to the unfolding process. The energy transfer efficiency (ET) and donor–acceptor distance (r/d), obtained from the FRET and NSET calculations, justify the refolding process over the entire binding isotherm. The refolding phenomenon is quite efficient as the percentage of refolding exceeds 90%. It was also found that α-helix bears a good inverse relationship with β-turns and random coils during this process. In the pre-refolding region, with increasing SDS concentration, a reduction is witnessed in the α-helical content with a simultaneous increase in the β-turns and random coils, which is just the reverse of the observation in refolding region. The SDS molecules extract the gemini surfactant molecules by mixed catanion formation. The extraction of surfactant molecules is more efficient for 12-8-12,2Br− than 12-4-12,2Br− due to its longer spacer chain length. A longer spacer increases the hydrophobicity of the micelles by tending to bend towards the tails, thus easing the formation of mixed assemblies. This study thereby illustrates an appropriate approach for implementing bioconjugated AuNPs in the form of an optical-based molecular ruler in order to demonstrate the refolding of a gemini surfactant-induced denatured protein. The study subsequently highlights the alterations in the structures of bioconjugates with the formation of different mixed assemblies in the solution during refolding process.Author contributions
All the authors have contributed to writing the manuscript. All authors have approved the final version of the manuscript.Conflicts of interest
The authors declare no competing financial interest.Acknowledgements
The Central Analytical Laboratory, Birla Institute of Technology and Science (BITS), Pilani, Hyderabad Campus, India, is thankfully acknowledged by the authors for the instrumentation facilities provided for different characterizations. S. K. S. also acknowledges the Department of Science and Technology (DST) FIST program, Government of India. R. A., S. H. and S. D. are thankful to Birla Institute of Technology & Science (BITS), Pilani – Hyderabad campus, for providing financial support. Also, Mr Dinabandhu Patra, a research scholar from the Department of Chemistry, BITS Pilani, Hyderabad campus, is acknowledged for cooperating in the ED-XRF analysis.References
- S. C. Wang and C. T. Lee, Biochemistry, 2007, 46, 14557–14566 CrossRef CAS PubMed.
- A. V. Latnikova, S. Y. Lin, G. Loglio, R. Miller and B. A. Noskov, J. Phys. Chem. C, 2008, 112, 6126–6131 CrossRef CAS.
- J. Chen, M. Chi, M. Lin, L. Lin and T. Wang, BioMed Res. Int., 2015, 2015, 1–9 CAS.
- S. Kumari, S. Halder, R. Aggrawal, V. K. Aswal, G. Sundar and S. K. Saha, J. Mol. Liq., 2020, 300, 112238 CrossRef CAS.
- V. Dahiya and T. K. Chaudhuri, Biochemistry, 2013, 52, 4517–4530 CrossRef CAS.
- H. Yamaguchi and M. Miyazaki, Biomolecules, 2014, 4, 235–251 CrossRef PubMed.
- D. Rozema and S. H. Gellman, Biochemistry, 1996, 35, 15760–15771 CrossRef CAS PubMed.
- D. L. Daugherty, D. Rozema, P. E. Hanson and S. H. Gellman, J. Biol. Chem., 1998, 273, 33961–33971 CrossRef CAS PubMed.
- U. Anand, C. Jash and S. Mukherjee, Phys. Chem. Chem. Phys., 2011, 13, 20418–20426 RSC.
- S. Tayyab, B. Ahmad, Y. Kumar and M. M. Khan, Int. J. Biol. Macromol., 2002, 30, 17–22 CrossRef CAS PubMed.
- Sonu, S. Halder, S. Kumari, R. Aggrawal, V. K. Aswal and S. K. Saha, J. Mol. Liq., 2017, 243, 369–379 CrossRef CAS.
- S. Bhuin, S. Halder, S. K. Saha and M. Chakravarty, RSC Adv., 2021, 11, 1679–1693 RSC.
- S. Halder, R. Aggrawal, S. Jana and S. K. Saha, J. Photochem. Photobiol., B, 2021, 225, 112351 CrossRef CAS PubMed.
- P. Bharmoria, K. S. Rao, T. J. Trivedi and A. Kumar, J. Phys. Chem. B, 2014, 118, 115–124 CrossRef CAS PubMed.
- J. K. Choi, J. Ho, S. Curry, D. Qin, R. Bittman and J. A. Hamilton, J. Lipid Res., 2002, 43, 1000–1010 CrossRef CAS PubMed.
- T. Zhou, M. Ao, G. Xu, T. Liu and J. Zhang, J. Colloid Interface Sci., 2013, 389, 175–181 CrossRef CAS PubMed.
- P. B. Kandagal, S. Ashoka, J. Seetharamappa, S. M. T. Shaikh, Y. Jadegoud and O. B. Ijare, J. Pharm. Biomed. Anal., 2006, 41, 393–399 CrossRef CAS PubMed.
- S. Halder, S. Kumari, S. Kumar, V. K. Aswal and S. K. Saha, ACS Omega, 2018, 3, 11192–11204 CrossRef CAS PubMed.
- S. Halder, R. Aggrawal, V. K. Aswal, D. Ray and S. K. Saha, J. Mol. Liq., 2021, 322, 114532 CrossRef CAS.
- S. Halder, R. Aggrawal and S. K. Saha, Colloids Surf., A, 2022, 644, 128862 CrossRef CAS.
- E. L. Gelamo, R. Itri, A. Alonso, J. V. Da Silva and M. Tabak, J. Colloid Interface Sci., 2004, 277, 471–482 CrossRef CAS PubMed.
- Y. Moriyama, Y. Kawasaka and K. Takeda, J. Colloid Interface Sci., 2003, 257, 41–46 CrossRef CAS PubMed.
- Y. Moriyama, Y. Sato and K. Takeda, J. Colloid Interface Sci., 1993, 156, 420–424 CrossRef CAS.
- R. C. Lu, A. N. Cao, L. H. Lai, B. Y. Zhu, G. X. Zhao and J. X. Xiao, Colloids Surf., B, 2005, 41, 139–143 CrossRef CAS PubMed.
- J. X. Xiao, U. Sivars and F. Tjerneld, J. Chromatogr. B: Biomed. Sci. Appl., 2000, 743, 327–338 CrossRef CAS.
- D. Saha, D. Ray, S. Kumar, J. Kohlbrecher and V. K. Aswal, Soft Matter, 2021, 17, 6972–6984 RSC.
- G. X. Zhao and B. Y. Zhu, J. Dispersion Sci. Technol., 1995, 16, 305–332 CrossRef CAS.
- D. Saha, D. Ray, J. Kohlbrecher and V. K. Aswal, ACS Omega, 2018, 3, 8260–8270 CrossRef CAS PubMed.
- Y. Shang, H. Liu, Y. Hu and J. M. Prausnitz, Colloids Surf., A, 2007, 294, 203–211 CrossRef CAS.
- Y. Shang, Y. Xu, H. Liu and Y. Hu, J. Dispersion Sci. Technol., 2006, 27, 105–108 CrossRef CAS.
- Y. Shang, Q. Chen and H. Liu, J. Dispersion Sci. Technol., 2007, 28, 854–859 CrossRef CAS.
- Y. Wang, G. Bai, E. F. Marques and H. Yan, J. Phys. Chem. B, 2006, 110, 5294–5300 CrossRef CAS PubMed.
- D. Jurašin, I. Weber and N. Filipović-Vinceković, J. Dispersion Sci. Technol., 2009, 30, 622–633 CrossRef.
- F. Höök, M. Rodahl, B. Kasemo and P. Brzezinski, Proc. Natl. Acad. Sci., 1998, 95, 12271–12276 CrossRef PubMed.
- M. Wahlgren, Trends Biotechnol., 1991, 9, 201–208 CrossRef CAS PubMed.
- J. Yao, Y. He, L. Li, P. Li and M. Yang, Ind. Eng. Chem. Res., 2019, 58, 21201–21207 CrossRef CAS.
- L. Shang, Y. Wang, J. Jiang and S. Dong, Langmuir, 2007, 23, 2714–2721 CrossRef CAS PubMed.
- T. Sen, K. K. Haldar and A. Patra, J. Phys. Chem. C, 2008, 112, 17945–17951 CrossRef CAS.
- A. Selva sharma and M. Ilanchelian, J. Phys. Chem. B, 2015, 119, 9461–9476 CrossRef CAS PubMed.
- C. Salinas, M. V. Amé and A. G. Bracamonte, RSC Adv., 2020, 10, 20620–20637 RSC.
- K. Nakanishi, T. Sakiyama and K. Imamura, J. Biosci. Bioeng., 2001, 91, 233–244 CrossRef CAS PubMed.
- A. Sahraei, F. Mohammadi, R. Boukherroub and S. Szunerits, Langmuir, 2020, 36, 10321–10330 CrossRef CAS PubMed.
- F. Aldeek, M. Safi, N. Zhan, G. Palui and H. Mattoussi, ACS Nano, 2013, 7, 10197–10210 CrossRef CAS PubMed.
- P. Srivastava, S. K. Hira, D. N. Srivastava, U. Gupta, P. Sen, R. A. Singh and P. P. Manna, ACS Biomater. Sci. Eng., 2017, 3, 3376–3385 CrossRef CAS PubMed.
- D. Dutta, A. Chattopadhyay and S. S. Ghosh, ACS Biomater. Sci. Eng., 2016, 2, 2090–2098 CrossRef CAS PubMed.
- R. Zhong, Y. Liu, P. Zhang, J. Liu, G. Zhao and F. Zhang, ACS Appl. Mater. Interfaces, 2014, 6, 19465–19470 CrossRef CAS PubMed.
- C. Bhan, R. Mandlewala, A. Gebregeorgis and D. Raghavan, Langmuir, 2012, 28, 17043–17052 CrossRef CAS PubMed.
- J. R. Lakowicz, Principles of Fluorescence Spectroscopy, Kluwer Academic, New York, 1999 Search PubMed.
- C. S. Yun, A. Javier, T. Jennings, M. Fisher, S. Hira, S. Peterson, B. Hopkins, N. O. Reich and G. F. Strouse, J. Am. Chem. Soc., 2005, 127, 3115–3119 CrossRef CAS PubMed.
- Z. Li, N. Xue, H. Ma, Z. Cheng and X. Miao, Talanta, 2018, 181, 346–351 CrossRef CAS PubMed.
- H. Ma, N. Xue, Z. Li, K. Xing and X. Miao, Sens. Actuators, B, 2018, 266, 221–227 CrossRef CAS.
- B. N. J. Persson and N. D. Lang, Phys. Rev. B: Condens. Matter Mater. Phys., 1982, 26, 5409–5415 CrossRef CAS.
- T. Pons, I. L. Medintz, K. E. Sapsford, S. Higashiya, A. F. Grimes, D. S. English and H. Mattoussi, Nano Lett., 2007, 7, 3157–3164 CrossRef CAS PubMed.
- M. Ghosh, S. Kundu, A. Pyne and N. Sarkar, J. Phys. Chem. C, 2020, 124, 3905–3914 CrossRef CAS.
- A. K. Tiwari, Sonu and S. K. Saha, J. Phys. Chem. B, 2014, 118, 3582–3592 CrossRef CAS PubMed.
- Sonu, S. Kumari and S. K. Saha, J. Phys. Chem. B, 2015, 119, 9751–9763 CrossRef CAS PubMed.
- F. M. Menger and C. A. Littau, J. Am. Chem. Soc., 1993, 115, 10083–10090 CrossRef CAS.
- X. Wang, J. Wang, Y. Wang, H. Yan, P. Li and R. K. Thomas, Langmuir, 2004, 20, 53–56 CrossRef CAS PubMed.
- Sonu, S. Kumari and S. K. Saha, J. Phys. Chem. B, 2015, 119, 9751–9763 CrossRef CAS PubMed.
- S. Muthusubramanian and S. K. Saha, J. Lumin., 2012, 132, 2166–2177 CrossRef CAS.
- L. Wu, C. Huang, B. P. Emery, A. C. Sedgwick, S. D. Bull, X. P. He, H. Tian, J. Yoon, J. L. Sessler and T. D. James, Chem. Soc. Rev., 2020, 49, 5110–5139 RSC.
- T. L. Jennings, M. P. Singh and G. F. Strouse, J. Am. Chem. Soc., 2006, 128, 5462–5467 CrossRef CAS PubMed.
- T. L. Jennings, J. C. Schlatterer, M. P. Singh, N. L. Greenbaum and G. F. Strouse, Nano Lett., 2006, 6, 1318–1324 CrossRef CAS PubMed.
- T. Sen, K. K. Haldar and A. Patra, J. Phys. Chem. C, 2010, 114, 11409–11413 CrossRef CAS.
- R. Yasuda, T. Masaike, K. Adachi, H. Moji, H. Itoh and K. Kinosita, Proc. Natl. Acad. Sci., 2003, 100, 9314–9318 CrossRef CAS PubMed.
- N. Gull, S. Chodankar, V. K. Aswal, P. Sen, R. H. Khan and K.-u. -Din, Colloids Surf., B, 2009, 69, 122–128 CrossRef CAS PubMed.
- N. Gull, P. Sen, R. H. Khan and K.-U. -Din, Langmuir, 2009, 25, 11686–11691 CrossRef CAS PubMed.
- K.-u. -Din, W. Fatma, Z. A. Khan and A. A. Dar, J. Phys. Chem. B, 2007, 111, 8860–8867 CrossRef PubMed.
- J. L. Burt, C. Gutiérrez-Wing, M. Miki-Yoshida and M. José-Yacamán, Langmuir, 2004, 20, 11778–11783 CrossRef CAS PubMed.
- L. Liu, H. Z. Zheng, Z. J. Zhang, Y. M. Huang, S. M. Chen and Y. F. Hu, Spectrochim. Acta, Part A, 2008, 69, 701–705 CrossRef PubMed.
- L. Wang, J. Li, J. Pan, X. Jiang, Y. Ji, Y. Li, Y. Qu, Y. Zhao, X. Wu and C. Chen, J. Am. Chem. Soc., 2013, 135, 17359–17368 CrossRef CAS PubMed.
- X. Zhao, R. Liu, Z. Chi, Y. Teng and P. Qin, J. Phys. Chem. B, 2010, 114, 5625–5631 CrossRef CAS PubMed.
- M. Iosin, F. Toderas, P. L. Baldeck and S. Astilean, J. Mol. Struct., 2009, 924–926, 196–200 CrossRef CAS.
- M. Iosin, V. Canpean and S. Astilean, J. Photochem. Photobiol., A, 2011, 217, 395–401 CrossRef CAS.
- K. M. Bromley, A. J. Patil, A. W. Perriman, G. Stubbs and S. Mann, J. Mater. Chem., 2008, 18, 4796–4801 RSC.
- M. J. Meziani, P. Pathak, B. A. Harruff, R. Hurezeanu and Y. P. Sun, Langmuir, 2005, 21, 2008–2011 CrossRef CAS PubMed.
- A. M. Dennis, D. C. Sotto, B. C. Mei, I. L. Medintz, H. Mattoussi and G. Bao, Bioconjugate Chem., 2010, 21, 1160–1170 CrossRef CAS PubMed.
- A. Micsonai, F. Wien, L. Kernya, Y. H. Lee, Y. Goto, M. Réfrégiers and J. Kardos, Proc. Natl. Acad. Sci., 2015, 112, E3095–E3103 CrossRef CAS PubMed.
- A. Micsonai, F. Wien, É. Bulyáki, J. Kun, É. Moussong, Y. H. Lee, Y. Goto, M. Réfrégiers and J. Kardos, Nucleic Acids Res., 2018, 46, W315–W322 CrossRef CAS PubMed.
- J. S. Mandeville and H. A. Tajmir-Riahi, Biomacromolecules, 2010, 11, 465–472 CrossRef CAS PubMed.
- N. N. Mamedova, N. A. Kotov, A. L. Rogach and J. Studer, Nano Lett., 2001, 1, 281–286 CrossRef CAS.
- P. Joshi, S. Chakraborty, S. Dey, V. Shanker, Z. A. Ansari, S. P. Singh and P. Chakrabarti, J. Colloid Interface Sci., 2011, 355, 402–409 CrossRef CAS PubMed.
- S. J. Strickler and R. A. Berg, J. Chem. Phys., 1962, 37, 814–822 CrossRef CAS.
- P. C. Ray, Z. Fan, R. A. Crouch, S. S. Sinha and A. Pramanik, Chem. Soc. Rev., 2014, 43, 6370–6404 RSC.
- B. Klajnert, L. Stanisławska, M. Bryszewska and B. Pałecz, Biochim. Biophys. Acta, Proteins Proteomics, 2003, 1648, 115–126 CrossRef CAS PubMed.
- N. J. Turro, X. G. Lei, K. P. Ananthapadmanabhan and M. Aronson, Langmuir, 1995, 11, 2525–2533 CrossRef CAS.
- S. F. Santos, D. Zanette, H. Fischer and R. Itri, J. Colloid Interface Sci., 2003, 262, 400–408 CrossRef CAS PubMed.
- H. H. Mantsch and D. Chapman, Infrared Spectroscopy of Biomolecules, Wiley-Liss, New York, 1996 Search PubMed.
- W. K. Surewicz, H. H. Mantsch and D. Chapman, Biochemistry, 1993, 32, 389–394 CrossRef CAS PubMed.
- Y. Y. Lou, K. L. Zhou, J. H. Shi and D. Q. Pan, J. Photochem. Photobiol., B, 2017, 173, 589–597 CrossRef CAS PubMed.
- Y. Y. Lou, K. L. Zhou, D. Q. Pan, J. Le Shen and J. H. Shi, J. Photochem. Photobiol., B, 2017, 167, 158–167 CrossRef CAS PubMed.
- C. Hu, R. Li, H. Yang and J. Wang, J. Colloid Interface Sci., 2011, 356, 605–613 CrossRef CAS PubMed.
- R. Zana, Curr. Opin. Colloid Interface Sci., 1996, 1, 566–571 CrossRef CAS.
- R. Zana and Y. Talmon, Nature, 1993, 362, 228–230 CrossRef CAS.
- R. Zana, J. Colloid Interface Sci., 2002, 248, 203–220 CrossRef CAS PubMed.
Footnotes |
| † Electronic supplementary information (ESI) available. See https://doi.org/10.1039/d2ra02618j |
| ‡ The authors have contributed equally. |
| This journal is © The Royal Society of Chemistry 2022 |