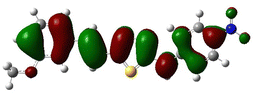Open Access Article
Open Access ArticleSynthesis and characterization of new 1,3,4-thiadiazole derivatives: study of their antibacterial activity and CT-DNA binding
Hakan S. Sayinera,
Mehmet I. Yilmazer b,
Aisha. T. Abdelsalamc,
Mohamed A. Ganimc,
Cengiz Balogluc,
Yasemin Celik Altunogluc,
Mahmut Gür
b,
Aisha. T. Abdelsalamc,
Mohamed A. Ganimc,
Cengiz Balogluc,
Yasemin Celik Altunogluc,
Mahmut Gür d,
Murat Saracoglu*b,
Mohamed S. Attia
d,
Murat Saracoglu*b,
Mohamed S. Attia *e,
Safwat A. Mahmoud*f,
Ekram H. Mohamed
*e,
Safwat A. Mahmoud*f,
Ekram H. Mohamed g,
Rabah Boukherroubh,
Nora Hamad Al-Shaalan
g,
Rabah Boukherroubh,
Nora Hamad Al-Shaalan i,
Sarah Alharthi
i,
Sarah Alharthi j,
Fatma Kandemirli*k and
Mohammed A. Amin
j,
Fatma Kandemirli*k and
Mohammed A. Amin *j
*j
aDepartment of Infectious Diseases, Faculty of Medicine, Adiyaman University, Adiyaman, Turkey
bFaculty of Education, Erciyes University, 38039, Kayseri, Turkey. E-mail: muratsaracoglu@gmail.com
cDepartment of Genetic & Bioengineering, Faculty of Engineering & Architecture, Kastamonu University, 37150, Kastamonu, Turkey
dDepartment of Forest Industrial Engineering, Faculty of Forestry, Kastamonu University, 37150, Kastamonu, Turkey
eChemistry Department, Faculty of Science, Ain Shams University, Abbassia 11566, Cairo, Egypt. E-mail: mohd_mostafa@sci.asu.edu.eg
fPhysics Department, Faculty of Science, Northern Border University, Arar, Saudi Arabia
gDepartment of Analytical Chemistry, Faculty of Pharmacy, The British University in Egypt, 11837, El-Sherouk City, Egypt
hUniv. Lille, CNRS, Centrale Lille, Univ. Polytechnique Hauts-de-France, UMR 8520 – IEMN, F59000 Lille, France
iDepartment of Chemistry, College of Science, Princess Nourah Bint Abdulrahman University, P.O. Box 84428, Riyadh 11671, Saudi Arabia
jDepartment of Chemistry, College of Science, Taif University, P.O. Box 11099, Taif 21944, Saudi Arabia. E-mail: mohamed@tu.edu.sa
kDepartment of Biomedical Engineering, Faculty of Engineering and Architecture, Kastamonu University, 37150, Kastamonu, Turkey. E-mail: fkandemirli@yahoo.com
First published on 17th October 2022
Abstract
1,3,4-Thiadiazole molecules (1–4) were synthesized by the reaction of phenylthiosemicarbazide and methoxy cinnamic acid molecules in the presence of phosphorus oxychloride, and characterized with UV, FT-IR, 13C-NMR, and 1H-NMR methods. DFT calculations (b3lyp/6-311++G(d,p)) were performed to investigate the structures' geometry and physiochemical properties. Their antibacterial activity was screened for various bacteria strains such as Enterobacter aerogenes, Escherichia coli ATCC 13048, Salmonella kentucky, Pseudomonas aeruginosa, Klebsiella pneumoniae, Proteus and Gram positive such as Staphylococcus aureus ATCC 25923, Listeria monocytogenes ATCC 7644, Enterococcus faecium, Enterococcus durans, Staphylococcus aureus ATCC, Serratia marcescens, Staphylococcus hominis, Staphylococcus epidermidis, alfa Streptococcus haemolyticus, Enterococcus faecium and found to have an inhibitory effect on Klebsiella pneumoniae and Staphylococcus hominis, while molecules 1, 3 and 4 had an inhibitory effect on Staphylococcus epidermidis and alpha Streptococcus haemolyticus. The experimental results were supported by the docking study using the Kinase ThiM from Klebsiella pneumoniae. All the investigated compounds showed an inhibitory effect for the Staphylococcus epidermidis protein. In addition, the mechanism of the 1–4 molecule interaction with calf thymus-DNA (CT-DNA) was investigated by UV-vis spectroscopic methods.
1 Introduction
Heterocyclic molecules are cyclic compounds comprising carbon and other elements such as oxygen, nitrogen and sulphur. The heterocyclic molecules containing a single heteroatom are pyrrole, furan and thiophene, while those consisting of more than one heteroatom are azole, pyrrole, thiazole, thiadiazole, oxadiazole, triazene etc.1 There are many different heterocyclic molecules that have therapeutic significance. Lately, thiadiazole derivatives have become an important field of research due to their broad-spectrum activities. Thiadiazoles belong to the classes of nitrogen–sulfur containing heterocycles with extensive application as structural units of biologically active molecules and are useful intermediates in medicinal chemistry. There are four possible structures of thiadiazole such as 1,2,5-thiadiazole, 1,3,4-thiadiazole, 1,2,3-thiadiazole and 1,2,4-thiadiazole, but 1,3,4-thiadiazole is the most versatile owing to its pharmacological and biological activities (Fig. 1).2 During the past years, substituted 1,3,4-thiadiazole derivatives have received significant attention and have been increasingly investigated due to their broad spectrum of pharmacological properties.2 It is supposed that 1,3,4-thiadiazole derivatives exhibit various biological activities due to the presence of![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N–C–S– moiety.2 Other authors assume that the biological activities of 1,3,4-thiadiazole derivatives are due to the strong aromaticity of the ring, which also provides great in vivo stability to this five-membered ring system and low toxicity for higher vertebrates, including human beings.3
N–C–S– moiety.2 Other authors assume that the biological activities of 1,3,4-thiadiazole derivatives are due to the strong aromaticity of the ring, which also provides great in vivo stability to this five-membered ring system and low toxicity for higher vertebrates, including human beings.3
Thiadiazole contains both a sulphur atom and two nitrogen atoms as part of its aromatic five-membered ring and has a “hydrogen bonding domain” and “two-electron donor system” functions as anhydrase.3
Among the thiadiazole isomers, our attention was focused on 1,3,4-thiadiazole. The importance of 1,3,4-thiadiazole originates from its biological activity, featuring antimicrobial,4,5 anti-tuberculosis,6 anti-inflammatory,7,8 carbonic anhydrase inhibitor,9 anticonvulsants,10,11 antihypertensive,12,13 antioxidant,14 anticancer,15,16 and antifungal17 properties. 1,3,4-Thiadiazole derivatives are important precursors in various chemical reactions for the synthesis of biologically-active molecules. Some examples of drug molecules that comprise 1,3,4-thiadiazole group in their structure are: acetazolamide and methazolamide as carbonic anhydrase inhibitors.9 It was reported that the thiadiazole group could act as the bio-isosteric substitute of the thiazole moiety.18 The biological activity of thiadiazole derivatives is due to their strong aromaticity. For higher vertebrates including humans, thiadiazole showed little or no toxicity effect.19
Farghaly et al.20 synthesized a series of 1,3,4-thiadiazoles, investigated their antimicrobial activity, and studied their structure activity relationship (SAR) towards some microorganisms. The results of this study revealed that this class of molecules holds promising activity. Bhatia et al.21 prepared various imidazo-thiadiazole derivatives, and screened their antimicrobial activity. The results revealed that these molecules are active against Shigella flexneri, Staphylococcus aureus (S. aureus) and Candida albicans bacterial strains. Kaur et al.17 synthesized biphenyl imidazo[2,1-b][1,3,4]thiadiazole derivatives and found that these molecules exhibit moderate to good activity when compared with standard antibiotics ampicillin and amphotericin B, and screened the two derivatives for anticancer activity. Gür et al.14 synthesized a series of 1,3,4-thiadiazole derivatives and reported that some of them had effective antioxidant properties.
In this study, 1,3,4-thiadiazole derivatives were synthesized and their antibacterial activity was assessed on both Gram-positive and Gram-negative bacteria. In addition, their antifungal property was assessed for Candida albicans ATCC 26555. Besides, interaction of 1–4 molecules and calf thymus DNA (CT-DNA) in physiological buffer (pH = 7.4) was studied by UV-vis absorption to understand the mechanism of interaction of molecules studied with nucleic acids.
2 Experimental section
2.1 Materials and methods
Solvents are distilled and dried before use. Infrared spectra were recorded on FT-IR Fourier Transform Infrared Spectroscopy. 13C and 1H-NMR spectra were acquired on Bruker AVANCE III 400 MHz NMR Spectrometer. Melting points were uncorrected and recorded on SMP30 melting point apparatus. UV-vis absorption spectra were performed by Thermo Scientific MULTISKAN GO spectrophotometer. Quartz cuvettes with 1 cm light path were used in the measurements.2.2 Synthesis
2.2.1.1 5-[(E)-2-(3-Methoxyphenyl)]vinyl-N-[4′-nitrophenyl]-1,3,4-thiadiazole-2-amine (1). Melting point (m.p.): 252 °C. Rf (formic acid
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethylacetate
ethylacetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) xylene, 1
xylene, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5): 0.60. IR ν (cm−1): 3262 (N–H), 3167 (Ar C–H), 1598 (Ar C
5): 0.60. IR ν (cm−1): 3262 (N–H), 3167 (Ar C–H), 1598 (Ar C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1623 (alkene C
C), 1623 (alkene C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1255 (C–N), 1575 (C
C), 1255 (C–N), 1575 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1187 (C–S). 1H-NMR (ppm, DMSO-d6): δ = 9.94 (N–H), δ = 8.43–7.00 (m, aromatic protons), δ = 6.95, 6.96 (alkenic protons, overlapped), δ = 3.87 (s, CH3O protons). 13C-NMR (ppm, DMSO-d6): δ = 55.9 (1C, –OCH3), δ = 164.0 and 159.4 (2C, thiadiazole carbons), δ = 119.8–157.4 (10C, aromatic carbons), δ = 111.4 and 111.9 (alkenic carbons). UV-vis spectrum: λmax = 241, 368 nm. Elemental analysis (%) for C17H14N4O3S, experimental (calc.): C = 57.29 (57.62); H = 3.83 (3.98); N = 14.94 (15.81); S = 9.09 (9.05).
N), 1187 (C–S). 1H-NMR (ppm, DMSO-d6): δ = 9.94 (N–H), δ = 8.43–7.00 (m, aromatic protons), δ = 6.95, 6.96 (alkenic protons, overlapped), δ = 3.87 (s, CH3O protons). 13C-NMR (ppm, DMSO-d6): δ = 55.9 (1C, –OCH3), δ = 164.0 and 159.4 (2C, thiadiazole carbons), δ = 119.8–157.4 (10C, aromatic carbons), δ = 111.4 and 111.9 (alkenic carbons). UV-vis spectrum: λmax = 241, 368 nm. Elemental analysis (%) for C17H14N4O3S, experimental (calc.): C = 57.29 (57.62); H = 3.83 (3.98); N = 14.94 (15.81); S = 9.09 (9.05).
2.2.1.2 5-[(E)-2-(2-Methoxyphenyl)]vinyl-N-[4′-nitrophenyl]-1,3,4-thiadiazole-2-amine (2). M.p.: 247–249 °C. Rf (formic acid
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethylacetate
ethylacetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) xylene, 1
xylene, 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5): 0.66. IR ν (cm−1): 3262 (N–H), 3161 (Ar C–H), 1596 (Ar C
5): 0.66. IR ν (cm−1): 3262 (N–H), 3161 (Ar C–H), 1596 (Ar C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1617 (alkene C
C), 1617 (alkene C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1245 (C–N), 1568 (C
C), 1245 (C–N), 1568 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1188 (C–S). 1H-NMR (ppm, DMSO-d6): δ = 9.95 (N–H), δ = 8.41–7.02 (m, aromatic protons), δ = 6.88, 6.90 (alkenic protons, overlapped), δ = 3.86 (s, CH3O protons). 13C-NMR (ppm, DMSO-d6): δ = 55.58 (1C, –OCH3), δ = 164.2 and 160.1 (2C, thiadiazole carbons), δ = 115.4–158.9 (10C, aromatic carbons), δ = 111.5 and 112.4 (alkenic carbons). UV-vis spectrum bands: λmax = 243, 267, 374 nm. Elemental analysis (%) for C17H14N4O3S, experimental (calc.): C = 57.45 (57.62); H = 3.83 (3.98); N = 14.91 (15.81); S = 8.78 (9.05).
N), 1188 (C–S). 1H-NMR (ppm, DMSO-d6): δ = 9.95 (N–H), δ = 8.41–7.02 (m, aromatic protons), δ = 6.88, 6.90 (alkenic protons, overlapped), δ = 3.86 (s, CH3O protons). 13C-NMR (ppm, DMSO-d6): δ = 55.58 (1C, –OCH3), δ = 164.2 and 160.1 (2C, thiadiazole carbons), δ = 115.4–158.9 (10C, aromatic carbons), δ = 111.5 and 112.4 (alkenic carbons). UV-vis spectrum bands: λmax = 243, 267, 374 nm. Elemental analysis (%) for C17H14N4O3S, experimental (calc.): C = 57.45 (57.62); H = 3.83 (3.98); N = 14.91 (15.81); S = 8.78 (9.05).
2.2.1.3 5-[(E)-2-(3-Methoxyphenyl)]vinyl-N-[4′-methylphenyl]-1,3,4-thiadiazole-2-amine (3). M.p.: 172–176 °C. Rf (formic acid
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethylacetate
ethylacetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) xylene: 1
xylene: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5): 0.62. IR ν (cm−1): 3247 (N–H), 3114 (Ar C–H), 1596 (Ar C
5): 0.62. IR ν (cm−1): 3247 (N–H), 3114 (Ar C–H), 1596 (Ar C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1235 (C–N), 1548 (C
C), 1235 (C–N), 1548 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1183 (C–S), 2953, 2917, 2834 (aliphatic, C–H). 1H-NMR (ppm, DMSO-d6): δ = 10.45 (N–H), δ = 7.72–7.00 (m, aromatic protons), δ = 6.98, 6.97 (overlapped, alkenic protons), δ = 3.88 (s, CH3O protons), δ = 2.27 (s, CH3 protons). 13C-NMR (ppm, DMSO-d6): δ = 56.1 (1C, –OCH3), δ = 163.5 and 158.4 (2C, thiadiazole carbons), δ = 120.0–157.4 (10C, aromatic carbons), δ = 112.0 and 118.2 (alkenic carbons). UV-vis spectrum bands λmax = 245, 276, 353 nm. Elemental analysis (%) for C18H17N3OS, experimental (calc.): C = 65.83 (66.85); H = 5.05 (5.30); N = 12.14 (12.99); S = 9.06 (9.91).
N), 1183 (C–S), 2953, 2917, 2834 (aliphatic, C–H). 1H-NMR (ppm, DMSO-d6): δ = 10.45 (N–H), δ = 7.72–7.00 (m, aromatic protons), δ = 6.98, 6.97 (overlapped, alkenic protons), δ = 3.88 (s, CH3O protons), δ = 2.27 (s, CH3 protons). 13C-NMR (ppm, DMSO-d6): δ = 56.1 (1C, –OCH3), δ = 163.5 and 158.4 (2C, thiadiazole carbons), δ = 120.0–157.4 (10C, aromatic carbons), δ = 112.0 and 118.2 (alkenic carbons). UV-vis spectrum bands λmax = 245, 276, 353 nm. Elemental analysis (%) for C18H17N3OS, experimental (calc.): C = 65.83 (66.85); H = 5.05 (5.30); N = 12.14 (12.99); S = 9.06 (9.91).
2.2.1.4 5-[(E)-2-(2-Methoxyphenyl)]vinyl-N-[4′-methylphenyl]-1,3,4-thiadiazole-2-amine (4). M.p.: 185 °C. Rf (formic acid
![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) ethylacetate
ethylacetate![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) xylene: 1
xylene: 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 4
4![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 5): 0.74. IR ν (cm−1): 3243 (N–H), 3121 (Ar C–H), 1566 (Ar C
5): 0.74. IR ν (cm−1): 3243 (N–H), 3121 (Ar C–H), 1566 (Ar C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1613 (alkene C
C), 1613 (alkene C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) C), 1237 (C–N), 1566 (C
C), 1237 (C–N), 1566 (C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N), 1190 (C–S), 2914, 2834 (aliphatic, C–H). 1H-NMR (ppm, DMSO-d6): δ = 10.47 (N–H), δ = 7.55–7.16 (m, aromatic protons), δ = 6.92, 6.90 (d, alkenic protons), δ = 3.80 (s, methoxy protons); δ = 2.27 (s, methyl protons). 13C-NMR (ppm, DMSO-d6): δ = 55.6 (1C, –OCH3), δ = 163.8 and 160.1 (2C, thiadiazole carbons), δ = 118.2–158.0 (10C, aromatic carbons), δ = 112.3 and 115.5 (alkenic carbons). UV-vis spectrum bands: λmax = 242, 274, 358 nm. Elemental analysis (%) for C18H17N3OS, experimental (calc.): C = 65.97 (66.85); H = 4.89 (5.30); N = 12.51 (12.99); S = 9.93 (9.91).
N), 1190 (C–S), 2914, 2834 (aliphatic, C–H). 1H-NMR (ppm, DMSO-d6): δ = 10.47 (N–H), δ = 7.55–7.16 (m, aromatic protons), δ = 6.92, 6.90 (d, alkenic protons), δ = 3.80 (s, methoxy protons); δ = 2.27 (s, methyl protons). 13C-NMR (ppm, DMSO-d6): δ = 55.6 (1C, –OCH3), δ = 163.8 and 160.1 (2C, thiadiazole carbons), δ = 118.2–158.0 (10C, aromatic carbons), δ = 112.3 and 115.5 (alkenic carbons). UV-vis spectrum bands: λmax = 242, 274, 358 nm. Elemental analysis (%) for C18H17N3OS, experimental (calc.): C = 65.97 (66.85); H = 4.89 (5.30); N = 12.51 (12.99); S = 9.93 (9.91).
2.3 Enterobacter aerogenes
The antifungal activity of 1,3,4-thiadiazoles was tested on Candida albicans ATCC 26555 and their antibacterial activity was evaluated on Gram-negative such as Enterobacter aerogenes Escherichia coli ATCC 13048, Salmonella kentucky, Pseudomonas aeruginosa, Klebsiella pneumoniae, Proteus and Gram-positive strains such as Staphylococcus aureus ATCC 25923, Listeria monocytogenes ATCC 7644, Enterococcus faecium, Enterococcus durans, Staphylococcus aureus ATCC, Serratia marcescens, Staphylococcus hominis, Staphylococcus epidermidis, alfa Streptococcus haemolyticus, Enterococcus faecium. Antifungal activity of 1,3,4-thiadiazoles was assessed on Candida albicans ATCC 26555. Standard and other strains were provided from Kastamonu University and Gazi University.2.4 Antimicrobial activity
All bacterial strains were incubated at 37 ± 2 °C for 24 h and inocula were prepared by transferring morphologically similar colonies of each organism into 0.9% sterile saline solution until the visible turbidity was equal to 0.5 McFarland standard corresponding approximately 108 cfu mL−1. 1000 mL of medium was autoclaved at 120 °C for 20 min, then placed in sterile Petri dishes. The prepared bacterial suspension was distributed over the sterile agar surface by rubbing the sterile swab. Before the discs were applied, the inoculated plates were allowed to dry at room temperature for 5–10 min.Sterile discs filled with solutions containing 1–4 molecules at a concentration of 2 ppm were placed on the surface of inoculated Petri dish. After placing the discs on each plate, it was lightly pressed to make contact with the agar surface and incubated at 37 °C for 24 h. Antibacterial activity against organisms was recorded in mm units after 24 h by measurement of the zone of inhibition. All tests were repeated three times each. After the operations were completed, the plates were sterilized in an autoclave at 120 °C for 20 min and then disposed.22,23
2.5 CT-DNA binding experiments
Absorption spectra measurements were conducted using various CT-DNA concentrations (0–90 μM) and keeping the concentration of molecules 1–4 constant (40 μM). The samples were mixed for 30 min at 37 °C.24 Nucleotide concentration was determined at 260 nm by UV-vis absorption spectroscopy using a molar absorption coefficient of 6600 M−1 cm−1.25Molecules 1–4 were prepared at a concentration of 500 μM in DMSO/Tris HCl (65%/35%). The (K) value, known as the binding constant of small molecules with DNA, can be determined by using the Benesi–Hildebrand eqn (1).26
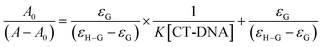 | (1) |
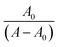 vs.
vs. 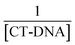 plots.
plots.
The intercept-to-slope ratios of the plot of [DNA] vs. 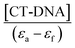 gives also the K value. Where εa (or εG) is the absorption coefficient of the molecule and εf (or εH–G) is the absorption of the complex obtained through the interaction of the molecule with CT-DNA.16
gives also the K value. Where εa (or εG) is the absorption coefficient of the molecule and εf (or εH–G) is the absorption of the complex obtained through the interaction of the molecule with CT-DNA.16
The interaction of thiadiazole derivatives with CT-DNA was examined by calculating the intrinsic binding constant (K) between molecules 1–4 and CT-DNA using the following eqn (2):
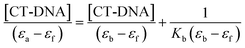 | (2) |
εf was calculated using the calibration curve generated from the absorbance of molecules 1–4 in aqueous solution, following Beer's law. εa was determined as the ratio between the molecules 1–4 concentration and the measured absorbance, Aobs/[molecules 1–4].
A plot of 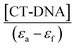 vs. [CT-DNA] gives a slope of
vs. [CT-DNA] gives a slope of 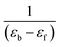 and a y-intercept equals to
and a y-intercept equals to 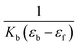 .13
.13
| IP = −EHOMO | (3) |
| EA = −ELUMO | (4) |
The chemical hardness, electron donating power (ω−), and electron acceptor power (ω+) were calculated using eqn (5)–(7):28
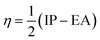 | (5) |
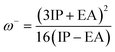 | (6) |
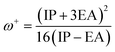 | (7) |
The molecular docking study was carried out by the Autodock Vina.29,30 The optimized structures of compounds 1–4 (b3lyp/6-311++G(d,p)) were used for the docking calculations. The crystal structure of Kinase ThiM from Klebsiella pneumoniae was downloaded from the protein data bank (PDB 6k28), the water molecules and heteroatoms were removed, the polar hydrogen atoms were added, and the Gasteiger charges were applied to the protein structure. All chains were deleted except chain A. The search space was determined using GetBox PyMOL plugin using the cocrystallized TZE molecule at the center, yielding a box with center at 22.5, 63.5, 35.4, and dimensions of 15.5, 14.6, 15.1. Docking automation was carried out using PyRx 0.8 (https://link.springer.com/protocol/10.1007/978-1-4939-2269-7_19). Figures were generated using UCSF Chimera 1.13.1 (https://onlinelibrary.wiley.com/doi/10.1002/jcc.20084) and PoseView (https://academic.oup.com/bioinformatics/article/22/14/1710/226866?login=false) (https://pubs.acs.org/doi/10.1021/ci049958u).
3 Results and discussion
3.1 Synthesis and characterization of molecules 1–4
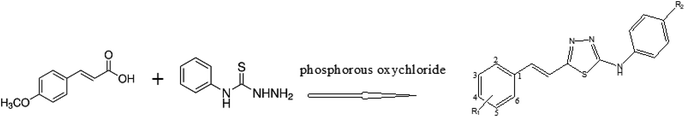
The synthesis of thiadiazole derivatives 1–4 was carried out using a mixture of methoxy cinnamic acid and phenylthiosemicarbazide derivatives in presence of phosphorous oxychloride (Fig. 2).| Compound | R1 | R2 |
|---|---|---|
| 1 | 5-OCH3 | NO2 |
| 2 | 6-OCH3 | NO2 |
| 3 | 5-OCH3 | CH3 |
| 4 | 6-OCH3 | CH3 |
13C-NMR, 1H-NMR, and FT-IR, spectroscopies and elemental analysis were performed to determine the structure of the molecules 1–4.
The FT-IR spectrum of molecules 1, 2, 3 and 4 shows bands in range 3262–3167 cm−1 due to the stretching of N–H and C–HAr groups and the bands in range 1575–1183 cm−1 are attributed to C![[double bond, length as m-dash]](https://www.rsc.org/images/entities/char_e001.gif) N, C–N and C–S stretching vibrations.
N, C–N and C–S stretching vibrations.
N–H proton between phenyl and thiadiazole ring resonated at δH 9.94, 9.95, 10.45 and 10.47 ppm, respectively for the molecules 1, 2, 3 and 4. The signals observed as multiple bands are between δ 8.43–7.00, 8.41–7.02, 7.27–7.00, and 7.55–7.16 ppm respectively for the molecules 1, 2, 3 and 4, due to aromatic protons.
In the 13C-NMR spectra, the two characteristic peaks of the two carbons of 1,3,4-thiadiazole ring are at 163.5–158.4, 164.23–160.1, and 164.0 and 159.4 ppm for molecules 1, 2 and 3, respectively. The two peaks at δ 163.8 and 160.1 ppm were the characteristic peaks of two carbons of 1,3,4-thiadiazole ring, and the other 13C-NMR (DMSO) spectrum data of molecule 4 showed 118.2–158.0 (10C, aromatic carbons), 112.3 and 115.5 (alkenic carbons), and 55.62 (1C, –OCH3) for molecule 1.
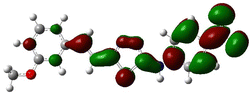
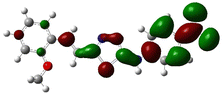
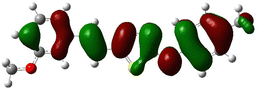
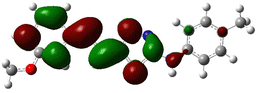
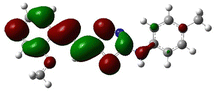
In order to investigate the structures' geometry, DFT calculations were performed. The DFT calculations at b3lyp/6-311++G(d,p) level in the gas phase showed that all the structures were linear as the dihedral angles C7–C8–C9–S13 and C12–N10–C21–C22 were close to 0° (Fig. 3). Fig. 4 depicts the frontier orbital calculations of compounds 1–4.31 For all molecules, the HOMO orbitals were distributed over the aromatic skeleton, while the LUMO orbitals of molecules 1 and 2 were positioned on the NO2-ph unit. The substituted –CH3 group changed the situation, where the LUMO orbitals were distributed over the thiadiazole ring. Table 1 summarizes the calculated HOMO, LUMO, and energy gap values for the investigated compounds. The calculated HOMO energies revealed that values were in the order of 1 > 2 > 3 > 4. These results indicate that compound 1 is the most reactive ligand towards the interactions with protein receptors.32,33 The calculated energy gap of the investigated molecules showed that compound 1 and 2 featured the lower values, indicating lower reactivity of the two molecules.
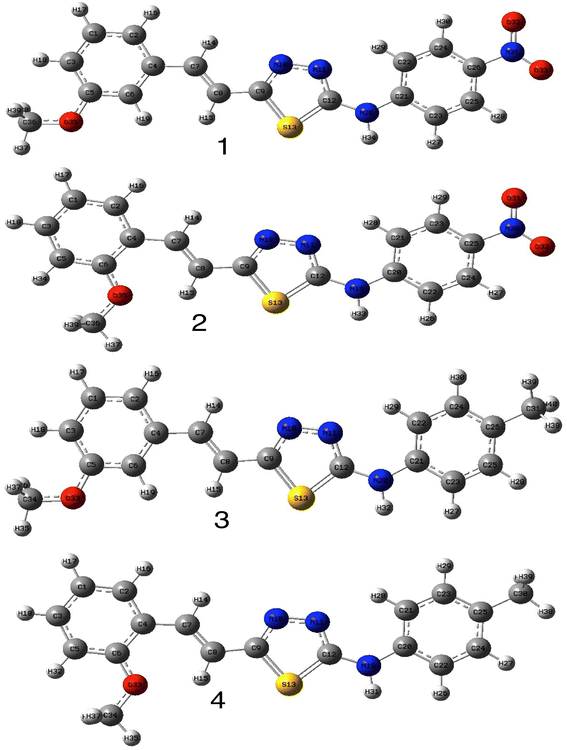 | ||
| Fig. 3 Optimized structures of compounds 1–4 at b3lyp/6-311++G(d,p) level of theory in the gas phase. | ||
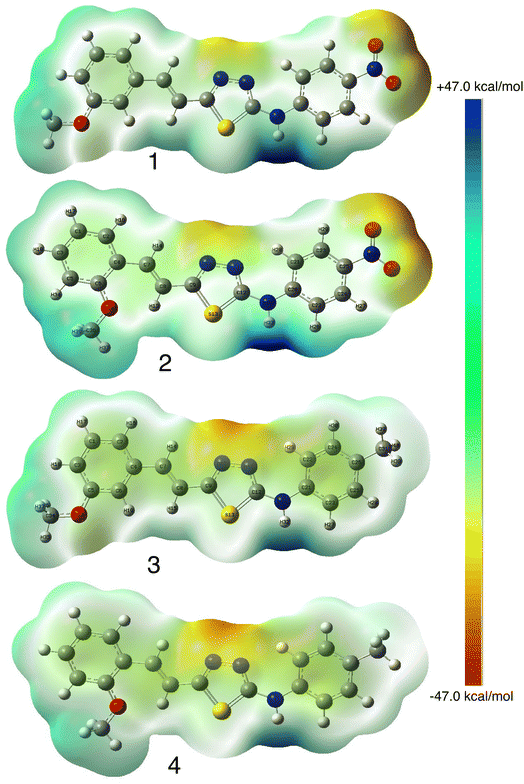
Molecular electrostatic surface potential (MESP) of molecules is an important factor to describe the active site of ligands.34–37 The MESP of 1–4 molecules were calculated using the optimized structures at b3lyp/6-311++G(d,p) level of theory to explore the nucleophilic and electrophilic sites on the ligands' surface. Fig. 4 indicates that the positive regions (nucleophilic sites) were positioned on the –NH group, while the negative regions (electrophilic sites) were situated over the thiadiazole and NO2 group in compounds 1 and 2, and the thiadiazole unit dominated the electrophilic attack in compounds 2 and 4. Ionization potential and electron affinity of the investigated molecules were used to assess the reactivity parameters of compounds 1–4. The calculated IP and EA of the investigated compounds showed that compound 1 has the highest affinity to donate and accept electrons from the receptors, while compound 4 is the weakest electron donor (Table 2). This result indicates the high polarizability of compound 1 as compared to the other ligands. The calculated ω− and ω+ values confirmed the high electron mobility of the molecules in the order 1 > 2 > 3 > 4. The calculated chemical hardness of the investigated molecules suggested that compound 1 and 2 have the lowest values, which indicates the facile charge transfer process.
| 1 | 2 | 3 | 4 | |
|---|---|---|---|---|
| EHOMO (eV) | −6.15 | −5.99 | −5.59 | −5.47 |
| ELUMO (eV) | −2.73 | −2.67 | −2.07 | −1.92 |
| Eg (eV) | 3.42 | 3.32 | 3.52 | 3.55 |
| IP (eV) | 6.15 | 5.99 | 5.59 | 5.47 |
| EA (eV) | 2.73 | 2.67 | 2.07 | 1.92 |
| η (eV) | 3.42 | 3.32 | 3.52 | 3.55 |
| ω− (eV) | 8.20 | 8.02 | 6.30 | 5.91 |
| ω+ (eV) | 2.58 | 2.56 | 2.01 | 1.90 |
3.2 Antimicrobial activity
In vitro biological screening tests for antimicrobial activity of the synthesized 1,3,4-thiodiazole molecules were performed against various Gram-positive and Gram-negative bacterial strains. The Gram-negative bacteria consist of Salmonella kentucky, Enterobacter aerogenes ATCC 13048, Klebsiella pneumoniae, Escherichia coli, Proteus, and Pseudomonas aeruginosa. The Gram-positive bacteria investigated are Enterococcus faecium, Listeria monocytogenes ATCC 7644, ATCC 25923, Staphylococcus aureus, Enterococcus durans, Serratia marcescens, Staphylococcus aureus ATCC, Staphylococcus epidermidis, alfa Streptococcus haemolyticus, Staphylococcus hominis, and Enterococcus faecium. Additionally, antifungal activity was tested using Candida albicans ATCC 26555. Antimicrobial studies were carried out by the disk diffusion method and the tests were performed in triplicate. No zone of inhibition was observed in the tests performed with the molecules 1–4 at 0.2 M against Gram-positive (Listeria monocytogenes ATCC 7644, S. aureus ATCC, monocytogenes E. faecium, S. aureus ATCC 25923, E. durans, S. marcescens, E. faecium), Gram negative (S. kentucky, E. aerogenes ATCC 13048, P. aeruginosa, K. pneumoniae, E. coli, Proteus) and the fungi C. albicans ATCC 26555 (Table 3).| No. | Type of bacteria | Compound 1 | Compound 2 | Compound 3 | Compound 4 |
|---|---|---|---|---|---|
| 1 | Klebsiella pneumoniae | 6 mm | 9 mm | 14 mm | 14 mm |
| 2 | Staphylococcus aureus ATCC | — | — | — | — |
| 3 | Staphylococcus aureus | — | — | — | — |
| 4 | Proteus | — | — | — | — |
| 5 | E. coli | — | — | — | — |
| 6 | Serratia marcescens | — | — | — | — |
| 7 | Staphylococcus hominis | 8 mm | 8 mm | 17 mm | 8 mm |
| 8 | Staphylococcus epidermidis | 6 mm | — | 19 mm | 6 mm |
| 9 | Alfa Streptococcus haemolyticus | 6 mm | — | 8 mm | 6 mm |
| 10 | Enterococcus faecium | — | — | — | — |
| 11 | Pseudomonas aeroginosa | — | — | — | — |
| 12 | Listeria monocytogenes ATCC 7644 | — | — | — | — |
| 13 | Enterococcus faecium | — | — | — | — |
| 14 | Enterococcus durans | — | — | — | — |
| 15 | Salmonella kentucky | — | — | — | — |
| 16 | Enterobacter aerogenes ATCC 13048 | — | — | — | — |
| 17 | Candida albicans ATCC 26555 | — | — | — | — |
Molecules 1–4 displayed inhibition zone on Gram-positive S. hominis, S. epidermidis, alfa S. haemolyticus and Gram-negative K. pneumoniae (Fig. 5).
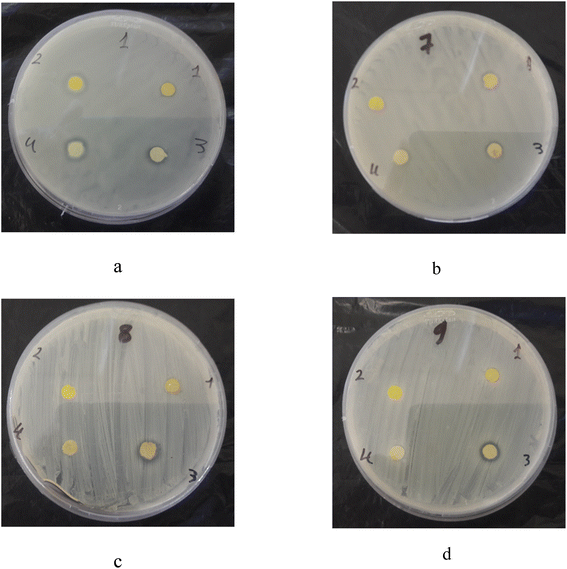 | ||
| Fig. 5 Inhibition zone of 1–4 compounds on (a) Klebsiella pneumoniae, (b) Staphylococcus hominis, (c) Staphylococcus epidermidis, (d) alfa Streptococcus haemolyticus. | ||
The difference between molecules 1 and 2 is that the methoxy group is in the ortho position in molecule 1 and the meta position in molecule 2. When the methoxy group was in the ortho position, an inhibition zone of approximately 8 mm was obtained for S. hominis and 6 mm for S. epidermidis, K. pneumoniae and alpha S. haemolyticus. When the methoxy group was in the meta position, an inhibition zone of about 8 and 9 mm on S. hominis and K. pneumoniae were recorded, respectively, while no effect on S. epidermidis and alpha S. haemolyticus was observed (Fig. 6).
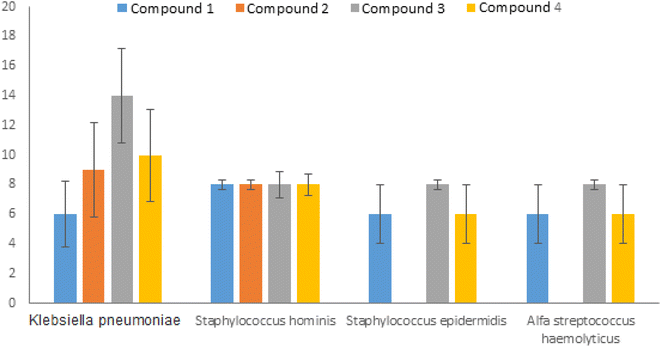 | ||
| Fig. 6 Inhibition zone for compounds on a: Klebsiella pneumoniae, Staphylococcus hominis, Staphylococcus epidermidis, and alfa Streptococcus haemolyticus. | ||
The difference between molecules 1 and 3 is that the methyl group, which is the electron donating group, is attached to the phenyl ring instead of the nitro group, which is the electron withdrawing group.
Molecule 3, to which electron donor groups were bound, exhibited the highest zone of inhibition on S. epidermis (19 mm), S. hominis (17 mm), K. pneumoniae (14 mm) and alfa S. haemolyticus (8 mm). El-Sheshtawy et al.37 synthesized the complex [TiCl2(bzac)(L4)] and reported that is more potent against S. typhimurium, E. aerogenes, S. epidermidis, S. aureus, and M. luteus with 13, 13, 15, 15, and 16.5 mm zone inhibition diameters, respectively. Raj Kaushal et al. also indicated that titanium(IV) complex can bind to calf thymus DNA (CT-DNA) via an intercalative mode.38
Molecule 4 revealed the biggest inhibition zone for K. pneumoniae (14 mm) followed by S. hominis (8 mm), S. epidermidis (6 mm) and alfa S. haemolyticus (6 mm).
In order to investigate the potential activity of the investigated compounds as inhibitors, the molecular docking of the four compounds was performed. The 5-(hydroxyethyl)-methylthiazole kinase (ThiM) of Klebsiella pneumoniae has a key role in bacterial thiamine and salvage pathway. It is essential in the pathway forming thiamine pyrophosphate (TPP) which is and essential co-factor in bacterial amino acid and carbohydrate metabolic pathways. It is considered an attractive antibacterial target due to its absence in humans. Hence, molecular docking study was carried out on ThiM (PDB 6k28) to investigate the compounds inhibitory activity to Klebsiella pneumoniae.
The molecular docking results were reported as 9 poses for each ligand with the corresponding ligand-protein binding affinity. Docking poses were chosen based on the alignment of the compounds' thiadiazole ring over the co-crystallized ligand thiazole ring. The docking results showed docking affinity values for the complex of −6.9, −6.9, −7.1, and −7 kcal mol−1 for compounds 1, 2, 3, and 4, respectively. This result indicates that compound 3 has the highest affinity towards the Klebsiella pneumoniae followed by 4, then 2 and 1, which is in accordance with the experimental results.
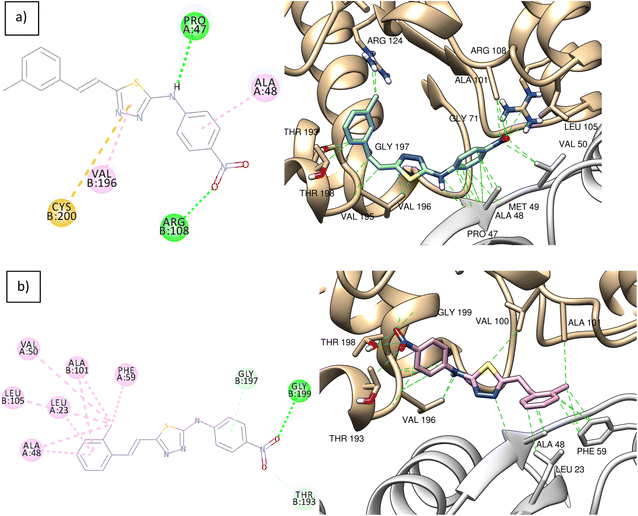
The ligand–protein interactions for compounds 1 and 2 are shown in Fig. 7. In Fig. 7a, Compound 1 is stabilized inside the binding site by a main conventional hydrogen bond between its nitro group and Arg 108 residue of chain B. Another hydrogen bond with Pro 47 of chain A is formed by the compound's amino group. Compound 1 also forms and additional pi–sulphur interaction with its thiadiazole pi system and sulphur group of Cys 200 residue of chain B. Compound 2 forms; a hydrogen bond by its nitro group with Gly 199 residue of chain B. In Fig. 7b, the compound is also stabilized with hydrophobic interactions formed between the compound's o-phenyl group with the protein's hydrophobic pocket formed by residues of Leu 23, Ala 48, Val 50, and phenylalanine 59 residues of chain A, and Ala 101 and Leu 105 residues of chain B.
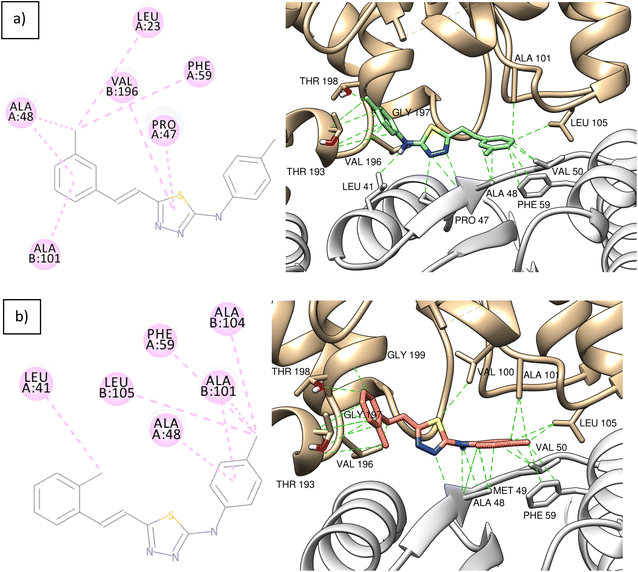
The ligand–protein interactions of compounds 3 and 4 are shown in Fig. 8. Both compounds are stabilized inside the binding site by hydrophobic interactions. In Fig. 8a, compound 3 forms hydrophobic interactions by its m-phenyl group with the hydrophobic pocket formed by residues of alanine 101 and leucine 105 of chain A, and proline 47, alanine 48, valine 50, and phenylalanine 59 of chain B. In Fig. 8b, compound 4 also forms hydrophobic interactions with the same hydrophobic pocket through its p-phenyl group.
3.3 CT-DNA binding
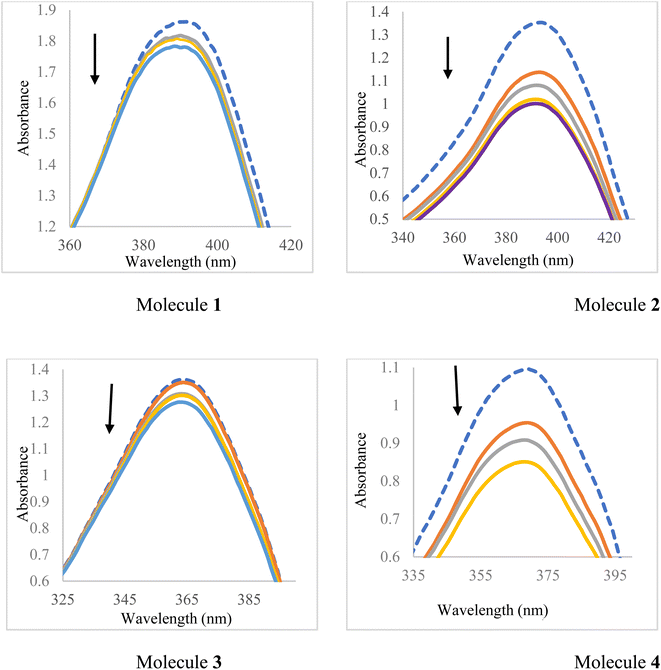
In this section, the binding of the synthesized ligand molecules (1–4) with CT-DNA was assessed by UV-Vis absorption. Generally, DNA plays an important role in the life process as it contains all the genetic information for cellular function. However, DNA molecules are prone to damage under a variety of conditions, such as interactions with certain molecules. This damage can lead to various pathological changes in living organisms since the DNA binding interaction of molecules is of interest for both therapeutic and scientific reasons.39 Absorption spectroscopy is an effective method to examine the binding mode of DNA with micromolecules. Because of compound binding to DNA, the absorbance spectrum shows hypochromism and hyperchromism, which involve a strong stacking interaction between an aromatic chromophore and the base pairs of DNA.40 Thus, in order to provide evidence for the possibility of binding of synthesized ligand molecules (1–4) with CT-DNA, spectroscopic titration of solutions of the CT-DNA with synthesized ligand molecules (1–4) have been performed and the binding constants have been calculated by absorption spectra using Benesi–Hildebrand equation. In general, the hyperchromism and hypochromism were regarded as spectral features for DNA double-helix structural change when DNA reacted with other molecules. The hyperchromism means the breakage of the secondary structure of DNA; the hypochromism originates from the stabilization of the DNA duplex by either the intercalation-binding mode or the electrostatic effect of small molecules.33 The absorption spectra of the constant concentration (40 μM) of the synthesized ligand molecules (1–4) upon addition of different concentration of CT-DNA are shown in Fig. 9. It is apparent from Fig. 9 that the absorption peak of the synthesized ligand molecules (1–4) around at 390 nm, 393 nm, 360 nm, 367 nm, respectively, showed gradual decrease with the increasing concentration of CT-DNA. This was attributed to the electrostatic interaction between the DNA and the small molecules. Slight hypsochromic shifts can be explained by the increasing of energy between π and π* due to repulsion between the non-bonding, electrons localized on the hetero atoms (N, S and O) in both CT-DNA base pairs and the synthetized molecules. The increasing of the energy gap between π and π* explained the low absorptivity of the synthesized molecules in the presence of CT-DNA.16 This explanation is agreed with the values of binding constant Kb of molecules 1–4 are 5.4 × 104, 5.5 × 104, 1.5 × 104 and 3 × 104, respectively. When the methyl group (electron donor group) attached to the phenyl ring instead of the electron withdrawing NO2 group, there is a decrease in the binding constant value. The more repulsion is occurred between the non-bonding electrons in case of the molecules with electron donating groups due to the electron density has been increased on the heteroatoms in the organic ligands.40–544 Conclusions
To sum up, 5-[(E)-2-(3-methoxyphenyl)]vinyl-N-[4′-nitrophenyl]-1,3,4-thiadiazole-2-amine (1), 5-[(E)-2-(2-methoxyphenyl)]vinyl-N-[4′-nitrophenyl]-1,3,4-thiadiazole-2-amine (2), 5-[(E)-2-(3-methoxyphenyl)]vinyl-N-[4′-methylphenyl]-1,3,4-thiadiazole-2-amine (3), and 5-[(E)-2-(2-methoxyphenyl)]vinyl-N-[4′-methylphenyl]-1,3,4-thiadiazole-2-amine (4) were synthesised. The DFT calculations have been used to explore the geometric and spectroscopic properties of the molecules. Derivatives 1–4 showed good antibacterial activity against Klebsiella pneumoniae and Staphylococcus hominis, where 1, 3 and 4 were more effective against Staphylococcus epidermidis and alfa Streptococcus haemolyticus. The docking study on Kinase ThiM from Klebsiella pneumoniae shows the formation of hydrogen bonding with the receptor residue, which confirms the inhibition effect of the ligands. The collected results revealed that molecules 1–4 interact with CT-DNA by intercalative mechanism. The binding ability study of the synthesized molecules with CT-DNA revealed that nitro groups attached to the phenyl group slightly increased the binding constant compared to methyl groups.Conflicts of interest
There are no conflicts to declare.Acknowledgements
The authors would like to thank Princess Nourah bint Abdulrahman University Researchers Supporting Project number (PNURSP2022R9), Princess Nourah bint Abdulrahman University, Riyadh, Saudi Arabia for supporting this project.References
- U. Kalidhar and A. Kaur, 1,3,4-Thiadiazole derivatives and their biological activities: A review, Res. J. Pharm., Biol. Chem. Sci., 2011, 2, 1091–1106 CAS.
- S. Georgeta, S. Oana, S. Eugenia and B. Sanda, 2-Amino-1,3,4-thiadiazole as a potential scaffold for promising antimicrobial agents, Drug Des., Dev. Ther., 2018, 12, 1545–1566 CrossRef PubMed.
- S. Haider, M. S. Alam and H. Hamid, 1,3,4-Thiadiazoles: A potent multi targeted pharmacological scaffold, Eur. J. Med. Chem., 2015, 92, 156–177 CrossRef CAS PubMed.
- M. Amir, A. Kumar, I. Ali and S. A. Khan, Synthesis of pharmaceutically important 1,3,4-thiadiazole and imidazolinone derivatives as antimicrobials, Indian J. Chem., 2009, 48B, 1288–1293 CAS.
- J. Salimon, N. Salih, A. Hameed, H. Ibraheem and E. Yousif, Synthesis and antibacterial activity of some new 1,3,4-oxadiazole and 1,3,4-thiadiazole derivatives, J. Appl. Sci. Res., 2010, 6(7), 866–870 CAS.
- E. E. Oruc, S. Rollas, F. Kandemirli, N. Shvets and A. Dimoglo, 1,3,4-Thiadiazole derivatives. Synthesis, structure elucidation and structure-antituberculosis activity relationship investigation, J. Med. Chem., 2004, 47(27), 6760–6767 CrossRef CAS PubMed.
- S. Schenone, C. Brullo and O. Bruno, et al., New 1,3,4-thiadiazole derivatives endowed with analgesic and anti-inflammatory activities, Bioorg. Med. Chem., 2006, 14(6), 1698–1705 CrossRef CAS PubMed.
- L. Labanauskas, V. Kalcas, E. Udrenaite, P. Gaidelis, A. Brukstus and A. Dauksas, Synthesis of 3-(3,4-dimethoxy phenyl)-1H-1,2,4-triazole-5-thiol and 2-amino-5-(3,4-dimethoxy phenyl)-1,3,4-thiadiazole derivatives exhibiting anti-inflammatory activity, Pharmazie, 2001, 56, 617–619 CAS.
- H. Khalilullah, M. U. Khan, D. Mahmood, J. Akhtar and G. O. Elhassan, 1,3,4-Thiadiazole: A biologically active scaffold, Int. J. Pharm. Pharm. Sci., 2014, 6, 8–15 CAS.
- S. R. Pattan, N. S. Desai, P. A. Rabara, A. A. Bukitgar and V. A. Wakale, Synthesis and antimicrobial evaluation of some 1,3,4-thiadiazole derivatives, Indian J. Pharm. Educ. Res., 2008, 42(4), 314–318 Search PubMed.
- A. Basso, M. Liu and C. Dai, et al., SCH 2047069, a novel oral kinesin spindle protein inhibitor, shows single-agent antitumor activity and enhances the efficacy of chemotherapeutics, Mol. Cancer Ther., 2010, 9(11), 2993–3002 CrossRef CAS PubMed.
- S. S. Taha, A. A. Ahmad, S. I. Mawlood and N. O. Ali, Synthesis of some series of 2-amino-1,3,4-thiadiazole derivatives with their pathogenic bacterial activity, J. Raparin Univ., 2017, 4(11), 63–78 Search PubMed.
- A. A. Kadi, E. S. Al-Abdullah, I. A. Shehata, E. E. Habib, T. M. Ibrahim and A. A. El-Emam, Synthesis, antimicrobial and anti-inflammatory activities of novel 5-(1-adamantyl)-1,3,4-thiadiazole derivatives, Eur. J. Med. Chem., 2010, 45(11), 5006–5011 CrossRef CAS PubMed.
- M. Gür, M. Muğlu, M. S. Çavuş, A. Güder, H. S. Sayiner and F. Kandemirli, Synthesis, characterization, quantum chemical calculations and evaluation of antioxidant properties of 1,3,4-thiadiazole derivatives including 2-and 3-methoxy cinnamic acids, J. Mol. Struct., 2017, 1134, 40–50 CrossRef.
- A. A. Othman, M. Kihel and S. Amara, 1,3,4-Oxadiazole, 1,3,4-thiadiazole and 1,2,4-triazole derivatives as potential antibacterial agents, Arabian J. Chem., 2014, 12, 1660–1675 CrossRef.
- J. Y. Chou, S. Y. Lai, S. L. Pan, G. M. Jow, J. W. Chern and J. H. Guh, Investigation of anticancer mechanism of thiadiazole-based compound in human nonsmall cell lung cancer A549 cells, Biochem. Pharmacol., 2003, 66, 115–124 CrossRef CAS PubMed.
- A. Kaur, R. Kumar and U. Kalidhar, Synthesis, spectral studies and biological activity of some novel biphenyl imidazo[2,1-b][1,3,4]thiadiazole derivatives, Res. J. Pharm., Biol. Chem. Sci., 2012, 3, 1084–1096 CAS.
- S. Georgeta, S. Oana, S. Eugenia and B. Sanda, Drug Des., Dev. Ther., 2018, 12, 1545–1566 CrossRef PubMed.
- N. Kushwaha, S. K. S. Kushwaha and A. K. Rai, Biological activities of thiadiazole Derivatives: A Review, Int. J. Chem. Res., 2012, 4, 517–531 CAS.
- T. A. Farghaly, M. A. Abdallah and M. R. A. Aziz, Synthesis and antimicrobial activity of some new 1,3,4-thiadiazole derivatives, Molecules, 2012, 17, 14625–14636 CrossRef CAS PubMed.
- R. Bhatia and A. Kaur, Synthesis, spectral studies and antimicrobial activity of some imidazo [2,1-b] [1,3,4] thiadiazole derivatives, Der Pharma Chem., 2014, 6, 114–120 CAS.
- O. E. Ozkan, G. Zengin, M. Akça, M. C. Baloğlu, Ç. Olgun, E. M. Altuner, S. Ateş, A. Aktümsek and H. Vurdu, DNA protection, antioxidant, antibacterial and enzyme inhibition activities of heartwood and sapwood extracts from juniper and olive woods, RSC Adv., 2015, 5, 72950–72958 RSC.
- N. Sharath, S. Halehatty, B. Naik, B. N. Kumar and J. Hoskeri, Antibacterial, molecular docking, DNA binding and photocleavage studies on novel heterocyclic pyrazoles, Br. J. Pharm. Res., 2011, 1, 46–65 CrossRef.
- P. Pakravan and S. Masoudian, Study on the interaction between isatin-β-thiosemicarbazone and Calf Thymus DNA by spectroscopic techniques, Iran. J. Pharm. Res., 2015, 14, 111–123 CAS.
- N. Hadjiliadis and E. Sletten, Metal complex–DNA interactions, Blackwell Publishing Ltd, 2009, pp. 138–129, https://onlinelibrary.wiley.com/doi/book/10.1002/9781444312089 Search PubMed.
- H. A. Benesi and J. H. Hildebrand, A spectrophotometric investigation of the interaction of iodine with aromatic hydrocarbons, J. Am. Chem. Soc., 1949, 71, 2703–2707 CrossRef CAS.
- M. J. Frisch, G. W. Trucks, H. B. Schlegel, G. E. Scuseria, M. A. Robb, J. R. Cheeseman, G. Scalmani, V. Barone, B. Mennucci and G. A. Petersson, Gaussian 09, Revision d1, Inc., Wallingford, CT, USA, 2009 Search PubMed.
- J. L. Gázquez, A. Cedillo and A. Vela, Electrodonating and Electroaccepting Powers, J. Phys. Chem. A, 2007, 111, 1966–1970 CrossRef PubMed.
- O. Trott and A. J. Olson, AutoDock Vina: Improving the speed and accuracy of docking with a new scoring function, efficient optimization, and multithreading, J. Comput. Chem., 2010, 31, 455–461 CAS.
- K. Schöning-Stierand, K. Diedrich, R. Fährrolfes, F. Flachsenberg, A. Meyder, E. Nittinger, R. Steinegger and M. Rarey, ProteinsPlus: interactive analysis of protein–ligand binding interfaces, Nucleic Acids Res., 2020, 48, W48–W53 CrossRef PubMed.
- R. Fährrolfes, S. Bietz, F. Flachsenberg, A. Meyder, E. Nittinger, T. Otto, A. Volkamer and M. Rarey, ProteinsPlus: a web portal for structure analysis of macromolecules, Nucleic Acids Res., 2017, 45, W337–W343 CrossRef PubMed.
- M. Er, B. Ergüven, H. Tahtaci, A. Onaran, T. Karakurt and A. Ece, Synthesis, characterization, preliminary SAR and molecular docking study of some novel substituted imidazo[2,1-b][1,3,4]thiadiazole derivatives as antifungal agents, Med. Chem. Res., 2017, 26, 615–630 CrossRef CAS.
- M. Er, A. Özer, Ş. Direkel, T. Karakurt and H. Tahtaci, Novel substituted benzothiazole and Imidazo[2,1-b][1,3,4]Thiadiazole derivatives: Synthesis, characterization, molecular docking study, and investigation of their in vitro antileishmanial and antibacterial activities, J. Mol. Struct., 2019, 1194, 284–296 CrossRef CAS.
- H. S. El-Sheshtawy and I. El-Mehasseb, Role of halogen and hydrogen bonds for stabilization of antithyroid drugs with hypohalous acids (HOX, X = I, Br, and Cl) adducts, J. Mol. Struct., 2017, 1147, 643–650 CrossRef CAS.
- Q.-L. Zhang, J.-G. Liu, H. Chao, G.-Q. Xue and L.-N. Ji, DNA binding and photocleavage studies of cobalt(III) polypyridyl complexes: [Co(phen)2IP]3+ and [Co(phen)2PIP]3+, J. Inorg. Biochem., 2001, 83(1), 49–55 CrossRef CAS PubMed.
- H. S. El-Sheshtawy, M. M. Ibrahim, I. El-Mehasseb and M. El-Kemary, Orthogonal hydrogen/halogen bonding in 1-(2-methoxyphenyl)-1H-imidazole-2(3H)-thione-I2 adduct: An experimental and theoretical study, Spectrochim. Acta, Part A, 2015, 143, 120–127 CrossRef CAS PubMed.
- H. S. El-Sheshtawy, H. M. A. Salman and M. El-Kemary, Halogen vs Hydrogen Bonding in Thiazoline-2-thione Stabilization with σ- and π-Electron Acceptors Adducts: Theoretical and Experimental study, Spectrochim. Acta, Part A, 2015, 137, 442–449 CrossRef CAS PubMed.
- R. Kaushal, S. Thakur and K. Nehra, ct-DNA Binding and Antibacterial Activity of Octahedral Titanium (IV) Heteroleptic (Benzoylacetone and Hydroxamic Acids) Complexes, Int. J. Med. Chem., 2016, 2361214 Search PubMed.
- H. S. El-Sheshtawy, M. M. Ibrahim, M. R. E. Aly and M. El-Kemary, Spectroscopic and structure investigation of the molecular complexes of tris(2-aminoethyl)amine with π-acceptors, J. Mol. Liq., 2016, 213, 82–91 CrossRef CAS.
- M. S. Attia, M. N. Ramsis, L. H. Khalil and S. G. Hashem, J. Fluoresc., 2012, 22, 779–788 CrossRef CAS PubMed.
- M. S. Attia, A. O. Youssef, Z. A. Khan and M. N. Abou-Omar, Talanta, 2018, 186, 36–43 CrossRef CAS PubMed.
- M. S. Attia, A. O. Youssef and A. A. Essawy, Anal. Methods, 2012, 4, 2323–2328 RSC.
- M. S. Attia, K. Ali, M. El-Kemary and W. M. Darwish, Talanta, 2019, 201, 185–193 CrossRef CAS PubMed.
- M. S. Attia, W. H. Mahmoud, A. O. Youssef and M. S. Mostafa, J. Fluoresc., 2011, 21, 2229–2235 CrossRef CAS PubMed.
- M. S. Attia, M. H. Khalil, M. S. A. Abdel-Mottaleb, M. B. Lukyanova, Yu. A. Alekseenko and B. Lukyanov, Int. J. Photoenergy, 2006, 1–9 CrossRef.
- M. S. Attia, W. H. Mahmoud, M. N. Ramsis, L. H. Khalil, A. M. Othman, S. G. Hashem and M. S. Mostafa, J. Fluoresc., 2011, 21, 1739–1748 CrossRef CAS PubMed.
- M. S. Attia, A. M. Othman, E. Elraghi and H. Y. Aboul-Enein, J. Fluoresc., 2011, 21, 739–745 CrossRef CAS PubMed.
- A. A. Elabd and M. S. Attia, J. Lumin., 2016, 169, 313–318 CrossRef CAS.
- M. S. Attia, S. A. Elsaadany, K. A. Ahmed, M. M. El-Molla and M. S. A. Abdel-Mottaleb, J. Fluoresc., 2015, 25, 119–125 CrossRef CAS PubMed.
- S. G. Hashem, M. M. Elsaady, H. G. Afify, M. El-Kemary and M. S. Attia, Talanta, 2019, 199, 89–96 CrossRef CAS PubMed.
- M. S. A. Abdel-Mottaleb, M. Saif, M. S. Attia, M. M. Abo-Aly and S. N. Mobarez, Photochem. Photobiol. Sci., 2018, 17, 221–230 CrossRef CAS PubMed.
- W. E. Omer, M. A. El-Kemary, M. M. Elsaady, A. A. Gouda and M. S. Attia, ACS Omega, 2020, 5, 5629–5637 CrossRef CAS PubMed.
- L. M. Abdullah, M. S. Attia and M. S. A. Abdel-Mottaleb, Egypt. J. Chem., 2019, 62, 247–255 Search PubMed.
- M. S. Attia, A. O. Youssef, A.-S. S. H. Elgazwy, S. M. Agami and S. I. Elewa, J. Fluoresc., 2014, 24, 759–765 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |