 Open Access Article
Open Access ArticleExosome-based drug delivery systems and their therapeutic applications
Jaewook Lee
 a,
Ji-Heon Lee
a,
Kushal Chakraborty
b,
Joon Hwang
ac and
Yong-Kyu Lee
*ad
a,
Ji-Heon Lee
a,
Kushal Chakraborty
b,
Joon Hwang
ac and
Yong-Kyu Lee
*ad
a4D Convergence Technology Institute (National Key Technology Institute in University), Korea National University of Transportation, Jeungpyeong, Chung-Buk 27909, Republic of Korea. E-mail: leeyk@ut.ac.kr; Tel: +82-43-841-5224
bDepartment of IT and Energy Convergence (BK21 FOUR), Korea National University of Transportation, Chungju, Chung-Buk 27469, Republic of Korea
cDepartment of Aeronautical & Mechanical Design Engineering, Korea National University of Transportation, Chungju, Chung-Buk 27469, Republic of Korea
dDepartment of Chemical and Biological Engineering, Korea National University of Transportation, Chungju, Chung-Buk 27469, Republic of Korea
First published on 23rd June 2022
Abstract
In the past few decades, scientists have actively worked on developing effective drug delivery systems (DDSs) as means to control life-threatening diseases and challenging illnesses. In order to develop such DDSs, nanobiotechnological strategies have been introduced, and many nanomaterial-based DDS platforms have been proposed. Among these nanomaterials, DDSs based on exosomes and hybrids of exosomes have been focused upon and developed due to their low toxicity, high bioactivity, and biocompatibility. In this review, we describe the processes involved in drug loading into exosomes and the surface modification of exosomes with treatment agents. Furthermore, we describe the synthesis methods of hybrid exosomes with organic or inorganic nanoparticles. Moreover, we focus on the effective therapeutic applications of exosome-based DDSs against various diseases. In conclusion, we show that exosomes and hybrids of exosomes show excellent drug carrier potential and capacity.
1 Introduction
Nanoparticles (NPs) have been utilized in various applications and in different fields ranging from cosmetics to nanobiomedicine and drug delivery systems (DDSs).1–4 NP-based DDSs have gained special attention, as they have been designed to effectively transport various therapeutic agents to target diseases of different organ systems.5–8 Both organic and inorganic NPs have been used in DDSs. Inorganic NPs composed of various materials, including gold, silver, iron oxide, and silicates, have been loaded with drugs and used for drug delivery to treat different diseases.9–13 Similarly, treatment agents have been encapsulated into organic NPs like polymeric NPs and liposomes in order to enhance the healing effect.14–19 Although these NPs have been described as potential drug carriers, a major and critical limitation to their application in the biomedical and biotechnological fields is their toxicity profile.20–23Recently, many scientists have shifted focus to exosomes as new potential drug carriers and attempted the development of exosome-based DDSs.24–29 Exosomes are a bioproduct that can be obtained from various cell lines such as immune cells, cancer cells, or stem cells as well as from body fluids, including blood and cerebrospinal fluid.30–36 Given their source, exosomes are less toxic. The surface of exosomes is composed of a lipid bilayer membrane with embedded tetraspanins, glycoproteins, and signaling receptors, encapsulating DNA and micro RNA.37–39 Interestingly, the characteristics of exosomes vary depending on their origin, so they can also be utilized in biopsy-based diagnosis as biomarkers.40–45 Moreover, several researchers have modified the surface or nucleic acid content of exosomes through cell engineering to expand their applications in nanobiotechnology, especially in the field of DDSs.46–50
Based on the valuable efforts of researchers, various types of exosomal DDSs have been developed and produced by several strategies using both physical and chemical processes and approaches. Many kinds of drugs have been either encapsulated into exosomes or surface-modified and have revealed substantial treatment effects in various diseases. Furthermore, an advanced structure called a hybrid exosome was also successfully developed to improve the therapeutic applications.51–55
Therefore, many start-up companies have been established in Korea and worldwide to work on exosome-based DDSs; some of these companies have already started clinical trials using exosomes and their engineered structure in various fields like diabetes treatment, bone treatment, skin regeneration, and anti-cancer therapy.56–60
In this review article, we discuss the methods for the preparation of exosome-based DDSs through encapsulation and loading of drugs into exosomes as well as the synthesis methods of hybrid exosomes through diverse approaches. We also describe the effects of treatment using exosome-based DDSs in different diseases.
2 Modification methods for exosome-based DDSs
Many studies have been conducted to develop various strategies for encapsulation or modification of drugs with exosomes in order to obtain high performance exosome-based DDS.61 Drug-loaded exosome systems have been prepared through physical processes or chemical modification or cell engineering methods. For the case of physical encapsulation methods including electroporation, sonication, extrusion, freeze/thaw method, they are facile methods and high molecular weight cargo loading is possible. Meanwhile they can disturb the exosome integrity and account for relatively lower loading efficacy. It meant the exosome carrier could get the damage through the physical stimulation, so recovery time should be required. On the other hands, the exosome-based drug carrier could be tuned without structure damage through chemical modification process and precise control could be possible. However, by product and side reaction should be avoid for effective nano bio application. The exosome-based DDS could be produced by cell engineering approaches, and in this case, advantage of various biomolecules transportation could be achieved. But the methodology is complicated and function of various biomolecules should be defined before modification. In addition, encapsulation methods should be selected depending on the property of the drug. Hydrophobic drugs can penetrate into the exosomes via mixing due to the hydrophobic interactions between the drug and the lipid bilayer of the exosomes. However, hydrophilic drugs cannot enter exosomes due to their inability to penetrate the lipid bilayer. Therefore, other loading or modification processes are required to force this penetration such as physical stimulations using pressure, sonication, or electrical force or chemical coupling reactions between the drugs and the surface of the exosomes.2.1 Encapsulation process of therapeutic agents into exosome
Many researchers have tried to load or encapsulate drugs into the exosomes via several physical methods including ultrasonication, electroporation, extrusion, agitation. However, the structure of exosomes can be damaged via such external stimulations. Therefore, the exosome encapsulation processes should be carefully conducted under well-established conditions.Haney et al. successfully loaded a catalase into an exosome through sonication, extrusion, and other methods.62 In a study, they used exosomal DDS for delivering a therapeutic agent for Parkinson's disease, where the loading efficiency was characterized by catalase enzymatic activity with H2O2. The results showed that sonication had the highest loading efficiency (26.1 ± 2%) compared to that of other processes, such as extrusion (22.2 ± 2%), freeze/thaw method (14.7 ± 1.1%), and incubation (4.9 ± 2%). Another study on sonication-induced encapsulation of poly(lactic-co-glycolic acid) (PLGA) into an exosome was carried out by Liu et al., where the exosome was isolated from A549 cells (human lung carcinoma cell line);63 an in situ encapsulation process was used, wherein the exosome and PLGA were mixed in microfluidic channels, and then coating was performed in the sonication area under a frequency of 80 kHz frequency and a power of 100 W (Fig. 1A). The final product was collected at the outlet channel, and 90.5% of EM-PLGA NPs showed a typical core–shell structure as per transmission electron microscopy (TEM) (Fig. 1B). Additionally, in this study, an EM-(dye/PLGA) hybrid structure was also developed. Interestingly, this cancer cell-derived exosome coating the core of an imaging dye accumulated into cancer cells in mice, by a homotypic targeting effect without off-target labeling, and allowed fluorescence imaging of the target organ.
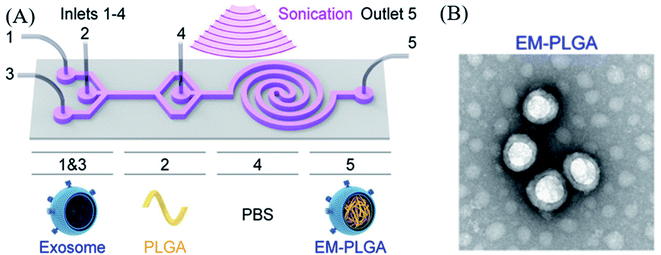 | ||
| Fig. 1 (A) Schematic illustration of the sonication-assisted encapsulation process for PLGA-loaded exosome structure (EM-PLGA) and (B) TEM image of EM-PLGA. Reprinted with permission.63 Copyright 2019, American Chemical Society. | ||
A study by Wang et al. focused on the synthesis of paclitaxel (PTX)-loaded exosome as an anti-cancer DDS.64 They isolated exosomes from RAW264.7 macrophage cells and then mixed them with PTX in a 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 6 ratio (m m−1). The mixture was sonicated at 20% amplitude, with 6 cycles of 30 s on/off for 3 min (with 2 min cooling time between cycles). The loading efficiency was measured by High Performance Liquid Chromatography (HPLC), and it was found to be 19.55 ± 2.48%. Compared to that of a simple agitation process, the loading efficiency of PTX was 4 times higher in case of sonication. The authors rationalized that this improved loading efficiency could be because the transient pores that are created during sonication allow the drug to diffuse into the exosomes. The tetraspanin quantity in the exosomes was clearly measured after sonication, and it confirmed that the exosome structure was maintained after sonication. In a different study, Yu et al. loaded erastin into an HFL-1 cell-derived (human lung fibroblast cell line) exosome through sonication, to use it as an anti-cancer drug.65 The authors evaluated the loading amount of the drug via HPLC, which was found to be 3.2 mg erastin per mg protein. Furthermore, in another study, insulin was encapsulated into an exosome through electroporation for use in diabetes treatment.66 The authors used exosomes derived from HepG2 (hepatocellular carcinoma), primary dermal fibroblasts (HDFa), and pancreatic islet cell tumor (RIN-m) to prepare the drug carrier. Electroporation was carried out at 200 V and 50 μF in order to encapsulate insulin. The results showed that the loading efficiency was estimated to be 50.75 ± 1.2%, 57.42 ± 5.47%, and 49.70 ± 4.32% for HepG2, HDFa, and RIN-m exosomes, respectively. Similarly, Tian et al. reported the encapsulation of doxorubicin (DOX) into an exosome via electroporation to produce an anti-cancer agent delivery platform.67 The exosome was collected from imDC (mouse immature dendritic cells) culture medium, and DOX was loaded into the imDC-derived exosome by electroporation at 350 V and 150 μF. During this process, some pores on the surface of the exosome were created by electrical stimulation, so DOX could enter more freely into the exosome. However, an additional 30 min of incubation of the product at 37 °C was needed to recover the surface of the exosome. The loading efficiency was estimated by the measurement of the fluorescence intensity of DOX with fluorescence spectroscopy (excitation: 480 nm; emission: 594 nm), and it was reported to be approximately 20%. Liang et al. also developed exosomal DDS via electroporation using an engineered exosome, 5-fluorouracil (5-FU), and miR-21i (miRNA) for cancer treatment (Fig. 2).68 In this study, the authors prepared the exosome that was engineered by Her 2 affibody to improve cancer cell targeting. Subsequently, 5-FU and miR-21i were co-encapsulated into the engineered exosome by electroporation at 1000 V for 10 ms. The results showed that the loading efficiency of 5-FU and miR-21i was approximately 3.1% and 0.5%, respectively. Although the loading efficiency was slightly low, co-delivering RNA and a drug by an exosome exhibited the potential for efficient and effective cancer treatment.
6 ratio (m m−1). The mixture was sonicated at 20% amplitude, with 6 cycles of 30 s on/off for 3 min (with 2 min cooling time between cycles). The loading efficiency was measured by High Performance Liquid Chromatography (HPLC), and it was found to be 19.55 ± 2.48%. Compared to that of a simple agitation process, the loading efficiency of PTX was 4 times higher in case of sonication. The authors rationalized that this improved loading efficiency could be because the transient pores that are created during sonication allow the drug to diffuse into the exosomes. The tetraspanin quantity in the exosomes was clearly measured after sonication, and it confirmed that the exosome structure was maintained after sonication. In a different study, Yu et al. loaded erastin into an HFL-1 cell-derived (human lung fibroblast cell line) exosome through sonication, to use it as an anti-cancer drug.65 The authors evaluated the loading amount of the drug via HPLC, which was found to be 3.2 mg erastin per mg protein. Furthermore, in another study, insulin was encapsulated into an exosome through electroporation for use in diabetes treatment.66 The authors used exosomes derived from HepG2 (hepatocellular carcinoma), primary dermal fibroblasts (HDFa), and pancreatic islet cell tumor (RIN-m) to prepare the drug carrier. Electroporation was carried out at 200 V and 50 μF in order to encapsulate insulin. The results showed that the loading efficiency was estimated to be 50.75 ± 1.2%, 57.42 ± 5.47%, and 49.70 ± 4.32% for HepG2, HDFa, and RIN-m exosomes, respectively. Similarly, Tian et al. reported the encapsulation of doxorubicin (DOX) into an exosome via electroporation to produce an anti-cancer agent delivery platform.67 The exosome was collected from imDC (mouse immature dendritic cells) culture medium, and DOX was loaded into the imDC-derived exosome by electroporation at 350 V and 150 μF. During this process, some pores on the surface of the exosome were created by electrical stimulation, so DOX could enter more freely into the exosome. However, an additional 30 min of incubation of the product at 37 °C was needed to recover the surface of the exosome. The loading efficiency was estimated by the measurement of the fluorescence intensity of DOX with fluorescence spectroscopy (excitation: 480 nm; emission: 594 nm), and it was reported to be approximately 20%. Liang et al. also developed exosomal DDS via electroporation using an engineered exosome, 5-fluorouracil (5-FU), and miR-21i (miRNA) for cancer treatment (Fig. 2).68 In this study, the authors prepared the exosome that was engineered by Her 2 affibody to improve cancer cell targeting. Subsequently, 5-FU and miR-21i were co-encapsulated into the engineered exosome by electroporation at 1000 V for 10 ms. The results showed that the loading efficiency of 5-FU and miR-21i was approximately 3.1% and 0.5%, respectively. Although the loading efficiency was slightly low, co-delivering RNA and a drug by an exosome exhibited the potential for efficient and effective cancer treatment.
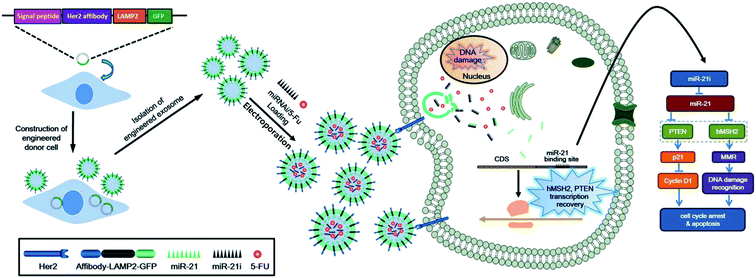 | ||
| Fig. 2 Preparation scheme of engineered exosome-based DDS and its treatment application. Reprinted with permission.68 Copyright 2020, BioMed Central. | ||
Li et al. synthesized an anti-cancer drug-loaded exosome through agitation and dialysis.69 In this study, DOX was encapsulated into exosomes derived from LIM1215 cells (colorectal cancer (CRC) cell line) by incubation, and the loading capacity was estimated by detection of fluorescence of DOX with microplate leader, where it was determined to be around 9.06%. Similarly, Zhuang et al. developed anti-inflammatory drug-loaded exosomes that were prepared by incubation and agitation.70 According to this study, curcumin was encapsulated into the EL4 cell-derived exosome by incubation for 5 min at 22 °C, followed by purification through sucrose gradient (8, 30, 45, and 60%, respectively), and then centrifugation for 1.5 h at 36![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000 rpm. In another study, PTX was loaded into LNCaP cells (prostate cancer cells) derived exosomes by incubation.71 In this case, Saari et al. mixed PTX in DMSO and isobutanol (1
000 rpm. In another study, PTX was loaded into LNCaP cells (prostate cancer cells) derived exosomes by incubation.71 In this case, Saari et al. mixed PTX in DMSO and isobutanol (1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 ratio) with the exosomes, and the mixture was incubated at 22 °C for 1 h. The loading efficiency was measured using Ultra Performance Liquid Chromatography (UPLC) and was estimated to be 9.2 ± 4.5%. In other studies, porphyrin was encapsulated by extrusion into exosomes derived from various cells, including MDA-MB231 (breast cancer cells), hESC (human embryonic stem cells), and hMSC (human mesenchymal stem cells).72
1 ratio) with the exosomes, and the mixture was incubated at 22 °C for 1 h. The loading efficiency was measured using Ultra Performance Liquid Chromatography (UPLC) and was estimated to be 9.2 ± 4.5%. In other studies, porphyrin was encapsulated by extrusion into exosomes derived from various cells, including MDA-MB231 (breast cancer cells), hESC (human embryonic stem cells), and hMSC (human mesenchymal stem cells).72
Porphyrin loading was conducted at 42 °C using a mini extruder with a polycarbonate membrane, which had 400 nm-sized pores, and extrusion was repeated 31 times in each group. In this study, the loading efficiency was associated with the degree of hydrophobicity.
Interestingly, not only chemical drug but also protein drug were modified with exosome via various approaches. And protein drug involved exosome DDSs were described in the next part.
In another study, Shi and colleagues prepared synthetic multivalent antibodies retargeted exosome (SMART-Exo) for breast cancer treatment.73 In this case, HER-2 Ab and CD3 Ab were attached on the surface of exosome and this SMART-Exo was isolated from anti-CD3–anti-HER2 bispecific scFv Ab transfected Expi293 cell, and the existence of two types Ab was confirmed by ELISA. The SMART-Exos could exhibit dually targeting T cell CD3 and breast cancer associated HER2 receptors and as a result, specific anti-tumor activity and immunotherapy were achieved.
Some research groups reported that the protein modification process with exosome was taken a place through the incident light. For example, Yim and co-worker introduced ‘exosomes for protein loading via optically reversible protein–protein interactions’ (EXPLORs) technique.74 Briefly, they prepared target protein and light sensitive protein conjugation structure which was cargo protein and CRY2 protein (cargo–CRY2). And such conjugated protein was co-cultured with HEK293T under blue light (460 nm) irradiation environment. During the incubation, CRY2 part of cargo–CRY2 was activated by blue light in the media and this activated part was conjugated with CIBN part of CIBN modified CD9 (CIBN-CD9) at the surface of the cell. Subsequently, endocytosis of conjugated protein was occurred, and then exosome structure was created in the cell. Finally, cargo protein encapsulated exosomes were released from the cell and collected for further application. And in this study, Cre recombinase, Bax and super-repressor IkB were encapsulated as cargo protein into the exosome through EXPLORs method and as results, these kinds of protein were effectively delivered to target cell, so it showed novel protein carrier function. The other case of incident light assisted protein delivery was introduced by Cheng and colleagues.75 Firstly, the target cargo protein was modified with photocleavable protein (PhoCl) and CD9 by genetically linking in the Expi293F cell which was transfected with expression constructs. And then, target protein included exosomes were isolated and collected by UC. Interestingly, such linked cargo protein with PhoCl in the exosome could be disassembled by incident UV light (408 nm), and finally cargo protein could be delivered and released into the target cell effectively.
2.2 Surface modification of exosomes with therapeutic agents
To prevent damage to exosomes during the drug encapsulation process via physical processes, some researchers have explored alternative strategies and modified the surface of exosomes with therapeutic agents using chemical reactions. However, it is essential that such reactions are conducted with precise and well-established methods to reduce side reactions and by products.Kim et al. studied surface modification of an exosome with an anti-cancer drug, where DOX was attached on to the surface of the exosome by the assistance of phenylboronic acid (BPA).76 First, BPA was chemically conjugated on the surface of the exosome through EDC/NHS coupling reaction, and then DOX was added to induce binding with BPA on the exosome. The DOX loading efficiency was estimated to be 36.7%, using fluorescence spectroscopy with excitation wavelength at 470 nm and emission at 595 nm.
In a previous study, the surface of an exosome was also functionalized with an anti-cancer drug by a streptavidin (strep)–biotin coupling reaction.77 According to the study by Kim et al., the surface of a serum-derived exosome was conjugated with biotin through an EDC/NHS (1-ethyl-3-[3-dimethylamino-propy]carbodiimide hydrochloride/N-hydroxysuccinimide) coupling reaction, and then strep was mixed with the biotin-exosome mixture to obtain the strep–biotin–exosome complex (strep–bio–exo). Meanwhile, DOX, as an anti-cancer drug, was intercalated into the double-stranded biotin-imotif/flare duplex (ds-i-motif-bio) in order to tune the drug releasing behavior, depending on the pH. Subsequently, strep–bio–exo was mixed with ds-i-motif-bio-DOX, and the two precursors were combined via strep–biotin interaction to produce a pH-sensitive DOX delivery system. The release of DOX from the exosomal complex structure was monitored, and it was shown that about 17.5 ± 1.5 and 21.0 ± 4.9% of DOX was released at an acidic pH after 1 and 3 h, respectively.
Zhou et al. synthesized an anti-PD-L1 (aPD1) modified exosome as an immunotherapy agent.78 First, the authors conjugated aPD1 with NHS-functionalized PEG (aPD1-PEG). Next, macrophage-derived exosome-mimetic nanovesicles (EMVs) were mixed with aPD1-PEG for 2 h. Interestingly, AB680 was also encapsulated into the EMVs by a mini extruder in order to inhibit CD73, which is related to the activation of tumor reactive T-cells and natural killer cells (Fig. 3). In this study, encapsulation efficiency of AB680 was estimated to be 61.0 ± 3.1%, and the graft ratio of aPDL1 on the surface of the EMVs was also determined to be approximately 5.47 ± 0.36% (w/w) under a 2.5 weight ratio of EMVs to aPDL1 (mg protein per mg, w/w).
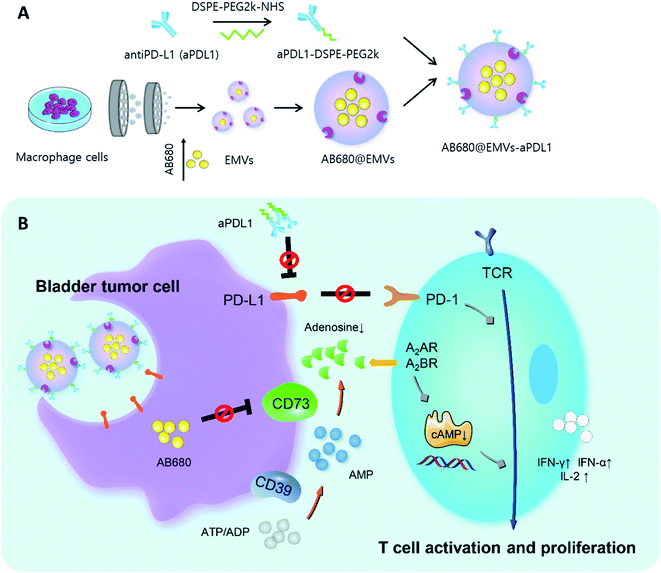 | ||
| Fig. 3 (A) Diagram representing synthesis of AB680-EMVs-aPDL1 and (B) its immunotherapy mechanism. Reprinted with permission.78 Copyright 2020, American Chemical Society. | ||
Examples of exosome-based DDSs are listed in Table 1.
| Exosome origin | Encapsulated agent | Loading methodology | Loading efficiency (LE)/loading amount (LA) | Application | Ref. |
|---|---|---|---|---|---|
| Raw 264.7 | Catalase | (1) Incubation, (2) incubation with saponin, (3) freeze–thaw cycles, (4) sonication, (5) extrusion | LE: (1) 4.9 ± 0.5%, (2) 18.5 ± 1.3%, (3) 14.7 ± 1.1%, (4) 26.1 ± 1.2%, (5) 22.2 ± 3.1% | Parkinson's disease treatment | 62 |
| A549 | Dye/PEG | Sonication under microfluidics | N/A | Cancer therapy and image | 63 |
| RAW 264.7 | PTX | Sonication | LE: 19.55 ± 2.48% | Cancer therapy | 64 |
| HFL-1 | Erastin | Sonication | LA: 3.2 mg erastin per mg protein | Cancer therapy | 65 |
| Macrophage | PTX | Sonication | LE: 33% | Cancer therapy | 88 |
| imDC | Curcumin | Sonication | LE: 70% | Parkinson's disease treatment | 107 |
| (1) HepG2, (2) HDFa, (3) RIN-m | Insulin | Electroporation | LE: (1) 50.75 ± 1.2%, (2) 57.42 ± 5.47%, (3) 49.70 ± 4.32% | Diabetes treatment | 66 |
| imDC | DOX | Electroporation | LE: 20% | Cancer therapy | 67 |
| MSC | DOX | Electroporation | LE: 35% | Cancer therapy | 87 |
| RAW 264.7 | Dex | Electroporation | LE: 11.12 ± 1.82% | Rheumatoid arthritis | 102 |
| Dendritic cell | miR-140 | Electroporation | LA: 0.03 μmol miR per μg exosome | Osteoarthritis | 104 |
| Dendritic cell | GAPDH siRNA | Electroporation | LE: 25% | Alzheimer's disease treatment | 107 |
| M2 macrophage | (1) BSP and (2) IL10pDNA | Electroporation and gene transfection | LE: (1) 29.03% and (2) 6.97% | Rheumatoid arthritis | 103 |
| Engineered HEK293T | (1) 5-FU and (2) miR-21i | Electroporation and incubation | LE: (1) 3.1% and (2) 0.5% | Cancer therapy | 68 |
| LIM1215 c | DOX | Incubation | LE: 9.06% | Cancer therapy | 69 |
| LNCaP l | PTX | Incubation | LE: 9.2 ± 4.5% | Cancer therapy | 71 |
| MSC | miR-155 inhibitor | Incubation with CaCl2 | LE: 60% | Diabetic wound treatment | 97 |
| EL 4 | Curcumin | Agitation | LA: 2.9 g cur per g exosome | Brain inflammatory diseases | 70 |
| THP-1 | (1) DOX and (2) Cho-miR159 | Agitation and shaking | (1) LA: 160 ng DOX per μg exosome | Cancer therapy | 90 |
| Autologous breast cancer cell | siS100A4/CBSA NP | Extrusion | LE: 86.70 ± 1.22% | Cancer therapy | 91 |
| HEK293T | Melatonin | Extrusion | LA: 97.1 ng mel per μg exosome | Atopic dermatitis | 99 |
| HEK293T | DOX | Surface modification with BPA | LE: 35% | Cancer therapy | 76 |
| Macrophage | (1) anti-PD-L1 and (2) AB680 | (1) Surface modification by PEGylation and (2) extrusion | LE: (1) 5.47 ± 0.36%. (2) 61.0 ± 3.1% | Cancer therapy | 78 |
| Serum | DOX | Surface modification with strep/biotin | N/A | Cancer therapy | 77 |
| HUVEC | KV11 peptide | Surface modification with CP05 linker | LE: 83.1% | Pathological retinal angiogenesis | 109 |
| Engineered Expi293 | Anti-CD3 and Anti-HER2 | Cell engineering | N/A | Cancer therapy | 73 |
| Engineered HEK293T | Cargo protein–CRY2 protein | Cell engineering and light irradiation | LA: 1.4 molecules per exosome | Protein-based therapy | 74 |
| Engineered Expi293 | Cargo protein–photocleavable protein | Cell engineering and light irradiation | N/A | Protein-based therapy | 75 |
3 Methods for synthesis of hybrid exosomes as drug carriers
Recently, hybrid exosomes have been developed, such as liposome–exosome fusion structure, inorganic NP–exosome core/shell, or NP-decorated exosome structure. These hybrid exosomes have been developed to obtain synergistic properties that can be utilized in multiple treatments and theragnostic applications. Several methods have been used for the production of hybrid systems including extrusion, sonication, freeze–thaw method. In this section, the preparation process for organic or inorganic nanoparticle/exosome hybrid structure has been discussed.3.1 Exosome–liposome nanoparticle hybrid structure
Many scientists have been interested in the development of exosome–liposome hybrid structures to improve DDSs, and they tried to prepare these hybrid structures through various physical processes. For example, Cheng et al. reported that engineered exosomes and thermosensitive liposomes were successfully combined via membrane fusion technology to be used in photothermal therapy and cancer immunotherapy.79 In this case, the engineered exosomes were isolated from CD47-overexpressed CT26 cells, and the thermosensitive liposomes (TSLs) were prepared by loading ICG/R837 (indocyanine green (ICG)/imiquimod (R837)) as a photothermal treatment agent. Subsequently, these two precursors were mixed at a ratio of 1![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) :
:![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 1 and fused via the freeze–thaw method. Briefly, the mixture was frozen at −80 °C for 15 min and incubated at 37 °C for 15 min in 3 cycles, and then the TSL–exosome hybrid was isolated. The photothermal activity of the TSL–exosome was characterized by NIR irradiation (808 nm, 2 W cm−2) for 10 min, and the temperature change was monitored up to 56.2 °C. At 42 °C, 90.7% of the encapsulated ICG and R837 were released after 6 h and 69.5% of the encapsulated ICG/R837 were released after 12 h. Another hybrid structure was introduced by Piffoux et al.; they produced a liposome–exosome hybrid DDS structure to improve cellular uptake.80 In this study, the authors obtained exosomes from MSCs, and the liposomes were prepared by the extrusion method. Then, the exosomes and liposomes were mixed to synthesize the hybrid structure (exo–lipo) via incubation with PEG assistance in a thermomixer for 2 h at 40 °C. In addition, mTHPC (anti-cancer photosensitizer) was also loaded into the exo–lipo with 90% encapsulation efficiency. The cellular uptake ratio of only mTHPC was shown to be approximately 20%, whereas mTHPC–exo–lipo showed a higher cellular uptake ratio of up to 70% in CT26 colon cancer cells. In a study by Rayamajhi et al., a macrophage-derived exosome and liposome hybrid structure (mExo–lipo) was introduced. This hybrid DDS was produced via membrane extrusion, and DOX was loaded into the mExo–lipo through extrusion.81 The DOX loading efficiency was reported to be from 82% to 99%, depending on the drug concentration (400, 200, 100, 50 mg ml−1). However, the researchers observed that mExo–lipo was aggregated at high DOX concentrations, and the best loading amount of DOX was determined to be 100 mg ml−1, by dynamic light scattering (DLS). The viability of 4T1 cells against free DOX and DOX–mExo–lipo was investigated, and the results showed higher cytotoxicity with DOX–mExo–lipo than with free DOX. Therefore, drug delivery was effectively performed using this system.
1 and fused via the freeze–thaw method. Briefly, the mixture was frozen at −80 °C for 15 min and incubated at 37 °C for 15 min in 3 cycles, and then the TSL–exosome hybrid was isolated. The photothermal activity of the TSL–exosome was characterized by NIR irradiation (808 nm, 2 W cm−2) for 10 min, and the temperature change was monitored up to 56.2 °C. At 42 °C, 90.7% of the encapsulated ICG and R837 were released after 6 h and 69.5% of the encapsulated ICG/R837 were released after 12 h. Another hybrid structure was introduced by Piffoux et al.; they produced a liposome–exosome hybrid DDS structure to improve cellular uptake.80 In this study, the authors obtained exosomes from MSCs, and the liposomes were prepared by the extrusion method. Then, the exosomes and liposomes were mixed to synthesize the hybrid structure (exo–lipo) via incubation with PEG assistance in a thermomixer for 2 h at 40 °C. In addition, mTHPC (anti-cancer photosensitizer) was also loaded into the exo–lipo with 90% encapsulation efficiency. The cellular uptake ratio of only mTHPC was shown to be approximately 20%, whereas mTHPC–exo–lipo showed a higher cellular uptake ratio of up to 70% in CT26 colon cancer cells. In a study by Rayamajhi et al., a macrophage-derived exosome and liposome hybrid structure (mExo–lipo) was introduced. This hybrid DDS was produced via membrane extrusion, and DOX was loaded into the mExo–lipo through extrusion.81 The DOX loading efficiency was reported to be from 82% to 99%, depending on the drug concentration (400, 200, 100, 50 mg ml−1). However, the researchers observed that mExo–lipo was aggregated at high DOX concentrations, and the best loading amount of DOX was determined to be 100 mg ml−1, by dynamic light scattering (DLS). The viability of 4T1 cells against free DOX and DOX–mExo–lipo was investigated, and the results showed higher cytotoxicity with DOX–mExo–lipo than with free DOX. Therefore, drug delivery was effectively performed using this system.
3.2 Exosome–inorganic NP hybrid structure
In this section, exosome–inorganic NP hybrid DDSs are discussed, including how they are formed in a core/shell or surface assembly structure. Inorganic NPs like metallic NPs, metal oxide NPs, and quantum dots (QDs) possess excellent physical properties, such as plasmonic, magnetic, and fluorescence properties; therefore, a synergistic treatment effect can be expected from these NPs.82For example, hollow gold NPs (HGNs) were encapsulated into exosomes derived from murine melanoma cells (B16–F10–exos) through various methods, such as incubation, freeze–thaw method, sonication, and electroporation, and the encapsulation efficiency for each process was estimated.83 In case of incubation, saponin, as a proper detergent to enhance encapsulation, increased the efficiency of encapsulation from 13.7 ± 9.9% (with no saponin) to 16.4 ± 5.1%. In contrast, the efficiencies of encapsulation by other methods like freeze–thaw (thermal shock I and II), sonication, and electroporation were approximately 18.20 ± 1.35, 9.11 ± 2.0, 19.34 ± 10, and 1%, respectively. The researchers rationalized that the exosome structure was damaged during the electroporation process, so its encapsulation efficiency might not have been monitored clearly. Furthermore, the researchers also conducted encapsulation of HGNs into exosomes via a cell-mediated process. Briefly, B16–F10 cells were incubated with PEGylated HGNs (PEG-HGNs) and the secreted HGNs–exosome structures were collected and characterized. Then, tetraspanin CD9 content in HGN–exosomes was measured by western blotting, and the core/shell structures were observed by TEM. This cell-mediated method yielded the highest encapsulation efficiency, which was estimated to be 50%. Interestingly, HGNs in the exosomes could generate heat by incident light stimulation due to its plasmonic property. Therefore, this hybrid material could be used as a photothermal treatment agent.
Khongkow et al. also introduced a gold NP–exosome hybrid structure that was prepared by the extrusion method to improve neuron targeting through enhanced blood–brain barrier (BBB) penetration.84 In this case, engineered exosomes that possessed neuron-specific rabies viral glycoprotein (RVG) and glycosylation-stabilized (GNSTM) peptides were produced by transfected HEK293T cells with the pcDNA GNSTM-3-RVG-10-Lamp2b-HA vector. The gold NPs were synthesized by a heat-assisted method. In order to prepare the gold-RVG/GNSTM-Exo, the gold NP solution and engineered exosome solution were mixed and extruded using a mini extruder with 400 nm, 200 nm, and 100 nm polycarbonate porous membranes. The physical properties of gold-RVG/GNSTM-Exo were characterized by DLS, and its hydrodynamic ratio was measured at 105 ± 10.1 nm; the targeting properties of these hybrids were examined by measuring the percentage of gold NPs that crossed the BBB co-culture model into the basal chamber of the Transwell™ System, by spectrophotometry over a certain period of incubation, and the results revealed that from 30 min to 20 h, the percentage increased from 5 to 20%. Furthermore, brain uptake of gold-RVG/GNSTM-Exo was characterized via mice experiments. According to in vivo analysis, gold-RVG/GNSTM-Exo-induced fluorescence image was clearly observed at the mouse brain site, indicating that this hybrid system could penetrate the BBB and targeted the brain.
In another study, a metal–organic frame (MOF) was coated with an exosome to be used as an anti-cancer agent.85 Cheng et al. produced a protein loaded MOF (ZIF-8) by a self-assembly process, and the exosomes were obtained from MDA-MB-231 cells (Fig. 4 I and II). These two components were then mixed to form a protein/ZIF-8-exosome structure by sonication and repeated extrusion (Fig. 4 III). The results showed that the protein loading efficiency of the ZIF-8 was estimated to be 94% (low initial protein concentration) and around 41% (high initial protein concentration). Furthermore, over 91% of protein/ZIF-8 was coated with exosomes. Interestingly, because the exosome in the hybrid structure originated from a cancer cell, this protein/ZIF-8-exosome could be automatically accumulated into the cancer site through homotypic effect without the need for targeting moieties on the surface of the exosome. Moreover, after injecting this protein/ZIF-8-exosome into tumor-bearing mice, tumor growth was effectively suppressed, which implied that this hybrid could be applied as an anti-cancer drug. In 2017, Popovtzer et al. created gold NP–exosome hybrid structure through surface modification approaches.86 Briefly, the exosomes were isolated from MSCs (human mesenchymal stem cells), and the surface of gold NPs was modified with glucose and PEG by EDC/NHS coupling reaction. Then, the hMSC-derived exosomes and the glucose-gold NPs were mixed and incubated for 3 h at 37 °C. This exosome–gold NP hybrid structure could be accumulated in the brain and successfully used as a neuroimaging agent.
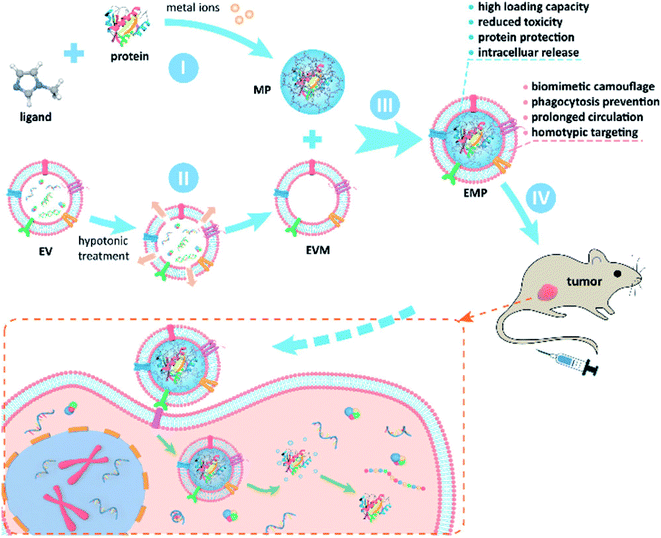 | ||
| Fig. 4 Schematic figure of synthesis of MOF–exosome hybrid structure and its anti-tumor activity. Reprinted with permission.85 Copyright 2020, American Chemical Society. | ||
Meanwhile, Wang et al. reported that inorganic NPs were attached on the surface of drug-loaded exosomes to be used as multiple treatment agents87 (Fig. 5). In this study, DOX was first encapsulated into a biotin labeled exosome (DOX–exo–bio) by electroporation. The surface of iron oxide was coated with polydopamine (PDA), and then modified with avidin and molecular beacon (Fe3O4–PDA–MB–avidin). Next, DOX–exo–bio and av–mb–IONP were mixed and incubated at 4 °C overnight, and during incubation, these two precursors attached via av–bio interaction to finally produce the hybrid system. This hybrid system could be used as a multi-functional DDS and it could be utilized in cancer therapy for chemo-thermal-gene treatment. Interestingly, this hybrid DDS could also accumulate at the tumor site through external magnetic forces, thus its therapeutic effect could be enhanced.
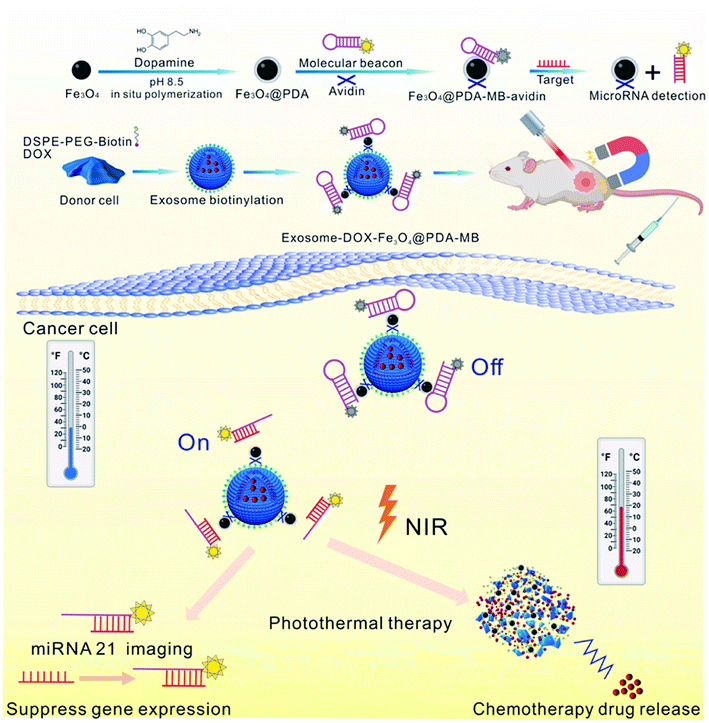 | ||
| Fig. 5 Schematic illustration of the preparation of exosome–DOX–Fe3O4–PDA–MB and its multiple therapeutic applications. Reprinted with permission.87 Copyright 2021, Elsevier. | ||
A series of examples of hybrid exosome-based DDS is presented in Table 2.
| Exosome origin | Combined NP | Assembly strategy | Loaded agent | Application | Ref. |
|---|---|---|---|---|---|
| CT26 | Liposome | Freeze–thaw | ICG/R837 | Cancer therapy | 79 |
| MSC | Liposome | Extrusion | mTHPC | Cancer therapy | 80 |
| Macrophage | Liposome | Extrusion | DOX | Cancer therapy | 81 |
| Fibroblast | Liposome | Freeze–thaw | DTX and GM-CSF | Cancer therapy | 92 |
| L-929 | Liposome | Extrusion | Clodronate | Pulmonary fibrosis treatment | 110 |
| BMSC | Liposome | Freeze–thaw | Polypyrrole NPs | Peripheral neuropathy treatment | 111 |
| Murine melanoma cell | Hollow gold NPs | Incubation, freeze–thaw method, sonication and electroporation | N/A | Photothermal cancer therapy and imaging | 83 |
| HEK293T | Gold NP | Extrusion | N/A | Brain imaging | 84 |
| MSC | Gold NP | Incubation with PEG | N/A | Brain imaging | 86 |
| Macrophage | Iron oxide NP | Surface modification with strep/biotin | DOX and miRNA21 | Cancer therapy | 87 |
| MSC | Iron oxide NP | Cell-mediated process | N/A | Cutaneous wound treatment | 98 |
| MDA-MB-231 | MOF | Sonication and extrusion | Protein drug | Cancer therapy | 85 |
| H22 and Bel7402 | MPS | Cell-mediated process | DOX | Cancer therapy | 93 |
4 Exosome-based DDSs in treatment of various diseases
In this part, therapeutic applications of exosome- and hybrid exosome-based DDSs are described. These DDSs could be used as treatment agents in various conditions, including cancer, skin damage, bone-related diseases.4.1 Cancer treatment
In order to enhance the efficiency of anti-cancer drug therapy, several types of chemical drugs have been loaded into exosomes, and these exosomal formulations have been continuously developed to improve biodistribution and pharmacokinetics of anti-cancer drugs while reducing their side effects.For example, Bagheri et al. prepared a DOX-encapsulated exosome to utilize it in anti-cancer treatment.88 The exosome was isolated from MSCs and DOX was encapsulated by electroporation; the system had 35% encapsulation efficiency. Moreover, the MSC-derived exosome was functionalized with the MUC1 aptamer through the EDC/NHS coupling reaction to introduce the cancer-targeting ligand on the exosome, thus improve its accumulation in target cancer cells. In order to evaluate the efficiency of MUC1apt–MExo–DOX in cancer therapy, a C26 carcinoma-bearing mouse model was treated with this hybrid. The results showed that the DDS effectively reduced the cancer volume compared to that of the control groups, and the survival ratio in MUC1apt–MExo–DOX-treated mice was around 100% after 30 days.
In 2018, Kim et al. evaluated the anti-cancer effect of PTX-loaded macrophage-derived exosome (exo–PTX) that was produced by sonication, with a loading capacity of approximately 33%.89 In an attempt to enhance targeted delivery, the surface of exo-PTX was modified with aminoethylanisamide–polyethylene glycol (AA–PEG), as it recognizes a sigma receptor that is overexpressed by lung cancer cells. In vivo analysis showed that the treatment with AA–PEG–exo–PTX system suppressed metastases on the lungs that was less than 5%. The results also showed excellent survival rate in mice treated with AA–PEG–exo–PTX compared to those who received other treatments such as exo–PTX, exosome alone. In another study, Gong et al. used exosomes derived from THP-1 cells (human leukemia monocytic cell line) (THP–1exo) as anti-cancer drug carriers to treat breast cancer.90 They functionalized THP-1exo with metalloproteinase 15 (A15) to enhance cancer targeting. Two different therapeutic agents, DOX and cholesterol-modified miRNA 159 (Cho-miR159), were loaded into A15-exo by agitation and shaking incubation. The loading amount of DOX was estimated to be 160 ng in 1 μg A15-exo, meanwhile the encapsulation efficiency of Cho-miR159 was approximately 5.33%. DOX–Cho-miR159–A15-exo was injected into a breast cancer xenograft mouse model, and its treatment capacity was studied. In vivo analysis showed that the cancer volume and weight were clearly reduced in mice treated with DOX–Cho-miR159–A15-exo compared to those of the control groups. Additionally, the survival period of the mice treated with this exosome-based DDS was longer than that of the other control groups. Therefore, A15-exo carriers could successfully reach the cancer site and chemotherapy and miR159 treatment were effective.
In addition to the studies on DDSs loaded with various chemical drugs for cancer therapy, attempts have also been made to deliver siRNA to tumor cells using exosome carriers. Zhao et al. encapsulated siS100A4 (siRNA)/cationic bovine serum albumin (CBSA) complex into a breast cancer cell-derived exosome as an anti-cancer agent.91 In this study, CBSA and siS100A4 were first mixed and incubated at 42 °C for 30 min to produce the siS100A4/CBSA NP structure. Then, siS100A4/CBSA NP and a cancer cell-derived exosome were mixed and incubated at 4 °C for 30 min and then at 37 °C for 1 h. Subsequently, this mixture was extruded using a mini extruder with 200 nm and 100 nm pore-diameter filters to prepare the siS100A4/CBSA-exosome. After siRNA was loaded into the exosome, the encapsulation efficiency was estimated to be 86.70 ± 1.22%. Mice with lung cancer were used as models to study the effect of siS100A4/CBSA-exosomes in cancer treatment. Mice treated with siS100A4/CBSA-exosomes had an accumulation of the complex in the lung tumors. The mean number of metastatic nodules in the lungs of the mice treated with siS100A4/CBSA-exosome was the lowest compared to that of the other control groups, which were treated with siS100A4/CBSA-liposome or siS100A4-CBSA.
Organic or inorganic NP–exosome hybrid DDSs have also been used in cancer treatment. For example, Qijun et al. synthesized a hybrid structure of a genetically-engineered exosome (gExo) and a TSL encapsulating an anti-tumor agent to be used in cancer therapy.92 In this study, gExo was isolated from fibroblasts overexpressing CD47 and granulocyte-macrophage colony-stimulating factor (GM-CSF), and DTX was loaded into TSL (DTX-TSL). The gExo and DTX-TSLs were fused by the freeze–thaw method (Fig. 6). The gETL can act as a therapeutic agent in two different ways. First, it can activate T-cells by GM-CSF from the exosomes (immunotherapy application). Second, DTX in the liposomes can treat the tumor through chemotherapy. Consequently, through these functions, this DDS platform showed excellent anti-cancer treatment performance.
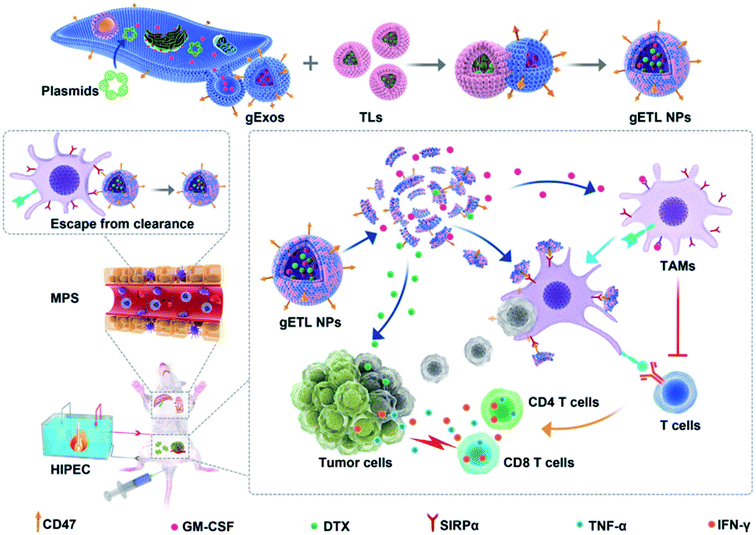 | ||
| Fig. 6 Synthetic scheme of gETL NPs (exosome/liposome hybrid structure) and their anti-tumor activity. Reprinted with permission.92 Copyright 2020, John Wiley and Sons. | ||
Yong et al. described another inorganic NP-exosome hybrid structure for delivery of anti-cancer agents.93 In this study, DOX was loaded into a mesoporous silica NP (DOX-MPS) and applied to cancer cells. DOX-MPSs could penetrate into the H22 or Bel7402 cells through endocytosis and turn into DOX-MPS/exosome core/shell structure in the cells. Finally, these hybrids were released from the cells and collected by centrifugation. Their anti-cancer effect was proven in H22 tumor-bearing mice and B16–F10 lung metastasis mice. After the injection of DOX-MPS/exosomes into both cancer xenograft mouse models, the tumor volume did not increase in either cancer group compared to that of the mice treated with DOX and DOX/exosomes. In addition, the colony number and size was smaller in DOX-MPS/exosomes treated mice groups, and these mice exhibited the highest survival. Therefore, these cancer cell-derived exosome coated-inorganic NPs could serve as excellent drug carrier.
4.2 Skin therapy
Exosomes loaded with therapeutic agents have also been used in skin therapy. Some cells such as MSCs are known to promote skin repair, thus MSC-derived exosomes can be used to induce skin regeneration.94 Additionally, some milk-derived exosomes could prevent skin aging and help in wound repairing.95 This shows that skin therapeutic agent-loaded exosomes can aid in the treatment of skin damage.Moran et al. conducted both in vivo and in vitro analyses of the treatment of ischemic wounds using exosomes containing transforming growth factor beta (TGF-β).96 This study explored the utility of a purified CD63, CD9, and ALG-2 interacting protein X (Alix)-positive exosome population (PEP), which was extracted from activated platelets, in promoting wound healing; the results confirmed that bioactive TGF-β was released to aid the wound healing process. The continuous delivery of PEP through fibrin sealant (TISSEEL) showed significant regeneration effects on overall ischemic wound healing, and the results described the potential ability to preserve the biological activity of TGF-β in freeze-dried exosomes applied to accelerate wound healing. The researchers developed a PEP bioactive hydrogel specialized in curing ischemic wounds in rabbits—a cell-free regeneration treatment method. The mechanisms governing this effect on ischemic wound healing were related to regulation of epithelial metastatic differentiation, collagen reconstruction, and overall skin tissue development through the TGF-β pathways. In a study by Gondaliya et al., it was reported that miRNA-loaded exosomes could treat a diabetic wound.97 The exosome was isolated from MSCs and miR-155 inhibitor was encapsulated within the exosome with CaCl2 assistance by incubation at 42 °C for 60 s and then on ice for 5 min. Diabetic wound healing is known to be delayed by miR-155 upregulation, so its inhibitor was loaded into the exosome to enhance the wound healing effect. The loading efficiency was estimated to be 60%. After the treatment of diabetic wound model mice with miR-155 inhibitor-encapsulated MSC-derived exosome, it was shown that collagen deposition, angiogenesis, and re-epithelialization were enhanced in these mice groups compared to those in the control groups.
In a study by Li et al., magnetic NP–exosome core/shell structures were applied for cutaneous wound treatment.98 The researchers incubated MSCs with iron oxide NPs for 16 h, and then the iron oxide NP–exosomes secreted from the MSCs were collected by centrifugation. Next, the iron oxide NP–exosomes were injected into surgically wounded mouse models. Interestingly, this NP–exosome hybrid could accumulate at the wound site by external magnetic forces, which allowed excellent wound healing. The results showed that the wound size in the group treated with iron oxide NP–exosomes was more quickly reduced than that in the control groups. Additionally, the collagen deposition area in the group treated with the hybrid exosome almost recovered back to normal, in contrast to that in the other groups treated with a vehicle or only MSC-derived exosome. Moreover, high blood vessel density was observed in the wounds of the mice treated with the hybrid NP–exosomes, which proves that angiogenesis was actively taking place. Therefore, these results established that iron oxide NP–exosome hybrids could be considered good wound healing agents.
In other study, Kim et al. produced melatonin encapsulated exosomes (mel-exosome) to treat atopic dermatitis (AD).99 To encapsulate melatonin into exosomes, melatonin was first incubated with HEK293 cells and extruded with 10, 5, and 1 μm polycarbonate membrane filters, and then mel-exosomes were isolated by ultracentrifugation at 100![[thin space (1/6-em)]](https://www.rsc.org/images/entities/char_2009.gif) 000×g for 1 h. The loading amount of melatonin was approximately 97.1 ng mel per μg exosome, which was determined using an ELISA kit. According to the in vitro analysis, mel-exosomes could suppress TNF-α and β-hexosaminidase release, which confirmed that the exosomes exhibited anti-inflammatory effects. Furthermore, AD-like mice were treated with mel-exosomes to evaluate their healing effect. The results showed that local inflammation, mast cell infiltration, and fibrosis were highly suppressed in the mice treated with mel-exosomes, and symptoms of AD such as erythema, edema, and dryness were alleviated in these mice. Furthermore, treatment with mel-exosomes caused the restoration IFN-γ and IL-4 levels, and the suppression of COX-2, TNF-α, and PAR-2 expression levels. These results prove that mel-exosome could be a candidate for AD treatment.
000×g for 1 h. The loading amount of melatonin was approximately 97.1 ng mel per μg exosome, which was determined using an ELISA kit. According to the in vitro analysis, mel-exosomes could suppress TNF-α and β-hexosaminidase release, which confirmed that the exosomes exhibited anti-inflammatory effects. Furthermore, AD-like mice were treated with mel-exosomes to evaluate their healing effect. The results showed that local inflammation, mast cell infiltration, and fibrosis were highly suppressed in the mice treated with mel-exosomes, and symptoms of AD such as erythema, edema, and dryness were alleviated in these mice. Furthermore, treatment with mel-exosomes caused the restoration IFN-γ and IL-4 levels, and the suppression of COX-2, TNF-α, and PAR-2 expression levels. These results prove that mel-exosome could be a candidate for AD treatment.
4.3 Bone-related disease treatment
Exosomal DDSs have been applied in the treatment of various bone-related diseases, including rheumatoid arthritis, osteoporosis. The therapeutic effect of cell-derived exosomes on such diseases have been tested previously, wherein they showed good therapeutic ability.100,101 In order to enhance the treatment effect, the structures of drug-containing exosomes have been investigated.Recently, Yan et al. studied rheumatoid arthritis (RA) treatment using dexamethasone sodium phosphate (Dex)-loaded exosomes (Exo/Dex).102 In this case, Dex was encapsulated into exosomes derived from raw 264.7 macrophage cell lines via electroporation, with a loading efficiency of approximately 11.12 ± 1.82%. The Exo/Dex was functionalized with folic acid–polyethylene glycol–cholesterol (FPC) to improve its targeted delivery. Since FPC-Exo/Dex could recognize the folic acid receptors in the damaged areas, this DDS could accumulate there and treat the area more effectively. Micro-CT analysis indicated that the ankle and toe joint in mice treated with FPC-Exo/Dex showed bone morphology similar to that of normal mice (Fig. 7a). Other parameters like bone mineral density (BMD), percent bone volume (BV/TV), bone surface density (BS/BV), trabecular thickness (Tb.Th), trabecular number (Tb.N), and trabecular spacing (Tb.Sp) also reflected that FPC-Exo/Dex treatment was the most effective compared to the effect in control groups (Fig. 7b–g). Therefore, FPC-Exo/Dex could be considered a potential candidate for RA treatment.
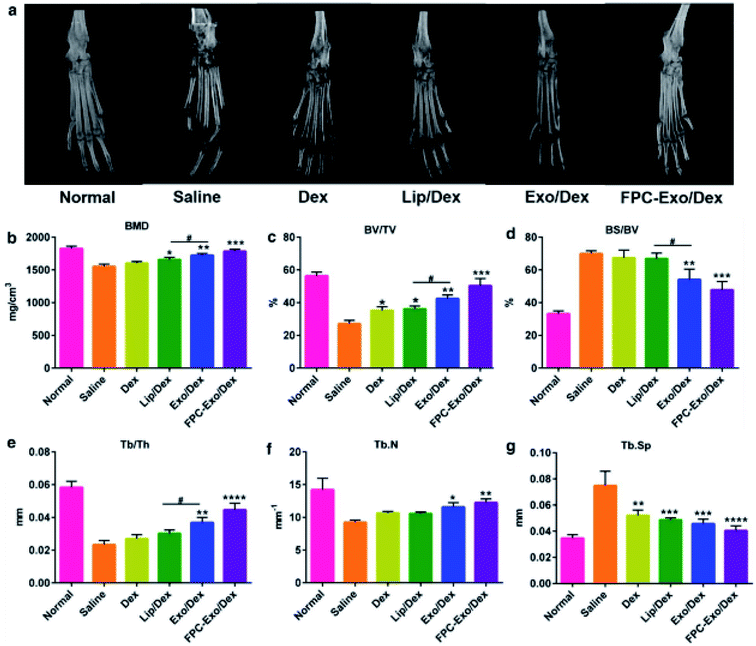 | ||
| Fig. 7 Micro-CT analysis of the hind limbs of the mice from different treatment groups. (a) Representative 3D reconstructed images from each treatment group. (b–g) Bone morphometric parameters of ROI within calcaneus bone (n = 3, *p < 0.05, **p < 0.01, ***p < 0.001, ****p < 0.0001 vs. mice treated with saline, #p < 0.05 is mice treated with Lip/Dex compared to Exo/Dex). Reprinted with permission.102 Copyright 2020, BioMed Central. | ||
Li et al. conducted further research on RA therapy using chemotherapeutic drugs and DNA co-loaded exosomes.103 In their study, IL-10 plasmid DNA (IL10pDNA) was first transfected into M2 macrophages and the secreted exosome that possessed IL10pDNA was obtained. Next, betamethasone sodium phosphate (BSP, a chemical drug) was encapsulated into the IL10pDNA–exosome by electroporation at 200 V for 5 ms. In this case, the loading efficiency was estimated to be 29.03% for BSP and 6.97% for IL-10 pDNA. The authors carried out an in vivo study using collagen-induced arthritis (CIA) mice model, where they injected BSP–IL10pDNA–exosome and monitored the effect. The results showed that the body weight in mice treated with BSP–IL10pDNA–exosome gradually increased to reach a level close to that in the healthy group. In contrast, mice in other control groups, like those treated with free BSP, naked pDNA, BSP/pDNA, showed body weight reduction. Besides, the paw of the mice showed swelling in the control groups especially in the saline group; however, the paw shape in mice treated with BSP–IL10pDNA–exosome group seemed similar to that in healthy mice. Furthermore, BSP–IL10pDNA–exosome was highly accumulated at the inflamed joint site and showed anti-inflammatory activity.
Liang et al. investigated osteoarthritis (OA) treatment using micro RNA encapsulated exosomes.104 In their study, miRNA-140 was encapsulated by electroporation into an exosome (miR-140-CAPexo) modified with chondrocyte-affinity peptide (CAP) and Lamp2b. In this structure, miR-140 can protect the chondrocytes and be a potential candidate for osteoarthritis treatment. The OA mice were injected with miR-140-CAPexo, and the results revealed that miR-140-CAPexo was highly accumulated in the cartilage tissues compared to miR-140-exo. In addition, the mice treated with miR-140-CAPexo exhibited smooth and flat cartilage, which was similar to that in healthy mice, while miR-140-exo-injected mice had small cavities at the junction of the cartilage and subchondral bone. Thus, the developed exosomal DDS could be considered as a candidate for OA treatment.
4.4 Treatment applications in other diseases
Exosome-based DDSs were developed to treat not only cancer and skin conditions, but also conditions like Alzheimer's disease and ocular disease.105,106 For example, Alvarez-Erviti et al. developed a treatment agent for Alzheimer's disease using siRNA-encapsulated exosomes.107 In this study, they delivered GAPDH siRNA to the brain in dendritic cells-derived exosomes; the loading of siRNA into the exosomes was conducted through electroporation at 400 V and 125 μF. The results of the in vivo analysis showed that the exosomal DDSs accumulated in many sites of the brain, and BACE1 mRNA levels and total β-amyloid 1-42 levels were significantly decreased, i.e., approximately 66 ± 15% and 55%, respectively after injection. On the other hand, Liu et al. introduced rabies virus glycoprotein and exosome-based core–shell structure (REXO) as a drug carrier for Parkinson's disease treatment.108 Curcumin and phenylboronic acid-poly(2-(dimethylamino)ethyl acrylate) NPs (CANP) were co-loaded into the REXO by sonication, with curcumin loading efficiency to be approximately 70%. Then, the surface of the carrier was modified with small interfering RNA targeting SNCA (REXO-C/ANP/S) to improve the penetration into BBB. REXO-C/ANP/S effectively accumulated in mice brains and enhanced neuronal recovery without any organ toxicity.In another study, Dong et al. reported the inhibition of pathological retinal angiogenesis using anti-angiogenic peptide-conjugated exosomes.109 In this study, the researchers prepared human umbilical vein endothelial cell (HUVEC)-derived exosomes (HUVEC-exo) and incubated them with CP05-linked KV11 peptide, as anti-angiogenic peptide, to prepare the conjugation structure. The loading efficiency was estimated to be 83.1%. According to the results, vessel regression was not induced in the avascular areas of the retina against oxygen stress, while retinal neovascularization was clearly reduced after KV11-HUVEC-Exo treatment (Fig. 8). The expression level of key proteins such as HIF1α and VEGF that are known to induce the neovascularization was significantly decreased after the treatment. Therefore, exosome-based DDSs could be potential therapeutic agents for ocular disease.
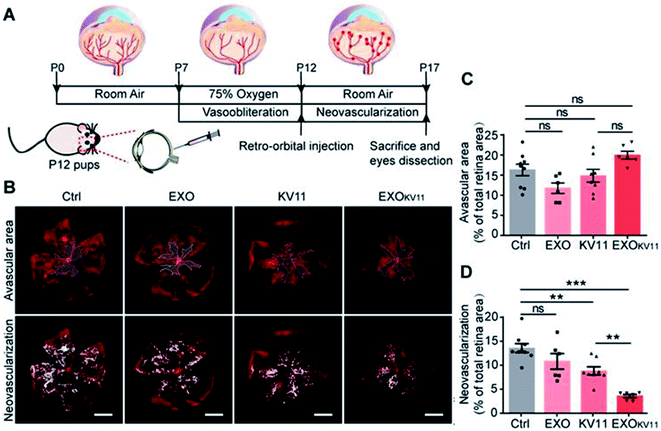 | ||
| Fig. 8 (A) Schematic illustration of the treatment of oxygen-induced retinopathy (OIR) mouse model, (B) confocal images of retina vasculature stained with IsoB4, (C) vascular area, and (D) neovascularization quantification. Reprinted with permission.109 Copyright 2021, Ivyspring International Publisher. | ||
Another study explored the application of an exosome–liposome hybrid system (exo–lipo) as a treatment agent for pulmonary fibrosis (lung disease).110 Sun et al. isolated the exosomes from L-929 cells (fibroblast cell line) and synthesized the clodronate (CLD)-loaded liposomes. Subsequently, the exosomes and CLD-liposomes were mixed and fused through the membrane extrusion method using a mini extruder with 400 and 200 nm polycarbonate membranes under 10 times cycle. The authors conducted in vivo experiments using pulmonary fibrosis-induced mice to evaluate the effect of exosome–CLD-liposome hybrid structure (EL-CLD). The results showed that EL-CLD accumulated at the lung site compared to exo–lipo, and EL-CLD was detected in the lung even 48 h after injection. Furthermore, collagen deposition increased, and thickened alveolar walls and alveolar air areas decreased after treatment with EL-CLD. Hence, this hybrid DDS can be considered as a potential candidate for anti-fibrotic treatment of pulmonary fibrosis. Furthermore, it was reported that exo–lipo system could be utilized in the treatment of peripheral neuropathy.111 Singh et al. produced an exosome from the bone marrow mesenchymal stromal cells (BMSCs) and prepared polypyrrole NPs (PpyNPs)-loaded liposome (PpyNP-lipo). In this case, PpyNPs possess electrical conductivity, so they can assist in nerve regeneration. These two precursors were mixed and assembled by the freeze–thaw process using liquid nitrogen (freeze) and 50 °C water bath (thaw) in a 10 times cycle. An in vivo experiment was performed using diabetic peripheral neuropathy mouse models with electrical stimulation assistance. After injecting exo–PpyNP-lipo, the nerve diameter was significantly restored compared to the diameter in other controls, where it was thicker than that in healthy mice. In addition, the diameter of the axon was also recovered by the exo–PpyNP-lipo treatment. Moreover, gastrocnemius muscle was also regenerated after the exosomal DDS treatment. Therefore, this hybrid DDS can be utilized as a novel agent for regenerative therapy.
5 Summary
Effective chemotherapy, gene therapy, and photothermal therapy can be carried using proper DDS platforms, and exosomes with their hybrid systems have been considered as alternative drug carriers. To achieve high performance DDSs based on exosomes and their hybrid structures, many methods of encapsulation and modification of therapeutic agents have been developed, and these DDSs have been utilized to treat various diseases.Exosome, itself could be utilized as novel drug carrier without target Ab labeling due to homotypic targeting effect and self-recognition. However, challenges with respect to loading efficiency and stability of the exosome structure still persist. On the other hands, hybrid exosomes showed excellent synergic effect including treatment and diagnosis at the same time, so it could perform theragnosis function. However, some disadvantages also could be raised such as the high cost for synthesis of hybrid exosomes and the complication of preparation process and thus facile synthesis method should be suggested for accessible applications. Moreover, the function of various types of exosome and its hybrid structure-based DDS should be tuned to fit the proper therapeutic applications.
By various approaches, exosome-based DDSs have been developed as mentioned it above, and recently, it was reported that exosome-based DDS showed more effective therapy performance than cell therapy. In addition, in the several countries including USA, China, clinical trials have been taken a place to help for the patient treatment. Also, many research groups and some start-up companies have developed exosome-based DDSs as 3rd generation anticancer therapeutic agents to apply for chemo-immunotherapy. However, still mass production of exosome and exosome-based DDS is very difficult for commercialization, and drug loading efficiency should be improved in order to achieved high therapeutic function. And these kinds of huddles should be overcome in order to contribute the human health care and commercialize. Furthermore, during the preparation of hybrid exosome or encapsulation of drug into the exosome via physical process and chemical synthesis, therapeutic agents, exosome carrier, and other active materials would be lost in each modification steps. And it could bring the low yield for production and poor reproducibility. Therefore, these kinds of challenge should be solved to grow the exosome-based DDS field.
Nevertheless, as mentioned it above, exosomes and their hybrid structures have showed a lot of advantages such as high targeting, non-toxicity, biocompatibility. The clinical application of these exosomes should thus be expanded to the treatment of various diseases. In conclusion, exosomes exhibit a promising potential for application in DDS, which will provide substantial benefit to public health and society.
Conflicts of interest
Authors declare no conflicts of interest.Acknowledgements
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education (No. 2021R1A6A1A03046418 and No. 2022R1I1A1A01060691) and This work was supported by the National Research Foundation of Korea (NRF) grant funded by the Korea government (MSIT) (No. 2021R1A2C2095113).References
- I. S. Raja, N. Duraipandi, M. S. Kiran and N. N. Fathima, RSC Adv., 2016, 6, 100916–100924 RSC.
- C. Xue, S. Hu, Z.-H. Gao, L. Wang, M.-X. Luo, X. Yu, B.-F. Li, Z. Shen and Z.-S. Wu, Nat. Commun., 2021, 12, 2928 CrossRef CAS PubMed.
- S. Li, Y. Zhang, J. Wang, Y. Zhao, T. Ji, X. Zhao, Y. Ding, X. Zhao, R. Zhao, F. Li, X. Yang, S. Liu, Z. Liu, J. Lai, A. K. Whittaker, G. J. Anderson, J. Wei and G. Nie, Nat. Biomed. Eng., 2017, 1, 667–679 CrossRef CAS PubMed.
- X. Zeng, J. Sun, S. Li, J. Shi, H. Gao, W. Sun Leong, Y. Wu, M. Li, C. Liu, P. Li, J. Kong, Y.-Z. Wu, G. Nie, Y. Fu and G. Zhang, Nat. Commun., 2020, 11, 567 CrossRef CAS PubMed.
- B. G. Carvalho, F. F. Vit, H. F. Carvalho, S. W. Han and L. G. de la Torre, J. Mater. Chem. B, 2021, 9, 1208–1237 RSC.
- C. Dhand, M. P. Prabhakaran, R. W. Beuerman, R. Lakshminarayanan, N. Dwivedi and S. Ramakrishna, RSC Adv., 2014, 4, 32673–32689 RSC.
- K. R. B. Singh, V. Nayak, J. Singh, A. K. Singh and R. P. Singh, RSC Adv., 2021, 11, 24722–24746 RSC.
- J. E. Kim, J. Lee, M. Jang, M. H. Kwak, J. Go, E. K. Kho, S. H. Song, J. E. Sung, J. Lee and D. Y. Hwang, Biomater. Sci., 2015, 3, 509–519 RSC.
- S. H. Kang, V. Revuri, S.-J. Lee, S. Cho, I.-K. Park, K. J. Cho, W. K. Bae and Y.-k. Lee, ACS Nano, 2017, 11, 10417–10429 CrossRef CAS PubMed.
- R. Kumar, K. Mondal, P. K. Panda, A. Kaushik, R. Abolhassani, R. Ahuja, H.-G. Rubahn and Y. K. Mishra, J. Mater. Chem. B, 2020, 8, 8992–9027 RSC.
- S. Zanganeh, G. Hutter, R. Spitler, O. Lenkov, M. Mahmoudi, A. Shaw, J. S. Pajarinen, H. Nejadnik, S. Goodman, M. Moseley, L. M. Coussens and H. E. Daldrup-Link, Nat. Nanotechnol., 2016, 11, 986–994 CrossRef CAS PubMed.
- Z. Ding, K. Sigdel, L. Yang, Y. Liu, M. Xuan, X. Wang, Z. Gu, J. Wu and H. Xie, J. Mater. Chem. B, 2020, 8, 8781–8793 RSC.
- Y. Chen, H. Zhang, X. Cai, J. Ji, S. He and G. Zhai, RSC Adv., 2016, 6, 92073–92091 RSC.
- J. S. Lee, P. Han, R. Chaudhury, S. Khan, S. Bickerton, M. D. McHugh, H. B. Park, A. L. Siefert, G. Rea, J. M. Carballido, D. A. Horwitz, J. Criscione, K. Perica, R. Samstein, R. Ragheb, D. Kim and T. M. Fahmy, Nat. Biomed. Eng., 2021, 5, 983–997 CrossRef CAS PubMed.
- J. M. An, S. M. S. Shahriar, Y. H. Hwang, S. R. Hwang, D. Y. Lee, S. Cho and Y.-k. Lee, ACS Appl. Mater. Interfaces, 2021, 13, 23314–23327 CrossRef CAS PubMed.
- O. Sedlacek, V. Bardoula, E. Vuorimaa-Laukkanen, L. Gedda, K. Edwards, A. Radulescu, G. A. Mun, Y. Guo, J. Zhou, H. Zhang, V. Nardello-Rataj, S. Filippov and R. Hoogenboom, Small, 2022, 2106251 CrossRef CAS PubMed.
- P. Ramamurthi, Z. Zhao, E. Burke, N. F. Steinmetz and M. Müllner, Adv. Healthcare Mater., 2022, 2200163 CrossRef PubMed.
- D. Shi, G. Mi, Y. Shen and T. J. Webster, Nanoscale, 2019, 11, 15057–15071 RSC.
- A. Babu, N. Amreddy, R. Muralidharan, G. Pathuri, H. Gali, A. Chen, Y. D. Zhao, A. Munshi and R. Ramesh, Sci. Rep., 2017, 7, 14674 CrossRef PubMed.
- X. Zhao, X. Fan, Z. Gong, X. Gao, Y. Wang and B. Ni, Microb. Ecol., 2022 DOI:10.1007/s00248-022-01972-3.
- A. Manuja, B. Kumar, R. Kumar, D. Chhabra, M. Ghosh, M. Manuja, B. Brar, Y. Pal, B. N. Tripathi and M. Prasad, Toxicol. Rep., 2021, 8, 1970–1978 CrossRef CAS PubMed.
- R. D. Brohi, L. Wang, H. S. Talpur, D. Wu, F. A. Khan, D. Bhattarai, Z.-U. Rehman, F. Farmanullah and L.-J. Huo, Front. Pharmacol., 2017, 8, 606 CrossRef PubMed.
- J. Lee, E. Y. Park and J. Lee, Bioprocess Biosyst. Eng., 2014, 37, 983–989 CrossRef CAS PubMed.
- Z. Xu, S. Zeng, Z. Gong and Y. Yan, Mol. Cancer, 2020, 19, 160 CrossRef CAS PubMed.
- M. Zhang, X. Zang, M. Wang, Z. Li, M. Qiao, H. Hu and D. Chen, J. Mater. Chem. B, 2019, 7, 2421–2433 RSC.
- Y. Liang, L. Duan, J. Lu and J. Xia, Theranostics, 2021, 11, 3183–3195 CrossRef CAS PubMed.
- M. Sancho-Albero, A. Medel-Martínez and P. Martín-Duque, RSC Adv., 2020, 10, 23975–23987 RSC.
- Y. Yang, Y. Hong, E. Cho, G. B. Kim and I.-S. Kim, J. Extracell. Vesicles, 2018, 7, 1440131 CrossRef PubMed.
- J. P. K. Armstrong, M. N. Holme and M. M. Stevens, ACS Nano, 2017, 11, 69–83 CrossRef CAS PubMed.
- P. Li, M. Kaslan, S. H. Lee, J. Yao and Z. Gao, Theranostics, 2017, 7, 789–804 CrossRef CAS PubMed.
- H. H. Jung, J.-Y. Kim, J. E. Lim and Y.-H. Im, Sci. Rep., 2020, 10, 14069 CrossRef PubMed.
- S. A. Choi, E. J. Koh, R. N. Kim, J. W. Byun, J. H. Phi, J. Yang, K.-C. Wang, A. K. Park, D. W. Hwang, J. Y. Lee and S.-K. Kim, Cancer Cell Int., 2020, 20, 558 CrossRef CAS PubMed.
- D. Choi, L. Montermini, H. Jeong, S. Sharma, B. Meehan and J. Rak, ACS Nano, 2019, 13, 10499–10511 CrossRef CAS PubMed.
- B. W. Sódar, Á. Kittel, K. Pálóczi, K. V. Vukman, X. Osteikoetxea, K. Szabó-Taylor, A. Németh, B. Sperlágh, T. Baranyai, Z. Giricz, Z. Wiener, L. Turiák, L. Drahos, É. Pállinger, K. Vékey, P. Ferdinandy, A. Falus and E. I. Buzás, Sci. Rep., 2016, 6, 24316 CrossRef PubMed.
- D. H. Kim, V. K. Kothandan, H. W. Kim, K. S. Kim, J. Y. Kim, H. J. Cho, Y.-k. Lee, D.-E. Lee and S. R. Hwang, Pharmaceutics, 2019, 11, 649 CrossRef CAS PubMed.
- A. Hoshino, H. S. Kim, L. Bojmar, K. E. Gyan, M. Cioffi, J. Hernandez, C. P. Zambirinis, G. Rodrigues, H. Molina, S. Heissel, M. T. Mark, L. Steiner, A. Benito-Martin, S. Lucotti, A. Di Giannatale, K. Offer, M. Nakajima, C. Williams, L. Nogués, F. A. Pelissier Vatter, A. Hashimoto, A. E. Davies, D. Freitas, C. M. Kenific, Y. Ararso, W. Buehring, P. Lauritzen, Y. Ogitani, K. Sugiura, N. Takahashi, M. Alečković, K. A. Bailey, J. S. Jolissant, H. Wang, A. Harris, L. M. Schaeffer, G. García-Santos, Z. Posner, V. P. Balachandran, Y. Khakoo, G. P. Raju, A. Scherz, I. Sagi, R. Scherz-Shouval, Y. Yarden, M. Oren, M. Malladi, M. Petriccione, K. C. De Braganca, M. Donzelli, C. Fischer, S. Vitolano, G. P. Wright, L. Ganshaw, M. Marrano, A. Ahmed, J. DeStefano, E. Danzer, M. H. A. Roehrl, N. J. Lacayo, T. C. Vincent, M. R. Weiser, M. S. Brady, P. A. Meyers, L. H. Wexler, S. R. Ambati, A. J. Chou, E. K. Slotkin, S. Modak, S. S. Roberts, E. M. Basu, D. Diolaiti, B. A. Krantz, F. Cardoso, A. L. Simpson, M. Berger, C. M. Rudin, D. M. Simeone, M. Jain, C. M. Ghajar, S. K. Batra, B. Z. Stanger, J. Bui, K. A. Brown, V. K. Rajasekhar, J. H. Healey, M. de Sousa, K. Kramer, S. Sheth, J. Baisch, V. Pascual, T. E. Heaton, M. P. La Quaglia, D. J. Pisapia, R. Schwartz, H. Zhang, Y. Liu, A. Shukla, L. Blavier, Y. A. DeClerck, M. LaBarge, M. J. Bissell, T. C. Caffrey, P. M. Grandgenett, M. A. Hollingsworth, J. Bromberg, B. Costa-Silva, H. Peinado, Y. Kang, B. A. Garcia, E. M. O'Reilly, D. Kelsen, T. M. Trippett, D. R. Jones, I. R. Matei, W. R. Jarnagin and D. Lyden, Cell, 2020, 182, 1044–1061 CrossRef CAS PubMed .e1018..
- J. Lu, J. Li, S. Liu, T. Wang, A. Ianni, E. Bober, T. Braun, R. Xiang and S. Yue, Oncotarget, 2017, 8, 62803 CrossRef PubMed.
- G. Raposo and W. Stoorvogel, J. Cell Biol., 2013, 200, 373–383 CrossRef CAS PubMed.
- Z. Andreu and M. Yáñez-Mó, Front. Immunol., 2014, 5, 442 Search PubMed.
- S. Li, L. Zhu, L. Zhu, X. Mei and W. Xu, Biosens. Bioelectron., 2022, 200, 113902 CrossRef CAS PubMed.
- D.-Y. Choi, J.-N. Park, S.-H. Paek, S.-C. Choi and S.-H. Paek, Biosens. Bioelectron., 2022, 198, 113828 CrossRef CAS PubMed.
- M. Lee, S. J. Park, G. Kim, C. Park, M.-H. Lee, J.-H. Ahn and T. Lee, Biosens. Bioelectron., 2022, 199, 113872 CrossRef CAS PubMed.
- J. Kowal, G. Arras, M. Colombo, M. Jouve, J. P. Morath, B. Primdal-Bengtson, F. Dingli, D. Loew, M. Tkach and C. Théry, Proc. Natl. Acad. Sci., 2016, 113, E968–E977 CrossRef CAS PubMed.
- M. Ramirez-Garrastacho, V. Berge, A. Linē and A. Llorente, Br. J. Cancer, 2022, 126, 492–501 CrossRef CAS PubMed.
- N. Ullah Khan, Z. Muhammad, X. Liu, J. Lin, Q. Zheng, H. Zhang, S. Malik, H. He and L. Shen, Nano Lett., 2021, 21, 5532–5539 CrossRef CAS PubMed.
- Q. Zhan, K. Yi, H. Qi, S. Li, X. Li, Q. Wang, Y. Wang, C. Liu, M. Qiu, X. Yuan, J. Zhao, X. Hou and C. Kang, Theranostics, 2020, 10, 7889–7905 CrossRef CAS PubMed.
- Y. Song, Y. Kim, S. Ha, S. Sheller-Miller, J. Yoo, C. Choi and C. H. Park, Am. J. Reprod. Immunol., 2021, 85, e13329 CrossRef PubMed.
- X. Luan, K. Sansanaphongpricha, I. Myers, H. Chen, H. Yuan and D. Sun, Acta Pharmacol. Sin., 2017, 38, 754–763 CrossRef CAS PubMed.
- J. U. Choi, I.-K. Park, Y.-K. Lee and S. R. Hwang, Int. J. Mol. Sci., 2020, 21, 7363 CrossRef CAS PubMed.
- X. Shi, Q. Cheng and Y. Zhang, Methods, 2020, 177, 95–102 CrossRef CAS PubMed.
- K. Popowski, H. Lutz, S. Hu, A. George, P.-U. Dinh and K. Cheng, J. Extracell. Vesicles, 2020, 9, 1785161 CrossRef CAS PubMed.
- M. Barok, M. Puhka, N. Yazdi and H. Joensuu, J. Extracell. Vesicles, 2021, 10, e12070 CAS.
- S. G. Antimisiaris, S. Mourtas and A. Marazioti, Pharmaceutics, 2018, 10, 218 CrossRef CAS PubMed.
- L. van der Koog, T. B. Gandek and A. Nagelkerke, Adv. Healthcare Mater., 2022, 11, 2100639 CrossRef CAS PubMed.
- M.-H. Chan, Z.-X. Chang, C.-Y. F. Huang, L. J. Lee, R.-S. Liu and M. Hsiao, Nanoscale Horiz., 2022, 7, 352–367 RSC.
- B.-C. Lee, I. Kang and K.-R. Yu, J. Clin. Med., 2021, 10, 711 CrossRef CAS PubMed.
- P. Santos and F. Almeida, Front. Immunol., 2021, 12, 711565 CrossRef CAS PubMed.
- A. Sanghani, P. Andriesei, K. N. Kafetzis, A. D. Tagalakis and C. Yu-Wai-Man, Acta Ophthalmol., 2022, 100, 243–252 CrossRef PubMed.
- M.-m. Shi, Q.-y. Yang, A. Monsel, J.-y. Yan, C.-x. Dai, J.-y. Zhao, G.-c. Shi, M. Zhou, X.-m. Zhu, S.-k. Li, P. Li, J. Wang, M. Li, J.-g. Lei, D. Xu, Y.-g. Zhu and J.-m. Qu, J. Extracell. Vesicles, 2021, 10, e12134 CAS.
- C. K. Das, B. C. Jena, I. Banerjee, S. Das, A. Parekh, S. K. Bhutia and M. Mandal, Mol. Pharm., 2019, 16, 24–40 CrossRef CAS PubMed.
- F. Mehryab, S. Rabbani, S. Shahhosseini, F. Shekari, Y. Fatahi, H. Baharvand and A. Haeri, Acta Biomater., 2020, 113, 42–62 CrossRef CAS PubMed.
- M. J. Haney, N. L. Klyachko, Y. Zhao, R. Gupta, E. G. Plotnikova, Z. He, T. Patel, A. Piroyan, M. Sokolsky, A. V. Kabanov and E. V. Batrakova, J. Controlled Release, 2015, 207, 18–30 CrossRef CAS PubMed.
- C. Liu, W. Zhang, Y. Li, J. Chang, F. Tian, F. Zhao, Y. Ma and J. Sun, Nano Lett., 2019, 19, 7836–7844 CrossRef CAS PubMed.
- P. Wang, H. Wang, Q. Huang, C. Peng, L. Yao, H. Chen, Z. Qiu, Y. Wu, L. Wang and W. Chen, Theranostics, 2019, 9, 1714–1727 CrossRef CAS PubMed.
- M. Yu, C. Gai, Z. Li, D. Ding, J. Zheng, W. Zhang, S. Lv and W. Li, Cancer Sci., 2019, 110, 3173–3182 CrossRef CAS PubMed.
- B. Rodríguez-Morales, M. Antunes-Ricardo and J. González-Valdez, Pharmaceutics, 2021, 13, 1870 CrossRef PubMed.
- Y. Tian, S. Li, J. Song, T. Ji, M. Zhu, G. J. Anderson, J. Wei and G. Nie, Biomaterials, 2014, 35, 2383–2390 CrossRef CAS PubMed.
- G. Liang, Y. Zhu, D. J. Ali, T. Tian, H. Xu, K. Si, B. Sun, B. Chen and Z. Xiao, J. Nanobiotechnol., 2020, 18, 10 CrossRef CAS PubMed.
- Y. Li, Y. Gao, C. Gong, Z. Wang, Q. Xia, F. Gu, C. Hu, L. Zhang, H. Guo and S. Gao, Nanomed. Nanotechnol. Biol. Med., 2018, 14, 1973–1985 CrossRef CAS PubMed.
- X. Zhuang, X. Xiang, W. Grizzle, D. Sun, S. Zhang, R. C. Axtell, S. Ju, J. Mu, L. Zhang, L. Steinman, D. Miller and H.-G. Zhang, Mol. Ther., 2011, 19, 1769–1779 CrossRef CAS PubMed.
- H. Saari, E. Lázaro-Ibáñez, T. Viitala, E. Vuorimaa-Laukkanen, P. Siljander and M. Yliperttula, J. Controlled Release, 2015, 220, 727–737 CrossRef CAS PubMed.
- G. Fuhrmann, A. Serio, M. Mazo, R. Nair and M. M. Stevens, J. Controlled Release, 2015, 205, 35–44 CrossRef CAS PubMed.
- X. Shi, Q. Cheng, T. Hou, M. Han, G. Smbatyan, J. E. Lang, A. L. Epstein, H.-J. Lenz and Y. Zhang, Mol. Ther., 2020, 28, 536–547 CrossRef CAS PubMed.
- N. Yim, S.-W. Ryu, K. Choi, K. R. Lee, S. Lee, H. Choi, J. Kim, M. R. Shaker, W. Sun, J.-H. Park, D. Kim, W. D. Heo and C. Choi, Nat. Commun., 2016, 7, 12277 CrossRef CAS PubMed.
- Q. Cheng, Z. Dai, X. Shi, X. Duan, Y. Wang, T. Hou and Y. Zhang, Biomaterials, 2021, 277, 121129 CrossRef CAS PubMed.
- H. Kim, S. J. Kang and W. J. Rhee, Biotechnol. Bioprocess Eng., 2021, 26, 78–85 CrossRef CAS.
- J. Y. Kim, J. Song, H. Jung and H. Mok, Appl. Biol. Chem., 2018, 61, 599–606 CrossRef CAS.
- Q. Zhou, W. Ding, Z. Qian, Q. Zhu, C. Sun, Q. Yu, Z. Tai and K. Xu, Mol. Pharm., 2021, 18, 4015–4028 CrossRef CAS PubMed.
- L. Cheng, X. Zhang, J. Tang, Q. Lv and J. Liu, Biomaterials, 2021, 275, 120964 CrossRef CAS PubMed.
- M. Piffoux, A. K. A. Silva, C. Wilhelm, F. Gazeau and D. Tareste, ACS Nano, 2018, 12, 6830–6842 CrossRef CAS PubMed.
- S. Rayamajhi, T. D. T. Nguyen, R. Marasini and S. Aryal, Acta Biomater., 2019, 94, 482–494 CrossRef CAS PubMed.
- T. Barjesteh, S. Mansur and Y. Bao, Molecules, 2021, 26, 1135 CrossRef CAS PubMed.
- M. Sancho-Albero, M. d. M. Encabo-Berzosa, M. Beltrán-Visiedo, L. Fernández-Messina, V. Sebastián, F. Sánchez-Madrid, M. Arruebo, J. Santamaría and P. Martín-Duque, Nanoscale, 2019, 11, 18825–18836 RSC.
- M. Khongkow, T. Yata, S. Boonrungsiman, U. R. Ruktanonchai, D. Graham and K. Namdee, Sci. Rep., 2019, 9, 8278 CrossRef PubMed.
- G. Cheng, W. Li, L. Ha, X. Han, S. Hao, Y. Wan, Z. Wang, F. Dong, X. Zou, Y. Mao and S.-Y. Zheng, J. Am. Chem. Soc., 2018, 140, 7282–7291 CrossRef CAS PubMed.
- O. Betzer, N. Perets, A. Angel, M. Motiei, T. Sadan, G. Yadid, D. Offen and R. Popovtzer, ACS Nano, 2017, 11, 10883–10893 CrossRef CAS PubMed.
- J. Wang, P. Chen, Y. Dong, H. Xie, Y. Wang, F. Soto, P. Ma, X. Feng, W. Du and B.-F. Liu, Biomaterials, 2021, 276, 121056 CrossRef CAS PubMed.
- E. Bagheri, K. Abnous, S. A. Farzad, S. M. Taghdisi, M. Ramezani and M. Alibolandi, Life Sci., 2020, 261, 118369 CrossRef CAS PubMed.
- M. S. Kim, M. J. Haney, Y. Zhao, D. Yuan, I. Deygen, N. L. Klyachko, A. V. Kabanov and E. V. Batrakova, Nanomed. Nanotechnol. Biol. Med., 2018, 14, 195–204 CrossRef CAS PubMed.
- C. Gong, J. Tian, Z. Wang, Y. Gao, X. Wu, X. Ding, L. Qiang, G. Li, Z. Han, Y. Yuan and S. Gao, J. Nanobiotechnol., 2019, 17, 93 CrossRef PubMed.
- L. Zhao, C. Gu, Y. Gan, L. Shao, H. Chen and H. Zhu, J. Controlled Release, 2020, 318, 1–15 CrossRef CAS PubMed.
- Q. Lv, L. Cheng, Y. Lu, X. Zhang, Y. Wang, J. Deng, J. Zhou, B. Liu and J. Liu, Adv. Sci., 2020, 7, 2000515 CrossRef CAS PubMed.
- T. Yong, X. Zhang, N. Bie, H. Zhang, X. Zhang, F. Li, A. Hakeem, J. Hu, L. Gan, H. A. Santos and X. Yang, Nat. Commun., 2019, 10, 3838 CrossRef PubMed.
- E. R. Bray, A. R. Oropallo, D. A. Grande, R. S. Kirsner and E. V. Badiavas, Pharmaceutics, 2021, 13, 1543 CrossRef CAS PubMed.
- G. Han, H. Kim, D. E. Kim, Y. Ahn, J. Kim, Y. J. Jang, K. Kim, Y. Yang and S. H. Kim, Pharmaceutics, 2022, 14, 307 CrossRef CAS PubMed.
- A. Shi, J. Li, X. Qiu, M. Sabbah, S. Boroumand, T. C.-T. Huang, C. Zhao, A. Terzic, A. Behfar and S. L. Moran, Theranostics, 2021, 11, 6616–6631 CrossRef CAS PubMed.
- P. Gondaliya, A. A. Sayyed, P. Bhat, M. Mali, N. Arya, A. Khairnar and K. Kalia, Mol. Pharm., 2022, 19, 1294–1308 CrossRef CAS PubMed.
- X. Li, Y. Wang, L. Shi, B. Li, J. Li, Z. Wei, H. Lv, L. Wu, H. Zhang, B. Yang, X. Xu and J. Jiang, J. Nanobiotechnol., 2020, 18, 113 CrossRef CAS PubMed.
- Y. S. Kim, G. Go, C.-W. Yun, J.-H. Yea, S. Yoon, S.-Y. Han, G. Lee, M.-Y. Lee and S. H. Lee, Biomolecules, 2021, 11, 1450 CrossRef CAS PubMed.
- Y. Zhu, Y. Wang, B. Zhao, X. Niu, B. Hu, Q. Li, J. Zhang, J. Ding, Y. Chen and Y. Wang, Stem Cell Res. Ther., 2017, 8, 64 CrossRef PubMed.
- H. Song, X. Li, Z. Zhao, J. Qian, Y. Wang, J. Cui, W. Weng, L. Cao, X. Chen, Y. Hu and J. Su, Nano Lett., 2019, 19, 3040–3048 CrossRef CAS PubMed.
- F. Yan, Z. Zhong, Y. Wang, Y. Feng, Z. Mei, H. Li, X. Chen, L. Cai and C. Li, J. Nanobiotechnol., 2020, 18, 115 CrossRef CAS PubMed.
- H. Li, Y. Feng, X. Zheng, M. Jia, Z. Mei, Y. Wang, Z. Zhang, M. Zhou and C. Li, J. Controlled Release, 2022, 341, 16–30 CrossRef CAS PubMed.
- Y. Liang, X. Xu, X. Li, J. Xiong, B. Li, L. Duan, D. Wang and J. Xia, ACS Appl. Mater. Interfaces, 2020, 12, 36938–36947 CrossRef CAS PubMed.
- M. Heidarzadeh, Y. Gürsoy-Özdemir, M. Kaya, A. Eslami Abriz, A. Zarebkohan, R. Rahbarghazi and E. Sokullu, Cell Biosci., 2021, 11, 142 CrossRef PubMed.
- N. Li, L. Zhao, Y. Wei, V. L. Ea, H. Nian and R. Wei, Stem Cell Res. Ther., 2019, 10, 278 CrossRef PubMed.
- L. Alvarez-Erviti, Y. Seow, H. Yin, C. Betts, S. Lakhal and M. J. A. Wood, Nat. Biotechnol., 2011, 29, 341–345 CrossRef CAS PubMed.
- L. Liu, Y. Li, H. Peng, R. Liu, W. Ji, Z. Shi, J. Shen, G. Ma and X. Zhang, Sci. Adv., 2020, 6, 3967 CrossRef PubMed.
- X. Dong, Y. Lei, Z. Yu, T. Wang, Y. Liu, G. Han, X. Zhang, Y. Li, Y. Song, H. Xu, M. Du, H. Yin, X. Wang and H. Yan, Theranostics, 2021, 11, 5107–5126 CrossRef CAS PubMed.
- L. Sun, M. Fan, D. Huang, B. Li, R. Xu, F. Gao and Y. Chen, Biomaterials, 2021, 271, 120761 CrossRef CAS PubMed.
- A. Singh, A. Raghav, P. A. Shiekh and A. Kumar, Bioact. Mater., 2021, 6, 2231–2249 CrossRef CAS PubMed.
| This journal is © The Royal Society of Chemistry 2022 |





